Advances and Applications of Plant Base Editing Technologies
Abstract
1. Introduction
2. Technical Advances
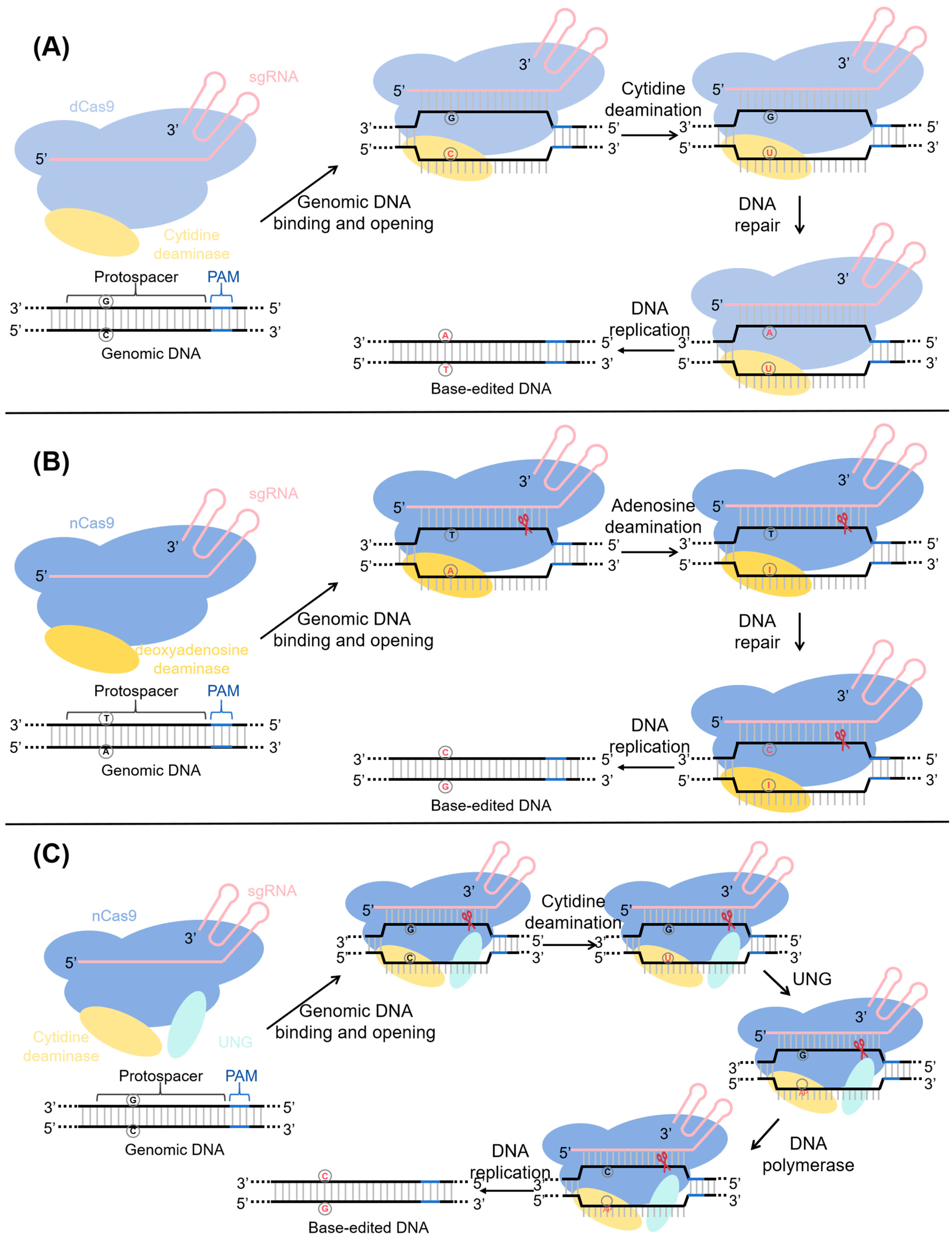
2.1. Cytosine Base Editing System (CBE)
2.2. Adenine Base Editing System (ABE)
2.3. The Glycosylase Base Editor (GBE)
3. Application of Base Editing Technologies in Plants
3.1. Optimization of Base Editing Technologies in Plants
3.2. Application of Base Editing Technology in Rice and Wheat
3.3. Application of Base Editing Technology in Other Crops
4. Prospects of Base Editing Technology in Crop Applications
4.1. Limitations of Base Editing Technology
4.1.1. Factors Affecting Editing Efficiency
4.1.2. Limitations of PAM Regions
4.1.3. Limitations of the Editing Window
4.1.4. Limited Types of Base Substitutions
4.1.5. Off-Target Effects
4.2. Summary and Outlook
Funding
Conflicts of Interest
References
- Durai, S.; Mani, M.; Kandavelou, K.; Wu, J.; Porteus, M.H.; Chandrasegaran, S. Zinc finger nucleases: Custom-designed molecular scissors for genome engineering of plant and mammalian cells. Nucleic Acids Res. 2005, 33, 5978–5990. [Google Scholar] [CrossRef] [PubMed]
- Joung, J.K.; Sander, J.D. TALENs: A widely applicable technology for targeted genome editing. Nat. Rev. Mol. Cell Biol. 2013, 14, 49–55. [Google Scholar] [CrossRef]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef]
- Lieber, M.R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 2010, 79, 181–211. [Google Scholar] [CrossRef]
- Zhao, H.Y.; Xu, D.Y. Selection and regulation of DNA double-strand break repair pathways. Sci. Sin. Vitae 2021, 51, 56–69. (In Chinese) [Google Scholar]
- Henikoff, S.; Comai, L. Single-nucleotide mutations for plant functional genomics. Annu. Rev. Plant Biol. 2003, 54, 375–401. [Google Scholar] [CrossRef]
- Huq, M.A.; Akter, S.; Nou, I.S.; Kim, H.T.; Jung, Y.J.; Kang, K.K. Identification of functional SNPs in genes and their effects on plant phenotypes. J. Plant Biotechnol. 2016, 43, 1–11. [Google Scholar] [CrossRef]
- Miura, K.; Ikeda, M.; Matsubara, A.; Song, X.J.; Ito, M.; Asano, K.; Matsuoka, M.; Kitano, H.; Ashikari, M. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 2010, 42, 545–549. [Google Scholar] [CrossRef]
- Su, Z.; Hao, C.; Wang, L.; Dong, Y.; Zhang, X. Identification and development of a functional marker of TaGW2 associated with grain weight in bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 2011, 122, 211–223. [Google Scholar] [CrossRef]
- Komor, A.C.; Kim, Y.B.; Packer, M.S.; Zuris, J.A.; Liu, D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016, 533, 420–424. [Google Scholar] [CrossRef] [PubMed]
- Komor, A.C.; Zhao, K.T.; Packer, M.S.; Gaudelli, N.M.; Waterbury, A.L.; Koblan, L.W.; Kim, Y.B.; Badran, A.H.; Liu, D.R. Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C:G-to-T:A base editors with higher efficiency and product purity. Sci. Adv. 2017, 3, eaao4774. [Google Scholar] [CrossRef] [PubMed]
- Koblan, L.W.; Doman, J.L.; Wilson, C.; Levy, J.M.; Tay, T.; Newby, G.A.; Maianti, J.P.; Raguram, A.; Liu, D.R. Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction. Nat. Biotechnol. 2018, 36, 843–846. [Google Scholar] [CrossRef]
- Gehrke, J.M.; Cervantes, O.; Clement, M.K.; Wu, Y.; Zeng, J.; Bauer, D.E.; Pinello, L.; Joung, J.K. An APOBEC3A-Cas9 base editor with minimized bystander and off-target activities. Nat. Biotechnol. 2018, 36, 977–982. [Google Scholar] [CrossRef]
- Thuronyi, B.W.; Koblan, L.W.; Levy, J.M.; Yeh, W.H.; Zheng, C.; Newby, G.A.; Wilson, C.; Bhaumik, M.; Shubina-Oleinik, O.; Holt, J.R.; et al. Continuous evolution of base editors with expanded target compatibility and improved activity. Nat. Biotechnol. 2019, 37, 1070–1079, Correction in Nat. Biotechnol. 2019, 37, 1091. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhu, B.; Ru, G.; Meng, H.; Yan, Y.; Hong, M.; Zhang, D.; Luan, C.; Zhang, S.; Wu, H.; et al. Re-engineering the adenine deaminase TadA-8e for efficient and specific CRISPR-based cytosine base editing. Nat. Biotechnol. 2023, 41, 663–672, Correction in Nat. Biotechnol. 2024, 42, 987. [Google Scholar] [CrossRef]
- Neugebauer, M.E.; Hsu, A.; Arbab, M.; Krasnow, N.A.; McElroy, A.N.; Pandey, S.; Doman, J.L.; Huang, T.P.; Raguram, A.; Banskota, S.; et al. Evolution of an adenine base editor into a small, efficient cytosine base editor with low off-target activity. Nat. Biotechnol. 2023, 41, 673–685. [Google Scholar] [CrossRef] [PubMed]
- Gaudelli, N.M.; Komor, A.C.; Rees, H.A.; Packer, M.S.; Badran, A.H.; Bryson, D.I.; Liu, D.R. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 2017, 551, 464–471. [Google Scholar] [CrossRef]
- Gaudelli, N.M.; Lam, D.K.; Rees, H.A.; Solá-Esteves, N.M.; Barrera, L.A.; Born, D.A.; Edwards, A.; Gehrke, J.M.; Lee, S.J.; Liquori, A.J.; et al. Directed evolution of adenine base editors with increased activity and therapeutic application. Nat. Biotechnol. 2020, 38, 892–900. [Google Scholar] [CrossRef]
- Richter, M.F.; Zhao, K.T.; Eton, E.; Lapinaite, A.; Newby, G.A.; Thuronyi, B.W.; Wilson, C.; Koblan, L.W.; Zeng, J.; Bauer, D.E.; et al. Phage-assisted evolution of an adenine base editor with improved Cas domain compatibility and activity. Nat. Biotechnol. 2020, 38, 883–891, Correction in Nat. Biotechnol. 2020, 38, 901. [Google Scholar] [CrossRef]
- Zhao, D.; Li, J.; Li, S.; Xin, X.; Hu, M.; Price, M.A.; Rosser, S.J.; Bi, C.; Zhang, X. Glycosylase base editors enable C-to-A and C-to-G base changes. Nat. Biotechnol. 2021, 39, 35–40, Correction in Nat. Biotechnol. 2021, 39, 115. [Google Scholar] [CrossRef]
- Kurt, I.C.; Zhou, R.; Iyer, S.; Garcia, S.P.; Miller, B.R.; Langner, L.M.; Grünewald, J.; Joung, J.K. CRISPR C-to-G base editors for inducing targeted DNA transversions in human cells. Nat. Biotechnol. 2021, 39, 41–46. [Google Scholar] [CrossRef]
- Sun, N.; Zhao, D.; Li, S.; Zhang, Z.; Bi, C.; Zhang, X. Reconstructed glycosylase base editors GBE2.0 with enhanced C-to-G base editing efficiency and purity. Mol. Ther. 2022, 30, 2452–2463. [Google Scholar] [CrossRef]
- Dong, X.; Yang, C.; Ma, Z.; Chen, M.; Zhang, X.; Bi, C. Enhancing glycosylase base-editor activity by fusion to transactivation modules. Cell Rep. 2022, 40, 111090. [Google Scholar] [CrossRef]
- Chen, L.; Park, J.E.; Paa, P.; Rajakumar, P.D.; Prekop, H.T.; Chew, Y.T.; Manivannan, S.N.; Chew, W.L. Programmable C:G to G:C genome editing with CRISPR-Cas9-directed base excision repair proteins. Nat. Commun. 2021, 12, 1384. [Google Scholar] [CrossRef]
- Zong, Y.; Wang, Y.; Li, C.; Zhang, R.; Chen, K.; Ran, Y.; Qiu, J.L.; Wang, D.; Gao, C. Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat. Biotechnol. 2017, 35, 438–440, Correction in Nat. Biotechnol. 2025, 43, 1011. [Google Scholar] [CrossRef]
- Zong, Y.; Song, Q.; Li, C.; Jin, S.; Zhang, D.; Wang, Y.; Qiu, J.L.; Gao, C. Efficient C-to-T base editing in plants using a fusion of nCas9 and human APOBEC3A. Nat. Biotechnol. 2018, 36, 950–953, Correction in Nat. Biotechnol. 2025, 43, 1011. [Google Scholar] [CrossRef] [PubMed]
- Zeng, D.; Liu, T.; Tan, J.; Zhang, Y.; Zheng, Z.; Wang, B.; Zhou, D.; Xie, X.; Guo, M.; Liu, Y.G.; et al. PhieCBEs: Plant High-Efficiency Cytidine Base Editors with Expanded Target Range. Mol. Plant 2020, 13, 1666–1669. [Google Scholar] [CrossRef] [PubMed]
- Nishimasu, H.; Shi, X.; Ishiguro, S.; Gao, L.; Hirano, S.; Okazaki, S.; Noda, T.; Abudayyeh, O.O.; Gootenberg, J.S.; Mori, H.; et al. Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science 2018, 361, 1259–1262. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zong, Y.; Wang, Y.; Jin, S.; Zhang, D.; Song, Q.; Zhang, R.; Gao, C. Expanded base editing in rice and wheat using a Cas9-adenosine deaminase fusion. Genome Biol. 2018, 19, 59. [Google Scholar] [CrossRef]
- Yan, F.; Kuang, Y.; Ren, B.; Wang, J.; Zhang, D.; Lin, H.; Yang, B.; Zhou, X.; Zhou, H. Highly Efficient A·T to G·C Base Editing by Cas9n-Guided tRNA Adenosine Deaminase in Rice. Mol. Plant 2018, 11, 631–634. [Google Scholar] [CrossRef]
- Wei, C.; Wang, C.; Jia, M.; Guo, H.X.; Luo, P.Y.; Wang, M.G.; Zhu, J.K.; Zhang, H. Efficient generation of homozygous substitutions in rice in one generation utilizing an rABE8e base editor. J. Integr. Plant Biol. 2021, 63, 1595–1599. [Google Scholar] [CrossRef]
- Yan, D.; Ren, B.; Liu, L.; Yan, F.; Li, S.; Wang, G.; Sun, W.; Zhou, X.; Zhou, H. High-efficiency and multiplex adenine base editing in plants using new TadA variants. Mol. Plant 2021, 14, 722–731. [Google Scholar] [CrossRef]
- Tan, J.; Zeng, D.; Zhao, Y.; Wang, Y.; Liu, T.; Li, S.; Xue, Y.; Luo, Y.; Xie, X.; Chen, L.; et al. PhieABEs: A PAM-less/free high-efficiency adenine base editor toolbox with wide target scope in plants. Plant Biotechnol. J. 2022, 20, 934–943. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Chen, L.; Zhu, B.; Wang, L.; Chen, C.; Hong, M.; Huang, Y.; Li, H.; Han, H.; Cai, B.; et al. Increasing the efficiency and targeting range of cytidine base editors through fusion of a single-stranded DNA-binding protein domain. Nat. Cell Biol. 2020, 22, 740–750. [Google Scholar] [CrossRef] [PubMed]
- Sretenovic, S.; Liu, S.; Li, G.; Cheng, Y.; Fan, T.; Xu, Y.; Zhou, J.; Zheng, X.; Coleman, G.; Zhang, Y.; et al. Exploring C-To-G Base Editing in Rice, Tomato, and Poplar. Front. Genome Ed. 2021, 3, 756766. [Google Scholar] [CrossRef]
- Tian, Y.; Shen, R.; Li, Z.; Yao, Q.; Zhang, X.; Zhong, D.; Tan, X.; Song, M.; Han, H.; Zhu, J.K.; et al. Efficient C-to-G editing in rice using an optimized base editor. Plant Biotechnol. J. 2022, 20, 1238–1240. [Google Scholar] [CrossRef]
- Fan, T.; Cheng, Y.; Wu, Y.; Liu, S.; Tang, X.; He, Y.; Liao, S.; Zheng, X.; Zhang, T.; Qi, Y.; et al. High performance TadA-8e derived cytosine and dual base editors with undetectable off-target effects in plants. Nat. Commun. 2024, 15, 5103. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Xiao, Z.; Luo, Z.; Zhou, S.; Tong, C.; Jin, S.; Liu, X.; Qin, R.; Xu, R.; Pan, L.; et al. Improving plant C-to-G base editors with a cold-adapted glycosylase and TadA-8e variants. Trends Biotechnol. 2025, 43, 1765–1787. [Google Scholar] [CrossRef]
- Li, J.; Sun, Y.; Du, J.; Zhao, Y.; Xia, L. Generation of Targeted Point Mutations in Rice by a Modified CRISPR/Cas9 System. Mol. Plant 2017, 10, 526–529. [Google Scholar] [CrossRef]
- Lu, Y.; Zhu, J.K. Precise Editing of a Target Base in the Rice Genome Using a Modified CRISPR/Cas9 System. Mol. Plant 2017, 10, 523–525. [Google Scholar] [CrossRef]
- Kuang, Y.; Li, S.; Ren, B.; Yan, F.; Spetz, C.; Li, X.; Zhou, X.; Zhou, H. Base-Editing-Mediated Artificial Evolution of OsALS1 In Planta to Develop Novel Herbicide-Tolerant Rice Germplasms. Mol. Plant 2020, 13, 565–572. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Kuang, Y.; Yan, F.; Li, S.; Ren, B.; Gosavi, G.; Spetz, C.; Li, X.; Wang, X.; Zhou, X.; et al. Developing a novel artificial rice germplasm for dinitroaniline herbicide resistance by base editing of OsTubA2. Plant Biotechnol. J. 2021, 19, 5–7. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Liu, J.; Chai, Z.; Chen, S.; Bai, Y.; Zong, Y.; Chen, K.; Li, J.; Jiang, L.; Gao, C. Generation of herbicide tolerance traits and a new selectable marker in wheat using base editing. Nat. Plants 2019, 5, 480–485. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Wu, Z.; Zheng, L.; Han, J.; Zhang, Y.; Li, J.; Wang, P. Generation of a high-efficiency adenine base editor with TadA8e for developing wheat dinitroaniline-resistant germplasm. Crop J. 2022, 10, 368–374. [Google Scholar] [CrossRef]
- Veillet, F.; Chauvin, L.; Kermarrec, M.P.; Sevestre, F.; Merrer, M.; Terret, Z.; Szydlowski, N.; Devaux, P.; Gallois, J.L.; Chauvin, J.E. The Solanum tuberosum GBSSI gene: A target for assessing gene and base editing in tetraploid potato. Plant Cell Rep. 2019, 38, 1065–1080. [Google Scholar] [CrossRef]
- Kang, B.C.; Yun, J.Y.; Kim, S.T.; Shin, Y.; Ryu, J.; Choi, M.; Woo, J.W.; Kim, J.S. Precision genome engineering through adenine base editing in plants. Nat. Plants 2018, 4, 427–431, Correction in Nat. Plants 2018, 4, 730. [Google Scholar] [CrossRef]
- Wu, J.; Chen, C.; Xian, G.; Liu, D.; Lin, L.; Yin, S.; Sun, Q.; Fang, Y.; Zhang, H.; Wang, Y. Engineering herbicide-resistant oilseed rape by CRISPR/Cas9-mediated cytosine base-editing. Plant Biotechnol. J. 2020, 18, 1857–1859. [Google Scholar] [CrossRef]
- Qin, L.; Li, J.; Wang, Q.; Xu, Z.; Sun, L.; Alariqi, M.; Manghwar, H.; Wang, G.; Li, B.; Ding, X.; et al. High-efficient and precise base editing of C•G to T•A in the allotetraploid cotton (Gossypium hirsutum) genome using a modified CRISPR/Cas9 system. Plant Biotechnol. J. 2020, 18, 45–56. [Google Scholar] [CrossRef]
- Wang, G.; Xu, Z.; Wang, F.; Huang, Y.; Xin, Y.; Liang, S.; Li, B.; Si, H.; Sun, L.; Wang, Q.; et al. Development of an efficient and precise adenine base editor (ABE) with expanded target range in allotetraploid cotton (Gossypium hirsutum). BMC Biol. 2022, 20, 45. [Google Scholar] [CrossRef]
- Li, Y.; Zhu, J.; Wu, H.; Liu, C.; Huang, C.; Lan, J.; Xie, C. Precise base editing of non-allelic acetolactate synthase genes confers sulfonylurea herbicide resistance in maize. Crop J. 2020, 8, 449–456. [Google Scholar] [CrossRef]
- Fu, X.; Wang, N.; Li, L.; Qiao, D.; Qi, X.; Liu, C.; Gao, Z.; Xie, C.; Zhu, J. Development of cytosine and adenine base editors for maize precision breeding. J. Integr. Plant Biol. 2025; early view. [Google Scholar] [CrossRef]
- Shimatani, Z.; Kashojiya, S.; Takayama, M.; Terada, R.; Arazoe, T.; Ishii, H.; Teramura, H.; Yamamoto, T.; Komatsu, H.; Miura, K.; et al. Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat. Biotechnol. 2017, 35, 441–443. [Google Scholar] [CrossRef] [PubMed]
- Veillet, F.; Perrot, L.; Chauvin, L.; Kermarrec, M.P.; Guyon-Debast, A.; Chauvin, J.E.; Nogué, F.; Mazier, M. Transgene-Free Genome Editing in Tomato and Potato Plants Using Agrobacterium-Mediated Delivery of a CRISPR/Cas9 Cytidine Base Editor. Int. J. Mol. Sci. 2019, 20, 402. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.; Jiang, L.; Cui, X.; Zhang, J.; Guo, S.; Li, M.; Zhang, H.; Ren, Y.; Gong, G.; Zong, M.; et al. Engineering herbicide-resistant watermelon variety through CRISPR/Cas9-mediated base-editing. Plant Cell Rep. 2018, 37, 1353–1356. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Yang, S.; Hao, H.; Du, W.; Pang, Y.; Zhao, S.; Zhao, X. Research Progress on the Application of Single Base Editing Technology. China Anim. Husb. Vet. Med. 2021, 48, 4403–4411. (In Chinese) [Google Scholar]
- Hua, K.; Han, P.; Zhu, J.K. Improvement of base editors and prime editors advances precision genome engineering in plants. Plant Physiol. 2022, 188, 1795–1810. [Google Scholar] [CrossRef]
- Liu, H.; Wang, K.; Jia, Z.; Gong, Q.; Lin, Z.; Du, L.; Pei, X.; Ye, X. Efficient induction of haploid plants in wheat by editing of TaMTL using an optimized Agrobacterium-mediated CRISPR system. J. Exp. Bot. 2020, 71, 1337–1349. [Google Scholar] [CrossRef]
- Nanasato, Y.; Kawabe, H.; Ueno, S.; Konagaya, K.I.; Endo, M.; Taniguchi, T. Improvement of genome editing efficiency by Cas9 codon optimization in Japanese cedar (Cryptomeria japonica D. Don). Plant Biotechnol. 2024, 41, 335–344. [Google Scholar] [CrossRef]
- Vats, S.; Kumawat, S.; Kumar, V.; Patil, G.B.; Joshi, T.; Sonah, H.; Sharma, T.R.; Deshmukh, R. Genome Editing in Plants: Exploration of Technological Advancements and Challenges. Cells 2019, 8, 1386. [Google Scholar] [CrossRef]
- Hu, J.H.; Miller, S.M.; Geurts, M.H.; Tang, W.; Chen, L.; Sun, N.; Zeina, C.M.; Gao, X.; Rees, H.A.; Lin, Z.; et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 2018, 556, 57–63. [Google Scholar] [CrossRef]
- Zhang, C.; Xu, W.; Wang, F.; Kang, G.; Yuan, S.; Lv, X.; Li, L.; Liu, Y.; Yang, J. Expanding the base editing scope to GA and relaxed NG PAM sites by improved xCas9 system. Plant Biotechnol. J. 2020, 18, 884–886. [Google Scholar] [CrossRef]
- Wang, M.; Wang, Z.; Mao, Y.; Lu, Y.; Yang, R.; Tao, X.; Zhu, J.K. Optimizing base editors for improved efficiency and expanded editing scope in rice. Plant Biotechnol. J. 2019, 17, 1697–1699. [Google Scholar] [CrossRef] [PubMed]
- Walton, R.T.; Christie, K.A.; Whittaker, M.N.; Kleinstiver, B.P. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science 2020, 368, 290–296. [Google Scholar] [CrossRef]
- Miller, S.M.; Wang, T.; Randolph, P.B.; Arbab, M.; Shen, M.W.; Huang, T.P.; Matuszek, Z.; Newby, G.A.; Rees, H.A.; Liu, D.R. Continuous evolution of SpCas9 variants compatible with non-G PAMs. Nat. Biotechnol. 2020, 38, 471–481. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, Y.; Wang, F.; Zhao, S.; Song, J.; Feng, F.; Zhao, J.; Yang, J. Expanding base editing scope to near-PAMless with engineered CRISPR/Cas9 variants in plants. Mol. Plant 2021, 14, 191–194. [Google Scholar] [CrossRef]
- Wang, X.; Ding, C.; Yu, W.; Wang, Y.; He, S.; Yang, B.; Xiong, Y.C.; Wei, J.; Li, J.; Liang, J.; et al. Cas12a Base Editors Induce Efficient and Specific Editing with Low DNA Damage Response. Cell Rep. 2020, 31, 107723. [Google Scholar] [CrossRef]
- Li, X.; Wang, Y.; Liu, Y.; Yang, B.; Wang, X.; Wei, J.; Lu, Z.; Zhang, Y.; Wu, J.; Huang, X.; et al. Base editing with a Cpf1-cytidine deaminase fusion. Nat. Biotechnol. 2018, 36, 324–327. [Google Scholar] [CrossRef] [PubMed]
- Kleinstiver, B.P.; Sousa, A.A.; Walton, R.T.; Tak, Y.E.; Hsu, J.Y.; Clement, K.; Welch, M.M.; Horng, J.E.; Malagon-Lopez, J.; Scarfò, I.; et al. Engineered CRISPR-Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nat. Biotechnol. 2019, 37, 276–282, Correction in Nat. Biotechnol. 2020, 38, 901. [Google Scholar] [CrossRef]
- Gaillochet, C.; Peña Fernández, A.; Goossens, V.; D’Halluin, K.; Drozdzecki, A.; Shafie, M.; Van Duyse, J.; Van Isterdael, G.; Gonzalez, C.; Vermeersch, M.; et al. Systematic optimization of Cas12a base editors in wheat and maize using the ITER platform. Genome Biol. 2023, 24, 6. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Zhang, Y.; Li, G.; Fang, H.; Sretenovic, S.; Fan, A.; Li, J.; Xu, J.; Que, Q.; Qi, Y. CRISPR-Cas12a base editors confer efficient multiplexed genome editing in rice. Plant Commun. 2023, 4, 100601. [Google Scholar] [CrossRef]
- Lv, P.; Su, F.; Chen, F.; Yan, C.; Xia, D.; Sun, H.; Li, S.; Duan, Z.; Ma, C.; Zhang, H.; et al. Genome editing in rice using CRISPR/Cas12i3. Plant Biotechnol. J. 2024, 22, 379–385. [Google Scholar] [CrossRef]
- Rees, H.A.; Liu, D.R. Base editing: Precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 2018, 19, 770–788, Correction in Nat. Rev. Genet. 2018, 19, 801. [Google Scholar] [CrossRef]
- Porto, E.M.; Komor, A.C.; Slaymaker, I.M.; Yeo, G.W. Base editing: Advances and therapeutic opportunities. Nat. Rev. Drug Discov. 2020, 19, 839–859. [Google Scholar] [CrossRef]
- Jeong, Y.K.; Lee, S.; Hwang, G.H.; Hong, S.A.; Park, S.E.; Kim, J.S.; Woo, J.S.; Bae, S. Adenine base editor engineering reduces editing of bystander cytosines. Nat. Biotechnol. 2021, 39, 1426–1433. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhang, S.; Xue, N.; Hong, M.; Zhang, X.; Zhang, D.; Yang, J.; Bai, S.; Huang, Y.; Meng, H.; et al. Engineering a precise adenine base editor with minimal bystander editing. Nat. Chem. Biol. 2023, 19, 101–110, Correction in Nat. Chem. Biol. 2024, 20, 1094. [Google Scholar] [CrossRef]
- Hao, W.; Cui, W.; Liu, Z.; Suo, F.; Wu, Y.; Han, L.; Zhou, Z. A New-Generation Base Editor with an Expanded Editing Window for Microbial Cell Evolution In Vivo Based on CRISPR—Cas12b Engineering. Adv. Sci. 2024, 11, e2309767. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Xiao, Y.L.; Tang, W. High-precision cytosine base editors by evolving nucleic-acid-recognition hotspots in deaminase. Nat. Biotechnol. 2025; early view. [Google Scholar] [CrossRef] [PubMed]
- Valdez, I.; O’Connor, I.; Patel, D.; Gierer, K.; Harrington, J.; Ellis, E.; Caponetti, S.A.; Sebra, R.P.; Valley, H.C.; Coote, K.; et al. A streamlined base editor engineering strategy to reduce bystander editing. Nat. Commun. 2025, 16, 8115. [Google Scholar] [CrossRef]
- Tong, H.; Wang, X.; Liu, Y.; Liu, N.; Li, Y.; Luo, J.; Ma, Q.; Wu, D.; Li, J.; Xu, C.; et al. Programmable A-to-Y base editing by fusing an adenine base editor with an N-methylpurine DNA glycosylase. Nat. Biotechnol. 2023, 41, 1080–1084. [Google Scholar] [CrossRef]
- Li, Y.; Li, S.; Li, C.; Zhang, C.; Yan, L.; Li, J.; He, Y.; Guo, Y.; Lin, Y.; Zhang, Y.; et al. Engineering a plant A-to-K base editor with improved performance by fusion with a transactivation module. Plant Commun. 2023, 4, 100667. [Google Scholar] [CrossRef]
- Wu, X.; Ren, B.; Liu, L.; Qiu, S.; Li, X.; Li, P.; Yan, F.; Lin, H.; Zhou, X.; Zhang, D.; et al. Adenine base editor incorporating the N-methylpurine DNA glycosylase MPGv3 enables efficient A-to-K base editing in rice. Plant Commun. 2023, 4, 100668. [Google Scholar] [CrossRef]
- Tong, H.; Wang, H.; Wang, X.; Liu, N.; Li, G.; Wu, D.; Li, Y.; Jin, M.; Li, H.; Wei, Y.; et al. Development of deaminase-free T-to-S base editor and C-to-G base editor by engineered human uracil DNA glycosylase. Nat. Commun. 2024, 15, 4897. [Google Scholar] [CrossRef]
- Anzalone, A.V.; Randolph, P.B.; Davis, J.R.; Sousa, A.A.; Koblan, L.W.; Levy, J.M.; Chen, P.J.; Wilson, C.; Newby, G.A.; Raguram, A.; et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 2019, 576, 149–157. [Google Scholar] [CrossRef]
- Lin, Q.; Zong, Y.; Xue, C.; Wang, S.; Jin, S.; Zhu, Z.; Wang, Y.; Anzalone, A.V.; Raguram, A.; Doman, J.L.; et al. Prime genome editing in rice and wheat. Nat. Biotechnol. 2020, 38, 582–585. [Google Scholar] [CrossRef] [PubMed]
- Hua, K.; Jiang, Y.; Tao, X.; Zhu, J.K. Precision genome engineering in rice using prime editing system. Plant Biotechnol. J. 2020, 18, 2167–2169. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Li, J.; Chen, J.; Yan, L.; Xia, L. Precise Modifications of Both Exogenous and Endogenous Genes in Rice by Prime Editing. Mol. Plant 2020, 13, 671–674. [Google Scholar] [CrossRef]
- Jiang, Y.; Chai, Y.; Qiao, D.; Wang, J.; Xin, C.; Sun, W.; Cao, Z.; Zhang, Y.; Zhou, Y.; Wang, X.C.; et al. Optimized prime editing efficiently generates glyphosate-resistant rice plants carrying homozygous TAP-IVS mutation in EPSPS. Mol. Plant 2022, 15, 1646–1649. [Google Scholar] [CrossRef]
- Qiao, D.; Wang, J.; Lu, M.H.; Xin, C.; Chai, Y.; Jiang, Y.; Sun, W.; Cao, Z.; Guo, S.; Wang, X.C.; et al. Optimized prime editing efficiently generates heritable mutations in maize. J. Integr. Plant Biol. 2023, 65, 900–906. [Google Scholar] [CrossRef]
- Gupta, A.; Liu, B.; Chen, Q.J.; Yang, B. High-efficiency prime editing enables new strategies for broad-spectrum resistance to bacterial blight of rice. Plant Biotechnol. J. 2023, 21, 1454–1464. [Google Scholar] [CrossRef] [PubMed]
- Ni, P.; Zhao, Y.; Zhou, X.; Liu, Z.; Huang, Z.; Ni, Z.; Sun, Q.; Zong, Y. Efficient and versatile multiplex prime editing in hexaploid wheat. Genome Biol. 2023, 24, 156. [Google Scholar] [CrossRef]
- Jin, S.; Zong, Y.; Gao, Q.; Zhu, Z.; Wang, Y.; Qin, P.; Liang, C.; Wang, D.; Qiu, J.L.; Zhang, F.; et al. Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science 2019, 364, 292–295. [Google Scholar] [CrossRef]
- Zhou, C.; Sun, Y.; Yan, R.; Liu, Y.; Zuo, E.; Gu, C.; Han, L.; Wei, Y.; Hu, X.; Zeng, R.; et al. Off-target RNA mutation induced by DNA base editing and its elimination by mutagenesis. Nature 2019, 571, 275–278. [Google Scholar] [CrossRef]
- Slesarenko, Y.S.; Lavrov, A.V.; Smirnikhina, S.A. Off-target effects of base editors: What we know and how we can reduce it. Curr. Genet. 2022, 68, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Guo, M.; Gao, Q.; Jia, H.; He, M.; Wang, Z.; Guo, L.; Liu, G.; Gao, Q.; Zhao, K.T. QBEmax is a sequence-permuted and internally protected base editor. Nat. Biotechnol. 2025; early view. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Zheng, K.; Yao, Y. China’s regulatory change toward genome-edited crops. Trends Biotechnol. 2024, 42, 801–806. [Google Scholar] [CrossRef]
- Wei, D.; Cheng, P.; Song, Z.; Liu, Y.; Xu, X.; Huang, X.; Wang, X.; Zhang, Y.; Shu, W.; Wei, Y. AI-guided Cas9 engineering provides an effective strategy to enhance base editing. Mol. Syst. Biol. 2025; advance online publication. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Watson, J.L.; Juergens, D.; Bennett, N.R.; Trippe, B.L.; Yim, J.; Eisenach, H.E.; Ahern, W.; Borst, A.J.; Ragotte, R.J.; Milles, L.F.; et al. De novo design of protein structure and function with RFdiffusion. Nature 2023, 620, 1089–1100. [Google Scholar] [CrossRef]
| Species | Base Editors | Target Gene | Editing Efficiency (%) | Transformation | Function | References |
|---|---|---|---|---|---|---|
| Rice | BE3 | OsSBEIIb, OsPDS | 20.00 | Agrobacterium | Starch structure | [39] |
| APOBEC1-XTEN-Cas9n | NRT1.1B, SLR1 | 2.70–13.30 | Agrobacterium | Nitrogen absorption efficiency, Plant height, Lodging resistance | [40] | |
| PABE | OsACC-T1, OsALS-T1, OsCDC48-T3, OsDEP1-T1, OsDEP1-T2, OsNRT1.1B-T1 | 15.80–59.10 | Agrobacterium | Herbicide resistance | [29] | |
| rBE9, rBE14 | OsALS1 | 15.20–23.80 | Agrobacterium, Bombardment | Herbicide resistance | [41] | |
| rBE14 | OsTubA2 | 12.70 | Agrobacterium | Herbicide resistance | [42] | |
| CGBE | OsIPA1, OsbZIP5, OsSIR1, OsALS1, NRT1.18 | 21.3 | Agrobacterium | Herbicide resistance, Nitrogen utilization rate | [36] | |
| Wheat | pnCas9-PBE | TaALS | 2.50 | Bombardment | Herbicide resistance | [43] |
| WhieABE8e | tubulin genes | 78.00 | Agrobacterium | Herbicide resistance | [44] | |
| Potato | CBE | StGBSSI | 90.00 | Agrobacterium | Amylose content | [45] |
| Arabidopsis, Rapeseed | pcABE7.10 | AtALS, BnPDS | 4.10–50.00 | Agrobacterium | Late flowering, Whitening | [46] |
| CBE | BrALS1 | 1.80 | Agrobacterium | Herbicide resistance | [47] | |
| Cotton | GhBE3 | GhCLA, GhPEBP | 37.32 | Agrobacterium | Whitening | [48] |
| GhABE7.10n | GhPEBP | 64.00 | Agrobacterium | Compact structure | [49] | |
| Corn | CBE | ZmALS1, ZmALS2 | 13.90 | Agrobacterium | Herbicide resistance | [50] |
| evoCDA1, TadA8.20 | ZmACC1, ZmACC2 | 79.00–100.00 | Agrobacterium | Herbicide resistance | [51] | |
| Tomatoes, Potatoes | Target-AID | DELLA, ETR1 | 18.30 | Agrobacterium | Leaf morphology | [52] |
| Target-AID | SlALS, StALS | 70.00 | Agrobacterium | Herbicide resistance | [53] | |
| Watermelon | BE3 | ClALS | 23.00 | Agrobacterium | Herbicide resistance | [54] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, H.; Li, J.; Sun, K.; Tang, H.; Huang, W.; Li, X.; Wang, S.; Ding, K.; Han, Z.; Li, Z.; et al. Advances and Applications of Plant Base Editing Technologies. Int. J. Mol. Sci. 2025, 26, 9452. https://doi.org/10.3390/ijms26199452
Peng H, Li J, Sun K, Tang H, Huang W, Li X, Wang S, Ding K, Han Z, Li Z, et al. Advances and Applications of Plant Base Editing Technologies. International Journal of Molecular Sciences. 2025; 26(19):9452. https://doi.org/10.3390/ijms26199452
Chicago/Turabian StylePeng, Hao, Jiajun Li, Kehui Sun, Huali Tang, Weihong Huang, Xi Li, Surong Wang, Ke Ding, Zhiyang Han, Zhikun Li, and et al. 2025. "Advances and Applications of Plant Base Editing Technologies" International Journal of Molecular Sciences 26, no. 19: 9452. https://doi.org/10.3390/ijms26199452
APA StylePeng, H., Li, J., Sun, K., Tang, H., Huang, W., Li, X., Wang, S., Ding, K., Han, Z., Li, Z., Xu, L., & Wang, K. (2025). Advances and Applications of Plant Base Editing Technologies. International Journal of Molecular Sciences, 26(19), 9452. https://doi.org/10.3390/ijms26199452








