Symmetric Dimeric Structure and Ligand Recognition of CutR, a LysR-Type Transcriptional Regulator from Mycobacterium sp. Strain JC1
Abstract
1. Introduction
2. Results
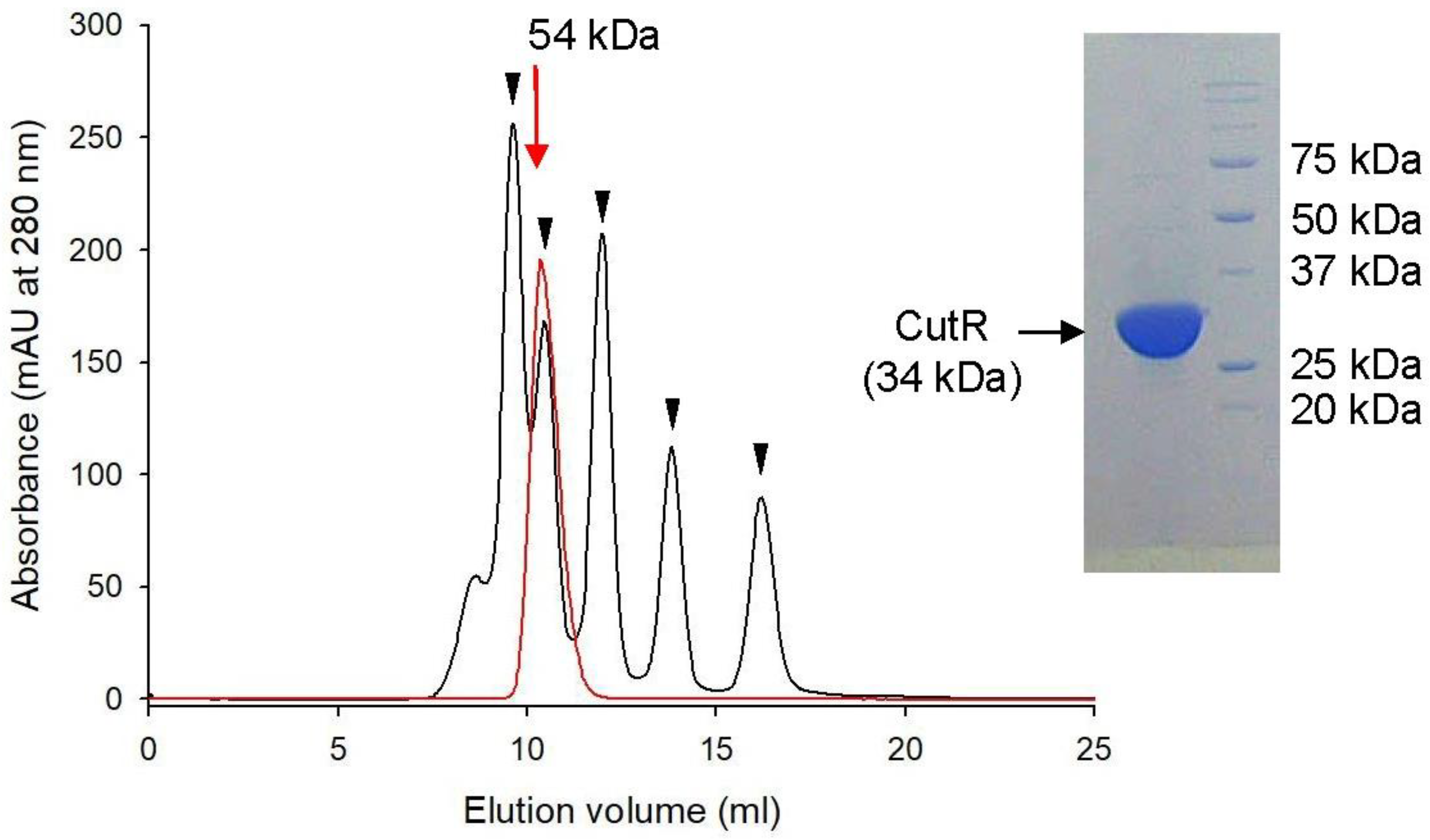
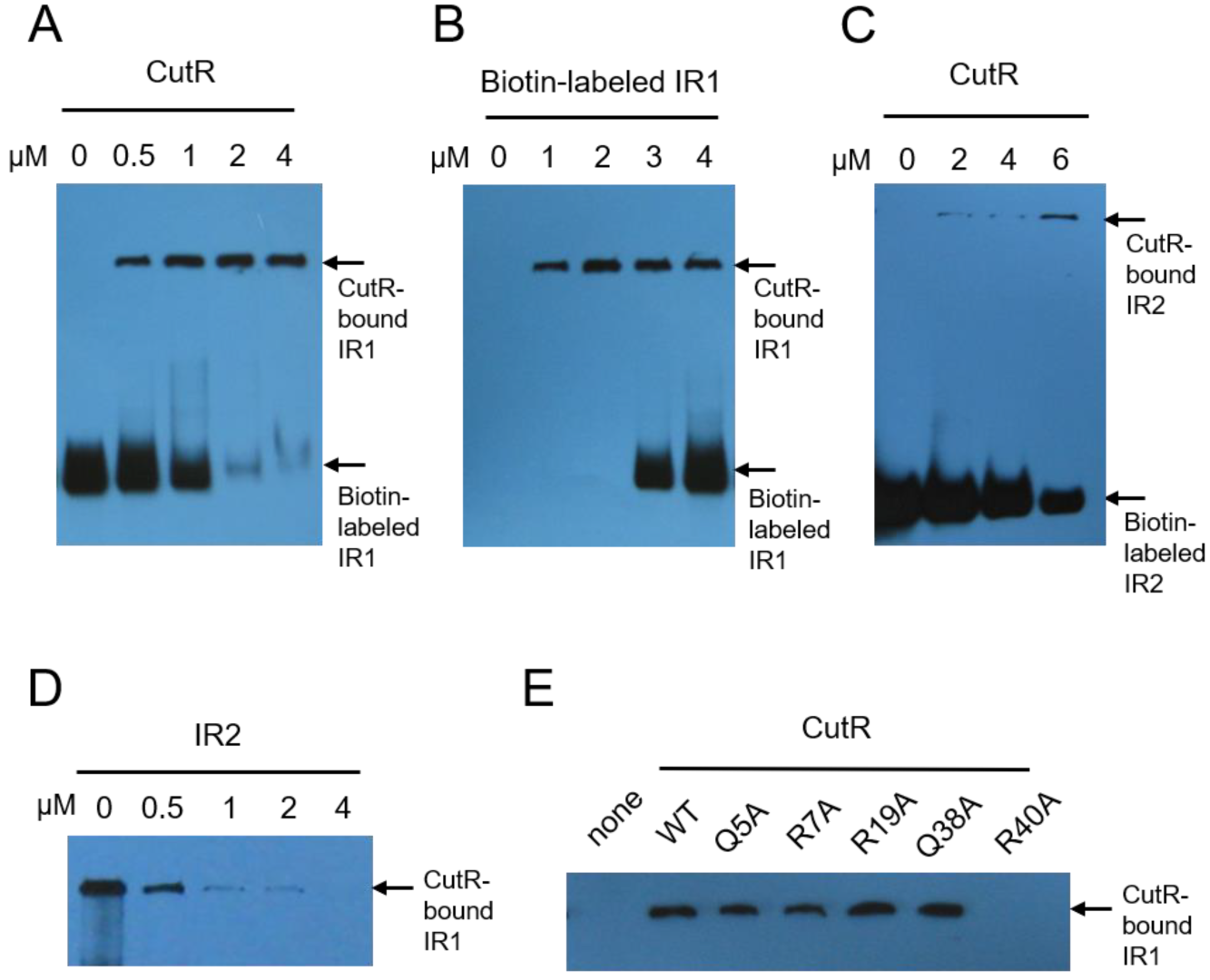
2.1. CutR Forms a Stable Dimer That Interacts with the Inverted Repeat Sequences Upstream of CutB
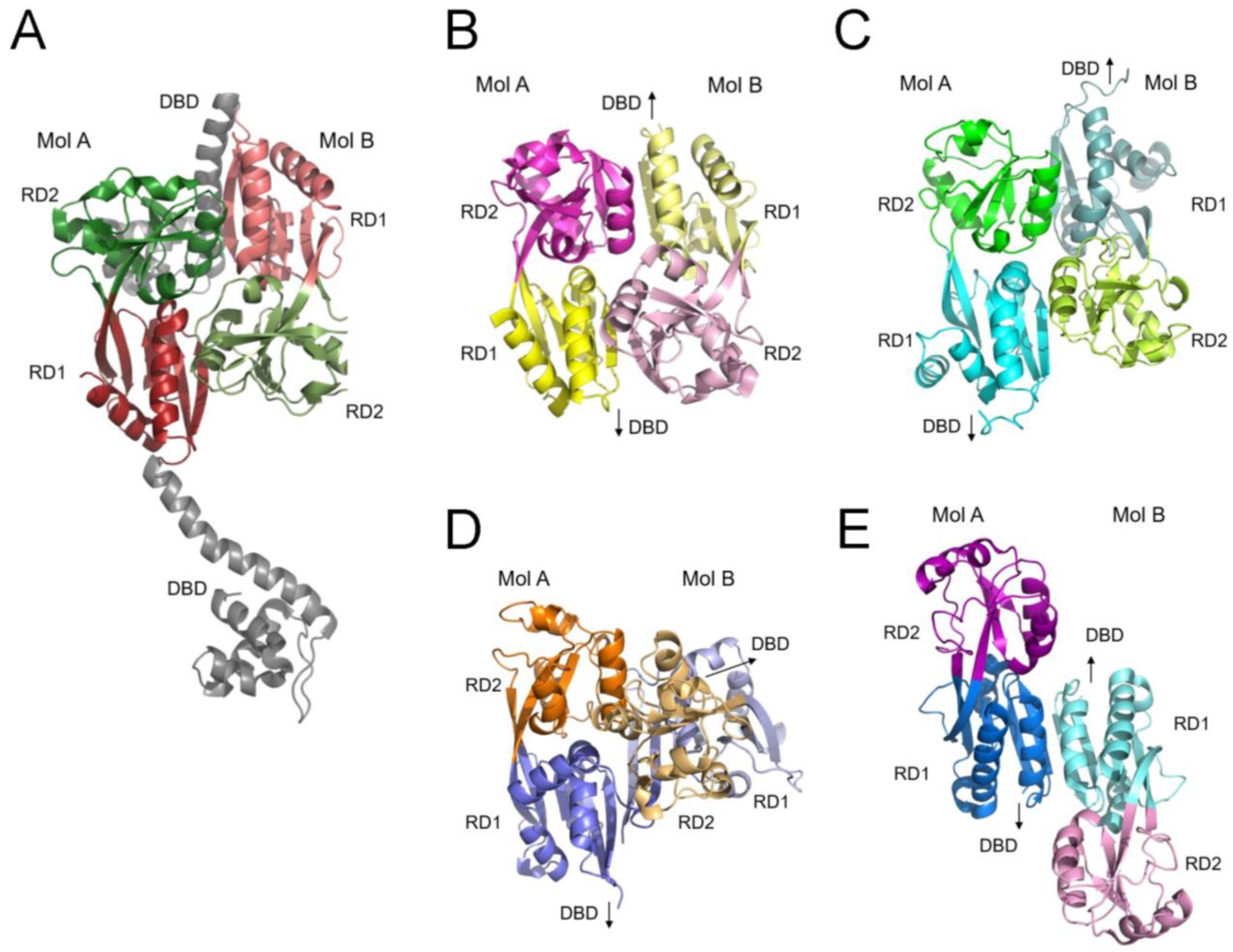
2.2. Crystal Structure of CutR Reveals a Two-Fold Symmetrical Dimer
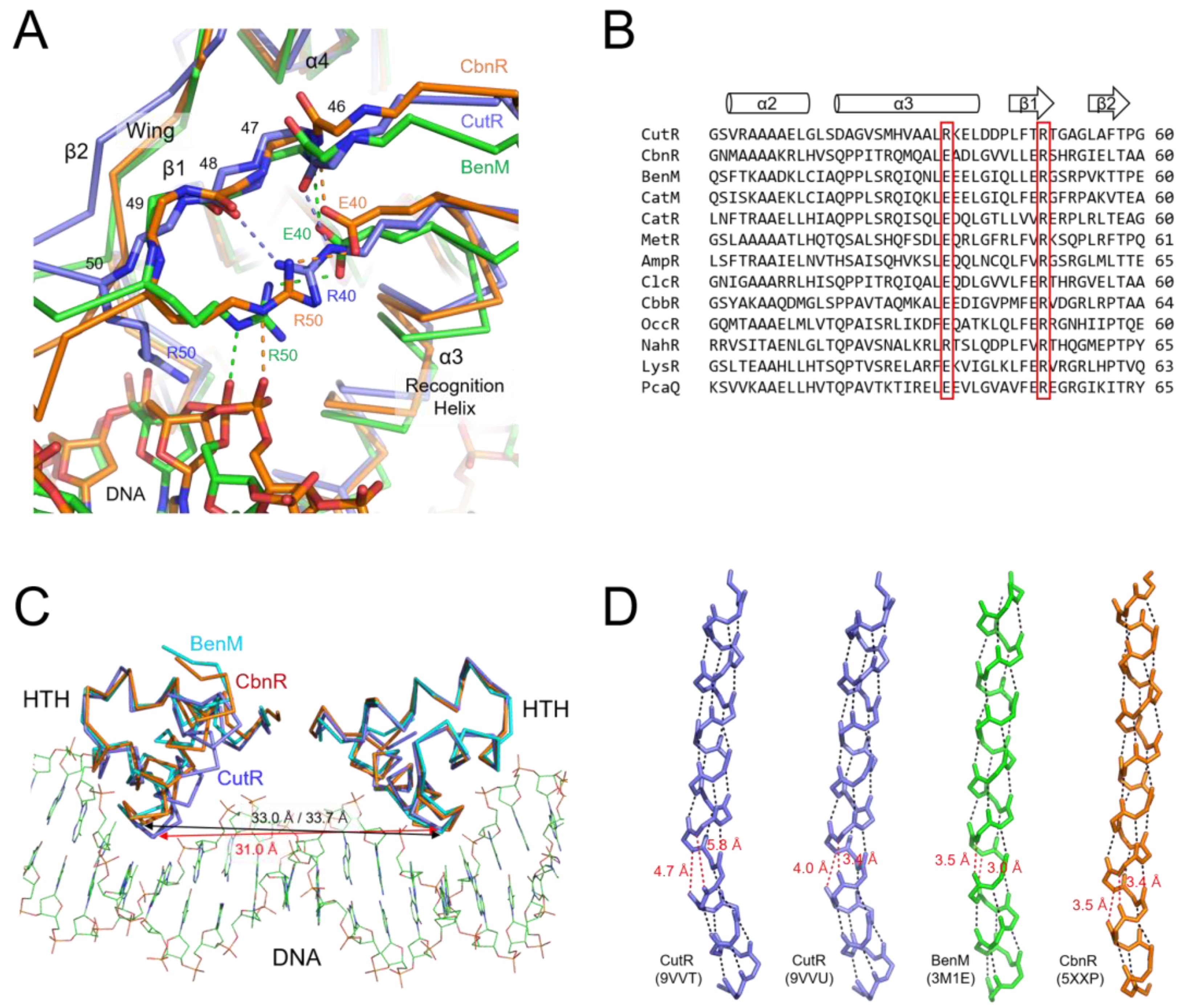
2.3. Putative Ligand-Binding Site
2.4. Structure of the Winged Helix-Turn-Helix Motif
3. Discussion
4. Materials and Methods
4.1. Cloning, Expression, and Purification
4.2. Crystallization and Data Collection
4.3. Structure Determination
4.4. Electrophoretic Mobility Shift Assay (EMSA)
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- To, K.; Cao, R.; Yegiazaryan, A.; Owens, J.; Venketaraman, V. General overview of nontuberculosis Mycobacteria opportunistic pathogens: Mycobacterium avium and Mycobacterium abscessus. J. Clin. Med. 2020, 9, 2541. [Google Scholar] [CrossRef] [PubMed]
- Conyers, L.E.; Saunders, B.M. Treatment for non-tuberculosis mycobacteria: Challenges and prospects. Front. Microbiol. 2024, 15, 1394220. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, L.; Qiao, W.; Luo, Y. Mycobacterium tuberculosis: Pathogenesis and therapeutic targets. MedComm 2023, 2, 353. [Google Scholar] [CrossRef] [PubMed]
- Warner, D.F.; Barczak, A.K.; Gutierrez, M.G.; Mizrahi, V. Mycobacterium tuberculosis biology, pathogenicity and interaction with the host. Nat. Rev. Microbiol. 2025, 4, e353. [Google Scholar] [CrossRef]
- Kumar, A.; Deshane, J.S.; Crossman, D.K.; Bolisetty, S.; Yan, B.; Kramnik, I.; Agarwal, A.; Steyn, A.J.C. Heme oxygenase-1-derived carbon monoxide induces the Mycobacterium tuberculosis dormancy regulon. J. Biol. Chem. 2008, 283, 18032–18039. [Google Scholar] [CrossRef]
- Park, S.W.; Hwang, E.H.; Park, H.; Kim, J.A.; Heo, J.; Lee, K.H.; Song, T.; Kim, E.; Ro, Y.T.; Kim, S.W.; et al. Growth of mycobacteria on carbon monoxide and methanol. J. Bacteriol. 2003, 185, 142–147. [Google Scholar] [CrossRef]
- Park, S.W.; Song, T.; Kim, S.Y.; Kim, E.; Oh, J.I.; Eom, C.Y.; Kim, Y.M. Carbon monoxide dehydrogenase in mycobacteria possesses a nitric oxide dehydrogenase activity. Biochem. Biophys. Res. Commun. 2007, 362, 449–453. [Google Scholar] [CrossRef]
- Dobbek, H.; Gremer, L.; Meyer, O.; Huber, R. Crystal structure and mechanism of CO dehydrogenase, a molybdo iron-sulfur flavoprotein containing S-selanylcysteine. Proc. Natl. Acad. Sci. USA 1999, 96, 8884–8889. [Google Scholar] [CrossRef]
- Kang, B.S.; Kim, Y.M. Cloning and molecular characterization of the genes for carbon monoxide dehydrogenase and localization of molybdopterin, flavin adenine dinucleotide, and iron-sulfur centers in the enzyme of Hydrogenophaga pseudoflava. J. Bacteriol. 1999, 181, 5581–5590. [Google Scholar] [CrossRef]
- Song, T.; Park, S.W.; Park, S.J.; Kim, J.H.; Yu, J.Y.; Oh, J.I.; Kim, Y.M. Cloning and expression analysis of the duplicated genes for carbon monoxide dehydrogenase of Mycobacterium sp. strain JC1 DSM 3803. Microbiology 2010, 156, 1009–1018. [Google Scholar] [CrossRef][Green Version]
- Oh, J.I.; Park, S.J.; Shin, S.J.; Ko, I.J.; Han, S.J.; Park, S.W.; Song, T.; Kim, Y.M. Identification of trans- and cis-control elements involved in regulation of the carbon monoxide dehydrogenase genes in Mycobacterium sp. strain JC1 DSM 3803. J. Bacteriol. 2010, 192, 3925–3933. [Google Scholar] [CrossRef]
- Maddocks, S.E.; Oyston, P.C.F. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology 2008, 154, 3609–3623. [Google Scholar] [CrossRef] [PubMed]
- Baugh, A.C.; Momany, C.; Neidle, E.L. Versatility and complexity: Common and uncommon facets of LysR-type transcriptional regulators. Annu. Rev. Microbiol. 2023, 77, 317–3319. [Google Scholar] [CrossRef] [PubMed]
- Lerche, M.; Dian, C.; Round, A.; Lönneborg, R.; Brzezinski, P.; Leonard, G.A. The solution configurations of inactive and activated DntR have implications for the sliding dimer mechanism of LysR transcription factors. Sci. Rep. 2016, 6, 19988. [Google Scholar] [CrossRef]
- Demeester, W.; De Paepe, B.; De Mey, M. Fundamentals and exceptions of the LysR-type transcriptional regulators. ACS Synth. Biol. 2024, 13, 3069–3092. [Google Scholar] [CrossRef] [PubMed]
- Maxon, M.E.; Wigboldus, J.; Brot, N.; Weissbach, H. Structure-function studies on Escherichia coli MetR protein, a putative prokaryotic leucine zipper protein. Proc. Natl. Acad. Sci. USA 1990, 87, 7076–7079. [Google Scholar] [CrossRef]
- Parsek, M.R.; Shinabarger, D.L.; Rothmel, R.K.; Chakrabarty, A.M. Roles of CatR and cis,cis-muconate in activation of the catBC operon, which is involved in benzoate degradation in Pseudomonas putida. J. Bacteriol. 1992, 174, 7798–7806. [Google Scholar] [CrossRef][Green Version]
- Akakura, R.; Winans, S.C. Mutations in the occQ operator that decrease OccR-induced DNA bending do not cause constitutive promoter activity. J. Biol. Chem. 2002, 277, 15773–15780. [Google Scholar] [CrossRef]
- Alanazi, A.M.; Neidle, E.L.; Momany, C. The DNA-binding domain of BenM reveals the structural basis for the recognition of a T-N11-A sequence motif by LysR-type transcriptional regulators. Acta Crystallogr. D Biol. Crystallogr. 2013, 69, 1995–2007. [Google Scholar] [CrossRef]
- Koentjoro, M.P.; Adachi, N.; Senda, M.; Ogawa, N.; Senda, T. Crystal structure of the DNA-binding domain of the LysR-type transcriptional regulator CbnR in the complex with a DNA fragment of the recognition-binding site in the promoter region. FEBS J. 2018, 285, 977–989. [Google Scholar] [CrossRef]
- Giannopoulou, E.A.; Senda, M.; Koentjoro, M.P.; Adachi, N.; Ogawa, N.; Senda, T. Crystal structure of the full-length LysR-type transcriptional regulator CbnR in complex with promoter DNA. FEBS J. 2021, 288, 4560–4575. [Google Scholar] [CrossRef] [PubMed]
- Muraoka, S.; Okumura, R.; Ogawa, N.; Nonaka, T.; Miyashita, K.; Senda, T. Crystal Structure of a Full-length LysR-type Transcriptional Regulator, CbnR: Unusual Combination of Two Subunit Forms and Molecular Bases for Causing and Changing DNA Bend. J. Mol. Biol. 2003, 328, 555–566. [Google Scholar] [CrossRef] [PubMed]
- Knapp, G.S.; Hu, J.C. The oligomerization of CynR in Escherichia coli. Protein Sci. 2009, 18, 2307–2315. [Google Scholar] [CrossRef] [PubMed]
- Stec, E.; Witkowska-Zimny, M.; Hryniewicz, M.M.; Neumann, P.; Wilkinson, A.J.; Brzozowski, A.M.; Verma, C.S.; Zaim, J.; Wysocki, S.D.; Bujacz, G. Structural basis of the sulphate starvation response in E. coli: Crystal structure and mutational analysis of the cofactor-binding domain of the Cbl transcriptional regulator. J. Mol. Biol. 2006, 364, 309–322. [Google Scholar] [CrossRef]
- van Keulen, G.; Girbal, L.; van den Bergh, E.R.E.; Dijkhuizen, L.; Meijer, W.G. The LysR-type transcriptional regulator CbbR controlling autotrophic CO2 fixation by Xanthobacter flavus. J. Bacteriol. 1998, 180, 1411–1417. [Google Scholar] [CrossRef]
- Sun, S.; Zhou, L.; Jin, K.; Jian, H.; He, Y. Quorum sensing systems differently regulate the production of phenazine-1-carboxylic acid in the rhizobacterium Pseudomonas aeruginosa PA1201. Sci. Rep. 2016, 6, 30352. [Google Scholar]
- Abdeksanue, A.S.; Hamed, M.M.; Schütz, C.; Röhrig, T.; Kany, A.M.; Schmelz, S.; Blankenfeldt, W.; Hirsch, A.K.H.; Hartmann, R.W.; Empting, M. Discovery and optimization of thiazole-based quorum sensing inhibitors as potent blockers of Pseudomonas aeruginosa pathogenicity. Eur. J. Med. Chem. 2024, 276, 116685. [Google Scholar] [CrossRef]
- Wang, L.; Winans, S.C. The sixty nucleotide OccR operator contains a subsite essential and sufficient for OccR binding and a second subsite required for ligand-responsive DNA bending. J. Mol. Biol. 1995, 253, 691–702. [Google Scholar] [CrossRef]
- MacLean, A.M.; Haerty, W.; Golding, G.B.; Finan, T.M. The LysR-type PcaQ protein regulates expression of a protocatechuate-inducible ABC-type transport system in Sinorhizobium meliloti. Microbiology 2011, 157, 2522–2533. [Google Scholar] [CrossRef]
- Wek, R.C.; Hatfield, G.W. Transcriptional activation at adjacent operators in the divergent-overlapping ilvY and ilvC promoters of Escherichia coli. J. Mol. Biol. 1988, 203, 643–663. [Google Scholar] [CrossRef]
- Lanzilotta, W.N.; Schuller, D.J.; Thorsteinsson, M.V.; Kerby, R.L.; Roberts, G.P.; Poulos, T.L. Structure of the CO sensing transcription activator CooA. Nat. Struct. Mol. Biol. 2000, 7, 876–880. [Google Scholar] [CrossRef] [PubMed]
- Dent, M.R.; Roberts, M.G.; Bowman, H.E.; Weaver, B.R.; McCaslin, D.R.; Burstyn, J.N. Quaternary structure and deoxyribonucleic acid-binding properties of the heme-dependent, CO-sensing transcriptional regulator PxRcoM. Biochemistry 2022, 61, 678–688. [Google Scholar] [CrossRef] [PubMed]
- Otwinowski, W.; Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997, 276, 307–326. [Google Scholar] [PubMed]
- Terwilliger, T.C.; Berendzen, J. Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 1999, 55, 849–861. [Google Scholar] [CrossRef]
- Terwilliger, T.C. Maximum-likelihood density modification. Acta Crystallogr. D Biol. Crystallogr. 2000, 56, 965–972. [Google Scholar] [CrossRef]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef]
- Winn, M.D.; Ballard, C.C.; Cowtan, K.D.; Dodson, E.J.; Emsley, P.; Evans, P.R.; Keegan, R.M.; Krissinel, E.B.; Leslie, A.G.; McCoy, A.; et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011, 67, 235–242. [Google Scholar] [CrossRef]
- Winn, M.D.; Murshudov, G.N.; Papiz, M.Z. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 2003, 374, 300–321. [Google Scholar]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef]
- Headd, J.J.; Echols, N.; Afonine, P.V.; Grosse-Kunstleve, R.W.; Chen, V.B.; Moriarty, N.W.; Richardson, D.C.; Richardson, J.S.; Adams, P.D. Use of knowledge-based restraints in phenix.refine to improve macromolecular refinement at low resolution. Acta Crystallogr. D Biol. Crystallogr. 2012, 68, 381–390. [Google Scholar] [CrossRef]



| Data Set | SeMet | Native |
|---|---|---|
| Experimental Data | ||
| X-Ray source | PAL 4A | PAL 7A |
| Wavelength (Å) | 0.97926 | 0.97933 |
| Space group | P212121 | P212121 |
| Unit cell parameters | ||
| a, b, c (Å) | 63.50, 92.08, 126.19 | 69.93, 81.81, 101.06 |
| α, β, γ (°) | 90.00, 90.00, 90.00 | 90.00, 90.00, 90.00 |
| Resolution limit (Å) Total reflections Unique reflections Redundancy Completeness (%) | 50–2.50 (2.59–2.50) a 378,265 26,311 14.4 (14.6) 100 (100) | 50–1.80 (1.83–1.80) 337,472 52,582 6.4 (3.8) 96.5 (76.4) |
| Rsymm b | 0.117 (0.492) | 0.062 (0.569) |
| Average I/σ (I) CC1/2 | 43.4 (6.2) - | 37.8 (3.0) 0.999 (0.927) |
| Refinement Details | ||
| Resolutions (Å) | 40.26–2.50 | 36.62–1.80 |
| Reflections (working) Reflections (test) | 23,187 1252 | 50,949 1949 |
| Rwork /Rfree c | 0.212/0.268 | 0.191/0.233 |
| Number of atoms Proteins Ligands Waters | 4889 0 44 | 4797 14 398 |
| RMSD | ||
| Bond length (Å) | 0.008 | 0.005 |
| Bond angle (o) Average B factors (Å2) | 1.115 | 0.736 |
| Mol. A (main/side) Mol. B (main/side) Water molecules Ramachandran plot Favoured (%) Allowed (%) Outliers (%) | 35.95 (35.73/36.21) 39.08 (39.02/39.14) 34.30 96.0 3.7 0.3 | 33.06 (30.22/36.31) 34.19 (37.05/31.72) 32.37 98.1 1.9 0.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cho, H.J.; Lee, K.Y.; Lee, H.-S.; Kang, B.S. Symmetric Dimeric Structure and Ligand Recognition of CutR, a LysR-Type Transcriptional Regulator from Mycobacterium sp. Strain JC1. Int. J. Mol. Sci. 2025, 26, 10533. https://doi.org/10.3390/ijms262110533
Cho HJ, Lee KY, Lee H-S, Kang BS. Symmetric Dimeric Structure and Ligand Recognition of CutR, a LysR-Type Transcriptional Regulator from Mycobacterium sp. Strain JC1. International Journal of Molecular Sciences. 2025; 26(21):10533. https://doi.org/10.3390/ijms262110533
Chicago/Turabian StyleCho, Hyo Je, Ka Young Lee, Hyun-Shik Lee, and Beom Sik Kang. 2025. "Symmetric Dimeric Structure and Ligand Recognition of CutR, a LysR-Type Transcriptional Regulator from Mycobacterium sp. Strain JC1" International Journal of Molecular Sciences 26, no. 21: 10533. https://doi.org/10.3390/ijms262110533
APA StyleCho, H. J., Lee, K. Y., Lee, H.-S., & Kang, B. S. (2025). Symmetric Dimeric Structure and Ligand Recognition of CutR, a LysR-Type Transcriptional Regulator from Mycobacterium sp. Strain JC1. International Journal of Molecular Sciences, 26(21), 10533. https://doi.org/10.3390/ijms262110533







