Genome-Wide Characterization of a Carbon Ion Beam-Induced Soybean Mutant Population Reveals Extensive Genetic Variation for Trait Improvement
Abstract
1. Introduction
2. Results
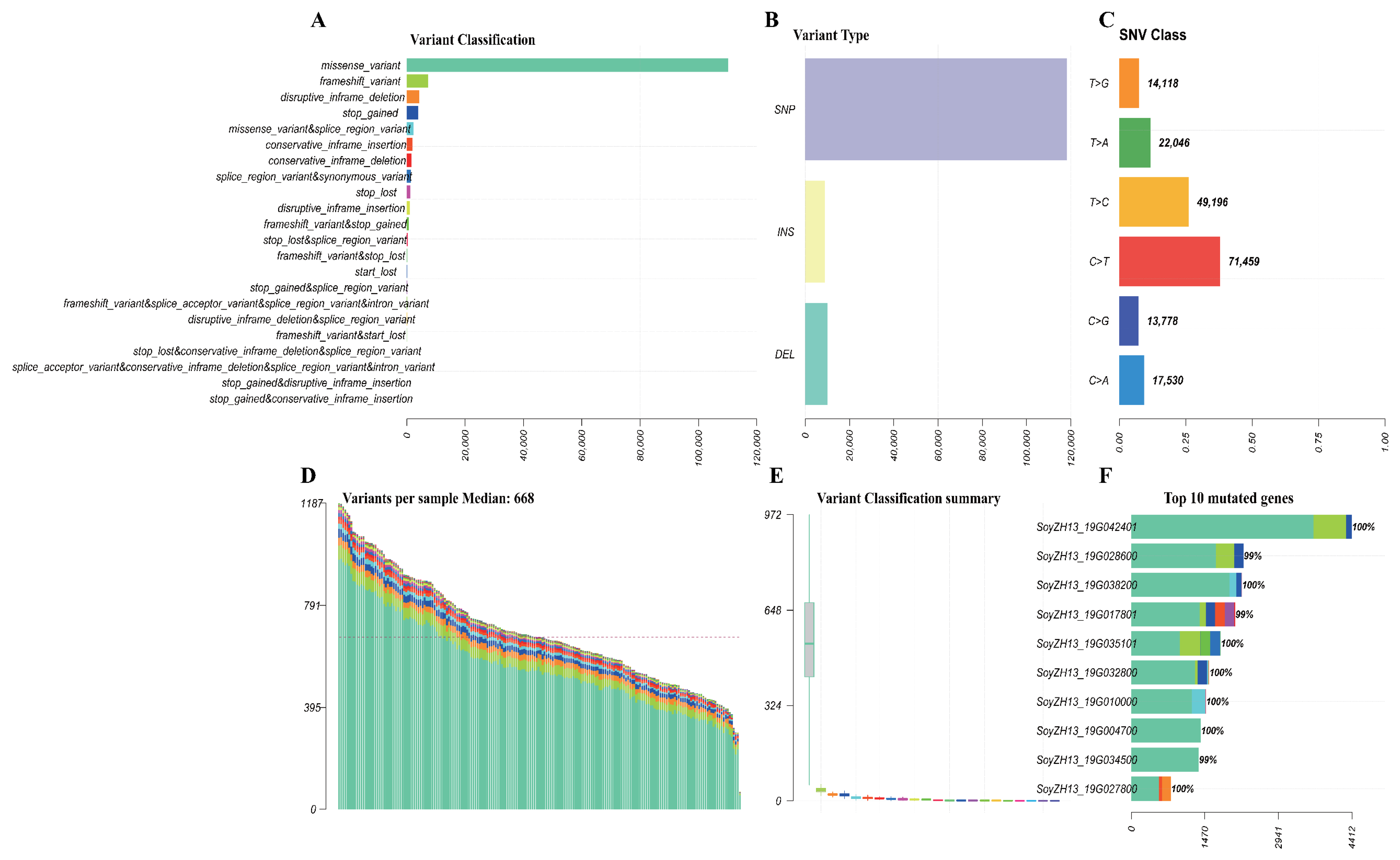
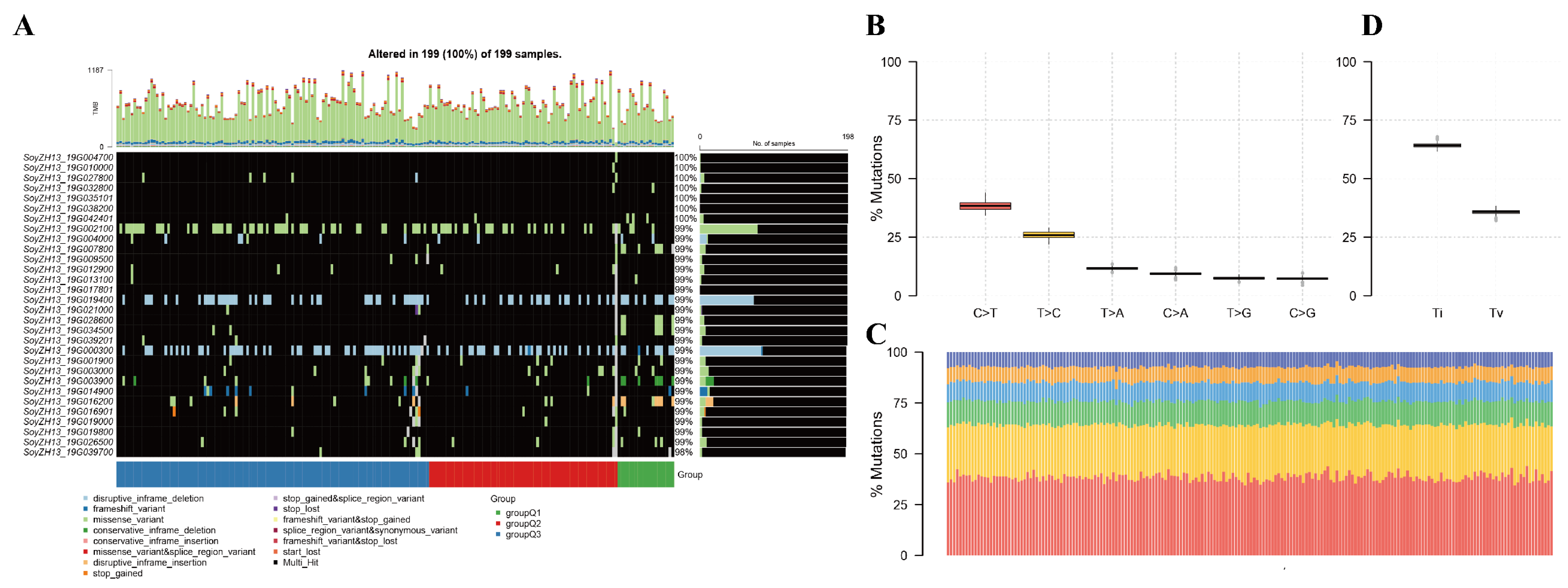
2.1. Soybean Population Sequencing and Variation
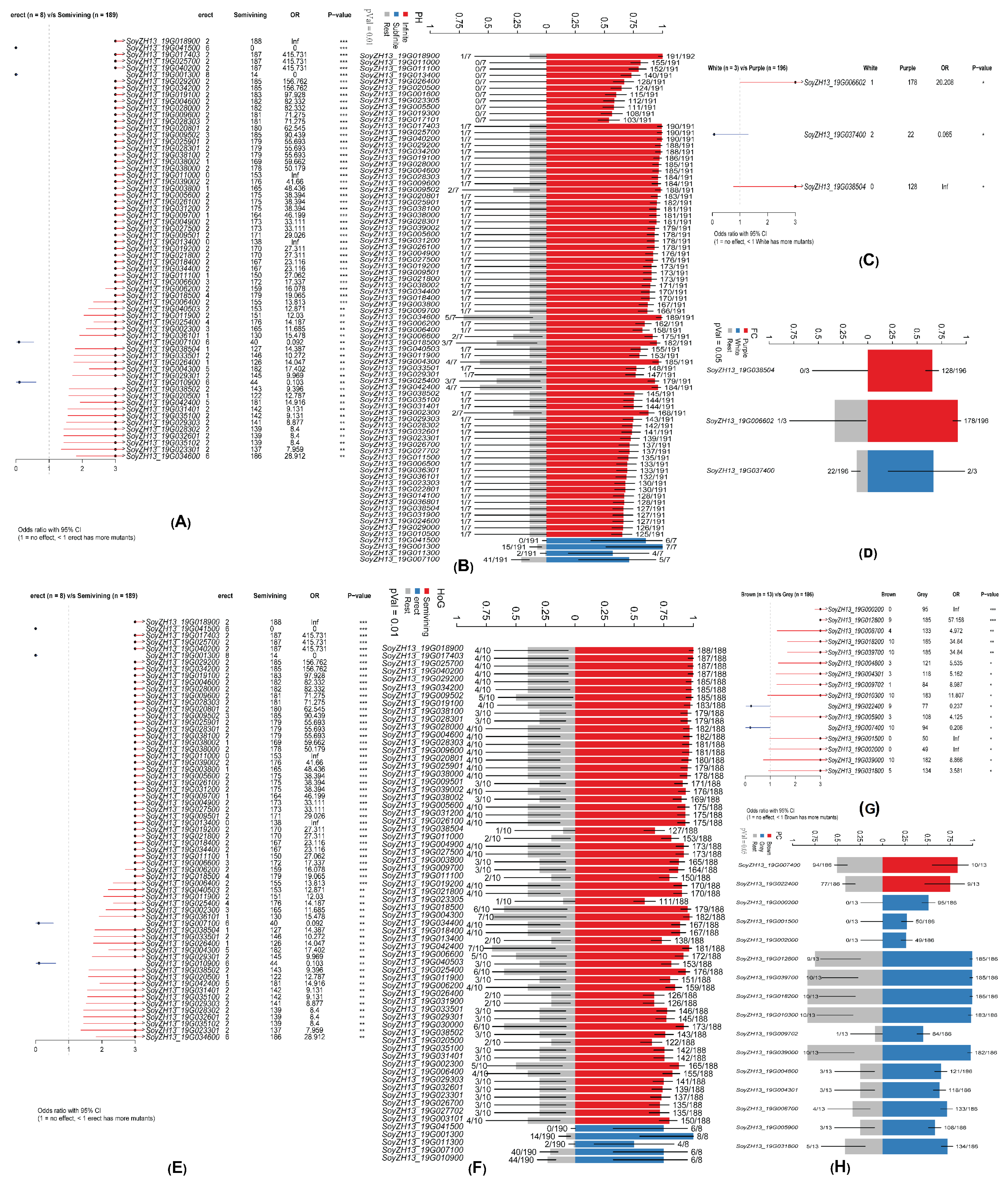
2.2. Phenotype Comparison and Enrichment Analysis
2.3. Population Structure Analysis
2.4. Morphological Traits Correlate with Seed Weight Variation
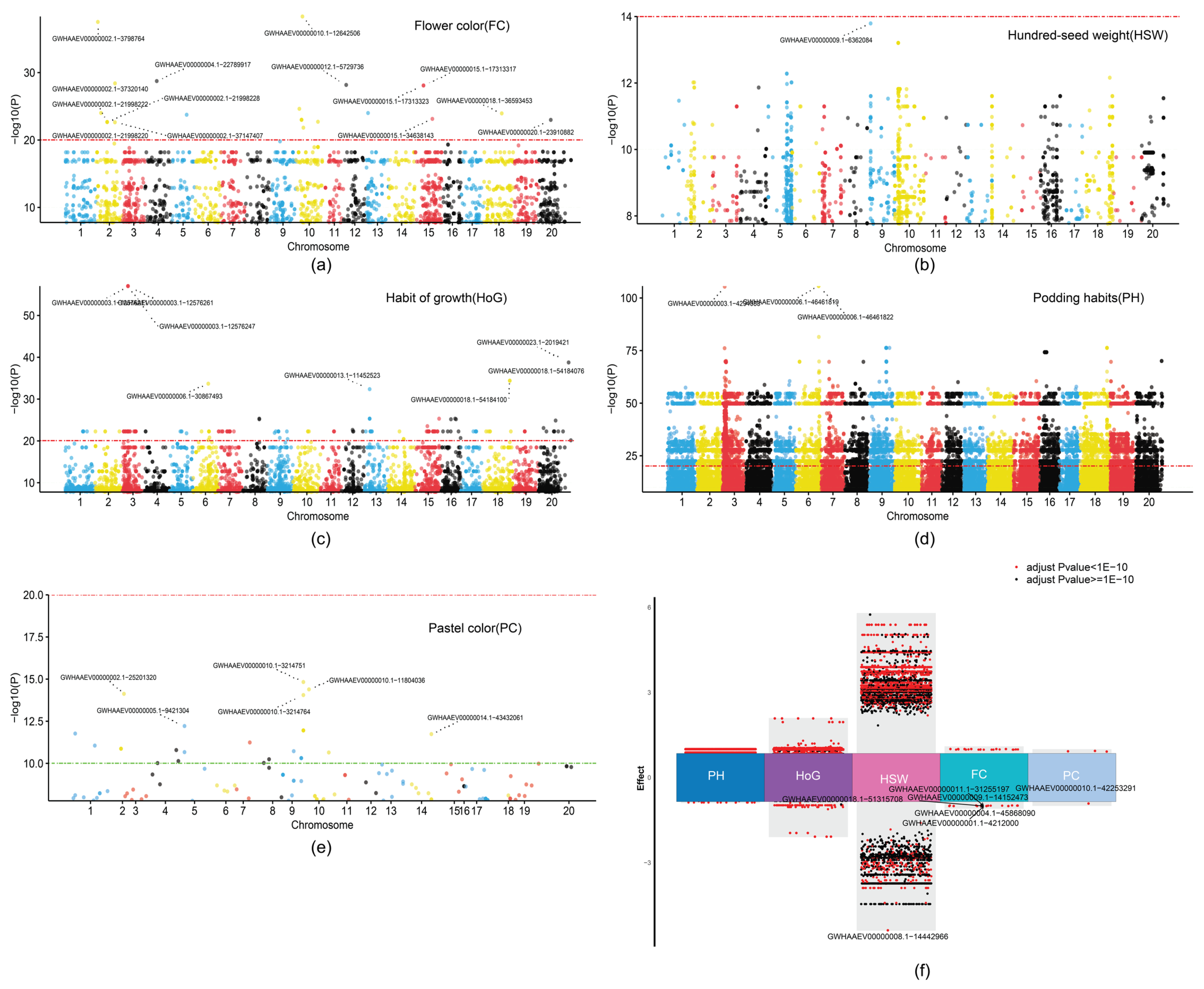
2.5. Genome-Wide Association Studies (GWAS) Reveal Distinct Genetic Architectures of Five Soybean Traits
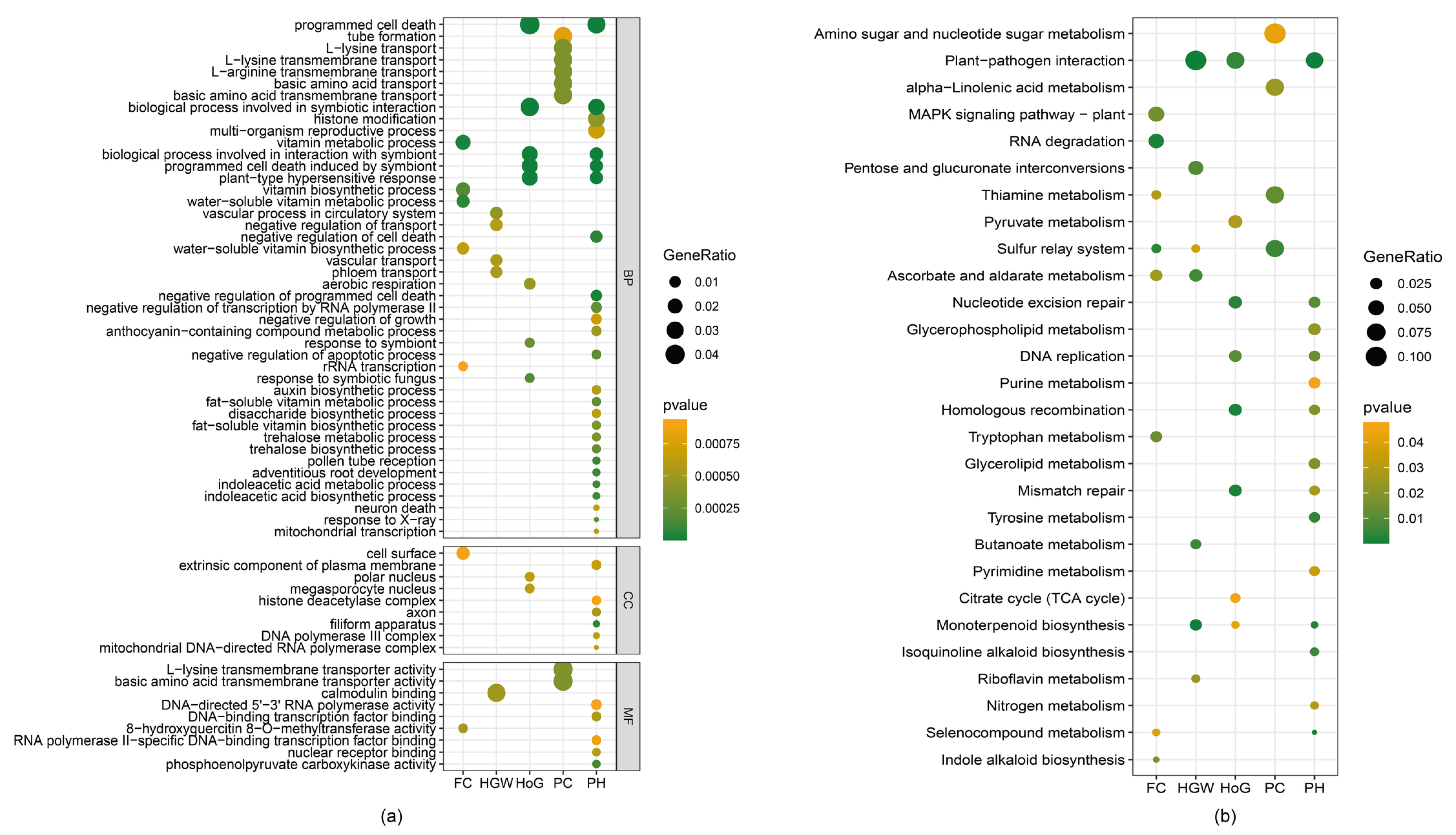
2.6. Gene Ontology Enrichment Reveals Trait-Specific Biological Processes
2.7. KEGG Pathway Analysis Identifies Key Metabolic Networks
2.8. Key Candidate Genes Underlying Trait Variation
3. Discussion
3.1. Carbon Ion Beam Mutagenesis Efficiency and Mutation Spectrum Analysis
3.2. Population Structure and Genetic Architecture Implications
3.3. Contrasting Genetic Architectures Reveal Evolutionary Constraints
3.4. Functional Pathway Analysis and Biological Interpretation
3.5. Breeding Applications and Strategic Implications
3.6. Future Research Directions and Validation Requirements
4. Materials and Methods
4.1. The Plant Material and Mutagenesis
4.2. Experimental Design and Field Conditions
4.3. DNA Extraction and Sequencing
4.4. Sequence Alignment and Variant Detection
4.5. Genetic Population Structure Analysis
4.6. Phenotyping and Trait Measurement
4.7. Genome-Wide Association Studies
4.8. Functional Enrichment Analysis
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lamlom, S.F.; Zhang, Y.; Su, B.; Wu, H.; Zhang, X.; Fu, J.; Zhang, B.; Qiu, L.-J. Map-based cloning of a novel qtl qbn-1 influencing branch number in soybean [Glycine max (L.) Merr.]. Crop J. 2020, 8, 793–801. [Google Scholar] [CrossRef]
- Ren, H.; Zhang, B.; Zhang, C.; Liu, X.; Wang, X.; Zhang, F.; Zhao, K.; Yuan, R.; Abdelghany, A.M.; Lamlom, S.F. Uncovering molecular mechanisms of soybean response to 12c6+ heavy ion irradiation through integrated transcriptomic and metabolomic profiling. Ecotoxicol. Environ. Saf. 2025, 289, 117689. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Liu, C.-K.; Tu, B.-J.; Li, Y.-S.; Zhang, Q.-Y.; Liu, X.-B. Effects of carbon ion beam irradiation on phenotypic variations and biochemical parameters in early generations of soybean plants. Agriculture 2021, 11, 98. [Google Scholar] [CrossRef]
- Feng, Z.; Du, Y.; Chen, J.; Chen, X.; Ren, W.; Wang, L.; Zhou, L. Comparison and characterization of phenotypic and genomic mutations induced by a carbon-ion beam and gamma-ray irradiation in soybean (Glycine max (L.) Merr.). Int. J. Mol. Sci. 2023, 24, 8825. [Google Scholar] [CrossRef]
- Zhao, K.; Hong, H.; Liu, X.; Wang, X.; Zhang, C.; Zhang, F.; Yuan, R.; Lamlom, S.F.; Ren, H.; Zhang, B. Multi-omics integration reveals heavy ion-induced enhancement of soybean isoflavone biosynthesis. Physiol. Plant. 2025, 177, e70508. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, M.; Li, F.; Liu, X.; Zhang, C.; Zhang, F.; Zhao, K.; Yuan, R.; Lamlom, S.F.; Ren, H. Genome-wide association study reveals key genetic loci controlling oil content in soybean seeds. Agronomy 2025, 15, 1889. [Google Scholar] [CrossRef]
- Jia, H.; Han, D.; Yan, X.; Zhang, L.; Liang, J.; Lu, W. Genome-wide association and rna-seq analyses reveal a potential candidate gene related to oil content in soybean seeds. Int. J. Mol. Sci. 2024, 25, 8134. [Google Scholar] [CrossRef]
- Zhang, F.; Xu, J.; Wang, W.; Liu, X.; He, D.; Zhang, B.; Liu, B.; Lamlom, S.F.; Abdelghany, A.M.; Hong, H. Genetic architecture of shade tolerance in soybean (Glycine max L. Merr.) revealed by genome-wide association study. Crop Sci. 2025, 65, e70107. [Google Scholar] [CrossRef]
- Zhang, C.; Zha, B.; Yuan, R.; Zhao, K.; Sun, J.; Liu, X.; Wang, X.; Zhang, F.; Zhang, B.; Lamlom, S.F. Identification of quantitative trait loci for node number, pod number, and seed number in soybean. Int. J. Mol. Sci. 2025, 26, 2300. [Google Scholar] [CrossRef]
- Awan, F.S.; Sadia, B.; Altaf, J.; Habib, M.; Hameed, K.; Hussain, S. Genetic variability through induced mutation. In Genetic Variation; IntechOpen: London, UK, 2021. [Google Scholar]
- Sinha, D.; Maurya, A.K.; Abdi, G.; Majeed, M.; Agarwal, R.; Mukherjee, R.; Ganguly, S.; Aziz, R.; Bhatia, M.; Majgaonkar, A. Integrated genomic selection for accelerating breeding programs of climate-smart cereals. Genes 2023, 14, 1484. [Google Scholar] [CrossRef]
- Saini, D.K.; Chopra, Y.; Singh, J.; Sandhu, K.S.; Kumar, A.; Bazzer, S.; Srivastava, P. Comprehensive evaluation of mapping complex traits in wheat using genome-wide association studies. Mol. Breed. 2022, 42, 1. [Google Scholar] [CrossRef]
- Xue, A.; Cui, Y. The research progress on crop genomics and genome-wide association studies: A review. Adv. Resour. Res. 2025, 5, 123–145. [Google Scholar]
- Tian, Z.; Nepomuceno, A.L.; Song, Q.; Stupar, R.M.; Liu, B.; Kong, F.; Ma, J.; Lee, S.-H.; Jackson, S.A. Soybean2035: A decadal vision for soybean functional genomics and breeding. Mol. Plant 2025, 18, 245–271. [Google Scholar] [CrossRef]
- Zhou, X.; Guo, T. Genomic tools in soybean breeding: Innovations and impacts. Legume Genom. Genet. 2024, 15, 126–139. [Google Scholar] [CrossRef]
- Razzaq, M.K.; Babur, M.N.; Awan, M.J.A.; Raza, G.; Mobeen, M.; Aslam, A.; Siddique, K.H. Revolutionizing soybean genomics: How crispr and advanced sequencing are unlocking new potential. Funct. Integr. Genom. 2024, 24, 153. [Google Scholar] [CrossRef]
- Zhou, C.; Wang, H.; Zhu, X.; Li, Y.; Zhang, B.; Tadege, M.; Wu, S.; Qi, Z.; Xia, Z. Functional genomics: From soybean to legume. Int. J. Mol. Sci. 2025, 26, 6323. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, Y.; Zhang, R.; Yuan, R.; Zhao, K.; Liu, X.; Wang, X.; Zhang, F.; Lamlom, S.F.; Zhang, B. Comprehensively characterize the soybean cam/cml gene family, as it provides resistance against both the soybean mosaic virus and cercospora sojina pathogens. Front. Plant Sci. 2025, 16, 1633325. [Google Scholar] [CrossRef] [PubMed]
- Ochar, K.; Lamlom, S.F. Identification of the genetic locus associated with the crinkled leaf phenotype in a soybean (Glycine max L.) mutant by bsa-seq technology. J. Integr. Agric. 2022, 21, 3524–3539. [Google Scholar] [CrossRef]
- Kim, S.H.; Kim, S.Y.; Ryu, J.; Jo, Y.D.; Choi, H.-I.; Kim, J.-B.; Kang, S.-Y. Suggested doses of proton ions and gamma-rays for mutation induction in 20 plant species. Int. J. Radiat. Biol. 2021, 97, 1624–1629. [Google Scholar] [CrossRef]
- Jo, Y.D.; Kim, J.-B. Frequency and spectrum of radiation-induced mutations revealed by whole-genome sequencing analyses of plants. Quantum Beam Sci. 2019, 3, 7. [Google Scholar] [CrossRef]
- Datta, S.; Jankowicz-Cieslak, J.; Nielen, S.; Ingelbrecht, I.; Till, B.J. Induction and recovery of copy number variation in banana through gamma irradiation and low-coverage whole-genome sequencing. Plant Biotechnol. J. 2018, 16, 1644–1653. [Google Scholar] [CrossRef]
- Quiroz, D.; Lensink, M.; Kliebenstein, D.J.; Monroe, J.G. Causes of mutation rate variability in plant genomes. Annu. Rev. Plant Biol. 2023, 74, 751–775. [Google Scholar] [CrossRef]
- Du, Y.; Feng, Z.; Wang, J.; Jin, W.; Wang, Z.; Guo, T.; Chen, Y.; Feng, H.; Yu, L.; Li, W.; et al. Frequency and spectrum of mutations induced by gamma rays revealed by phenotype screening and whole-genome re-sequencing in arabidopsis thaliana. Int. J. Mol. Sci. 2022, 23, 654. [Google Scholar] [CrossRef]
- Liu, J.; Zhao, G.; Geng, J.; Geng, Z.; Dou, H.; Liu, X.; An, Z.; Zhang, H.; Wang, Y. Genome-wide analysis of mutations induced by carbon ion beam irradiation in cotton. Front. Plant Sci. 2023, 14, 1056662. [Google Scholar] [CrossRef]
- Zhang, J.; Peng, Z.; Liu, Q.; Yang, G.; Zhou, L.; Li, W.; Wang, H.; Chen, Z.; Guo, T. Time course analysis of genome-wide identification of mutations induced by and genes expressed in response to carbon ion beam irradiation in rice (Oryza sativa L.). Genes 2021, 12, 1391. [Google Scholar] [CrossRef]
- Hoffmann, A.A.; Miller, A.D.; Weeks, A.R. Genetic mixing for population management: From genetic rescue to provenancing. Evol. Appl. 2021, 14, 634–652. [Google Scholar] [CrossRef]
- Sun, Y.; Wang, J.; Crouch, J.H.; Xu, Y. Efficiency of selective genotyping for genetic analysis of complex traits and potential applications in crop improvement. Mol. Breed. 2010, 26, 493–511. [Google Scholar] [CrossRef]
- García-Fernández, C.; Campa, A.; Garzón, A.S.; Miklas, P.; Ferreira, J.J. GWAS of pod morphological and color characters in common bean. BMC Plant Biol. 2021, 21, 184. [Google Scholar] [CrossRef]
- Crouch, D.J.; Bodmer, W.F. Polygenic inheritance, GWAS, polygenic risk scores, and the search for functional variants. Proc. Natl. Acad. Sci. USA 2020, 117, 18924–18933. [Google Scholar] [CrossRef]
- Wang, D.; Zhu, J.; Li, Z.; Paterson, A. Mapping qtls with epistatic effects and qtl× environment interactions by mixed linear model approaches. Theor. Appl. Genet. 1999, 99, 1255–1264. [Google Scholar] [CrossRef]
- Zha, B.; Zhang, C.; Yuan, R.; Zhao, K.; Sun, J.; Liu, X.; Wang, X.; Zhang, F.; Zhang, B.; Lamlom, S.F. Integrative qtl mapping and candidate gene analysis for main stem node number in soybean. BMC Plant Biol. 2025, 25, 422. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.; Zhao, K.; Zhang, C.; Lamlom, S.F.; Liu, X.; Wang, X.; Zhang, F.; Yuan, R.; Gao, Y.; Cao, B. Genetic analysis and qtl mapping of seed hardness trait in a soybean (Glycine max) recombinant inbred line (ril) population. Gene 2024, 905, 148238. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.; Qu, X.; Hong, H.; Sun, L.; Lamlom, S.F.; Liu, Z.; Lu, W. Multi-environment QTL mapping identifies major genetic loci influencing soybean main stem node architecture. PeerJ 2024, 12, e18539. [Google Scholar] [CrossRef]
- Kubis, A.; Bar-Even, A. Synthetic biology approaches for improving photosynthesis. J. Exp. Bot. 2019, 70, 1425–1433. [Google Scholar] [CrossRef]
- Li, X.; Zhang, X.; Zhu, L.; Bu, Y.; Wang, X.; Zhang, X.; Zhou, Y.; Wang, X.; Guo, N.; Qiu, L. Genome-wide association study of four yield-related traits at the R6 stage in soybean. BMC Genet. 2019, 20, 39. [Google Scholar] [CrossRef]
- Li, G.; Guo, X.; Sun, W.; Hou, L.; Wang, G.; Tian, R.; Wang, X.; Qu, C.; Zhao, C. Nitrogen application in pod zone improves yield and quality of two peanut cultivars by modulating nitrogen accumulation and metabolism. BMC Plant Biol. 2024, 24, 48. [Google Scholar] [CrossRef]
- Sun, X.; Sun, X.; Pan, X.; Zhang, H.; Wang, Y.; Ren, H.; Wang, F. Identification and fine mapping of a quantitative trait locus controlling the total flower and pod numbers in soybean. Agronomy 2022, 12, 790. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M. The genome analysis toolkit: A mapreduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Wong, K.; Keane, T.M.; Stalker, J.; Adams, D.J. Enhanced structural variant and breakpoint detection using SVMerge by integration of multiple detection methods and local assembly. Genome Biol. 2010, 11, R128. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, X.; Zhao, K.; Wang, X.; Zhang, C.; Zhang, F.; Yuan, R.; Lamlom, S.F.; Zhang, B.; Ren, H. Genome-Wide Characterization of a Carbon Ion Beam-Induced Soybean Mutant Population Reveals Extensive Genetic Variation for Trait Improvement. Int. J. Mol. Sci. 2025, 26, 9304. https://doi.org/10.3390/ijms26199304
Liu X, Zhao K, Wang X, Zhang C, Zhang F, Yuan R, Lamlom SF, Zhang B, Ren H. Genome-Wide Characterization of a Carbon Ion Beam-Induced Soybean Mutant Population Reveals Extensive Genetic Variation for Trait Improvement. International Journal of Molecular Sciences. 2025; 26(19):9304. https://doi.org/10.3390/ijms26199304
Chicago/Turabian StyleLiu, Xiulin, Kezhen Zhao, Xueyang Wang, Chunlei Zhang, Fengyi Zhang, Rongqiang Yuan, Sobhi F. Lamlom, Bixian Zhang, and Honglei Ren. 2025. "Genome-Wide Characterization of a Carbon Ion Beam-Induced Soybean Mutant Population Reveals Extensive Genetic Variation for Trait Improvement" International Journal of Molecular Sciences 26, no. 19: 9304. https://doi.org/10.3390/ijms26199304
APA StyleLiu, X., Zhao, K., Wang, X., Zhang, C., Zhang, F., Yuan, R., Lamlom, S. F., Zhang, B., & Ren, H. (2025). Genome-Wide Characterization of a Carbon Ion Beam-Induced Soybean Mutant Population Reveals Extensive Genetic Variation for Trait Improvement. International Journal of Molecular Sciences, 26(19), 9304. https://doi.org/10.3390/ijms26199304





