Abstract
Listeria monocytogenes is a ubiquitous foodborne pathogen whose occurrence in food and food-processing environments raises public-health concerns, particularly when isolates carry antimicrobial-resistance determinants. Next-generation sequencing (NGS) is increasingly used to detect resistance genes and to predict phenotypic resistance. Following the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA 2020) guidelines, PubMed, Web of Science, and Scopus were searched for original articles (2015–2024) that used second- and/or third-generation sequencing to characterize antibiotic resistance in L. monocytogenes from food and food-processing environments. After deduplication and screening, 58 studies were included from an initial 418 records. NGS reliably detected a set of recurrent resistance determinants across diverse sample types and geographies. The fosX locus (intrinsic fosfomycin-related marker) was effectively ubiquitous across studies, while acquired determinants were variably distributed: lin (35/58 studies, 60.34%), norB (33/58, 56.90%), and tetracycline genes overall in 20/58 (34.48%) with tetM as the most common (11/58, 18.97%). Reported concordance between the genotypes and phenotypes for acquired resistance was very high (>99% for most agents), with notable exceptions (e.g., ciprofloxacin and some fosfomycin cases). Common analysis pipelines and databases included ResFinder, CARD, BIGSdb-Lm, ABRicate, and ARIBA; most sequencing used Illumina short reads, with an increasing use of long-read or hybrid approaches. NGS is a powerful surveillance tool for detecting resistance determinants and for source-tracking, but its predictive value depends on integration with phenotypic testing, standardized reporting, and comprehensive, curated databases. Key gaps include inconsistent phenotype reporting, variable database coverage, and limited assessment of gene expression/regulatory effects.
1. Introduction
Listeria monocytogenes is a highly ubiquitous, multi-resistant foodborne pathogen found in soil, water, vegetation, and animal reservoirs [1]. This pathogen can grow over a wide range of temperatures and environmental pH and persist in food-processing environments, making it an extremely significant challenge for food safety [2]. L. monocytogenes is responsible for causing listeriosis. Human listeriosis has high rates of hospitalization and mortality, especially in vulnerable groups, and is on the rise in Europe despite efforts to control it [3,4]. Listeriosis therapy can be complicated because L. monocytogenes is an intracellular pathogen, so some antibiotics show efficacy in vitro, but in vivo, they only have a bacteriostatic effect [5]. In recent years, concerns about antibiotic-resistant L. monocytogenes in food and production environments have increased [2,6,7]. For example, a study by Rippa et al. (2024) [2] found alarmingly high levels of resistance—87.36% (n = 235) of 269 L. monocytogenes strains isolated from food and food-production environments were multidrug resistant (MDR). Similarly, in a Nigerian study by Kayode and Okoh (2023) [6], of 194 isolates from ready-to-eat (RTE) foods, >50% of strains resistant to sulfamethoxazole or trimethoprim and >40% resistant to amoxicillin, penicillin, or erythromycin were observed. These findings indicate that L. monocytogenes carrying resistance to clinically relevant antibiotics may be present in a variety of food sources (meat, dairy, seafood/vegetable RTE foods, etc.), raising public health concerns.
Importantly, L. monocytogenes has known intrinsic resistance traits that shape its antibiogram. The literature reports emerging acquired resistance in L. monocytogenes. A recent genomic study of 5339 French isolates (2012–2019) showed that only 2.23% contained acquired antimicrobial resistance (AMR) genes, and these were much more common in food-derived isolates (3.74%) than in clinical isolates (0.98%) [8]. Although most L. monocytogenes remain susceptible to standard treatment, the food chain contains a growing fraction of strains with genetic resistance traits [8].
The standard approach for antimicrobial resistance detection testing in microorganisms, including L. monocytogenes, is phenotypic testing. Typical methods include the following: the disk-diffusion method (Kirby–Bauer method) or the broth microdilution method to determine minimum inhibitory concentrations (MICs) against a panel of antibiotics [7]. Phenotypic tests characterize isolates as the following: “susceptible, standard dosing regimen (S)”, “susceptible, increased exposure (I)”, and “resistant (R)” for a specific antimicrobial agent [9]. These methods, while necessary for antimicrobial resistance diagnosis, have several drawbacks—they can be lengthy (requiring 1–2 days of culture), they may not reveal underlying genetic mechanisms, and their precision depends on standardized breakpoints that can miss low levels or newly occurring resistance [10]. They provide only a superficial picture of resistance and do not detect emerging resistance genotypes. Resistant isolates are identified in these methods by their growth, but the specific genes involved in encoding resistance to specific agents or mutations in these genes remain unknown [11].
In recent years, whole-genome sequencing (WGS), a next-generation sequencing (NGS) method, has become an increasingly important tool in the study of antibiotic resistance, including in L. monocytogenes [12]. WGS enables the high-resolution detection of resistance genes and mutations, including both chromosomal and mobile genetic elements, offering insights beyond the reach of traditional phenotypic methods [13]. Beyond identifying resistance genes, WGS allows researchers to assess their genomic context (e.g., plasmids and transposons), assign isolates to specific sequence types (STs) or clonal complexes (CCs), and trace their transmission in food-production chains [13].
This literature review aims to summarize original research published in the three main databases (PubMed, Web of Science, and Scopus) that applied whole-genome next-generation sequencing to investigate antibiotic resistance in L. monocytogenes isolated from food and food-processing environments.
2. Methods
The criteria following the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA 2020) were used to prepare a systematic review of original scientific articles.
The analysis involved searching three open-access research databases: PubMed, Web of Science, and Scopus (last search: 18 March 2025). These databases were used to find research articles specifically focused on using next-generation sequencing as a tool for predicting antibiotic resistance, by analyzing the presence of antibiotic resistance genes in L. monocytogenes isolates obtained from food in its production environment.
Inclusion criteria for the database search were (a) original research papers published between 2015 and 2024; (b) characterization of L. monocytogenes isolates from food and the food-production environment; and (c) the use of second- and/or third-generation sequencing technology.
The following criteria were used as exclusion criteria: (1) no/other source of strains than food and food-processing environment; (2) other than the original research article; (3) no analysis of L. monocytogenes strains; (4) no use of NGS for the analysis of L. monocytogenes; (5) different context for research than antibiotic resistance; (6) no analysis of antibiotic resistance genes; and (7) another reason. Search terms “Listeria monocytogenes” OR “L. monocytogenes” AND “next generation sequencing” AND “resistance” OR “Listeria monocytogenes” OR “L. monocytogenes” AND “whole-genome sequencing” AND “resistance”. After removing duplicate records from the databases, 418 results were obtained, of which 58 met the inclusion criteria. The database search and literature analysis identified key original research studies focusing on the use of next-generation sequencing in predicting antibiotic resistance for L. monocytogenes strains isolated from food and food-production environments (Table 1) [8,12,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69]. Data items: country, year of sample collection, sample type (food/processing environment), number of isolates, sequencing platform, bioinformatics pipeline, phenotypic resistance testing, and resistance genes detected (Table 1 and Table S1 and Figure S1).

Table 1.
The characterization and antibiotic resistance profile of sequenced L. monocytogenes strains from food and its production environment.
The entire selection process of the publications analyzed for this study is shown in Figure 1 and summarized in Table S1. Full search strategies for each database are provided in Supplementary Table S2. Duplicates were removed by P.W. and checked by P.A. and M.T. Data extraction was performed independently by P.W., P.A., and M.T. Extracted items were verified by A.Z. and W.C.-W. Discrepancies were resolved through discussion. Due to heterogeneity in study designs, sequencing methods, and reported outcomes, a meta-analysis was not performed. Results are synthesized narratively and presented in tables summarizing study characteristics and findings.
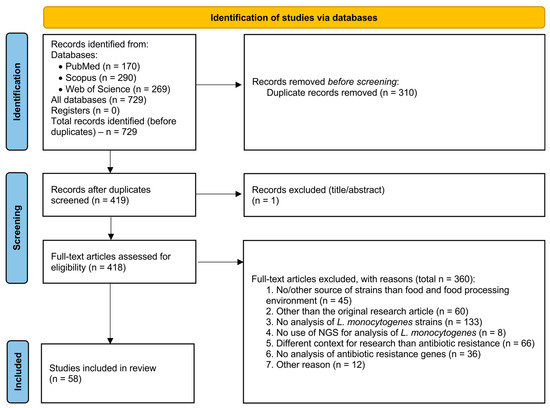
Figure 1.
A summary of the selection process for all publications reviewed for the study.
The authors (P.W., P.A., and M.T.) independently reviewed the titles and abstracts of the retrieved articles, while any disagreements were resolved through discussion with the next authors (A.Z. and W.C.-W.).
3. Results and Discussion
3.1. Profiling Resistance Genes as Phenotypic Predictors in L. monocytogenes Isolated from Food and Food-Production Environments
In recent years, there has been an increasing focus on the genetic markers of antibiotic resistance as potential predictors of phenotypic resistance. Among L. monocytogenes, there are both innate resistance mechanisms (such as natural resistance to cephalosporins or glycopeptides) and acquired resistance genes. Scientific studies have shown that the presence of specific genes strongly correlates with the phenotypic resistance profiles observed. It has been demonstrated that the phenotype of acquired antibiotic resistance in L. monocytogenes can be predicted from genome sequence analysis with an accuracy of over 99%. This indicates that strains carrying characteristic genes (e.g., tetM, lnuG, ermB, mphB, fexA, etc.) typically exhibit resistance to their respective antibiotics. Frequently transferred on plasmids or other mobile genetic elements, the tet and erm genes have also been documented in numerous studies of food-derived L. monocytogenes.
Due to the diverse antibiotic resistance profiles of L. monocytogenes strains isolated from food and food-production environments, molecular genotypic profiling is becoming a key part of microbial threat monitoring. Analyzing the presence of resistance genes in bacterial populations enables the early detection of rising resistance trends and the identification of potentially dangerous strains. Gene profiling can serve as a rapid marker of potential phenotypic resistance even before comprehensive phenotyping tests are completed. Whole-genome sequencing offers a complete view of a strain’s genome, allowing for the detection of all known resistance determinants, establishing strain relatedness, and tracing sources of contamination in the food chain. WGS analyses have proven useful in routine surveillance and epidemiological studies—enabling precise typing and source tracking of listeriosis outbreaks, as well as the identification of new vectors for resistance gene transmission in the production environment.
To highlight the benefits of next-generation sequencing in profiling antibiotic resistance genes, fifteen of the most common determinants were selected based on a literature review following established criteria. Figure 2 provides a summary of these genes, confirmed by numerous studies on L. monocytogenes strains isolated from food and production environments.
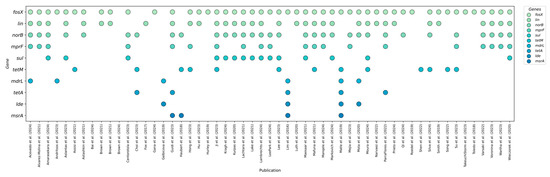
Figure 2.
The most frequently determined antibiotic resistance genes in L. monocytogenes from food and food-production environments that were analyzed in the studies [8,12,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69].
3.2. Prevalent Resistance Genes and Their Predictive Value
Based on a literature search following previously established criteria, the fifteen resistance genes most frequently detected in L. monocytogenes were identified through next-generation sequencing. Sequencing not only enables the simultaneous detection of all known genetic determinants of antibiotic resistance but also allows for the assessment of their prevalence in populations isolated from food and food-production environments. The articles shown in Figure 2 report the prevalence of genes such as fosX, lin, norB, mprF, sul, tetM, mdrL, tetA, aacA4, tet, msrA, lde, tetS, fexA, and mepA, which exhibited the highest detection rates in various studies. By compiling the results from these studies, we can identify which resistance mechanisms are most common in the food sector. An overview of the genetic profiles, including the number and percentage of strains with each gene in each study, provides a foundation for further risk analysis and the development of predictive models for phenotypic resistance based on NGS data.
Methodologically (Figure 3), detection relied on Illumina platforms (MiSeq, NextSeq, HiSeq, and NovaSeq) and was analyzed using tools such as ResFinder (Center for Genomic Epidemiology DTU; https://genepi.food.dtu.dk/resfinder (accessed on 21 July 2025)), Comprehensive Antibiotic Resistance Database/Resistance Gene Identifier (CARD/RGI; https://card.mcmaster.ca/, https://card.mcmaster.ca/analyze/rgi (accessed on 21 July 2025)), Bacterial Isolate Genome Sequence Database—Listeria monocytogenes (BIGSdb-Lm; https://bigsdb.pasteur.fr/listeria/ (accessed on 21 July 2025)), ABRicate (GitHub; https://github.com/tseemann/abricate (accessed on 21 July 2025)), or ARIBA (Antimicrobial Resistance Identification By Assembly, GitHub; https://github.com/sanger-pathogens/ariba (accessed on 21 July 2025)). Despite minor variations in read length and assembly pipelines, every study reported robust identification of antibiotic resistance genes, underscoring their stability across sequencing technologies and bioinformatic workflows.
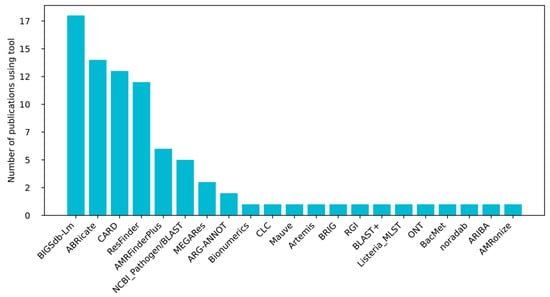
Figure 3.
The use of bioinformatic tools in the prediction of resistance genes in the analyzed studies.
The detection of antibiotic resistance genes in WGS studies typically relies on short-read Illumina sequencing, though long-read approaches are increasingly used. In most studies, L. monocytogenes genomes were generated on the Illumina MiSeq, HiSeq, NextSeq, NovaSeq, X10, and Nanopore platforms (150 bp paired-end reads, 250 bp paired-end reads, and 300 bp paired-end reads). Illumina reads provide high per-base accuracy (<0.1% error) but produce fragmented assemblies due to short read length [70]. In contrast, Oxford Nanopore long reads (10 kb+) can span repetitive regions and yield complete genomes, albeit with higher error rates. In practice, hybrid assembly (combining short and long reads) can recover full chromosomes and mobile elements. Regardless of platform, all studies report robust detection of AMR genes once the genome is assembled or even directly from reads [71].
Figure 4, Figure 5 and Figure 6 summarize the frequency and co-occurrence patterns of the top ten resistance genes across the surveyed publications. The co-occurrence matrix (Figure 4) displays the number of publications in which gene pairs were detected together; the strongest and most frequently reported pairings are fosX–lin, fosX–norB, and lin–norB. The gene-sample heatmap (Figure 5) shows the distribution of detections across sample categories and indicates that the greatest number of reports for fosX, lin, and norB originate from meat-related samples, while other sample types yield fewer but still notable detections. The bar chart of the top ten co-occurring gene pairs (Figure 6) ranks pairs by the number of publications reporting their simultaneous presence, with fosX–lin at the top, followed by fosX–norB, lin–norB, and additional frequent combinations involving mprF and sul. Collectively, these figures present a clear, publication-based picture of which resistance determinants and gene combinations are most commonly reported in L. monocytogenes from food and production environments, supporting targeted surveillance and comparative analyses of resistance prevalence.
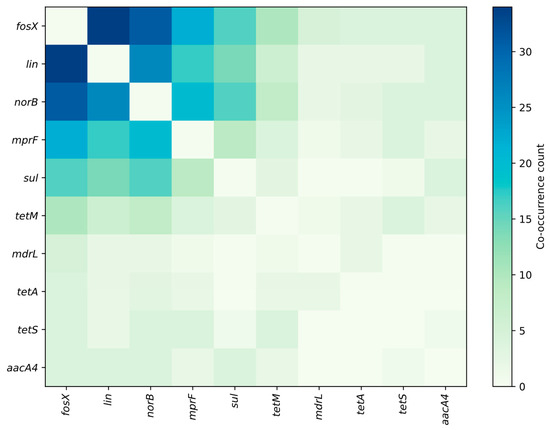
Figure 4.
Frequency matrices for the detection and co-occurrence of resistance genes (top 10 genes).
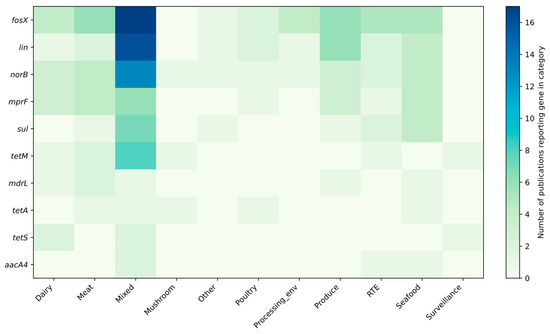
Figure 5.
Frequency of reporting resistance genes in individual sample categories in the analyzed studies (top 10 genes).
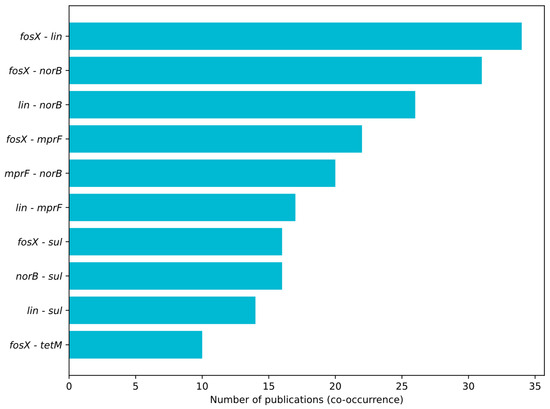
Figure 6.
Top 10 gene pairs with the highest co-occurrence—number of publications.
3.2.1. Fosfomycin Resistance
The fosX gene is the most frequently detected resistance gene among scientific studies analyzing the genome sequencing of L. monocytogenes strains isolated from food and food-production environments [8,12,14,15,16,17,18,19,20,21,22,23,26,28,30,31,32,33,34,35,36,37,38,40,41,42,43,44,45,46,47,48,49,51,52,53,54,55,57,58,59,60,61,62,63,64,65,66,68,69,72].
Across 50 independent studies spanning five continents, fosX was detected in every surveyed population—whether in traditional dairy products [17,43], ready-to-eat (RTE) meat [23,35], poultry [28], beef production chains [12,26], seafood [36,68], fresh produce [16,49], or environmental swabs from food-processing facilities [36,65].
The fosX gene (lmo1702 in strain EGD-e) encodes a Mn2+-dependent epoxide hydrolase that inactivates fosfomycin. It is part of the L. monocytogenes’ core genome and confers intrinsic resistance to fosfomycin [73,74]. Genomic analyses confirm that fosX is nearly ubiquitous across L. monocytogenes (and related Listeria sensu stricto) strains [74]. FosX-mediated resistance is encoded in the listerial core genome of all examined pathogenic and non-pathogenic Listeria, as demonstrated by Manqele et al. (2024) [12] and Parra-Flores et al. (2022) [51], who detected fosX in 100% of isolates from beef and ready-to-eat foods, respectively. Even in highly diverse sample sets—including 2431 isolates across Europe [8] and 322 Chinese isolates from meat, aquatic products, and RTE foods [33]—fosX prevalence consistently approached 100 %. No difference in fosX prevalence has been observed between food-derived and clinical isolates (both ~100%), underscoring its status as a core, chromosomal gene.
Standard antimicrobial resistance gene databases are used to search for fosX genes in L. monocytogenes sequences. Commonly used tools such as ResFinder (from CGE), the CARD database, and BIGSdb-Lm routinely include fosX for Listeria. Specifically, ResFinder identified fosX as the sole fosfomycin resistance gene in isolates from food and food-processing environments. Likewise, BIGSdb-Lm (Pasteur Institute’s Listeria scheme) annotates fosX as an intrinsic resistance locus. In a Polish study, BIGSdb-Lm scanning flagged fosX in all 91 isolates [36]. The study by Manqele et al. (2024) [12] screened their Illumina assemblies with ABRicate using ResFinder, CARD, and the NCBI AMR database; all three tools recovered fosX in every genome. Thus, whether using BLAST/ABRicate on ResFinder/CARD or querying the BIGSdb-Lm allele database, fosX is consistently recovered.
Although the fosX gene is present in almost 100% of L. monocytogenes isolates, data on phenotypic resistance to fosfomycin remain limited and inconclusive. Genotype–phenotype correlations are not well documented—fosX is common even when the strain is phenotypically susceptible to fosfomycin [8,74,75]. It is worth including this in the research analyses of phenotypic resistance to fosfomycin in strains containing this gene, as well as studies on changes in gene expression under the influence of various stress factors. However, the presence of fosX is considered an important marker of core resistance, which may be relevant for predictive analyses (genotype–phenotype) and the assessment of the natural tolerance range of the species [76]. Moreover, the gene fosX and phenotypic resistance to fosfomycin were detected [65,68]. Scortti et al. (2018) [74] described that, despite fosX gene expression, L. monocytogenes isolates were susceptible to the antibiotic fosfomycin due to epistasis, but only during infection.
3.2.2. Lincosamides Resistance
The resistance of L. monocytogenes to lincosamides is based, among other things, on mechanisms that modify the target site of the antibiotic, i.e., the ribosome. The major gene responsible for lincosamide resistance is the lin gene, which encodes the enzyme nucleotidyl transferase (LinA), which inactivates the antibiotic by adenylation of its molecule. The lin gene is responsible for the direct chemical modification of the structure of lincosamides, resulting in their neutralization [77,78,79]. In the analyzed studies, the lin gene was detected in 35 cases (35/58; 60.34%) [8,12,14,15,16,18,20,28,30,31,33,34,35,36,37,38,40,42,43,44,45,48,49,51,52,53,57,58,61,63,64,65,66,68]. Among the bioinformatic tools most commonly used for its detection, BIGSdb-Lm dominated and was used in 14 publications (14/35; 40.00%) [8,16,18,20,28,33,34,35,36,37,38,43,45,66]. It was followed by ABRicate, used in nine cases (9/35; 25.71%) [14,15,42,44,48,49,58,65,66], and ResFinder, present in six studies (7/35; 20.00%) [12,15,38,49,57,63]. The lin gene has been detected, among others, in L. monocytogenes isolates from fish and seafood [36,68], meat and meat products [12,35,44,45,47], vegetables and fruits [16,37,49,51], spices and plant products [31], food-production environments [14,34,36,65], poultry [28], and ready-to-eat products [61].
The erm (erythromycin r-methyltransferase) family of genes, particularly ermB, which encodes ribosomal methylases that modify 23S rRNA, play a key role here. This change prevents the antibiotic from binding to the ribosome, leading to MLSB (macrolides-lincosamides-streptogramins B) resistance [77,80,81]. The fact that the ermB gene is detected, among others, in L. monocytogenes isolates from the food-processing environment suggests a horizontal transfer of resistance, especially by transposons such as Tn917 or Tn1545 class elements [82]. The results of our analysis indicated that the ermB gene was detected in two cases (2/58; 3.45%). Both publications used the CARD program (2/2; 100.00%) as a tool for identifying resistance genes [8,60]. It has also been shown that the presence of ermB may correlate with the tetM gene, indicating co-selection of resistance to tetracyclines and lincosamides [83].
The vga(G) gene is another gene that determines phenotypic resistance to lincosamides [84]. In a study conducted by Gana et al. (2024) [26], the vga(G) gene, which encodes an ABC-F family ribosome protection protein, was detected in all 60 L. monocytogenes isolates analyzed. These results are consistent with observations from South Africa, where some antibiotics are available without a prescription and are used in animal husbandry for both therapeutic purposes and as growth promoters [26]. Similarly, Lee and Park (2024) [39] report the presence of the vga(G) gene in L. monocytogenes isolates, confirming the widespread distribution of lincosamide resistance determinants.
The next gene responsible for encoding resistance to lincosamides is lunG, discovered by Moura et al. (2024) [8]. This gene encodes the enzyme nucleotidyl transferase, which is responsible for inactivating antibiotics from this group through their adenylation. This process leads to a reduction in the therapeutic efficacy of lincosamides, which poses a significant threat from the point of view of clinical therapy [85]. Importantly, the same study also demonstrated the presence of other genes associated with antibiotic resistance, such as aphA, dfrD, ermB, mphB, and pbp, which encodes the mechanisms determining resistance to aminoglycosides, trimethoprim, macrolides, and β-lactams, among others. The presence of such a wide repertoire of resistance determinants indicates the high potential of the studied strains to inhibit the action of many classes of antibiotics, which significantly hinders their control and emphasizes the need to monitor the spread of these genes in clinical and non-clinical environments [8].
3.2.3. Resistance to Fluoroquinolones
The norB gene is responsible for encoding a chromosomally located multidrug efflux pump, classified within the Major Facilitator Superfamily (MFS), and is associated with resistance to fluoroquinolones, particularly conferring resistance to both hydrophobic quinolones, such as sparfloxacin and moxifloxacin, and hydrophilic quinolones, including norfloxacin and ciprofloxacin [86,87].
In the analyzed studies, the norB gene was detected in 33 cases (33/58; 56.90%) [8,12,14,15,16,18,20,23,24,28,29,33,34,35,36,37,38,39,40,43,44,45,47,48,51,54,57,60,63,64,65,66,68]. Among the bioinformatic tools most commonly used for its detection, BIGSdb-Lm dominated, used in 15 publications (15/33; 45.45%) [8,16,18,20,28,33,34,35,36,37,38,43,64,66,88], next, CARD (10/33; 30.30%) [12,15,51,54,57,60,63,64,65,68]. Following were ABRicate, used in seven cases (7/33; 21.21%) [14,15,44,48,63,65,66] and ResFinder used in six works (6/33; 18.19%) [12,15,38,48,57,63]. The following were used in isolated cases: MEGARes database [83], AMRFinderPlus [40], Listeria MLST database, AMRFinderPlus [39], Listeria MLST database [40], ARG-ANNOT [65]. The norB gene was detected, among others, in L. monocytogenes isolates from slaughterhouses, meat, and meat products [15,20,28,35,44], fish and seafood [20,33,66,68], vegetables and fruit [16,34,37,51], and food-production environments [34] as well as ready-to-eat products [8].
Other genes responsible for fluoroquinolone resistance are as follows: GYRA_23 [29,47], parC [28], and fepA [48].
The GYRA_23 gene, the presence of which has been linked to fluoroquinolone resistance, was detected in strains isolated from dairy products in studies conducted by Mejía et al. (2023) [47] and Haubert et al. (2018) [29]. However, analysis using different databases revealed discrepancies in its identification—the gene was not shown in the CARD database, while it was identified by MEGARes [47].
A study by Guidi et al. (2023) [28] demonstrated the presence of genes determining quinolone resistance in selected populations of L. monocytogenes; in addition to the common AMR genes, the authors observed the presence of the parC gene (encoding the C subunit of topoisomerase II), responsible for quinolone resistance, in all isolates studied.
In contrast, Mota et al. (2020) [48] identified the fepA gene as an important factor in L. monocytogenes’ resistance to fluoroquinolones. The mechanism is mainly related to the activity of pumps that pump the antibiotic out of the cell—the overexpression of fepA enables efficient removal of the drug, reducing its intracellular concentration [89]. The genomic analyses also searched for sequences in the NCBI BLAST database, which allowed us to identify additional genes responsible for resistance, including fepA, which—despite their description in the literature—are not always listed in commonly used databases such as ResFinder, CARD, NCBI AMRFinderPlus, or MEGARes.
3.2.4. Antimicrobial Peptides Resistance
The mprF gene encodes lysylphosphatidylglycerol synthetase, an enzyme that uses phosphatidylglycerol as its substrate. This reaction leads to the formation of lysyl-phosphatidylglycerol and diphosphatidylglycerol, both of which are essential structural components of the cell wall. As a result, bacteria exhibit resistance primarily to cationic antimicrobial peptides (CAMPs), such as defensins and cathelicidins, as well as to clinically used peptide antibiotics, particularly daptomycin [90,91].
The presence of the mprF gene was confirmed in 22 out of 58 studies (37.93%) [8,12,14,15,16,23,28,30,33,36,38,39,43,44,47,51,60,63,64,65,66,68,69]. For its identification, various bioinformatic tools were applied, with the most frequently used being the BIGSdb-Lm database—10/22 (45.45%) [16,33,36,38,43,64,65,66,88,92] and CARD—10/22 (45.45%) [15,47,51,60,63,64,65,68,69,93]. The other tools included ABRicate—6/22 (27.27%) [14,15,44,48,59,65], and ResFinder—5/22 (22.73%) [15,38,44,51,63]. Less frequently, AMRFinderPlus was used—3/22 (13.64%) [15,39,51], NCBI (BLASTN)—2/22 (9.09%) [44,60], while RGI [93] and ARIBA [47] appeared only once each, in 1 out of 22 studies (4.55%).
The sources of isolation in these studies covered a wide spectrum of samples from food and production environments. The isolates came from three fruit packing plants [14], a meat-processing factory [15], leafy, root, and seed vegetables [16], ready-to-eat meat products [88], broilers [92], raw meat, RTE foods and production environments [33,36,51], fish and seafood products [38], vacuum-packed salmon [43], meat samples and pork-production environments [44], beef products and their preparations [12], cheese [47], cow’s milk [60], dairy farms [63], fruits and berries [65], fish and the fish processing environment [66], bivalves [68], and traditional pork products and their production environment [69].
Other genes associated with peptide antibiotic resistance were the CdsA (cardiolipin synthase A) gene [94] and the PmrF (phosphoethanolamine transferase) gene [95], detected in only one study (1/58; 1.72%) [60]. The same study also reported an A-Ala-D-Ala fragment, indicating a mechanism of resistance to glycopeptides. The I-Sanger platform, developed by Shanghai Majorbio BioTech Co., Ltd. in Shanghai, China, was used to perform bioinformatics analyses [60].
3.2.5. Sulfonamides Resistance
Genes from the sul family encode modified variants of the dihydropteroate synthase (DHPS) enzyme, which plays a key role in folate biosynthesis. Normally, sulfonamides inhibit DHPS activity through competitive bidding, but sul gene products have low affinity for sulfonamides while maintaining enzymatic activity, resulting in complete resistance to this group of drugs [96,97].
In the 58 publications analyzed, genes associated with sulfonamide resistance (sul) were detected in 15 cases (15/58; 25.86%) [8,16,18,23,33,34,35,36,37,38,39,40,43,45,66]. Among the bioinformatics tools most commonly used for its detection, BIGSdb-Lm dominated, used in 13 publications (13/15; 86.67%) [8,16,18,33,34,39,40,43,45,60,66,68,88]. Each of the other programs mentioned—ResFinder, ABRicate, CARD, and AMRFinderPlus—were each labeled in one study (1/15; 6.67% each). Specific attributions are as follows: ResFinder [38], ABRicate [66], CARD, and AMRFinderPlus [39].
3.2.6. Tetracycline Resistance
Tetracycline resistance is the most frequently observed antimicrobial resistance trait among acquired antibiotic resistance determinants in L. monocytogenes isolates, occurring in both clinical and environmental sources, including food-derived strains [83,98,99]. The primary mechanism underlying this resistance involves the acquisition of genes such as tetM, tetS, and tetA, which are commonly associated with mobile genetic elements, particularly conjugative transposons from the Tn916–1545 family and plasmids facilitating horizontal gene transfer across bacterial species [99]. Among these, tetM is the most prevalent tetracycline resistance genotype identified in L. monocytogenes strains. It encodes a ribosomal protection protein [8,44] and has been detected in over 70% of tetracycline-resistant isolates from both environmental and clinical sources, highlighting its dominant role in the spread of resistance [83,99]. Whole genome sequencing enables not only the accurate identification of genes such as tetM, tetS, and tetA, but also provides insight into their genetic context, such as their localization on plasmids, integration within resistance islands, and proximity to other resistance or virulence determinants [51,100]. Our analysis showed that the most commonly used tools for detecting tetracycline resistance genes in L. monocytogenes strains were as follows: NCBI BLASTN, CLC Genomics Workbench, BIGSdb-Lm, MEGARes database, ResFinder, CARD, and ARIBA for rapid sequence screening against curated databases of known resistance gene variants.
In the 58 publications analyzed, genes associated with tetracycline resistance were detected in 20 studies (20/58; 34.48%) [8,19,22,24,25,29,30,33,40,41,44,46,47,51,56,59,60,61,67,92]. The most frequently identified gene was tetM, present in 13 studies (13/58; 22.41%) [8,19,24,29,30,33,40,44,46,56,59,60,67]. Next in frequency were tetA (6/58; 10.34%) [24,28,41,46,51,60], and tetS (5/58; 8.62%) [33,44,47,60,67]. TetR was reported less frequently (2/58; 3.45%) [22,61]. There were also single or very rare tet variants, including tetC [51,60], tetD, tetT, tet(42), and various tetA and tetB alleles (e.g., tetA(60), tetA(48), tetB(60), tetB(P))—most of these variants were listed in a detailed allele list published by Su et al. (2023) [60]; tetK was reported in Shen et al. (2022) [56].
An analysis of the bioinformatic tools used for tet gene detection in publications revealed diverse approaches: BIGSdb-Lm was used in 3 out of 20 studies (3/20; 15.0%) [8,28,33], ResFinder (including use via ARIBA/ABRicate) in four studies (4/20; 20.0%) [19,47,59,67], while ABRicate was reported in four studies (4/20; 20.0%) [44,56,59,67]. The CARD program (including RGI/CARD or references to the CARD database within pipelines) appeared in six papers (6/20; 30.0%) [30,41,47,51,60,67]. AMRFinderPlus was identified in two studies (2/20; 10.0%) [51,61], while NCBI BLASTN/NCBI Pathogen Detection appeared in two studies (2/20; 10.0%) [22,44]. CLC Genomics Workbench appeared in one study [24]. The remaining publications used combinations of databases (MEGARes, BacMet, ARIBA, etc.) or described gene aggregate results in a non-standard way (e.g., studies containing extensive lists of allelic variants).
3.2.7. Aminoglycosides Resistance
In the analyzed studies, the aacA4 gene, associated with resistance to aminoglycosides, was detected in four cases (4/58; 6.90%) [8,33,35,66]. The aacA4 gene encodes the aac(6′)-Ib7 enzyme, a key factor in mediating bacterial resistance to aminoglycosides, including tobramycin, kanamycin, and neomycin [101]. This enzyme modifies aminoglycoside molecules by acetylating the amino group at the 6′ position, a change that prevents the antibiotic from binding to the 30S ribosomal subunit. As a result, protein synthesis is disrupted, and the bacteria acquire resistance [102]. Among the bioinformatics tools most commonly used for its detection, BIGSdb-Lm dominated, being used in all cases (4/4; 100.00%) [8,33,35,66]. In addition, Abricate Software was also used in one publication (1/4; 25.00%; [66]). The aacA4 gene was most frequently detected among L. monocytogenes isolates from raw meat and ready-to-eat products [33,35,66].
Other resistance mechanisms have also been reported for aminoglycosides. Only in 1 of the 58 studies analyzed (1/58; 1.72%) were the presence of the ant(9)-Ia, ant(6)-Ia, and aph(3′)-IIIa genes detected, which encode enzymes that modify aminoglycoside molecules and lead to their inactivation. Bioinformatic analyses were performed using the I-Sanger platform (Shanghai Majorbio BioTech Co., Ltd., Shanghai, China) [60].
In a study conducted by Yan et al. (2019) [67], involving the isolation of L. monocytogenes strains from various food products in China, the aadE, ant9, and aph3 genes were identified, which encode the aminoglycoside O-nucleotidyl transferase enzyme. The presence of these genes indicates a potential mechanism for the resistance of the tested strains to streptomycin. In contrast, Shen et al. (2022) [56] identified, among others, the aph(4)-Ia gene, encoding aminoglycoside-inactivating aminoglycoside phosphotransferase, and the ermC gene, responsible for 23S rRNA methylation and causing the MLS_B resistance phenotype (resistance to macrolides, lincosamides, and type B streptogramins, including resistance to erythromycin).
3.2.8. Efflux Proteins/Multidrug Resistance
The mdrL gene was detected in six cases (6/58; 10.34%) [14,17,27,41,46,48], while the mepA, lde, and msrA genes appeared in three, four, and four publications, respectively, (mepA: Choi et al., 2023 [24]; Haubert et al., 2018 [29]; Su et al., 2023 [60]; lde: Gelbicova et al., 2019 [27]; Lim et al., 2016 [41]; Matle et al., 2019 [46]; Mota et al., 2020 [48]; msrA: Guidi et al., 2023 [28]; Haubert et al., 2018 [29]; Lim et al., 2016 [41]; Matle et al., 2019 [46]).
Among the bioinformatics tools used for detection of these genes, BIGSdb-Lm was actually used only in a minority of the relevant studies (mdrL: 1/6, 16.7%—Gelbicova et al., 2019 [27]; lde: 1/4, 25.0%—Gelbicova et al., 2019 [27]; msrA: 1/4, 25.0%—Guidi et al., 2023 [28]). ABRicate was used in two of the mdrL-positive studies (2/6, 33.3%—Méndez Acevedo et al., 2024 [14]; Mota et al., 2020 [48]). The CARD (or CARD-based RID/AMR callers) appears in single mdrL-positive studies (1/6, 16.7%—Lim et al., 2016 [41]) and was also the tool reported for detecting mepA in Su et al., 2023 [60] (mepA: 1/3, 33.3%). Other databases/tools reported across these papers include MEGARes, BacMet, Bionumerics, and the CLC Genomics Workbench.
3.2.9. Other Genes Detected in the Analyzed Studies
In the analyzed publications, the ACC-3 gene (encoding aminoglycoside-modifying enzymes [101]) was reported in only one study (1/58; 1.72%) [60]. In addition, the SRT-1 gene, which is also responsible for resistance to this class of antibiotics, was detected. In addition, the same study detected the presence of the Erm(34), ErmH, and ErmB genes, which encode rRNA methyltransferases [103]. The ErmB gene in particular was identified in most strains exhibiting phenotypic resistance to erythromycin, as confirmed, among others, in the NDDQ-065-1 strain described by Su et al. (2023) [60]. In addition, the presence of the LpeA gene, which also determines resistance to macrolides, was detected. The study also identified genes conferring resistance to other classes of antibiotics. These include the TaeA gene, which encodes resistance to pleuromutilins [104], and the Vatl gene, which confers resistance to streptogramins [105]. Bioinformatic analyses were performed using the I-Sanger platform (Shanghai Majorbio BioTech Co., Ltd., Shanghai, China) [60].
A study by Yan et al. (2019) [67] showed that L. monocytogenes isolates exhibit multidrug resistance (MDR) and contain the cat gene, which confers resistance to chloramphenicol, and the erm(B) gene, which encodes resistance to macrolide antibiotics. In this study, the chloramphenicol resistance marker, cat, was consistently identified in isolates positive for the tet(S) gene, indicating possible co-occurrence and links between resistance determinants in the L. monocytogenes genome [67]. An analysis conducted by Brown et al. (2024) [22], covering L. monocytogenes strains isolated from both dairy products and the environment of food-processing plants, revealed the presence of, among others, the emrE and emrC genes. These genes are responsible for encoding efflux pumps belonging to the MFS (Major Facilitator Superfamily), which participate in the active removal of antimicrobial compounds from the cell, thereby contributing to the increased tolerance of bacteria to antibiotics and biocides. The detection of these resistance determinants was confirmed using the NCBI BLASTN tool [22].
In the genome of L. monocytogenes food isolates (in dairy products), the TUFAB_7 gene, associated with resistance to efamycin, was also detected [47]. However, analysis using different databases revealed discrepancies—the gene was not identified in the CARD database, but its presence was confirmed using the MEGARes database [47].
A study by Su et al. (2023) [60] demonstrated the presence of the abeS gene, associated with resistance to macrolides through an efflux mechanism, which reduces the effectiveness of these antibiotics [106]. The same study also identified other resistance genes (arlR, bcrA, dfrG, fdP, kdpE, liaR, liaS, lmrC, lmrD, lnuB, IsaE, mdtL, patB, rphB, rpoB, and folp), which determine, among other things, the modification of the target site of action of drugs, the activation of efflux pumps, and the regulation of the response to antibiotic stress. Importantly, the lmrB gene, also associated with macrolide resistance, was previously described by Lim et al. (2016) [41], confirming its wider prevalence and role in the development of multidrug resistance. Such a wide repertoire of determinants highlights the high potential of the studied strains to develop multidrug resistance, posing a serious challenge for antimicrobial therapies [41,60].
Choi et al. (2023) [24] described L. monocytogenes isolates from fungi in which numerous genes associated with β-lactam antibiotic resistance were found, such as pbp, pbpX, pbpH, ponA, pbpB, and pbpF. In addition, genes associated with clindamycin resistance (emrY, emrB) were identified, as well as multidrug resistance determinants, including mdrL and norM, responsible for the action of efflux pumps. The results of the genomic analysis suggest that the L. monocytogenes SMFM2019-FV16 strain may be resistant to penicillin, tetracycline, and clindamycin.
Hauber et al. (2018) [29] highlighted the importance of single mutations in shaping the resistance profile of L. monocytogenes. It has been shown that changes in the efTu gene (C4881_02615) lead to bacterial resistance to elfamycin. Importantly, a comparative analysis showed that the reference genome of the L. monocytogenes EGD-e strain, which is used as a reference in numerous studies, lacks three gene-encoding efflux pumps identified in other isolates. This means that their presence is not a universal feature of the entire species but rather indicates the adaptive differentiation of individual strains in response to environmental and antibiotic stress [29].
Matle et al. (2019) [46] showed that all L. monocytogenes isolates in this study contained the mecC gene, which encodes a mechanism of resistance to β-lactam antibiotics [107]—the primary group of drugs used to treat listeriosis in humans, often in combination with aminoglycosides. In addition, the lmrB gene, responsible for resistance to lincomycin through an efflux pump mechanism, was detected. The presence of these genes indicates that L. monocytogenes possesses both genetic determinants of resistance to therapeutically critical antibiotics and mechanisms that enable tolerance to other antimicrobial agents. This accumulation of resistance genes highlights the potential clinical threat and the importance of genomic monitoring of environmental and food strains for their resistance profile [46].
Ji et al. (2023) [33] identified ten genes responsible for antibiotic resistance, including lmo0441 (penicillin-binding protein—PBP-like; associated with resistance to cephalosporins) [33]. Similar observations were reported by Kragh et al. (2024) [34], who detected the lmo0441 gene in all analyzed isolates, indicating its role in determining resistance to cephalosporins. Similarly, Astoshkin et al. (2021) [20] confirmed the presence of the lmo0441 gene, proving its widespread occurrence in various populations of L. monocytogenes. These results indicate that lmo0441 may be a key determinant of cephalosporin resistance within the species [20].
4. Epidemiological and Health Consequences of Detected Resistance Patterns
The detection of resistance genes in L. monocytogenes using NGS methods is an important element of contemporary epidemiological research and public health surveillance. The results of genomic analyses reveal both the scale of the spread of resistant strains in the food environment and the potential clinical consequences for patients infected with this microorganism [83,108]. Research confirms that resistance genes are often transferred by plasmids and mobile elements, which promotes their rapid spread among both environmental bacteria and clinical pathogens [98,109]. From a clinical perspective, ampicillin or penicillin in combination with gentamicin remains the standard treatment, but reported cases of aminoglycoside resistance limit the effectiveness of treatment [110]. Infections caused by resistant strains may be more severe, with a higher risk of recurrence and higher mortality, especially among immunocompromised patients, pregnant women, and newborns [111].
At the population level, the detection of resistance patterns in L. monocytogenes indicates growing challenges in controlling this bacterium throughout the food chain. Strains resistant to tetracyclines, macrolides, and fluoroquinolones are increasingly isolated in food-production environments, which increases the risk of their transmission to consumers [112]. Geographical diversity is also significant. Regions with the intensive use of antibiotics in animal husbandry have a higher incidence of antibiotic resistance [113]. Furthermore, resistant strains can persist for long periods in food-production facilities, increasing their epidemic potential. NGS not only enables the identification of resistance patterns but also allows the precise linking of clinical infection sources to specific production lines, which significantly supports preventive measures [114].
5. Conclusions and Limitations of the Research
Whole-genome sequencing (WGS) is an effective tool for detecting both innate and acquired resistance determinants in L. monocytogenes originating from food and processing environments. The studies analyzed showed that the fosX gene is virtually ubiquitous as part of the core genome, while other resistance genes show variable frequencies depending on the sample type and region of origin of the strain. Phenotype prediction based on sequence is high in most cases, but the observed discrepancies highlight the need to supplement genotypic analyses with phenotypic testing before making clinical or regulatory decisions.
A review of the literature identifies several key methodological limitations. The most frequently highlighted problem is the heterogeneity of data reporting—many studies do not provide complete phenotypic results, which prevents direct verification of the accuracy of genotype–phenotype predictions. Differences in the sequencing platforms used and depth of coverage affect data quality and may lead to the overlooking of less-common resistance determinants, especially those located on mobile genetic elements. Furthermore, the mere presence of a gene in the genome does not necessarily correlate with the phenotype–gene expression and regulation, as well as point mutations in target genes (e.g., penicillin-binding proteins) that can significantly affect antibiotic susceptibility, and standard bioinformatics procedures often do not take this into account. Models combining genotype and phenotype data are also prone to systematic errors resulting from unrepresentative samples or poor generalization between bacterial populations. The lack of standardized reporting protocols and unified databases limits the comparability of studies.
In view of the above, it is recommended to implement uniform reporting standards (including the mandatory sharing of phenotypic results), develop and validate consistent bioinformatics tools, and build extensive, consolidated databases. Where possible, long-read sequencing or hybrid approaches should be used to better determine the genomic context of resistance determinants. Future research should focus on studying the expression and regulatory mechanisms of common resistance loci, explaining the reasons for genotype–phenotype discordance, and evaluating the practical aspects of implementing NGS in food safety systems.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms262010112/s1.
Author Contributions
Conceptualization, P.W.; methodology, P.W.; validation, P.W.; formal analysis, P.W.; investigation, P.W.; resources, P.W.; data curation, P.W., P.A., and M.T.; writing—original draft preparation, P.W., P.A., and M.T.; writing—review and editing, P.W., A.Z., and W.C.-W.; visualization, P.W.; funding acquisition, P.W. and A.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Science Centre (Poland), grant number 2024/53/N/NZ9/00833 (PRELUDIUM-23). The APC was funded by the Minister of Science under the Regional Initiative of Excellence Program.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in this study are included in the article and Supplementary Material. Further inquiries can be directed to the corresponding author.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Manyi-Loh, C.E.; Okoh, A.I.; Lues, R. Occurrence and Multidrug Resistance in Strains of Listeria monocytogenes Recovered from the Anaerobic Co-Digestion Sludge Contained in a Single Stage Steel Biodigester: Implications for Antimicrobial Stewardship. Microorganisms 2023, 11, 725. [Google Scholar] [CrossRef]
- Rippa, A.; Bilei, S.; Peruzy, M.F.; Marrocco, M.G.; Leggeri, P.; Bossù, T.; Murru, N. Antimicrobial Resistance of Listeria monocytogenes Strains Isolated in Food and Food-Processing Environments in Italy. Antibiotics 2024, 13, 525. [Google Scholar] [CrossRef]
- Castello, A.; Alio, V.; Torresi, M.; Centorotola, G.; Chiaverini, A.; Pomilio, F.; Arrigo, I.; Giammanco, A.; Fasciana, T.; Ortoffi, M.F.; et al. Molecular Characterization and Antimicrobial Resistance Evaluation of Listeria monocytogenes Strains from Food and Human Samples. Pathogens 2025, 14, 294. [Google Scholar] [CrossRef]
- European Food Safety Authority (EFSA); European Centre for Disease Prevention and Control (ECDC). The European Union One Health 2023 Zoonoses Report. Eur. Food Saf. Auth. 2024, 22, 62–73. [Google Scholar] [CrossRef]
- Pagliano, P.; Arslan, F.; Ascione, T. Epidemiology and Treatment of the Commonest Form of Listeriosis: Meningitis and Bacteraemia. Infez. Med. 2017, 25, 210–216. [Google Scholar]
- Kayode, A.J.; Okoh, A.I. Antimicrobial-Resistant Listeria monocytogenes in Ready-to-Eat Foods: Implications for Food Safety and Risk Assessment. Foods 2023, 12, 1346. [Google Scholar] [CrossRef] [PubMed]
- Wiśniewski, P.; Zakrzewski, A.J.; Zadernowska, A.; Chajęcka-Wierzchowska, W. Antimicrobial Resistance and Virulence Characterization of Listeria monocytogenes Strains Isolated from Food and Food Processing Environments. Pathogens 2022, 11, 1099. [Google Scholar] [CrossRef]
- Moura, A.; Leclercq, A.; Vales, G.; Tessaud-Rita, N.; Bracq-Dieye, H.; Thouvenot, P.; Madec, Y.; Charlier, C.; Lecuit, M. Phenotypic and Genotypic Antimicrobial Resistance of Listeria monocytogenes: An Observational Study in France. Lancet Reg. Health. Eur. 2024, 37, 100800. [Google Scholar] [CrossRef]
- European Committee on Antimicrobial Susceptibility Testing to Clinical Colleagues: On Recent Changes in Clinical Microbiology Susceptibility Reports—New Interpretation of Susceptibility Categories S, I and R. 2021. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Guidance_documents/To_clinical_colleagues_on_recent_changes_in_clinical_microbiology_susceptibility_reports_9_July2021.pdf (accessed on 9 August 2025).
- Ellington, M.J.; Ekelund, O.; Aarestrup, F.M.; Canton, R.; Doumith, M.; Giske, C.; Grundman, H.; Hasman, H.; Holden, M.T.G.; Hopkins, K.L.; et al. The Role of Whole Genome Sequencing in Antimicrobial Susceptibility Testing of Bacteria: Report from the EUCAST Subcommittee. Clin. Microbiol. Infect. 2017, 23, 2–22. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Xin, X.; Cao, X.; Nasifu, L.; Nie, Z.; He, B. Phenotypic and Genotypic Perspectives on Detection Methods for Bacterial Antimicrobial Resistance in a One Health Context: Research Progress and Prospects. Arch. Microbiol. 2024, 206, 409. [Google Scholar] [CrossRef]
- Manqele, A.; Adesiyun, A.; Mafuna, T.; Pierneef, R.; Moerane, R.; Gcebe, N. Virulence Potential and Antimicrobial Resistance of Listeria monocytogenes Isolates Obtained from Beef and Beef-Based Products Deciphered Using Whole-Genome Sequencing. Microorganisms 2024, 12, 1166. [Google Scholar] [CrossRef] [PubMed]
- Lakicevic, B.; Jankovic, V.; Pietzka, A.; Ruppitsch, W. Wholegenome Sequencing as the Gold Standard Approach for Control of Listeria monocytogenes in the Food Chain. J. Food Prot. 2023, 86, 100003. [Google Scholar] [CrossRef] [PubMed]
- Méndez Acevedo, M.; Rolon, M.L.; Johnson, B.B.; Burns, L.H.; Stacy, J.; Aurand-Cravens, A.; LaBorde, L.; Kovac, J. Sanitizer Resistance and Persistence of Listeria monocytogenes Isolates in Tree Fruit Packing Facilities. J. Food Prot. 2024, 87, 100354. [Google Scholar] [CrossRef]
- Alvarez-Molina, A.; Cobo-Díaz, J.F.; López, M.; Prieto, M.; de Toro, M.; Alvarez-Ordóñez, A. Unraveling the Emergence and Population Diversity of Listeria monocytogenes in a Newly Built Meat Facility through Whole Genome Sequencing. Int. J. Food Microbiol. 2021, 340, 109043. [Google Scholar] [CrossRef]
- Amarasekara, N.R.; Swamy, A.S.; Paudel, S.K.; Jiang, W.; Li, K.; Shen, C.; Zhang, Y. Hypervirulent Clonal Complex (CC) of Listeria monocytogenes in Fresh Produce from Urban Communities. Front. Microbiol. 2024, 15, 1307610. [Google Scholar] [CrossRef]
- Andritsos, N.D.; Mataragas, M. Characterization and Antibiotic Resistance of Listeria monocytogenes Strains Isolated from Greek Myzithra Soft Whey Cheese and Related Food Processing Surfaces over Two-and-a-Half Years of Safety Monitoring in a Cheese Processing Facility. Foods 2023, 12, 1200. [Google Scholar] [CrossRef]
- Aslantaş, Ö.; Büyükaltay, K.; Keskin, O.; Güllü Yücetepe, A.; Adigüzel, A. Whole-Genome Sequencing-Based Characterization of Listeria monocytogenes from Food and Animal Clinical Cases. Kafkas. Univ. Vet. Fak. Derg. 2023, 29, 221–230. [Google Scholar] [CrossRef]
- Assisi, C.; Forauer, E.; Oliver, H.F.; Etter, A.J. Genomic and Transcriptomic Analysis of Biofilm Formation in Persistent and Transient Listeria monocytogenes Isolates from the Retail Deli Environment Does Not Yield Insight into Persistence Mechanisms. Foodborne Pathog. Dis. 2021, 18, 179–188. [Google Scholar] [CrossRef]
- Astashkin, E.I.; Alekseeva, E.A.; Borzenkov, V.N.; Kislichkina, A.A.; Mukhina, T.N.; Platonov, M.E.; Svetoch, E.A.; Shepelin, A.P.; Fursova, N.K. Molecular-Genetic Characteristics of Polyresistant Listeria monocytogenes Strains and Identification of New Sequence Types. Mol. Genet. Microbiol. Virol. 2021, 36, 159–169. [Google Scholar] [CrossRef]
- Bai, Y.; Li, J.; Huang, M.; Yan, S.; Li, F.; Xu, J.; Peng, Z.; Wang, X.; Ma, J.; Sun, J.; et al. Prevalence and Characterization of Foodborne Pathogens Isolated from Fresh-Cut Fruits and Vegetables in Beijing, China. Int. J. Food Microbiol. 2024, 421, 110804. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.R.B.; Bland, R.; McIntyre, L.; Shyng, S.; Weisberg, A.J.; Riutta, E.R.; Chang, J.H.; Kovacevic, J. Genomic Characterization of Listeria monocytogenes Recovered from Dairy Facilities in British Columbia, Canada from 2007 to 2017. Front. Microbiol. 2024, 15, 1304734. [Google Scholar] [CrossRef]
- Centorotola, G.; Guidi, F.; D’aurizio, G.; Salini, R.; Di Domenico, M.; Ottaviani, D.; Petruzzelli, A.; Fisichella, S.; Duranti, A.; Tonucci, F.; et al. Intensive Environmental Surveillance Plan for Listeria monocytogenes in Food Producing Plants and Retail Stores of Central Italy: Prevalence and Genetic Diversity. Foods 2021, 10, 1944. [Google Scholar] [CrossRef]
- Choi, S.; Choi, Y.; Seo, Y.; Lee, S.; Yoon, Y. Identification of Genetic Variations Related to Pathogenicity by Whole Genome Sequencing of Listeria monocytogenes SMFM2019-FV16 Isolated from Enoki Mushroom. J. Food Saf. 2023, 43, 13076. [Google Scholar] [CrossRef]
- Fox, E.M.; Casey, A.; Jordan, K.; Coffey, A.; Gahan, C.G.M.; McAuliffe, O. Whole Genome Sequence Analysis; an Improved Technology That Identifies Underlying Genotypic Differences between Closely Related Listeria monocytogenes Strains. Innov. Food Sci. Emerg. Technol. 2017, 44, 89–96. [Google Scholar] [CrossRef]
- Gana, J.; Gcebe, N.; Pierneef, R.E.; Chen, Y.; Moerane, R.; Adesiyun, A.A. Whole Genome Sequence Analysis of Listeria monocytogenes Isolates Obtained from the Beef Production Chain in Gauteng Province, South Africa. Microorganisms 2024, 12, 1003. [Google Scholar] [CrossRef] [PubMed]
- Gelbicova, T.; Florianova, M.; Tomastikova, Z.; Pospisilova, L.; Kolackova, I.; Karpiskova, R. Prediction of Persistence of Listeria monocytogenes ST451 in a Rabbit Meat Processing Plant in the Czech Republic. J. Food Prot. 2019, 82, 1350–1356. [Google Scholar] [CrossRef]
- Guidi, F.; Centorotola, G.; Chiaverini, A.; Iannetti, L.; Schirone, M.; Visciano, P.; Cornacchia, A.; Scattolini, S.; Pomilio, F.; D’Alterio, N.; et al. The Slaughterhouse as Hotspot of CC1 and CC6 Listeria monocytogenes Strains with Hypervirulent Profiles in an Integrated Poultry Chain of Italy. Microorganisms 2023, 11, 1543. [Google Scholar] [CrossRef] [PubMed]
- Haubert, L.; Kremer, F.S.; da Silva, W.P. Whole-Genome Sequencing Identification of a Multidrug-Resistant Listeria monocytogenes Serotype 1/2a Isolated from Fresh Mixed Sausage in Southern Brazil. Infect. Genet. Evol. 2018, 65, 127–130. [Google Scholar] [CrossRef] [PubMed]
- Hong, H.; Yang, S.M.; Kim, E.; Kim, H.J.; Park, S.H. Comprehensive Metagenomic Analysis of Stress-Resistant and -Sensitive Listeria monocytogenes. Appl. Microbiol. Biotechnol. 2023, 107, 6047–6056. [Google Scholar] [CrossRef]
- Hu, L.; Brown, E.W.; Zhang, G. Diversity of Antimicrobial Resistance, Stress Resistance, and Virulence Factors of Salmonella, Shiga Toxin-Producing Escherichia coli, and Listeria monocytogenes from Produce, Spices, and Tree Nuts by Whole Genome Sequencing. Front. Sustain. Food Syst. 2023, 7, 360–368. [Google Scholar] [CrossRef]
- Hurley, D.; Luque-Sastre, L.; Parker, C.T.; Huynh, S.; Eshwar, A.K.; Nguyen, S.V.; Andrews, N.; Moura, A.; Fox, E.M.; Jordan, K.; et al. Whole-Genome Sequencing-Based Characterization of 100 Listeria monocytogenes Isolates Collected from Food Processing Environments over a Four-Year Period. mSphere 2019, 4, e00252-19. [Google Scholar] [CrossRef]
- Ji, S.; Song, Z.; Luo, L.; Wang, Y.; Li, L.; Mao, P.; Ye, C.; Wang, Y. Whole-Genome Sequencing Reveals Genomic Characterization of Listeria monocytogenes from Food in China. Front. Microbiol. 2023, 13, 1049843. [Google Scholar] [CrossRef] [PubMed]
- Kragh, M.L.; Ivanova, M.; Dalgaard, P.; Leekitcharoenphon, P.; Hansen, L.T. Sensitivity of 240 Listeria monocytogenes Isolates to Common Industrial Biocides Is More Dependent on the Presence of Residual Organic Matter or Biofilm than on Genetic Determinants. Food Control 2024, 158, 110244. [Google Scholar] [CrossRef]
- Kurpas, M.; Osek, J.; Moura, A.; Leclercq, A.; Lecuit, M.; Wieczorek, K. Genomic Characterization of Listeria monocytogenes Isolated from Ready-to-Eat Meat and Meat Processing Environments in Poland. Front. Microbiol. 2020, 11, 1412. [Google Scholar] [CrossRef]
- Lachtara, B.; Osek, J.; Wieczorek, K. Molecular Typing of Listeria monocytogenes IVb Serogroup Isolated from Food and Food Production Environments in Poland. Pathogens 2021, 10, 482. [Google Scholar] [CrossRef]
- Lake, F.B.; van Overbeek, L.S.; Baars, J.J.P.; Koomen, J.; Abee, T.; den Besten, H.M.W. Genomic Characteristics of Listeria monocytogenes Isolated during Mushroom (Agaricus bisporus) Production and Processing. Int. J. Food Microbiol. 2021, 360, 109438. [Google Scholar] [CrossRef]
- Lambrechts, K.; Gouws, P.; Rip, D. Genetic Diversity of Listeria monocytogenes from Seafood Products, Its Processing Environment, and Clinical Origin in the Western Cape, South Africa Using Whole Genome Sequencing. AIMS Microb. 2024, 10, 608–643. [Google Scholar] [CrossRef]
- Lee, M.R.; Park, K.T. Whole-Genome Analysis of CC224 Listeria monocytogenes Strain IJPL9-1, Clonally Related to the Listeriosis Outbreak Strain in 2018, Isolated from Pork in Korea. Microbiol. Biotechnol. Lett. 2024, 52, 328–330. [Google Scholar] [CrossRef]
- Lee, S.H.; Lee, S.; Park, S.H.; Koo, O.K. Whole-Genome Sequencing of Listeria monocytogenes Isolated from the First Listeriosis Foodborne Outbreak in South Korea. Front. Microbiol. 2023, 14, 1182090. [Google Scholar] [CrossRef]
- Lim, S.Y.; Yap, K.-P.; Thong, K.L. Comparative Genomics Analyses Revealed Two Virulent Listeria monocytogenes Strains Isolated from Ready-to-Eat Food. Gut Pathog. 2016, 8, 65. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lüth, S.; Halbedel, S.; Rosner, B.; Wilking, H.; Holzer, A.; Roedel, A.; Dieckmann, R.; Vincze, S.; Prager, R.; Flieger, A.; et al. Backtracking and Forward Checking of Human Listeriosis Clusters Identified a Multiclonal Outbreak Linked to Listeria monocytogenes in Meat Products of a Single Producer. Emerg. Microbes Infect. 2020, 9, 1600–1608. [Google Scholar] [CrossRef]
- Mäesaar, M.; Mamede, R.; Elias, T.; Roasto, M. Retrospective Use of Whole-Genome Sequencing Expands the Multicountry Outbreak Cluster of Listeria monocytogenes St1247. Int. J. Genom. 2021, 2021, 6636138. [Google Scholar] [CrossRef]
- Mafuna, T.; Matle, I.; Magwedere, K.; Pierneef, R.E.; Reva, O.N. Whole Genome-Based Characterization of Listeria monocytogenes Isolates Recovered from the Food Chain in South Africa. Front. Microbiol. 2021, 12, 669287. [Google Scholar] [CrossRef]
- Markovich, Y.; Palacios-Gorba, C.; Gomis, J.; Gómez-Martín, Á.; Ortolá, S.; Quereda, J.J. Phenotypic and Genotypic Antimicrobial Resistance of Listeria spp. in Spain. Vet. Microbiol. 2024, 293, 110086. [Google Scholar] [CrossRef]
- Matle, I.; Pierneef, R.; Mbatha, K.R.; Magwedere, K.; Madoroba, E. Genomic Diversity of Common Sequence Types of Listeria monocytogenes Isolated from Ready-to-Eat Products of Animal Origin in South Africa. Genes 2019, 10, 1007. [Google Scholar] [CrossRef]
- Mejía, L.; Espinosa-Mata, E.; Freire, A.L.; Zapata, S.; González-Candelas, F. Listeria monocytogenes, a Silent Foodborne Pathogen in Ecuador. Front. Microbiol. 2023, 14, 1278860. [Google Scholar] [CrossRef]
- Mota, M.I.; Vázquez, S.; Cornejo, C.; D’Alessandro, B.; Braga, V.; Caetano, A.; Betancor, L.; Varela, G. Does Shiga Toxin-Producing Escherichia coli and Listeria monocytogenes Contribute Significantly to the Burden of Antimicrobial Resistance in Uruguay? Front. Vet. Sci. 2020, 7, 583930. [Google Scholar] [CrossRef] [PubMed]
- Narvaez, S.A.; Shen, Z.; Yan, L.; Stenger, B.L.S.; Goodman, L.B.; Lim, A.; Nissly, R.H.; Nair, M.S.; Zhang, S.; Sanchez, S. Optimized Conditions for Listeria, Salmonella and Escherichia Whole Genome Sequencing Using the Illumina iSeq100 Platform with Point-and-Click Bioinformatic Analysis. PLoS ONE 2022, 17, e0277659. [Google Scholar] [CrossRef] [PubMed]
- Brown, P.; Chen, Y.; Parsons, C.; Brown, E.; Loessner, M.J.; Shen, Y.; Kathariou, S. Whole Genome Sequence Analysis of Phage-Resistant Listeria monocytogenes Serotype 1/2a Strains from Turkey Processing Plants. Pathogens 2021, 10, 199. [Google Scholar] [CrossRef] [PubMed]
- Parra-Flores, J.; Holý, O.; Bustamante, F.; Lepuschitz, S.; Pietzka, A.; Contreras-Fernández, A.; Castillo, C.; Ovalle, C.; Alarcón-Lavín, M.P.; Cruz-Córdova, A.; et al. Virulence and Antibiotic Resistance Genes in Listeria monocytogenes Strains Isolated from Ready-to-Eat Foods in Chile. Front. Microbiol. 2022, 12, 796040. [Google Scholar] [CrossRef] [PubMed]
- Brown, P.; Chen, Y.; Siletzky, R.; Parsons, C.; Jaykus, L.-A.; Eifert, J.; Ryser, E.; Logue, C.M.; Stam, C.; Brown, E.; et al. Harnessing Whole Genome Sequence Data for Facility-Specific Signatures for Listeria monocytogenes: A Case Study with Turkey Processing Plants in the United States. Front. Sustain. Food Syst. 2021, 5, 742353. [Google Scholar] [CrossRef]
- Daza Prieto, B.; Pietzka, A.; Martinovic, A.; Ruppitsch, W.; Zuber Bogdanovic, I. Surveillance and Genetic Characterization of Listeria monocytogenes in the Food Chain in Montenegro during the Period 2014–2022. Front. Microbiol. 2024, 15, 1418333. [Google Scholar] [CrossRef]
- Qi, Y.; Cao, Q.; Zhao, X.; Tian, C.; Li, T.; Shi, W.; Wei, H.; Song, C.; Xue, H.; Gou, H. Comparative Genomic Analysis of Pathogenic Factors of Listeria spp. Using Whole-Genome Sequencing. BMC Genom. 2024, 25, 935. [Google Scholar] [CrossRef]
- Roedel, A.; Dieckmann, R.; Brendebach, H.; Hammerl, J.A.; Kleta, S.; Noll, M.; Al Dahouk, S.; Vinczea, S. Biocide-Tolerant Listeria monocytogenes Isolates from German Food Production Plants Do Not Show Crossresistance to Clinically Relevant Antibiotics. Appl. Environ. Microbiol. 2019, 85, e01253-19. [Google Scholar] [CrossRef]
- Shen, J.; Zhang, G.; Yang, J.; Zhao, L.; Jiang, Y.; Guo, D.; Wang, X.; Zhi, S.; Xu, X.; Dong, Q.; et al. Prevalence, Antibiotic Resistance, and Molecular Epidemiology of Listeria monocytogenes Isolated from Imported Foods in China during 2018 to 2020. Int. J. Food Microbiol. 2022, 382, 109916. [Google Scholar] [CrossRef] [PubMed]
- Silva, A.; Silva, V.; Gomes, J.P.; Coelho, A.; Batista, R.; Saraiva, C.; Esteves, A.; Martins, Â.; Contente, D.; Diaz-Formoso, L.; et al. Listeria monocytogenes from Food Products and Food Associated Environments: Antimicrobial Resistance, Genetic Clustering and Biofilm Insights. Antibiotics 2024, 13, 447. [Google Scholar] [CrossRef]
- Smith, A.; Hearn, J.; Taylor, C.; Wheelhouse, N.; Kaczmarek, M.; Moorhouse, E.; Singleton, I. Listeria monocytogenes Isolates from Ready to Eat Plant Produce Are Diverse and Have Virulence Potential. Int. J. Food Microbiol. 2019, 299, 23–32. [Google Scholar] [CrossRef]
- Song, Z.; Ji, S.; Wang, Y.; Luo, L.; Wang, Y.; Mao, P.; Li, L.; Jiang, H.; Ye, C. The Population Structure and Genetic Diversity of Listeria monocytogenes ST9 Strains Based on Genomic Analysis. Front. Microbiol. 2022, 13, 982220. [Google Scholar] [CrossRef] [PubMed]
- Su, R.; Wen, Y.; Prabakusuma, A.S.; Tang, X.; Huang, A.; Li, L. Prevalence, Antibiotic Resistance and Virulence Feature of Listeria monocytogenes Isolated from Bovine Milk in Yunnan, Southwest China. Int. Dairy J. 2023, 144, 105703. [Google Scholar] [CrossRef]
- Takeuchi-Storm, N.; Hansen, L.T.; Nielsen, N.L.; Andersen, J.K. Presence and Persistence of Listeria monocytogenes in the Danish Ready-to-Eat Food Production Environment. Hygiene 2023, 3, 18–32. [Google Scholar] [CrossRef]
- Toledo, V.; den Bakker, H.C.; Hormazábal, J.C.; González-Rocha, G.; Bello-Toledo, H.; Toro, M.; Moreno-Switt, A.I. Genomic Diversity of Listeria monocytogenes Isolated from Clinical and Non-Clinical Samples in Chile. Genes 2018, 9, 396. [Google Scholar] [CrossRef] [PubMed]
- Varsaki, A.; Ortiz, S.; Santorum, P.; López, P.; López-Alonso, V.; Hernández, M.; Abad, D.; Rodríguez-Grande, J.; Ocampo-Sosa, A.A.; Martínez-Suárez, J.V. Prevalence and Population Diversity of Listeria monocytogenes Isolated from Dairy Cattle Farms in the Cantabria Region of Spain. Animals 2022, 12, 2477. [Google Scholar] [CrossRef] [PubMed]
- Voronina, O.L.; Kunda, M.S.; Ryzhova, N.N.; Aksenova, E.I.; Kustova, M.A.; Karpova, T.I.; Melkumyan, A.R.; Klimova, E.A.; Gruzdeva, O.A.; Tartakovsky, I.S. Listeria monocytogenes ST37 Distribution in the Moscow Region and Properties of Clinical and Foodborne Isolates. Life 2023, 13, 2167. [Google Scholar] [CrossRef]
- Wartha, S.; Bretschneider, N.; Dangel, A.; Hobmaier, B.; Hörmansdorfer, S.; Huber, I.; Murr, L.; Pavlovic, M.; Sprenger, A.; Wenning, M.; et al. Genetic Characterization of Listeria from Food of Non-Animal Origin Products and from Producing and Processing Companies in Bavaria, Germany. Foods 2023, 12, 1120. [Google Scholar] [CrossRef]
- Wieczorek, K.; Bomba, A.; Osek, J. Whole-Genome Sequencing-Based Characterization of Listeria monocytogenes from Fish and Fish Production Environments in Poland. Int. J. Mol. Sci. 2020, 21, 9419. [Google Scholar] [CrossRef]
- Yan, S.; Li, M.; Luque-Sastre, L.; Wang, W.; Hu, Y.; Peng, Z.; Dong, Y.; Gan, X.; Nguyen, S.; Anes, J.; et al. Susceptibility (Re)-Testing of a Large Collection of Listeria monocytogenes from Foods in China from 2012 to 2015 and WGS Characterization of Resistant Isolates. J. Antimicrob. Chemother. 2019, 74, 1786–1794. [Google Scholar] [CrossRef]
- Zakrzewski, A.J.; Gajewska, J.; Chajęcka-Wierzchowska, W.; Zadernowska, A. Insights into the Genetic Diversity of Listeria monocytogenes from Bivalves. Sci. Total Environ. 2024, 908, 168481. [Google Scholar] [CrossRef]
- Zuber, I.; Lakicevic, B.; Pietzka, A.; Milanov, D.; Djordjevic, V.; Karabasil, N.; Teodorovic, V.; Ruppitsch, W.; Dimitrijevic, M. Molecular Characterization of Listeria monocytogenes Isolates from a Small-Scale Meat Processor in Montenegro, 2011–2014. Food Microbiol. 2019, 79, 116–122. [Google Scholar] [CrossRef]
- Schirmer, M.; D’Amore, R.; Ijaz, U.Z.; Hall, N.; Quince, C. Illumina Error Profiles: Resolving Fine-Scale Variation in Metagenomic Sequencing Data. BMC Bioinform. 2016, 17, 125. [Google Scholar] [CrossRef] [PubMed]
- Commichaux, S.; Javkar, K.; Ramachandran, P.; Nagarajan, N.; Bertrand, D.; Chen, Y.; Reed, E.; Gonzalez-Escalona, N.; Strain, E.; Rand, H.; et al. Evaluating the Accuracy of Listeria monocytogenes Assemblies from Quasimetagenomic Samples Using Long and Short Reads. BMC Genom. 2021, 22, 389. [Google Scholar] [CrossRef]
- Lee, Y.; Son, B.; Cha, Y.; Ryu, S. Characterization and Genomic Analysis of PALS2, a Novel Staphylococcus Jumbo Bacteriophage. Front. Microbiol. 2021, 12, 622755. [Google Scholar] [CrossRef]
- Rigsby, R.E.; Fillgrove, K.L.; Beihoffer, L.A.; Armstrong, R.N. Fosfomycin Resistance Proteins: A Nexus of Glutathione Transferases and Epoxide Hydrolases in a Metalloenzyme Superfamily. Methods Enzymol. 2005, 401, 367–379. [Google Scholar]
- Scortti, M.; Han, L.; Alvarez, S.; Leclercq, A.; Moura, A.; Lecuit, M.; Vazquez-Boland, J. Epistatic Control of Intrinsic Resistance by Virulence Genes in Listeria. PLoS Genet. 2018, 14, e1007525, Erratum in: PLoS Genet. 2018, 14, e1007727. [Google Scholar] [CrossRef]
- Wiśniewski, P.; Chajęcka-Wierzchowska, W.; Zadernowska, A. High-Pressure Processing—Impacts on the Virulence and Antibiotic Resistance of Listeria monocytogenes Isolated from Food and Food Processing Environments. Foods 2023, 12, 3899. [Google Scholar] [CrossRef]
- Kinde, M.Z.; Kerisew, B.; Eshetu, T.; Gessese, A.T. Genomic Analysis of Listeria monocytogenes Strains from Dairy Products in Ethiopia. Front. Bioinform. 2025, 5, 1572241. [Google Scholar] [CrossRef] [PubMed]
- Leclercq, R. Mechanisms of Resistance to Macrolides and Lincosamides: Nature of the Resistance Elements and Their Clinical Implications. Clin. Infect. Dis. 2002, 34, 482–492. [Google Scholar] [CrossRef] [PubMed]
- Petinaki, E.; Guérin-Faublée, V.; Pichereau, V.; Villers, C.; Achard, A.; Malbruny, B.; Leclercq, R. Lincomycin Resistance Gene Lnu (D) in Streptococcus uberis. Antimicrob. Agents Chemother. 2008, 52, 626–630. [Google Scholar] [CrossRef] [PubMed]
- Morar, M.; Bhullar, K.; Hughes, D.W.; Junop, M.; Wright, G.D. Structure and Mechanism of the Lincosamide Antibiotic Adenylyltransferase LinB. Structure 2009, 17, 1649–1659. [Google Scholar] [CrossRef]
- Vester, B.; Douthwaite, S. Macrolide Resistance Conferred by Base Substitutions in 23S rRNA. Antimicrob. Agents Chemother. 2001, 45, 1–12. [Google Scholar] [CrossRef]
- Roberts, M.C.; Facinelli, B.; Giovanetti, E.; Varaldo, P.E. Transferable Erythromycin Resistance in Listeria spp. Isolated from Food. Appl. Environ. Microbiol. 1996, 62, 269–270. [Google Scholar] [CrossRef]
- Wilson, A.; Gray, J.; Chandry, P.S.; Fox, E.M. Phenotypic and Genotypic Analysis of Antimicrobial Resistance among Listeria monocytogenes Isolated from Australian Food Production Chains. Genes 2018, 9, 80. [Google Scholar] [CrossRef]
- Haubert, L.; Mendonça, M.; Lopes, G.V.; de Itapema Cardoso, M.R.; da Silva, W.P. Listeria monocytogenes Isolates from Food and Food Environment Harbouring tetM and ermB Resistance Genes. Lett. Appl. Microbiol. 2016, 62, 23–29. [Google Scholar] [CrossRef]
- Novotna, G.; Janata, J. A New Evolutionary Variant of the Streptogramin a Resistance Protein, Vga(A)LC, from Staphylococcus haemolyticus with Shifted Substrate Specificity towards Lincosamides. Antimicrob. Agents Chemother. 2006, 50, 4070–4076. [Google Scholar] [CrossRef]
- Zhu, X.-Q.; Wang, X.-M.; Li, H.; Shang, Y.-H.; Pan, Y.-S.; Wu, C.-M.; Wang, Y.; Du, X.-D.; Shen, J.-Z. Novel Lnu (G) Gene Conferring Resistance to Lincomycin by Nucleotidylation, Located on Tn 6260 from Enterococcus faecalis E531. J. Antimicrob. Chemother. 2016, 72, 993–997. [Google Scholar] [CrossRef]
- Ding, Y.; Onodera, Y.; Lee, J.C.; Hooper, D.C. NorB, an Efflux Pump in Staphylococcus aureus Strain MW2, Contributes to Bacterial Fitness in Abscesses. J. Bacteriol. 2008, 190, 7123–7129. [Google Scholar] [CrossRef] [PubMed]
- Stephen, J.; Salam, F.; Lekshmi, M.; Kumar, S.H.; Varela, M.F. The Major Facilitator Superfamily and Antimicrobial Resistance Efflux Pumps of the ESKAPEE Pathogen Staphylococcus aureus. Antibiotics 2023, 12, 343. [Google Scholar] [CrossRef] [PubMed]
- Centorotola, G.; Ziba, M.W.; Cornacchia, A.; Chiaverini, A.; Torresi, M.; Guidi, F.; Cammà, C.; Bowa, B.; Mtonga, S.; Magambwa, P.; et al. Listeria monocytogenes in Ready to Eat Meat Products from Zambia: Phenotypical and Genomic Characterization of Isolates. Front. Microbiol. 2023, 14, 1228726. [Google Scholar] [CrossRef]
- Guérin, F.; Galimand, M.; Tuambilangana, F.; Courvalin, P.; Cattoir, V. Overexpression of the Novel MATE Fluoroquinolone Efflux Pump FepA in Listeria monocytogenes Is Driven by Inactivation of Its Local Repressor FepR. PLoS ONE 2014, 9, e106340. [Google Scholar] [CrossRef]
- Nowak, J.; Visnovsky, S.B.; Cruz, C.D.; Fletcher, G.C.; van Vliet, A.H.M.; Hedderley, D.; Butler, R.; Flint, S.; Palmer, J.; Pitman, A.R. Inactivation of the Gene Encoding the Cationic Antimicrobial Peptide Resistance Factor MprF Increases Biofilm Formation but Reduces Invasiveness of Listeria monocytogenes. J. Appl. Microbiol. 2021, 130, 464–477. [Google Scholar] [CrossRef]
- Bao, Y.; Sakinc, T.; Laverde, D.; Wobser, D.; Benachour, A.; Theilacker, C.; Hartke, A.; Huebner, J. Role of mprF1 and mprF2 in the Pathogenicity of Enterococcus faecalis. PLoS ONE 2012, 7, e38458. [Google Scholar] [CrossRef]
- Guidi, F.; Lorenzetti, C.; Centorotola, G.; Torresi, M.; Cammà, C.; Chiaverini, A.; Pomilio, F.; Blasi, G. Atypical Serogroup IVb-v1 of Listeria monocytogenes Assigned to New ST2801, Widely Spread and Persistent in the Environment of a Pork-Meat Producing Plant of Central Italy. Front. Microbiol. 2022, 13, 930895. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Moon, J.-S.; Lee, Y.J.; Kim, H.-Y. Whole-Genome Sequencing-Based Characterization of Listeria monocytogenes Isolated from Cattle and Pig Slaughterhouses. Infect. Genet. Evol. 2025, 130, 105737. [Google Scholar] [CrossRef]
- Tan, B.K.; Bogdanov, M.; Zhao, J.; Dowhan, W.; Raetz, C.R.H.; Guan, Z. Discovery of a Cardiolipin Synthase Utilizing Phosphatidylethanolamine and Phosphatidylglycerol as Substrates. Proc. Natl. Acad. Sci. USA 2012, 109, 16504–16509. [Google Scholar] [CrossRef]
- Lee, H.; Hsu, F.-F.; Turk, J.; Groisman, E.A. The PmrA-Regulated pmrC Gene Mediates Phosphoethanolamine Modification of Lipid A and Polymyxin Resistance in Salmonella enterica. J. Bacteriol. 2004, 186, 4124–4133. [Google Scholar] [CrossRef]
- Razavi, M.; Marathe, N.P.; Gillings, M.R.; Flach, C.-F.; Kristiansson, E.; Joakim Larsson, D.G. Discovery of the Fourth Mobile Sulfonamide Resistance Gene. Microbiome 2017, 5, 160. [Google Scholar] [CrossRef]
- Sköld, O. Sulfonamide Resistance: Mechanisms and Trends. Drug Resist. Updat. 2000, 3, 155–160. [Google Scholar] [CrossRef] [PubMed]
- Adamski, P.; Zakrzewski, A.; Wiśniewski, P.; Chajęcka-Wierzchowska, W.; Zadernowska, A. High-Pressure Processing (HPP) Alters Tetracycline Resistance in Listeria monocytogenes: A Phenotypic and Genotypic Study. NFS J. 2025, 38, 100223. [Google Scholar] [CrossRef]
- Bertsch, D.; Uruty, A.; Anderegg, J.; Lacroix, C.; Perreten, V.; Meile, L. Tn6198, a Novel Transposon Containing the Trimethoprim Resistance Gene dfrG Embedded into a Tn916 Element in Listeria monocytogenes. J. Antimicrob. Chemother. 2013, 68, 986–991. [Google Scholar] [CrossRef]
- Oniciuc, E.A.; Likotrafiti, E.; Alvarez-Molina, A.; Prieto, M.; Santos, J.A.; Alvarez-Ordóñez, A. The Present and Future of Whole Genome Sequencing (WGS) and Whole Metagenome Sequencing (WMS) for Surveillance of Antimicrobial Resistant Microorganisms and Antimicrobial Resistance Genes across the Food Chain. Genes 2018, 9, 268. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, M.S.; Tolmasky, M.E. Aminoglycoside Modifying Enzymes. Drug Resist. Updat. 2010, 13, 151–171. [Google Scholar] [CrossRef] [PubMed]
- Rather, P.N.; Munayyer, H.; Mann, P.A.; Hare, R.S.; Miller, G.H.; Shaw, K.J. Genetic Analysis of Bacterial Acetyltransferases: Identification of Amino Acids Determining the Specificities of the Aminoglycoside 6′-N-Acetyltransferase Lb and Ila Proteins. J. Bacteriol. 1992, 174, 3196–3203. [Google Scholar] [CrossRef][Green Version]
- Dzyubak, E.; Yap, M.-N.F. The Expression of Antibiotic Resistance Methyltransferase Correlates with mRNA Stability Independently of Ribosome Stalling. Antimicrob. Agents Chemother. 2016, 60, 7178–7188. [Google Scholar] [CrossRef]
- Pawlowski, A.C.; Wang, W.; Koteva, K.; Barton, H.A.; McArthur, A.G.; Wright, G.D. A Diverse Intrinsic Antibiotic Resistome from a Cave Bacterium. Nat. Commun. 2016, 7, 13803. [Google Scholar] [CrossRef]
- Li, Q.; Pellegrino, J.; Lee, D.J.; Tran, A.A.; Chaires, H.A.; Wang, R.; Park, J.E.; Ji, K.; Chow, D.; Zhang, N.; et al. Synthetic Group A Streptogramin Antibiotics That Overcome Vat Resistance. Nature 2020, 586, 145–150. [Google Scholar] [CrossRef]
- Srinivasan, V.B.; Rajamohan, G.; Gebreyes, W.A. Role of AbeS, a Novel Efflux Pump of the SMR Family of Transporters, in Resistance to Antimicrobial Agents in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2009, 53, 5312–5316. [Google Scholar] [CrossRef]
- Idrees, M.M.; Saeed, K.; Shahid, M.A.; Akhtar, M.; Qammar, K.; Hassan, J.; Khaliq, T.; Saeed, A. Prevalence of mecA- and mecC-Associated Methicillin-Resistant Staphylococcus aureus in Clinical Specimens, Punjab, Pakistan. Biomedicines 2023, 11, 878. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Lv, M.; Yang, K.; Zhao, G.; Fu, Y. A Case Report of Diagnosis and Dynamic Monitoring of Listeria monocytogenes Meningitis with NGS. Open Life Sci. 2023, 18, 20220738. [Google Scholar] [CrossRef]
- Shi, Q.; Zhang, X.; Wang, D.; Zhang, W.; Jin, X.; Sun, Y.; Huang, A. Genetic Diversity, Antimicrobial Resistance, and Virulence Profiles of Listeria monocytogenes Isolates from Nantong, China (2020–2023). Foodborne Pathog. Dis. 2025. [Google Scholar] [CrossRef] [PubMed]
- Andriyanov, P.A.; Zhurilov, P.A.; Liskova, E.A.; Karpova, T.I.; Sokolova, E.V.; Yushina, Y.K.; Zaiko, E.V.; Bataeva, D.S.; Voronina, O.L.; Psareva, E.K.; et al. Antimicrobial Resistance of Listeria monocytogenes Strains Isolated from Humans, Animals, and Food Products in Russia in 1950–1980, 2000–2005, and 2018–2021. Antibiotics 2021, 10, 1206. [Google Scholar] [CrossRef] [PubMed]
- Shamloo, E.; Hosseini, H.; Abdi Moghadam, Z.; Halberg Larsen, M.; Haslberger, A.; Alebouyeh, M. Importance of Listeria monocytogenes in Food Safety: A Review of Its Prevalence, Detection, and Antibiotic Resistance. Iran. J. Vet. Res. 2019, 20, 241–254. [Google Scholar]
- Sołtysiuk, M.; Przyborowska, P.; Wiszniewska-Łaszczych, A.; Tobolski, D. Prevalence and Antimicrobial Resistance Profile of Listeria spp. Isolated from Raw Fish. BMC Vet. Res. 2025, 21, 333. [Google Scholar] [CrossRef] [PubMed]
- Wiśniewski, P.; Trymers, M.; Chajęcka-Wierzchowska, W.; Tkacz, K.; Zadernowska, A.; Modzelewska-Kapituła, M. Antimicrobial Resistance in the Context of Animal Production and Meat Products in Poland—A Critical Review and Future Perspective. Pathogens 2024, 13, 1123. [Google Scholar] [CrossRef] [PubMed]
- Bradford, D.; Lodhi, K.; Yuan, J.; Graham, D.; Graham, J.; Maldani, M.; White, E.; Arhin, A.; Kassem, M. Comparative Genomics of Foodborne Pathogens: Diversity, Virulence, and Epidemiological Relevance. Am. J. Bot. 2025, 846–855. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).