Abstract
C. albicans has recently been described as a secondary colonizer associated with periodontal infections. This study aimed to determine the expression patterns of ALS and SAP family genes in C. albicans strains isolated from patients with periodontal disease (n = 268), and a control group of healthy individuals without any clinical signs of periodontal disease (n = 100) was included. C. albicans and the ALS and SAP genes were identified using polymerase chain reaction (PCR). An in vitro infection model was used with the strains using the human gingival fibroblast cell line. RNA was extracted using a QIAcube robotic workstation (Qiagen). A QuantiTect Reverse Transcription Kit (Qiagen) was used for first-strand cDNA synthesis. ALS and SAP gene expression in the strains was determined using real-time PCR. A total of 82.5% (n = 66) of the C. albicans strains were isolated from patients with moderate periodontitis, 10% (n = 8) from patients with chronic periodontitis, and 7.5% (n = 6) from patients with gingivitis. In the group of healthy individuals, C. albicans was identified in 9% (9/100). Overall, the most frequently expressed ALS genes in the strains from the three diagnoses were ALS1 (77/80), ALS3 (67/80), ALS4 (67/80), ALS6 (77/80), ALS7 (62/80), and ALS9 (73/80), while the most frequently expressed SAP genes were SAP1 (76/80), SAP6 (57/80), SAP9 (78/80), and SAP10 (77/80). The overall frequencies of expression of the ALS4, ALS9, SAP2, SAP3, SAP6, and SAP genes in the strains were statistically different across the three diagnoses. We identified different profiles of expression of the ALS and SAP genes in the strains of C. albicans that contribute directly to the degree of periodontal disease. Therefore, our findings may contribute to improving our knowledge of the molecular mechanisms of C. albicans in the pathogenesis of periodontal disease.
1. Introduction
Candida albicans is an opportunistic fungal pathogen associated with a wide range and various types of infections, such as oral, vaginal, skin, and systemic candidiasis [], as well as other skin infections and infections associated with medical devices such as catheters [,], primarily in immunocompromised patients []. C. albicans has recently been described as a secondary colonizer associated with periodontal infections [,].
Periodontitis is a chronic inflammatory disease primarily caused by the accumulation of bacterial plaques around the tooth surface. In chronic conditions, this leads to the formation of periodontal pockets, destruction of the supporting tissues of the teeth and alveolar bone, and consequent tooth loss []. In the United States of America (USA), periodontitis affects more than 40% of adults, and worldwide, the incidence is 11% [].
Numerous anaerobic bacteria are responsible for periodontal disease, including Prevotella intermedia, Porphyromonas gingivalis, Treponema denticola, Aggregatibacter actinomycetemcomitans, Tannerella forsythia, and Fusobacterium nucleatum [,,]. However, enterobacteria, such as Enterobacter cloacae, Klebsiella oxytoca, Klebsiella pneumoniae [], and Escherichia coli [], and the fungus C. albicans [] have also been described as secondary pathogens associated with periodontitis.
The pathogenicity of C. albicans is largely mediated by a repertoire of virulence genes that favor the acuteness and/or chronicity of infections. These genes include the HWP1 gene (hyphal wall protein 1) [], adhesion genes of the ALS family (agglutinin-like sequence) [] that favor adhesion to host epithelial cells and biofilm formation, SAP family genes (secreted aspartyl protease) that increase tissue degradation and evasion of the host’s immune response [,,], the LIP gene family (lipases) [] that participates in the digestion of lipids from the cell membranes for nutrient acquisition, and PHL genes (phospholipases) that promote phospholipid degradation in the membrane of host epithelial cells, favoring invasion [].
The analysis and expression of the different molecular markers of virulence of C. albicans causing vaginal and oral infections have been characterized in our previous study [,] and by other authors [,]. However, information on the role of C. albicans as a secondary pathogen associated with periodontal disease is limited.
Therefore, the aim of this study was to analyze the expression patterns of adhesion- and protease-encoding genes in C. albicans isolates from patients with gingivitis, moderate periodontitis, and chronic periodontitis. To achieve this, we used an in vitro infection model using a human gingival fibroblast (HGF) cell line, enabling the characterization of the different molecular arrangements of expression of the genotype encoding for adhesins and proteases in C. albicans strains relevant to periodontal disease pathogenesis.
2. Results
2.1. Origin of the Strains
C. albicans was identified in 24.6% (n = 80) of the 268 patients with periodontal disease, and other Gram-negative species, such as Escherichia coli (n = 50) and Klebsiella spp. (n = 13), were also identified as well as Gram-positive bacteria, such as Staphylococcus aureus (50/80). A total of 82.5% (n = 66) of C. albicans strains were isolated from patients with moderate periodontitis, 10% (n = 8) were isolated from patients with chronic periodontitis, and 7.5% (n = 6) were isolated from patients with gingivitis (Table 1). The association of C. albicans/E. coli (n = 3) and C. albicans/S. aureus (n = 2) was identified in patients with moderate periodontitis, and C. albicans/Klebsiella spp. (n = 1) was detected in a patient with chronic periodontitis. In the group of healthy individuals, C. albicans was identified in 9% (9/100), and no other species of enterobacteria or associated Gram-positive bacteria were detected.

Table 1.
Distribution of ALS genes in C. albicans strains associated with periodontal disease.
2.2. Frequency of ALS Genes in the Strains
Generally, the distribution of the presence of ALS genes in the strains was independent of the degree of periodontal disease (Table 1). ALS1, ALS4, ALS6, ALS7, and ALS9 genes were identified in 100% (6/6) of the strains from patients with gingivitis, whereas in strains from patients with moderate periodontitis, the most frequent genes were ALS4 (65/66), ALS6, and ALS7 (64/66) in each case and ALS9 (63/66). On the other hand, ALS1, ALS4, and ALS7 were detected in 100% (8/8) of the strains associated with chronic periodontitis. In the strains from the group of healthy individuals (n = 9), the ALS1, ALS3, ALS4, and ALS9 genes were detected in 88.8% (8/9), ALS2 and ALS5 were detected in 33.3% (3/9), and ALS6 and ALS7 were detected in 77.7% (7/9).
2.3. Distribution of SAP Genes in the Strains
Ten genes of the SAP family (SAP1–SAP10; Table 2) were detected in 100% of the strains isolated from patients with gingivitis (6/6), whereas only seven SAP genes (SAP1, SAP4, SAP6, SAP7, SAP8, SAP9, and SAP10) were identified in 100% (8/8) of the strains isolated from patients with chronic periodontitis. In contrast, only four SAP genes (SAP1, SAP6, SAP9, and SAP10) were found in 100% (66/66) of the strains recovered from moderate periodontitis. In the strains from the group of healthy individuals, the genes SAP1, SAP4, SAP6, SAP8, SAP9, and SAP10 were detected in 100% (9/9); SAP2, SAP3, and SAP7 were detected in 88.8% (8/9); and SAP5 was detected in 77.7% (7/9). The distribution of the presence of SAP genes in the strains was independent of the degree of periodontal disease (Table 2).

Table 2.
Distribution of SAP genes in C. albicans strains associated with periodontal disease.
2.4. ALS Gene Expression in the Strains
To simulate C. albicans adhesion during periodontal disease, an in vitro infection model was used in the human gingival fibroblast (HGF) cell line, which was infected with C. albicans strains. Subsequently, the levels of mRNA expressed by the ALS genes in the strains of C. albicans were studied after the reverse transcription of cDNA by real-time PCR.
The C. albicans strains that adhere to the HGF cell line present a high expression of ALS genes (Table 3). Overall, the genes ALS3, ALS4, ALS6, and ALS9 are expressed more frequently in the strains of gingivitis and moderate periodontitis in relation to the strains of chronic periodontitis.

Table 3.
Analysis of mRNA expression of ALS genes in strains associated with periodontal disease. Note: Significant p-values (<0.05) are shown in bold.
ALS6 and ALS9 were expressed in 100% (6/6) of the strains isolated from patients with gingivitis, while ALS1 (5/6), ALS3 (5/6), ALS4 (5/6), and ALS7 (4/6) were highly expressed.
Regarding the strains recovered from patients with moderate periodontitis, the most frequently expressed genes were ALS1 (64/66), ALS3 (58/66), ALS4 (58/66), ALS6 (64/66), ALS7 (52/66), and ALS9 (63/66).
On the other hand, in the strains recovered from patients with chronic periodontitis, the most frequently expressed genes were ALS1 (8/100), ALS6 (7/8), and ALS7 (6/8). A significant association was observed between the expression frequencies of the ALS3, ALS4, and ALS9 genes of the strains in relation to the degree of periodontal disease (Table 3).
2.5. SAP Gene Expression in the Strains
To simulate cellular damage by C. albicans during periodontal disease, after infection of the human gingival fibroblast (HGF) cell line, the levels of mRNA expressed by the SAP genes in the strains of C. albicans were studied after the reverse transcription of cDNA by PCR in real time.
The most frequently expressed SAP family genes in the strains from patients with gingivitis were SAP1 (6/6), SAP9 (5/6), and SAP10 (6/6; Table 4).

Table 4.
Analysis of mRNA expression of SAP genes in strains associated with periodontal disease. Note: Significant p-values (<0.05) are shown in bold.
On the other hand, the most frequently expressed genes in the strains from patients with moderate periodontitis were SAP9 (65/66), SAP10 (63/66), SAP1 (62/66), SAP6 (51/66), and SAP2 (49/66).
Of the strains isolated from patients with chronic periodontitis, 100% (8/8) expressed the SAP1, SAP9, and SAP10 genes. A significant association was found between the expression frequencies of the SAP2, SAP3, SAP6, and SAP7 genes in the strains of moderate periodontitis in relation to the strains of gingivitis and chronic periodontitis (Table 4).
2.6. Unsupervised Hierarchical Clustering
Unsupervised hierarchical clustering analysis showed a wide distribution and diversity in the expression of adhesion (ALS) and protease (SAP) markers in C. albicans strains. The overall cladogram of the strains presented two groups (I and II) based on the expression of virulence genes related to the clinical diagnosis (Figure 1). Similarly, the cladogram of expression of the different gene members of the ALS and SAP families showed two groups (A and B), where group B was the most abundant, with the expression of 15 genes (ALS and SAP), whereas group A was represented only by the expression of three genes (SAP3, SAP4, and ALS2). Group I consisted of 54 strains (range of the no. of strains: 121–142) that were distributed into two subgroups, where the distribution of the expression of the ALS and SAP genes in the strains mainly associated with moderate periodontitis was higher than that in group II. In contrast, group II comprised 26 strains (range of the no. of strains: 140–14), distributed in several subgroups, and associated with the three diagnoses (gingivitis, moderate, and chronic periodontitis). Notably, several subgroups of group I had strains with the same expression profiles for the virulence genes ALS and SAP. In this subgroup, six strains (no. of strains: 213, 210, 187, 185, 55, and 153), all from moderate periodontitis, exhibited simultaneous expression of 17 of the 18 ALS and SAP genes analyzed. In addition, three other strains (no. of strains: 222, 212, and 219) from moderate periodontitis and gingivitis presented the same expression profile, consisting of 16 ALS and SAP genes. In one subgroup of group II, a pair of strains from patients with moderate periodontitis (no. of strains: 140 and 141) with the same profile of expressed virulence genes (12 ALS and SAP genes) was found. Notably, ALS2 was the only gene not expressed in any strains studied (n = 80).
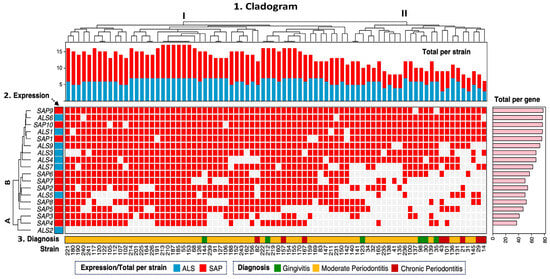
Figure 1.
Hierarchical clustering of C. albicans strains. The heat map is segmented into three panels. Top panel: (1) Overall cladogram of the distribution of virulence genotype expression across group I and II strains. Middle panel: (2) ALS and SAP gene expression across strains. A and B expression groups of the ALS and SAP genes in the strains. Bottom panel: (3) Diagnosis and origin of strains. The right panel (pink) shows the absolute frequency of expression by virulence genotype (ALS and SAP). Gene expression across strains is represented in red, and absence is represented in white.
3. Discussion
Periodontitis is a chronic, multifactorial disease that affects 62% of adults [] and is primarily caused by anaerobic bacteria []. The yeast C. albicans has also been described as a secondary pathogen associated with periodontitis []. In this study, C. albicans was more frequently isolated from patients with moderate periodontitis (66/80) than from gingivitis (6/80) or chronic periodontitis (8/80). Similar detection rates of C. albicans have been described: 20% in patients with periodontitis [] and 25.6% in middle-aged and older patients with periodontitis [].
Among the 268 patients studied, we also identified other bacteria commonly associated with periodontal disease, such as Escherichia coli (n = 50), Klebsiella spp. (n = 13), and Staphylococcus aureus (n = 50) [,,]. These findings suggest that C. albicans may coexist with Gram-negative and Gram-positive bacteria to form polymicrobial biofilms, thereby intensifying local inflammation and contributing to the persistence of periodontal lesions. This hypothesis aligns with prior evidence of synergistic interactions between C. albicans and Porphyromonas gingivalis during co-infection [].
Despite this growing body of evidence, the contribution of C. albicans ALS and SAP gene expression to periodontitis pathogenesis remains poorly characterized. Our group previously analyzed the distribution of gene association profiles of the lipase and phospholipase families related to antifungal resistance in the same periodontal strains of C. albicans []. Building on that work, the present study analyzed the different molecular expression patterns of ALS (ALS1, ALS2, ALS3, ALS4, ALS5, ALS6, ALS7, and ALS9) and SAP (SAP1-SAP10) family genes in C. albicans strains isolated from patients with periodontal disease in an in vitro infection model using an HGF cell line.
The presence of anaerobic red complex bacteria and the expression of virulence markers of Porphyromonas gingivalis (fimbriae, hemolysins, henaglutinin, and capsule), Tannerella forsythia (proteases, glycosidases, and outer membrane vesicles), and Treponema denticola (dentilisin, dentipain, main sheath protein, and motility and chemotaxis) [] during periodontal disease may facilitate colonization by secondary pathogens associated with periodontitis, such as C. albicans. The collective expression of virulence markers of anaerobic red complex bacteria with the ALS and SAP genes of C. albicans may synergistically increase the pathogenicity of periodontal disease.
The ALS (agglutinin-like sequence) family genes of C. albicans encode large glycoproteins on the fungal cell surface and are involved in the adhesion process to host surfaces []. A wide distribution of ALS genes was detected globally in the strains across the three diagnoses, where ALS1 (79/80), ALS3 (70/80), ALS4 (79/80), ALS6 (77/80), ALS7 (78/80), and ALS9 (76/80) were found in most strains from gingivitis, moderate periodontitis, and chronic periodontitis. The elevated detection frequency of ALS1 and ALS3 genes in the strains associated with the three diagnoses may favor not only the adhesion and colonization of the gingival epithelium during periodontal disease but also the acuity and/or chronicity of the disease. This is supported by studies showing that ALS1 and ALS3 have been implicated in biofilm formation that promotes evasion of the host immune response and protection against the action of antifungals [] and facilitates the formation of multispecies biofilms with other periodontal pathogens []. It has also been reported that the ALS6, ALS7, and ALS9 genes, prevalent in most of the studied strains, can participate in biofilm formation, similar to the ALS1 and ALS3 genes, as demonstrated in an als3ΔΔ/als1ΔΔ3 mutant strain []. Furthermore, C. albicans can coaggregate with the commensal bacteria S. gordonii, S. mitis, S. oralis, and S. sanguinis as early colonizers, which, in turn, collaborate with secondary colonizers, such as Fusobacterium nucleatum, promoting coaggregation and additional adhesion of late colonizers responsible for the disease, such as Porphyromonas gingivalis, Tannerella forsythia, and Treponema denticola []. In this way, the presence of C. albicans could favor the formation of interspecies biofilms during periodontal infection.
Compared to our previous findings in C. albicans vaginal isolates, the periodontal strains showed higher detection frequencies of ALS1, ALS2, ALS3, ALS4, ALS6, ALS7, and ALS9 in strains from the three diagnoses than those previously described by our working group in a study of C. albicans strains isolated from vaginal candidiasis []. In the in vitro infection model using the HGF cell line, the most frequently expressed genes in strains from gingivitis were ALS1 (5/6), ALS3 (5/6), ALS4 (5/6), ALS6 (6/6), and ALS9 (6/6). On the other hand, in strains associated with moderate periodontitis, high percentages of expression were detected for ALS1 (64/66), ALS3 (58/66), ALS4 (58/66), ALS6 (64/66), ALS7 (52/66), and ALS9 (63/66). Among the strains isolated from patients with chronic periodontitis, the most frequently expressed genes were ALS1 (8/8), ALS6 (7/8), and ALS7 (6/8).
We found high percentages of expression of ALS genes in periodontal strains of C. albicans, which are similar to the high percentages of expression of ALS1–ALS7 genes described in a group of C. albicans strains (n = 37) isolated from patients with cystic fibrosis []. The high expression of the ALS1 and ALS3 genes detected in our strains suggests their potential capacity to promote biofilm formation during periodontal disease. The ALS1 and ALS3 genes can be expressed in the hyphal form, which may favor tissue invasion and the severity of periodontal disease. High prevalence of expression of ALS1, ALS2, ALS3, and ALS9 has also been described in strains from vaginal candidiasis using an infection model in a reconstituted human vaginal epithelium (RHVE) []. Furthermore, high percentages of expression for ALS1, ALS2, ALS3, ALS4, ALS5, and ALS9 strains of C. albicans from oral candidiasis during the infection of a reconstituted human oral epithelium have been reported [].
We also detected all 10 SAP genes (SAP1–SAP10) in gingivitis strains (6/6), while in moderate periodontitis, SAP1, SAP6, SAP9, and SAP10 were detected in 100% (66/66), and in chronic periodontitis, SAP1, SAP4, SAP6, SAP7, SAP8, and SAP10 were present in all strains. These high frequencies indicate that SAP genes are key virulence determinants involved in adhesion, tissue degradation, and immune evasion [,,].
Although SAP gene expression has been widely studied in oral and vaginal candidiasis [,], SAP expression in C. albicans strains associated with periodontitis has not been extensively studied.
Using our HGF-based infection model, we observed high expression levels of SAP1 (76/80), SAP9 (78/80), and SAP10 (77/80) across strains from all clinical groups. In contrast, SAP2 (49/66), SAP3 (38/66), SAP4 (32/66), SAP5 (41/66), SAP6 (51/66), SAP7 (46/66), and SAP8 (44/66) were predominantly expressed in strains from moderate periodontitis, with lower frequencies in strains from gingivitis and chronic periodontitis. This expression pattern suggests that SAP2, SAP3, SAP6, and SAP7 may contribute specifically to the acute phases of disease progression. For instance, Sap2 has proteolytic activity that degrades host proteins in the oral cavity [], whereas Sap3 is mainly expressed in the yeast form and participates in the adhesion and colonization processes during the initial stage of infection []. Sap4–Sap6 favor the formation of hyphae [] and, consequently, tissue invasion [,]. Sap7 could participate in hypha formation, Sap8 is associated with vaginal and oral infections [], and Sap9 and Sap10 are involved in biofilm production in C. albicans strains []. The simultaneous expression of the 10 members of the SAP gene family in periodontal C. albicans strains was similar to that of an in vitro model of RHVE infection with C. albicans strains from vaginal candidiasis [] and those in strains from oral candidiasis [].
Interestingly, our reference strains of C. albicans, cv24, cv31, and cv42 from cervical and vaginal infections, have similar expression percentages of the ALS and SAP genes [,] to those found in this study, which leads us to assume that C. albicans strains from different types of candidiasis are capable of presenting distinct molecular arrangements of virolome expression.
The expression frequencies of the ALS3, ALS4, ALS5, ALS9, SAP4, SAP5, and SAP6 genes were lower in the strains isolated from chronic periodontitis compared to those from moderate periodontitis and gingivitis, which suggests that, in a more severe stage of the disease where there is a high destruction of tissues, these adhesion and protease genes are not essential.
Systematic, unsupervised hierarchical clustering revealed two major groups (I and II) based on gene expression profiles. Group I strains presented greater expression of the virulence genotypes (ALS and SAP) than group II strains. Notably, six strains from moderate periodontitis (213, 210, 187, 185, 55, 153) expressed 17 out of the 18 ALS and SAP genes analyzed. Three other strains (222, 212, 219) from moderate periodontitis (n = 2) and gingivitis (n = 1) expressed 16 genes. In contrast, two strains (140, 141) expressed only 12 genes.
Overall, 58 strains displayed distinct virulence gene expression profiles, highlighting the molecular diversity and adaptability of C. albicans in the periodontal environment. However, strains from different clinical diagnoses sharing identical profiles suggest that they may belong to the same C. albicans serotype. Therefore, whole-genome sequencing of the genomes of these strains in a subsequent study may provide more detailed information on the serotypes, the molecular composition of virulence, resistance to antifungals, and their relationship with the level of periodontal disease. Future studies should incorporate equivalent numbers of isolates from gingivitis, moderate periodontitis, and chronic periodontitis to improve the robustness of the virulome expression signatures. We acknowledge the uneven distribution of participants across diagnostic groups, which is determined by two main factors. This is a descriptive, observational study based on individuals who attended our dental clinic, where the prevalence of gingivitis cases is inherently lower than that of periodontitis. While this may create the appearance of a sampling imbalance, it reflects the natural distribution of both the condition and the pathogen within the studied population. These efforts will contribute to better vaccine design and optimization of antifungal treatment strategies for C. albicans-associated periodontal disease.
4. Materials and Methods
4.1. Patient Selection
This study was conducted at the Endoperiodontology Clinic of the Iztacala Faculty of Advanced Studies, National Autonomous University of Mexico (UNAM). A group of patients with periodontal disease (n = 268) and a control group of healthy individuals without any clinical signs of periodontal disease (n = 100) were included. All the patients enrolled in this study signed an informed consent form. This study was approved by the Ethics and Biosafety Committee of the Iztacala School of Advanced Studies, UNAM (approval No. 00012). Patients who were smokers, who were undergoing periodontal cleaning, or who had received antibiotic treatment in the previous six months were excluded from this study []. Patients with cancer treatment or treatment for any other progressive immune disease were excluded from this study. The three diagnoses of periodontal disease were established by endoperiodontology specialists (FES Iztacala, UNAM) using a Williams Fox periodontal probe (Hu-Friedy Mfg. Co., LLC, Chicago, IL, USA). The diagnostic criteria for periodontitis were loss of periodontal tissue attachment, periodontal pocket formation, alveolar bone loss, gingival recession, and tooth mobility. Patients with moderate periodontal disease presented with a tooth attachment loss of 2–4 mm, and those with chronic periodontal disease presented with a tooth attachment loss greater than or equal to 5 mm []. Patients with gingivitis presented exclusively with irritation, redness, and tissue inflammation; sensitivity; bleeding when brushing their teeth; and gums that were soft to the touch. Samples were collected from the periodontal pockets of the incisors, canines, premolars, or molars of patients with moderate or chronic periodontitis using sterile paper points. Patients with gingivitis did not present with periodontal pockets, so samples from the gingival surfaces were taken with sterile swabs. The samples were placed in brain–heart infusion (BHI; MCD LAB, Tlalnepantla, Mexico State) nutrient broth and transported to the Clinical Laboratory of CUSI (Clínica Universitaria de la Salud Integral), FES Iztacala, UNAM, for microbiological culture. The BHI cultures were incubated at 37 °C for 24 h. They were then plated on Sabouraud agar (MCD LAB, Tlalnepantla, Mexico City), eosin methylene blue (EMB; MCD LAB, Tlalnepantla, Edo. Mexico), and S110 (MCD LAB, Tlalnepantla, Edo. de México) and incubated at 37 °C for 24 h. The K. pneumoniae was identified by PCR based on the 16S–23S rDNA internal transcribed spacer [], E. coli strains were identified by standard IMViC biochemical tests (Indole, Methyl Red, Voges-Proskauer and Citrate; MCD LAB, Tlalnepantla, Edo. Mexico), and S. aureus strains were identified by bacteriological and biochemical tests that included mannitol, coagulase, and polymerase chain reaction (PCR) by amplification of the 23S rRNA, nuc, spax region, and coa genes [].
4.2. DNA Extraction
After incubation in BHI (MCD LAB, Tlalnepantla, Mexico State) at 37 °C for 24 h for samples taken from periodontal pockets of patients with moderate periodontitis and chronic periodontitis and from gums of patients with gingivitis and healthy control individuals, the samples were streaked by the cross-streak method using sterile loops on Sabouraud agar at 37 °C for 24 h. The Candida albicans species of pure cultures on Sabouraud agar were identified by the germ tube test in BHI (MCD LAB, Tlalnepantla, Mexico State) broth containing 10% horse serum and confirmed by the polymerase chain reaction (PCR) method, for which DNA extraction from the strains was performed by the boiling method as previously described []. From the fungal culture on Sabouraud agar, six colonies of approximately 2 mm in diameter of each strain were picked using a sterile loop and placed in Eppendorf tubes containing 1.5 mL of sterile deionized water. The tubes were boiled for 20 min and subsequently incubated on ice for 10 min. Finally, they were centrifuged at 10,000 rpm for 10 min, and the supernatant containing the DNA was separated and stored in another Eppendorf tube at −20 °C until used for PCR.
4.3. Identification of C. albicans
Using the DNA obtained from each strain by the boiling method, C. albicans was identified by PCR through amplification of internal transcribed spacers (ITS) 1 and 2 of the rRNA gene (Table 5), as previously described []. For each singleplex PCR assay, a final volume of 25 μL was used per reaction mixture: 12.5 μL of Taq DNA Polymerase Master Mix RED (Ampliqon, Copenhagen, Denmark), 1 μL of forward primer, 1 μL of reverse primer (10 pmol, Integrated DNA Technologies, San Diego, CA, USA: Table 5), 3 μL of DNA template (100 ng), and 7.5 μL of nuclease-free water. The PCR amplification conditions were as follows: initial denaturation at 96 °C for 5 min, followed by 40 cycles of 94 °C for 30 s, 58 °C for 30 s, and 72 °C for 30 s. Finally, the final extension was at 72 °C for 15 min. The strain used as a positive control was C. albicans ATCC32354. All PCR assays were performed in triplicate. The amplicons were stained with Midori Green (Nippon Genetics, Düren, Germany, EUROPE) after electrophoresis in 2% agarose gels, which were photographed under UV illumination with GEL LOGIC 100 (Kodak, Carestream Molecular Imaging, Rochester, NY, USA) equipment.

Table 5.
Oligonucleotides used for the identification of C. albicans and virulence genes.
4.4. Identification of ALS Genes in Strains
ALS family genes were detected using uniplex PCR, as previously described [] (Table 5). The final volume of the reaction mixture was 25 μL: 12.5 μL of Taq DNA Polymerase Master Mix RED (Ampliqon, Copenhagen, Denmark), 1 μL of forward primer, 1 μL of reverse primer (10 pmol, Integrated DNA Technologies, San Diego, CA, USA; Table 5), 3 μL of DNA template (100 ng), and 7.5 μL of nuclease-free water. DNA amplification was performed under the following conditions: initial denaturation at 94 °C for 5 min, followed by 40 cycles at 94 °C for 30 s, 58 °C for 30 s, and 72 °C for 30 s. Finally, an extension was performed at 72 °C for 7 min. C. albicans strains cv24, cv31, and cv42 from cervicovaginal infections belonging to the strain library of the CUSI Clinical Laboratory [,], FES Iztacala, and strain C. albicans ATCC32354 were used as positive controls. All PCR assays were performed in triplicate.
4.5. Identification of SAP Genes in Strains
SAP family genes were detected using uniplex PCR, as previously described [] (Table 5). The final volume of the reaction mixture was 25 μL: 12.5 μL of Taq DNA Polymerase Master Mix RED (Ampliqon, Copenhagen, Denmark), 1 μL of forward primer, 1 μL of reverse primer (10 pmol, Integrated DNA Technologies, San Diego, CA, USA: Table 5), 3 μL of DNA template (100 ng), and 7.5 μL of nuclease-free water. The amplification conditions of the SAP genes were as follows: initial denaturation at 95 °C for 15 min, followed by 45 cycles at 95 °C for 15 s, 58 °C for 30 s, and 60 °C for 60 s. Finally, an extension was performed at 72 °C for 5 min. C. albicans strains cv24, cv31, and cv42 from cervical and vaginal infections as well as strain C. albicans ATCC32354 were used as positive controls. All PCR assays were performed in triplicate.
4.6. Preparation of Gingival Fibroblasts
The HGF cell line (ATCC, CRL-2104) was seeded at a density of 5 × 103 cells per cm2 and cultured in 75 cm2 Corning cell culture flasks in a water-saturated atmosphere at 37 °C and 5% CO2. Fibroblasts were maintained in Dulbecco’s modified high glucose Eagle’s medium (Sigma Aldrich, Saint Louis, MO, USA), supplemented with 10% fetal bovine serum (previously inactivated at 56 °C for 30 min; GIBCO BRL, Gaithersburg, MD, USA), containing 10 U of penicillin/25 μg of streptomycin/mL (Sigma Aldrich). Fibroblasts were cultured to confluence at a density of 2.5 × 105 cells/mL, washed twice with phosphate-buffered saline, and dissociated with 0.25% trypsin and 1 mM EDTA for 5 min at 37 °C and 5% CO2. The cells were subsequently collected and placed in triplicate into the plate wells. Finally, the plate was incubated in the incubator at 37 °C with 5% CO2 for 48 h until a confluence of 1.8 × 105 cells/well was obtained.
4.7. Dilution of C. albicans Yeasts
One colony of each C. albicans strain was inoculated in triplicate into 2 mL of brain–heart infusion broth (MCD Lab, Edo. de México, Mexico) and incubated at 37 °C for 12 h with constant shaking. Dilutions of each culture were prepared at a 1:5 ratio using phosphate-buffered saline to obtain an optical density of 0.125 at 600 nm (OD600) using a Beckman DU-7400 spectrophotometer (Laguna Hills, CA, USA), equivalent to a concentration of 2 × 106 cells/mL.
4.8. In Vitro Infection of Fibroblasts
To simulate periodontal infection, we used a previously described in vitro infection model [,]. Fifty microliters at a concentration of 2 × 106 cells/mL of each C. albicans strain culture was added to a monolayer (1.8 × 105 cells/well) of HGFs (ATCC, CRL-2104). Triplicate 24-well plates were incubated with 1 mL of F12K (Sigma Aldrich; Merck KGaA, Darmstadt, Germany) and 10% fetal bovine serum at 37 °C for 48 h in a 5% CO2 atmosphere with saturated humidity. C. albicans strains cv24, cv31, and cv42 and C. albicans ATCC32354 were used as controls, and yeast adherence to fibroblasts was verified under a light microscope to ensure fungal viability.
4.9. C. albicans RNA Extraction and Reverse Transcription to cDNA
Triplicate C. albicans strains were harvested from the surface of the HGF cell line (ATCC, CRL-2104), resuspended in 1000 μL of RNA Protect Bacteria reagent (Qiagen, Hilden, Germany), and centrifuged at 8000 rpm for 5 min to obtain fungal cell pellets. C. albicans RNA was extracted using the QIAcube automated workstation (Qiagen, Hilden, Germany) with the RNeasy Mini commercial kit (Qiagen), which included a 200 μL treatment with buffer Y1 (Qiagen, Hilden, Germany) containing lyticase (50 U/107 cells) to disrupt the fungal cell wall. The concentration and purity of total RNA were measured using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). A QuantiTect Reverse Transcription Kit (Qiagen, Hilden, Germany) was used for first-strand cDNA synthesis.
4.10. Determination of Virulence Gene Expression in Strains by Real-Time PCR
To determine the expression of ALS and SAP family genes in C. albicans strains, we used the Corbette® Real-Time PCR device (Rotor-Gene Q 5plex; Qiagen, Hilden, Germany). The final volume per reaction mixture was 25 µL: 12.5 µL of the SYBR Green Master Mix, QuantiNova SYBR Green PCR (Qiagen, Hilden, Germany), 1 µL of each forward and reverse oligonucleotide (10 pmol, Integrated DNA Technologies, San Diego, CA, USA; Table 5), 1 µL of cDNA (100 ng/µL), and 9.5 µL of nuclease-free H2O. The real-time PCR conditions were as follows: one HotStart activation cycle at 95 °C for 2 min, 40 denaturation cycles at 95 °C for 5 s, and a 60 °C annealing/extension combination for 10 s. For each quantitative real-time PCR assay, a standard curve was constructed from three cDNA dilutions (100, 200, and 300 ng/µL) of a strain containing the gene of interest (positive control). Rotor-Gene Q 5plex HRM System software version 2.3.1.49 (Qiagen) was used to calculate the threshold cycle (CT) limit for each strain. Thus, using the CT values of each strain obtained with respect to the CT of the control strain standard curve, the arithmetic mean was obtained, and the percentage of expression of each gene was determined. For each real-time PCR run, the melting point (dissociation curve of specific products) and ACT1 and RPP2B genes were included as housekeeping genes. C. albicans strains cv24, cv31, and cv42 from cervical and vaginal infections [] and strain ATCC32354 were used as positive controls, and a negative control (no temperature control) was also used. All real-time PCR assays were performed in triplicate.
4.11. Statistical Analysis
The chi-square test was applied using SPSS statistical software (version 20.0; SPSS Inc., Chicago, IL, USA) (p < 0.05) to establish the association between the frequency of the eight adhesion genes of the ALS family and ten proteinase genes of the SAP family of the strains related to the three diagnoses (gingivitis, moderate periodontitis, and chronic periodontitis) as well as the association between the frequency of expression of the ALS and SAP genes in the C. albicans strains related to the three diagnoses (gingivitis, moderate periodontitis, and chronic periodontitis).
4.12. Unsupervised Hierarchical Clustering
To investigate the relationship between C. albicans gene expression and periodontal stage, unsupervised hierarchical clustering was performed using Gower’s similarity coefficient []. The analysis was based on a categorical data matrix constructed in R (v3.6.1) with the cluster package (v2.1.0), which integrated RT-PCR detected presence or absence of ALS and SAP virulence genes together with the clinical stage of periodontal disease (gingivitis, moderate periodontitis, chronic periodontitis). The distance between each strain was calculated based on the overall similarity coefficient, which was used to estimate the maximum possible absolute discrepancy between each matched pair of strains. After calculating the distances, we clustered mutually exclusive groups using Ward’s method in R []. The strains were visualized using a genotype expression distribution diagram, and a dendrogram was constructed using Complex heatmap (v3.6.2, R core).
5. Conclusions
The findings of this study highlight the widespread expression of the C. albicans virolome as a secondary pathogen associated with periodontal disease. The molecular properties involved in adhesion, biofilm formation, tissue degradation, and host immune evasion detected in C. albicans strains could contribute to the severity of periodontitis. Therefore, screening and medical treatment regimens are key against this opportunistic pathogen in patients with periodontal disease.
Author Contributions
Conceptualization, G.L.P.-C., A.C.-K. and E.M.-P.; Methodology, A.C.-K., A.M.F.-P., M.R.-D.l.C., G.L.P.-C. and E.M.-P.; Software, H.M.-G.; Validation, G.L.P.-C. and E.M.-P.; Formal analysis, G.L.P.-C., E.M.-P. and F.V.-P.; Investigation, G.L.P.-C., A.C.-K. and E.M.-P.; Resources, G.L.P.-C. and E.M.-P.; Data curation, A.C.-K., G.L.P.-C. and E.M.-P.; Writing—original draft preparation, G.L.P.-C. and E.M.-P.; Writing—review and editing, G.L.P.-C., E.M.-P. and F.V.-P.; Visualization, G.L.P.-C. and E.M.-P.; Supervision, G.L.P.-C. and E.M.-P.; Project administration, G.L.P.-C. and E.M.-P.; Funding acquisition, G.L.P.-C. and E.M.-P. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
This study was approved by the Ethics and Biosafety Committee of the Iztacala Faculty of Advanced Studies, UNAM (Approval No. 00012, 2016/03/18).
Informed Consent Statement
Informed consent was obtained from all subjects involved in this study.
Data Availability Statement
The original contributions presented in this study are included in the article.
Acknowledgments
We want to thank the Posgrado en Ciencias Biológicas of Universidad Nacional Autónoma de México (UNAM) at Facultad de Estudios Superiores Iztacala.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Gulati, M.; Nobile, C.J. Candida albicans biofilms: Development, regulation, and molecular mechanisms. Microbes Infect. 2016, 18, 310–321. [Google Scholar] [CrossRef]
- Yang, C.; Mo, F.; Zhang, J.; Zhang, P.; Li, Q.; Zhang, J.A. Catheter-Related Candida albicans Infection Model in Mouse. J. Vis. Exp. 2024, 205, e65307. [Google Scholar] [CrossRef]
- Duzgol, M.; Boncuoglu, E.; Kiymet, E.; Akaslan-Kara, A.; Erdem, M.; Odaman-Al, I.; Demirag, B.; Zihni, C.; Hilkay-Karapinar, T.; Oymak, Y.; et al. Evaluation for Metastatic Candida Focus and Mortality at Candida-associated Catheter-related Bloodstream Infections at the Pediatric Hematology-oncology Patients. J. Pediatr. Hematol. Oncol. 2022, 44, e643–e648. [Google Scholar] [CrossRef]
- Shahabudin, S.; Azmi, N.S.; Lani, M.N.; Mukhtar, M.; Hossain, M.S. Candida albicans skin infection in diabetic patients: An updated review of pathogenesis and management. Mycoses 2024, 67, e13753. [Google Scholar] [CrossRef] [PubMed]
- Jabri, B.; Iken, M.; Achmit, M.; Rida, S.; Ennibi, O.K. Occurrence of Candida albicans in Periodontitis. Int. J. Dent. 2021, 2021, 5589664. [Google Scholar] [CrossRef] [PubMed]
- Oka, I.; Shigeishi, H.; Ohta, K. Co-Infection of Oral Candida albicans and Porphyromonas gingivalis Is Associated with Active Periodontitis in Middle-Aged and Older Japanese People. Medicina 2022, 58, 723. [Google Scholar] [CrossRef]
- Kwon, T.; Lamster, I.B.; Levin, L. Current Concepts in the Management of Periodontitis. Int. Dent. J. 2021, 71, 462–476. [Google Scholar] [CrossRef] [PubMed]
- Eke, P.I.; Thornton-Evans, G.O.; Wei, L.; Borgnakke, W.S.; Dye, B.A.; Genco, R.J. Periodontitis in US Adults: National Health and Nutrition Examination Survey 2009–2014. J. Am. Dent. Assoc. 2018, 149, 576–588.e6. [Google Scholar] [CrossRef]
- Torrungruang, K.; Jitpakdeebordin, S.; Charatkulangkun, O.; Gleebbua, Y. Porphyromonas gingivalis, Aggregatibacter actinomycetemcomitans, and Treponema denticola/Prevotella intermedia Co-Infection Are Associated with Severe Periodontitis in a Thai Population. PLoS ONE 2015, 10, e0136646. [Google Scholar] [CrossRef]
- Carrouel, F.; Kanoute, A.; Lvovschi, V.E.; Bourgeois, D. Periodontal pathogens of the interdental microbiota in a 3 months pregnant population with an intact periodontium. Front. Microbiol. 2023, 14, 1275180. [Google Scholar] [CrossRef]
- Yekani, M.; Dastgir, M.; Fattahi, S.; Shahi, S.; Maleki Dizaj, S.; Memar, M.Y. Microbiological and molecular aspects of periodontitis pathogenesis: An infection-induced inflammatory condition. Front. Cell. Infect. Microbiol. 2025, 15, 1533658. [Google Scholar] [CrossRef] [PubMed]
- Jepsen, K.; Falk, W.; Brune, F.; Cosgarea, R.; Fimmers, R.; Bekeredjian-Ding, I.; Jepsen, S. Prevalence and Antibiotic Susceptibility Trends of Selected Enterobacteriaceae, Enterococci, and Candida albicans in the Subgingival Microbiota of German Periodontitis Patients: A Retrospective Surveillance Study. Antibiotics 2022, 11, 385. [Google Scholar] [CrossRef] [PubMed]
- Monroy-Pérez, E.; Hernández-Jaimes, T.; Morales-Espinosa, R.; Delgado, G.; Martínez-Gregorio, H.; García-Cortés, L.R.; Herrera-Gabriel, J.P.; De Lira-Silva, A.; Vaca-Paniagua, F.; Paniagua-Contreras, G.L. Analysis of in vitro expression of virulence genes related to antibiotic and disinfectant resistance in Escherichia coli as an emerging periodontal pathogen. Front. Cell. Infect. Microbiol. 2024, 14, 1412007. [Google Scholar] [CrossRef]
- Slazhneva, E.; Tikhomirova, E.; Tsarev, V.; Orekhova, L.; Loboda, E.; Atrushkevich, V. Candida species detection in patients with chronic periodontitis: A systematic review and meta-analysis. Clin. Exp. Dent. Res. 2022, 8, 1354–1375. [Google Scholar] [CrossRef]
- Martorano-Fernandes, L.; Goodwine, J.S.; Ricomini-Filho, A.P.; Nobile, C.J.; Del Bel Cury, A.A. Candida albicans Adhesins Als1 and Hwp1 Modulate Interactions with Streptococcus mutans. Microorganisms 2023, 11, 1391. [Google Scholar] [CrossRef]
- Hoyer, L.L.; Cota, E. Candida albicans Agglutinin-Like Sequence (Als) Family Vignettes: A Review of Als Protein Structure and Function. Front. Microbiol. 2016, 7, 280. [Google Scholar] [CrossRef]
- Kadry, A.A.; El-Ganiny, A.M.; El-Baz, A.M. Relationship between Sap prevalence and biofilm formation among resistant clinical isolates of Candida albicans. Afr. Health. Sci. 2018, 18, 1166–1174. [Google Scholar] [CrossRef]
- Kumar, R.; Rojas, I.G.; Edgerton, M. Candida albicans Sap6 Initiates Oral Mucosal Inflammation via the Protease Activated Receptor PAR2. Front. Immunol. 2022, 13, 912748. [Google Scholar] [CrossRef]
- Pawar, M.Y.; Hatolkar, S.M.; Misra, R.N. Phenotypic and molecular detection of virulence factor genes SAP4 and PLB in Candida albicans isolates from the Western part of India. Med. J. Armed Forces India 2022, 78, 271–276. [Google Scholar] [CrossRef]
- Hube, B.; Stehr, F.; Bossenz, M.; Mazur, A.; Kretschmar, M.; Schäfer, W. Secreted lipases of Candida albicans: Cloning, characterisation and expression analysis of a new gene family with at least ten members. Arch. Microbiol. 2000, 174, 362–374. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Zhou, Y.; Wang, R.; Xie, F.; Zhai, Z. Expression, purification, and characterization of phospholipase B1 from Candida albicans in Escherichia coli. 3 Biotech 2020, 10, 538. [Google Scholar] [CrossRef]
- Monroy-Pérez, E.; Sáinz-Espuñes, T.; Paniagua-Contreras, G.; Negrete-Abascal, E.; Rodríguez-Moctezuma, J.R.; Vaca, S. Frequency and expression of ALS and HWP1 genotypes in Candida albicans strains isolated from Mexican patients suffering from vaginal candidosis. Mycoses 2012, 55, e151–e157. [Google Scholar] [CrossRef]
- Monroy-Pérez, E.; Paniagua-Contreras, G.L.; Rodríguez-Purata, P.; Vaca-Paniagua, F.; Vázquez-Villaseñor, M.; Díaz-Velásquez, C.; Uribe-García, A.; Vaca, S. High Virulence and Antifungal Resistance in Clinical Strains of Candida albicans. Can. J. Infect. Dis. Med. Microbiol. 2016, 2016, 5930489. [Google Scholar] [CrossRef] [PubMed]
- Green, C.B.; Cheng, G.; Chandra, J.; Mukherjee, P.; Ghannoum, M.A.; Hoyer, L.L. RT-PCR detection of Candida albicans ALS gene expression in the reconstituted human epithelium (RHE) model of oral candidiasis and in model biofilms. Microbiology 2004, 150, 267–275. [Google Scholar] [CrossRef][Green Version]
- Bonfim-Mendonça, P.S.; Tobaldini-Valério, F.K.; Capoci, I.R.; Faria, D.R.; Sakita, K.M.; Arita, G.S.; Negri, M.; Kioshima, É.S.; Svidzinski, T.I. Different expression levels of ALS and SAP genes contribute to recurrent vulvovaginal candidiasis by Candida albicans. Future Microbiol. 2021, 16, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Trindade, D.; Carvalho, R.; Machado, V.; Chambrone, L.; Mendes, J.J.; Botelho, J. Prevalence of periodontitis in dentate people between 2011 and 2020: A systematic review and meta-analysis of epidemiological studies. J. Clin. Periodontol. 2023, 50, 604–626. [Google Scholar] [CrossRef] [PubMed]
- Jabri, B.; Iken, M.; Ait-Ou-Amar, S.; Rida, S.; Bouziane, A.; Ennibi, O.K. Candida albicans and Candida dubliniensis in Periodontitis in Adolescents and Young Adults. Int. J. Microbiol. 2022, 2022, 4625368. [Google Scholar] [CrossRef]
- Veras, E.L.; Castro Dos Santos, N.; Souza, J.G.S.; Figueiredo, L.C.; Retamal-Valdes, B.; Barão, V.A.R.; Shibli, J.; Bertolini, M.; Faveri, M.; Teles, F.; et al. Newly identified pathogens in periodontitis: Evidence from an association and an elimination study. J. Oral Microbiol. 2023, 15, 2213111. [Google Scholar] [CrossRef]
- Fritschi, B.Z.; Albert-Kiszely, A.; Persson, G.R. Staphylococcus aureus and other bacteria in untreated periodontitis. J. Dent. Res. 2008, 87, 589–593. [Google Scholar] [CrossRef]
- Monroy-Pérez, E.; Rodríguez-Bedolla, R.M.; Garzón, J.; Vaca-Paniagua, F.; Arturo-Rojas Jiménez, E.; Paniagua-Contreras, G.L. Marked virulence and azole resistance in Candida albicans isolated from patients with periodontal disease. Microb. Pathog. 2020, 148, 104436. [Google Scholar] [CrossRef]
- Zhao, X.; Oh, S.H.; Cheng, G.; Green, C.B.; Nuessen, J.A.; Yeater, K.; Leng, R.P.; Brown, A.J.P.; Hoyer, L.L. ALS3 and ALS8 represent a single locus that encodes a Candida albicans adhesin; functional comparisons between Als3p and Als1p. Microbiology 2004, 150, 2415–2428. [Google Scholar] [CrossRef]
- Roudbarmohammadi, S.; Roudbary, M.; Bakhshi, B.; Katiraee, F.; Mohammadi, R.; Falahati, M. ALS1 and ALS3 gene expression and biofilm formation in Candida albicans isolated from vulvovaginal candidiasis. Adv. Biomed. Res. 2016, 5, 105. [Google Scholar] [CrossRef]
- Satala, D.; González-González, M.; Smolarz, M.; Surowiec, M.; Kulig, K.; Wronowska, E.; Zawrotniak, M.; Kozik, A.; Rapala-Kozik, M.; Karkowska-Kuleta, J. The Role of Candida albicans Virulence Factors in the Formation of Multispecies Biofilms With Bacterial Periodontal Pathogens. Front. Cell Infect. Microbiol. 2022, 11, 765942. [Google Scholar] [CrossRef]
- Ponde, N.O.; Lortal, L.; Ramage, G.; Naglik, J.R.; Richardson, J.-P. Candida albicans biofilms and polymicrobial interactions. Crit. Rev. Microbiol. 2021, 47, 91–111. [Google Scholar] [CrossRef]
- Ríos-López, A.L.; Garza-Velásquez, M.F.; González, G.M.; Becerril-García, M.A.; Flores-Maldonado, O. Prevalence, virulence factors and antifungal susceptibility of oral isolates of Candida albicans from patients with cystic fibrosis in Mexico. Rev. Iberoam. Micol. 2024, 41, 31–36. [Google Scholar] [CrossRef]
- Cheng, G.; Wozniak, K.; Wallig, M.A.; Fidel Jr, P.L.; Trupin, S.R.; Hoyer, L.L. Comparison between Candida albicans agglutinin-like sequence gene expression patterns in human clinical specimens and models of vaginal candidiasis. Infect. Immun. 2005, 73, 1656–1663. [Google Scholar] [CrossRef] [PubMed]
- Borg-von Zepelin, M.; Meyer, I.; Thomssen, R.; Würzner, R.; Sanglard, D.; Telenti, A.; Monod, M. HIV-Protease inhibitors reduce cell adherence of Candida albicans strains by inhibition of yeast secreted aspartic proteases. J. Investig. Dermatol. 1999, 113, 747–751. [Google Scholar] [CrossRef] [PubMed]
- Borg-von Zepelin, M.; Beggah, S.; Boggian, K.; Sanglard, D.; Monod, M. The expression of the secreted aspartyl proteinases Sap4 to Sap6 from Candida albicans in murine macrophages. Mol. Microbiol. 1998, 28, 543–554. [Google Scholar] [CrossRef]
- Ibrahim, A.S.; Filler, S.G.; Sanglard, D.; Edwards, J.E.J.; Hube, B. Secreted aspartyl proteinases and interactions of Candida albicans with human endothelial cells. Infect. Immun. 1998, 66, 3003–3005. [Google Scholar] [CrossRef] [PubMed]
- Naglik, J.R.; Moyes, D.; Makwana, J.; Kanzaria, P.; Tsichlaki, E.; Weindl, G.; Hube, B. Quantitative expression of the Candida albicans secreted aspartyl proteinase gene family in human oral and vaginal candidiasis. Microbiology 2008, 154, 3266–3280. [Google Scholar] [CrossRef]
- Lian, C.H.; Liu, W.D. Differential expression of Candida albicans secreted aspartyl proteinase in human vulvovaginal candidiasis. Mycoses 2007, 50, 383–390. [Google Scholar] [CrossRef] [PubMed]
- Modrzewska, B.; Kurnatowski, P.; Khalid, K. Comparison of proteolytic activity of Candida sp. strains depending on their origin. J. Mycol. Med. 2016, 26, 138–147. [Google Scholar] [CrossRef]
- Pichová, I.; Pavlíčková, L.; Dostál, J.; Dolejší, E.; Hrušková-Heidingsfeldová, O.; Weber, J.; Ruml, T.; Souček, M. Secreted aspartic proteases of Candida albicans, Candida tropicalis, Candida parapsilosis and Candida lusitaniae: Inhibition with peptidomimetic inhibitors. Eur. J. Biochem. 2001, 268, 2669–2677. [Google Scholar] [CrossRef]
- Naglik, J.R.; Challacombe, S.J.; Hube, B. Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol. Mol. Biol. Rev. 2003, 67, 400–428. [Google Scholar] [CrossRef]
- Felk, A.; Kretschmar, M.; Albrecht, A.; Schaller, M.; Beinhauer, S.; Nichterlein, T.; Sanglard, D.; Korting, H.C.; Schäfer, W.; Hube, B. Candida albicans hyphal formation and the expression of the Efg1-regulated proteinases Sap4 to Sap6 are required for the invasion of parenchymal organs. Infect. Immun. 2002, 70, 3689–3700. [Google Scholar] [CrossRef]
- Sanglard, D.; Hube, B.; Monod, M.; Odds, F.C.; Gow, N.A. A triple deletion of the secreted aspartyl proteinase genes SAP4, SAP5, and SAP6 of Candida albicans causes attenuated virulence. Infect. Immun. 1997, 65, 3539–3546. [Google Scholar] [CrossRef]
- Monroy-Pérez, E.; Paniagua-Contreras, G.L.; Vaca-Paniagua, F.; Negrete-Abascal, E.; Vaca-Pacheco, S. SAP Expression in Candida albicans Strains Isolated from Mexican Patients with Vaginal Candidosis. Int. J. Clin. Med. 2013, 4, 25–31. [Google Scholar] [CrossRef]
- Tavanti, A.; Pardini, G.; Campa, D.; Davini, P.; Lupetti, A.; Senesi, S. Differential expression of secretory aspartyl proteinase genes (SAP1-10) in oral Candida albicans isolates with distinct karyotypes. J. Clin. Microbiol. 2004, 42, 4726–4734. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Koukos, G.; Sakellari, D.; Arsenakis, M.; Tsalikis, L.; Slini, T.; Konstantinidis, A. Prevalence of Staphylococcus aureus and methicillin resistant Staphylococcus aureus (MRSA) in the oral cavity. Arch. Oral Biol. 2015, 60, 1410–1415. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, C.; Zheng, W.; Zhang, X.; Yu, J.; Gao, Q.; Hou, Y.; Huang, X. PCR detection of Klebsiella pneumoniae in infant formula based on 16S-23S internal transcribed spacer. Int. J. Food. Microbiol. 2008, 125, 230–235. [Google Scholar] [CrossRef]
- Paniagua-Contreras, G.; Monroy-Pérez, E.; Gutiérrez-Lucas, R.; Sainz-Espuñes, T.; Bustos-Martínez, J.; Vaca, S. Genotypic characterization of methicillin-resistant Staphylococcus aureus strains isolated from the anterior nares and catheter of ambulatory hemodialysis patients in Mexico. Folia Microbiol. 2014, 59, 295–302. [Google Scholar] [CrossRef]
- Ooka, T.; Terajima, J.; Kusumoto, M.; Iguchi, A.; Kurokawa, K.; Ogura, Y.; Asadulghani, M.; Nakayama, K.; Murase, K.; Ohnishi, M.; et al. Development of a multiplex PCR-based rapid typing method for enterohemorrhagic Escherichia coli O157 strains. J. Clin. Microbiol. 2009, 47, 2888–2894. [Google Scholar] [CrossRef]
- Luo, G.; Mitchell, T.G. Rapid identification of pathogenic fungi directly from cultures by using Multiplex PCR. J. Clin. Microbiol. 2002, 40, 2860–2865. [Google Scholar] [CrossRef] [PubMed]
- Schaller, M.; Korting, H.C.; Schäfer, W.; Bastert, J.; Chen, W.; Hube, B. Secreted aspartic proteinase (Sap) activity contributes to tissue damage in a model of human oral candidosis. Mol. Microbiol. 1999, 34, 169–180. [Google Scholar] [CrossRef] [PubMed]
- Gower, J.C. A General Coefficient of Similarity and Some of Its Properties. Biometrics 1971, 27, 857–871. [Google Scholar] [CrossRef]
- Ward, J.H., Jr. Hierarchical Grouping to Optimize an Objective Function. J. Am. Stat. Assoc. 1963, 58, 236–244. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).