Linezolid in the Focus of Antimicrobial Resistance of Enterococcus Species: A Global Overview of Genomic Studies
Abstract
1. Introduction
2. Literature Search and Inclusion Criteria
3. LNZ Resistance in Enterococci
3.1. Historical Perspectives, Global Prevalence, Clonal Spread, and Mechanisms
3.2. Mutational Mechanisms Conferring LNZ Resistance in Enterococci
3.2.1. 23S rRNA Mutations
3.2.2. Mutations in Ribosomal Protein Genes
3.3. Non-Mutational Mechanisms Conferring LNZ Resistance in Enterococci
3.3.1. LNZ Resistance in Enterococci via Target Modification Mechanisms
3.3.2. LNZ Resistance in Enterococci via Target Protection Mechanisms
3.4. Tedizolid—Resistance Mechanisms and Cross-Resistance with LNZ
3.5. Future Perspectives on WGS for Detecting and Investigating LNZ Resistance in Enterococci
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hashemian, S.M.; Farhadi, T.; Ganjparvar, M. Linezolid: A Review of Its Properties, Function, and Use in Critical Care. Drug Des. Dev. Ther. 2018, 12, 1759–1767. [Google Scholar] [CrossRef]
- Bibel, B.; Raskar, T.; Couvillion, M.; Lee, M.; Kleinman, J.I.; Takeuchi-Tomita, N.; Churchman, L.S.; Fraser, J.S.; Fujimori, D.G. Context-Specific Inhibition of Mitochondrial Ribosomes by Phenicol and Oxazolidinone Antibiotics. Nucleic Acids Res. 2025, 53, gkaf046. [Google Scholar] [CrossRef]
- Vishnu, V.Y.; Modi, M.; Goyal, M.K.; Lal, V. Linezolid Induced Reversible Peripheral Neuropathy. Am. J. Ther. 2016, 23, e1839–e1841. [Google Scholar] [CrossRef]
- Soriano, A.; Miró, O.; Mensa, J. Mitochondrial Toxicity Associated with Linezolid. N. Engl. J. Med. 2005, 353, 2305–2306. [Google Scholar] [CrossRef]
- De Vriese, A.S.; Van Coster, R.; Smet, J.; Seneca, S.; Lovering, A.; Van Haute, L.L.; Vanopdenbosch, L.J.; Martin, J.-J.; Ceuterick-de Groote, C.; Vandecasteele, S.; et al. Linezolid-Induced Inhibition of Mitochondrial Protein Synthesis. Clin. Infect. Dis. 2006, 42, 1111–1117. [Google Scholar] [CrossRef]
- Ramesh, V.; Gattu, S.; Maqsood, M.; Rao, V. Linezolid-Induced Lactic Acidosis. BMJ Case Rep. 2024, 17, e259335. [Google Scholar] [CrossRef] [PubMed]
- Kato, H.; Hagihara, M.; Asai, N.; Koizumi, Y.; Yamagishi, Y.; Mikamo, H. A Systematic Review and Meta-Analysis of Myelosuppression in Pediatric Patients Treated with Linezolid for Gram-Positive Bacterial Infections. J. Infect. Chemother. 2021, 27, 1143–1150. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A.; Mendes, R.E.; Streit, J.M.; Hogan, P.A.; Flamm, R.K. Five-Year Summary of In Vitro Activity and Resistance Mechanisms of Linezolid against Clinically Important Gram-Positive Cocci in the United States from the LEADER Surveillance Program (2011 to 2015). Antimicrob. Agents Chemother. 2017, 61, e00609-17. [Google Scholar] [CrossRef]
- Luna, V.A.; King, D.S.; Gulledge, J.; Cannons, A.C.; Amuso, P.T.; Cattani, J. Susceptibility of Bacillus anthracis, Bacillus cereus, Bacillus mycoides, Bacillus pseudomycoides and Bacillus thuringiensis to 24 Antimicrobials Using Sensititre(R) Automated Microbroth Dilution and Etest(R) Agar Gradient Diffusion Methods. J. Antimicrob. Chemother. 2007, 60, 555–567. [Google Scholar] [CrossRef] [PubMed]
- Barberis, C.M.; Sandoval, E.; Rodriguez, C.H.; Ramírez, M.S.; Famiglietti, A.; Almuzara, M.; Vay, C. Comparison Between Disk Diffusion and Agar Dilution Methods to Determine In Vitro Susceptibility of Corynebacterium spp. Clinical Isolates and Update of Their Susceptibility. J. Glob. Antimicrob. Resist. 2018, 14, 246–252. [Google Scholar] [CrossRef]
- Noll, M.; Kleta, S.; Al Dahouk, S. Antibiotic Susceptibility of 259 Listeria monocytogenes Strains Isolated from Food, Food-Processing Plants and Human Samples in Germany. J. Infect. Public. Health 2018, 11, 572–577. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, A.; Trittler, R.; Bohnert, J.A.; Kümmerer, K.; Pagès, J.-M.; Kern, W.V. Intracellular Accumulation of Linezolid in Escherichia coli, Citrobacter freundii and Enterobacter aerogenes: Role of Enhanced Efflux Pump Activity and Inactivation. J. Antimicrob. Chemother. 2007, 59, 1261–1264. [Google Scholar] [CrossRef]
- Birmingham, M.C.; Rayner, C.R.; Meagher, A.K.; Flavin, S.M.; Batts, D.H.; Schentag, J.J. Linezolid for the Treatment of Multidrug-Resistant, Gram-Positive Infections: Experience from a Compassionate-Use Program. Clin. Infect. Dis. 2003, 36, 159–168. [Google Scholar] [CrossRef]
- Faella, F.; Pagliano, P.; Fusco, U.; Attanasio, V.; Conte, M. Combined Treatment with Ceftriaxone and Linezolid of Pneumococcal Meningitis: A Case Series Including Penicillin-Resistant Strains. Clin. Microbiol. Infect. 2006, 12, 391–394. [Google Scholar] [CrossRef]
- Thwaites, G.; Nguyen, N.V. Linezolid for Drug-Resistant Tuberculosis. N. Engl. J. Med. 2022, 387, 842–843. [Google Scholar] [CrossRef]
- Garazzino, S.; Krzysztofiak, A.; Esposito, S.; Castagnola, E.; Plebani, A.; Galli, L.; Cellini, M.; Lipreri, R.; Scolfaro, C.; Bertaina, C.; et al. Use of Linezolid in Infants and Children: A Retrospective Multicentre Study of the Italian Society for Paediatric Infectious Diseases. J. Antimicrob. Chemother. 2011, 66, 2393–2397. [Google Scholar] [CrossRef]
- Shi, Y.; Wu, H.-L.; Wu, Y.-H.; Li, S.; Zhang, L.-Y.; Xu, S.-S.; Huang, H.-Y.; Zhang, C.-H.; Yu, X.-B.; Cai, K.; et al. Safety and Clinical Efficacy of Linezolid in Children: A Systematic Review and Meta-Analysis. World J. Pediatr. 2023, 19, 129–138. [Google Scholar] [CrossRef]
- Dubin, K.; Pamer, E.G. Enterococci and Their Interactions with the Intestinal Microbiome. Microbiol. Spectr. 2017, 5, 1–16. [Google Scholar] [CrossRef]
- Komiyama, E.Y.; Lepesqueur, L.S.S.; Yassuda, C.G.; Samaranayake, L.P.; Parahitiyawa, N.B.; Balducci, I.; Koga-Ito, C.Y. Enterococcus Species in the Oral Cavity: Prevalence, Virulence Factors and Antimicrobial Susceptibility. PLoS ONE 2016, 11, e0163001. [Google Scholar] [CrossRef] [PubMed]
- Ghasemi, E.; Mansouri, S.; Shahabinejad, N. Vaginal Colonization and Susceptibility to Antibiotics of Enterococci During Late Pregnancy in Kerman City, Iran. Arch. Clin. Infect. Dis. 2016, 11, e35428. [Google Scholar] [CrossRef]
- Byappanahalli, M.N.; Nevers, M.B.; Korajkic, A.; Staley, Z.R.; Harwood, V.J. Enterococci in the Environment. Microbiol. Mol. Biol. Rev. 2012, 76, 685–706. [Google Scholar] [CrossRef]
- Ben Braïek, O.; Smaoui, S. Enterococci: Between Emerging Pathogens and Potential Probiotics. BioMed Res. Int. 2019, 2019, 5938210. [Google Scholar] [CrossRef] [PubMed]
- Fiore, E.; Van Tyne, D.; Gilmore, M.S. Pathogenicity of Enterococci. Microbiol. Spectr. 2019, 7, 1–23. [Google Scholar] [CrossRef]
- Lebreton, F.; Willems, R.J.L.; Gilmore, M.S. Enterococcus Diversity, Origins in Nature, and Gut Colonization. In Enterococci: From Commensals to Leading Causes of Drug Resistant Infection; Gilmore, M.S., Clewell, D.B., Ike, Y., Shankar, N., Eds.; Massachusetts Eye and Ear Infirmary: Boston, MA, USA, 2014. [Google Scholar]
- Ch’ng, J.-H.; Chong, K.K.L.; Lam, L.N.; Wong, J.J.; Kline, K.A. Biofilm-Associated Infection by Enterococci. Nat. Rev. Microbiol. 2019, 17, 82–94. [Google Scholar] [CrossRef]
- Dadashi, M.; Sharifian, P.; Bostanshirin, N.; Hajikhani, B.; Bostanghadiri, N.; Khosravi-Dehaghi, N.; van Belkum, A.; Darban-Sarokhalil, D. The Global Prevalence of Daptomycin, Tigecycline, and Linezolid-Resistant Enterococcus faecalis and Enterococcus faecium Strains from Human Clinical Samples: A Systematic Review and Meta-Analysis. Front. Med. 2021, 8, 720647. [Google Scholar] [CrossRef]
- Hegstad, K.; Mikalsen, T.; Coque, T.M.; Werner, G.; Sundsfjord, A. Mobile Genetic Elements and Their Contribution to the Emergence of Antimicrobial Resistant Enterococcus faecalis and Enterococcus faecium. Clin. Microbiol. Infect. 2010, 16, 541–554. [Google Scholar] [CrossRef]
- Arredondo-Alonso, S.; Top, J.; McNally, A.; Puranen, S.; Pesonen, M.; Pensar, J.; Marttinen, P.; Braat, J.C.; Rogers, M.R.C.; van Schaik, W.; et al. Plasmids Shaped the Recent Emergence of the Major Nosocomial Pathogen Enterococcus faecium. MBio 2020, 11, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, Y.; Suzuki, M.; Kobayashi, S.; Hirahara, Y.; Kurushima, J.; Hirakawa, H.; Nomura, T.; Tanimoto, K.; Tomita, H. Enterococcal Linear Plasmids Adapt to Enterococcus faecium and Spread within Multidrug-Resistant Clades. Antimicrob. Agents Chemother. 2023, 67, e01619-22. [Google Scholar] [CrossRef]
- Miller, W.R.; Arias, C.A. ESKAPE Pathogens: Antimicrobial Resistance, Epidemiology, Clinical Impact and Therapeutics. Nat. Rev. Microbiol. 2024, 22, 598–616. [Google Scholar] [CrossRef]
- Iqbal, F.; Alocious, A.; Joy, S.C.; Stanly, E.A.R.; Rajesh, V.; Unnikrishnan, M.K.; Steinke, D.; Chandra, P. Vancomycin-Resistant Enterococci: A Rising Challenge to Global Health. Clin. Epidemiol. Glob. Health 2024, 28, 101663. [Google Scholar] [CrossRef]
- WHO. Publishes List of Bacteria for Which New Antibiotics Are Urgently Needed. Available online: https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (accessed on 23 March 2025).
- WHO. Bacterial Priority Pathogens List, 2024: Bacterial Pathogens of Public Health Importance to Guide Research, Development and Strategies to Prevent and Control Antimicrobial Resistance. Available online: https://www.who.int/publications/i/item/9789240093461 (accessed on 17 August 2025).
- Wada, Y.; Afolabi, H.A.; Al-Mhanna, S.B.; Bello, K.E.; Irekeola, A.A.; Wada, M.; Ahmed, N.; Harun, A.; Yean, C.Y.; Mohamad Nasir, N.S.; et al. Global Occurrence of Linezolid-Resistant Enterococcus (LRE): The First Systematic Review and Meta-Analysis. Microbe 2024, 2, 100041. [Google Scholar] [CrossRef]
- Bi, R.; Qin, T.; Fan, W.; Ma, P.; Gu, B. The Emerging Problem of Linezolid-Resistant Enterococci. J. Glob. Antimicrob. Resist. 2018, 13, 11–19. [Google Scholar] [CrossRef]
- Kainer, M.A.; Devasia, R.A.; Jones, T.F.; Simmons, B.P.; Melton, K.; Chow, S.; Broyles, J.; Moore, K.L.; Craig, A.S.; Schaffner, W. Response to Emerging Infection Leading to Outbreak of Linezolid-Resistant Enterococci. Emerg. Infect. Dis. 2007, 13, 1024–1030. [Google Scholar] [CrossRef]
- Jia, S.; Xu, X.; Qu, M.; Pei, Y.; Sun, S.; Liu, Y.; Dong, W.; Hu, Y.; Zhu, B.; Gao, G.F.; et al. Longitudinal Trends and Drivers of Antimicrobial Resistance in Campylobacter Worldwide (1954–2023). Zoonoses 2025, 5, 989. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, X.; Jia, S.; Qu, M.; Pei, Y.; Qiu, S.; Zhang, J.; Liu, Y.; Ma, S.; Lyu, N.; et al. A Global Atlas and Drivers of Antimicrobial Resistance in Salmonella during 1900-2023. Nat. Commun. 2025, 16, 4611. [Google Scholar] [CrossRef]
- Waskito, L.A.; Rezkitha, Y.A.A.; Vilaichone, R.; Wibawa, I.D.N.; Mustika, S.; Sugihartono, T.; Miftahussurur, M. Antimicrobial Resistance Profile by Metagenomic and Metatranscriptomic Approach in Clinical Practice: Opportunity and Challenge. Antibiotics 2022, 11, 654. [Google Scholar] [CrossRef]
- Fines, M.; Leclercq, R. Activity of Linezolid against Gram-Positive Cocci Possessing Genes Conferring Resistance to Protein Synthesis Inhibitors. J. Antimicrob. Chemother. 2000, 45, 797–802. [Google Scholar] [CrossRef] [PubMed]
- Brumfitt, W.; Hamilton-Miller, J.M.T. In-Vitro Microbiological Activities of DuP 105 and DuP 721, Novel Synthetic Oxazolidinones. J. Antimicrob. Chemother. 1988, 21, 711–720. [Google Scholar] [CrossRef] [PubMed]
- Meka, V.G.; Gold, H.S. Antimicrobial Resistance to Linezolid. Clin. Infect. Dis. 2004, 39, 1010–1015. [Google Scholar] [CrossRef] [PubMed]
- Development of Linezolidresistant. Enterococcus faecium in Two Compassionate Use Program Patients Treated with Linezolid [Abstract 848]—New York University Health Sciences. Available online: https://search.hsl.med.nyu.edu (accessed on 24 March 2025).
- Klare, I.; Konstabel, C.; Mueller-Bertling, S.; Werner, G.; Strommenger, B.; Kettlitz, C.; Borgmann, S.; Schulte, B.; Jonas, D.; Serr, A.; et al. Spread of Ampicillin/Vancomycin-Resistant Enterococcus faecium of the Epidemic-Virulent Clonal Complex-17 Carrying the Genes esp and hyl in German Hospitals. Eur. J. Clin. Microbiol. Infect. Dis. 2005, 24, 815–825. [Google Scholar] [CrossRef]
- Theilacker, C.; Jonas, D.; Huebner, J.; Bertz, H.; Kern, W.V. Outcomes of Invasive Infection Due to Vancomycin-Resistant Enterococcus faecium during a Recent Outbreak. Infection 2009, 37, 540–543. [Google Scholar] [CrossRef]
- Nasir, S.A.R.; Zeeshan, M.; Ghanchi, N.; Saeed, N.; Ghayas, H.; Zaka, S.; Ashraf, J.; Jabeen, K.; Farooqi, J.; Hasan, Z.; et al. Linezolid-Resistant Enterococcus faecium Clinical Isolates from Pakistan: A Genomic Analysis. BMC Microbiol. 2024, 24, 347. [Google Scholar] [CrossRef]
- Abbo, L.; Shukla, B.S.; Giles, A.; Aragon, L.; Jimenez, A.; Camargo, J.F.; Simkins, J.; Sposato, K.; Tran, T.T.; Diaz, L.; et al. Linezolid- and Vancomycin-Resistant Enterococcus faecium in Solid Organ Transplant Recipients: Infection Control and Antimicrobial Stewardship Using Whole Genome Sequencing. Clin. Infect. Dis. 2019, 69, 259–265. [Google Scholar] [CrossRef] [PubMed]
- Almeida-Santos, A.C.; Novais, C.; Peixe, L.; Freitas, A.R. Vancomycin-Resistant Enterococcus faecium: A Current Perspective on Resilience, Adaptation, and the Urgent Need for Novel Strategies. J. Glob. Antimicrob. Resist. 2025, 41, 233–252. [Google Scholar] [CrossRef]
- Mortelé, O.; Van Kleef–van Koeveringe, S.; Vandamme, S.; Jansens, H.; Goossens, H.; Matheeussen, V. Epidemiology and Genetic Diversity of Linezolid-Resistant Enterococcus Clinical Isolates in Belgium from 2013 to 2021. J. Glob. Antimicrob. Resist. 2024, 38, 21–26. [Google Scholar] [CrossRef]
- Liu, P.; Zeng, B.; Wu, X.; Zheng, F.; Zhang, Y.; Liao, X. Risk Exploration and Prediction Model Construction for Linezolid-Resistant Enterococcus faecalis Based on Big Data in a Province in Southern China. Eur. J. Clin. Microbiol. Infect. Dis. 2024, 43, 259–268. [Google Scholar] [CrossRef] [PubMed]
- Kent, A.G.; Spicer, L.M.; Campbell, D.; Breaker, E.; McAllister, G.A.; Ewing, T.O.; Longo, C.; Balbuena, R.; Burroughs, M.; Burgin, A.; et al. Sentinel Surveillance Reveals Phylogenetic Diversity and Detection of Linear Plasmids Harboring vanA and optrA among Enterococci Collected in the United States. Antimicrob. Agents Chemother. 2024, 68, e00591-24. [Google Scholar] [CrossRef]
- Cinthi, M.; Coccitto, S.N.; Simoni, S.; Gherardi, G.; Palamara, A.T.; Di Lodovico, S.; Di Giulio, M.; Du, X.-D.; Vignaroli, C.; Brenciani, A.; et al. The optrA, cfr(D) and vanA Genes Are Co-Located on Linear Plasmids in Linezolid- and Vancomycin-Resistant Enterococcal Clinical Isolates in Italy. J. Antimicrob. Chemother. 2025, 80, 1362–1370. [Google Scholar] [CrossRef]
- Zahedi Bialvaei, A.; Rahbar, M.; Yousefi, M.; Asgharzadeh, M.; Samadi Kafil, H. Linezolid: A Promising Option in the Treatment of Gram-Positives. J. Antimicrob. Chemother. 2017, 72, 354–364. [Google Scholar] [CrossRef] [PubMed]
- Gemmell, C.G. Virulence Factor Expression by Gram-Positive Cocci Exposed to Subinhibitory Concentrations of Linezolid. J. Antimicrob. Chemother. 2002, 50, 665–672. [Google Scholar] [CrossRef]
- Long, K.S.; Vester, B. Resistance to Linezolid Caused by Modifications at Its Binding Site on the Ribosome. Antimicrob. Agents Chemother. 2012, 56, 603–612. [Google Scholar] [CrossRef]
- Turner, A.M.; Lee, J.Y.H.; Gorrie, C.L.; Howden, B.P.; Carter, G.P. Genomic Insights into Last-Line Antimicrobial Resistance in Multidrug-Resistant Staphylococcus and Vancomycin-Resistant Enterococcus. Front. Microbiol. 2021, 12, 637656. [Google Scholar] [CrossRef]
- Long, K.S.; Munck, C.; Andersen, T.M.B.; Schaub, M.A.; Hobbie, S.N.; Böttger, E.C.; Vester, B. Mutations in 23S rRNA at the Peptidyl Transferase Center and Their Relationship to Linezolid Binding and Cross-Resistance. Antimicrob. Agents Chemother. 2010, 54, 4705–4713. [Google Scholar] [CrossRef]
- Fleischmann, R.D.; Adams, M.D.; White, O.; Clayton, R.A.; Kirkness, E.F.; Kerlavage, A.R.; Bult, C.J.; Tomb, J.-F.; Dougherty, B.A.; Merrick, J.M.; et al. Whole-Genome Random Sequencing and Assembly of Haemophilus Influenzae Rd. Science 1995, 269, 496–512. [Google Scholar] [CrossRef]
- Beg, A.Z.; Khan, A.U. Exploring Bacterial Resistome and Resistance Dessemination: An Approach of Whole Genome Sequencing. Future Med. Chem. 2019, 11, 247–260. [Google Scholar] [CrossRef]
- Fuller, C.W.; Middendorf, L.R.; Benner, S.A.; Church, G.M.; Harris, T.; Huang, X.; Jovanovich, S.B.; Nelson, J.R.; Schloss, J.A.; Schwartz, D.C.; et al. The Challenges of Sequencing by Synthesis. Nat. Biotechnol. 2009, 27, 1013–1023. [Google Scholar] [CrossRef]
- Su, M.; Satola, S.W.; Read, T.D. Genome-Based Prediction of Bacterial Antibiotic Resistance. J. Clin. Microbiol. 2019, 57, e01405-18. [Google Scholar] [CrossRef] [PubMed]
- Florensa, A.F.; Kaas, R.S.; Clausen, P.T.L.C.; Aytan-Aktug, D.; Aarestrup, F.M. ResFinder—An Open Online Resource for Identification of Antimicrobial Resistance Genes in Next-Generation Sequencing Data and Prediction of Phenotypes from Genotypes. Microb. Genom. 2022, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Eyre, D.W.; De Silva, D.; Cole, K.; Peters, J.; Cole, M.J.; Grad, Y.H.; Demczuk, W.; Martin, I.; Mulvey, M.R.; Crook, D.W.; et al. WGS to Predict Antibiotic MICs for Neisseria gonorrhoeae. J. Antimicrob. Chemother. 2017, 72, 1937–1947. [Google Scholar] [CrossRef]
- Sinclair, A.; Arnold, C.; Woodford, N. Rapid Detection and Estimation by Pyrosequencing of 23S rRNA Genes with a Single Nucleotide Polymorphism Conferring Linezolid Resistance in Enterococci. Antimicrob. Agents Chemother. 2003, 47, 3620–3622. [Google Scholar] [CrossRef] [PubMed]
- Beukers, A.G.; Hasman, H.; Hegstad, K.; Van Hal, S.J. Recommendations to Address the Difficulties Encountered When Determining Linezolid Resistance from Whole-Genome Sequencing Data. Antimicrob. Agents Chemother. 2018, 62, e00613-18. [Google Scholar] [CrossRef]
- Hasman, H.; Clausen, P.T.L.C.; Kaya, H.; Hansen, F.; Knudsen, J.D.; Wang, M.; Holzknecht, B.J.; Samulioniené, J.; Røder, B.L.; Frimodt-Møller, N.; et al. LRE-Finder, a Web Tool for Detection of the 23S rRNA Mutations and the optrA, cfr, cfr(B) and poxtA Genes Encoding Linezolid Resistance in Enterococci from Whole-Genome Sequences. J. Antimicrob. Chemother. 2019, 74, 1473–1476. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and Accurate Short Read Alignment with Burrows–Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Mowlaboccus, S.; Daley, D.A.; Coombs, G.W. Genomic Characterisation of Linezolid-Resistant Enterococcus faecalis from Western Australia 2016–2021. Pathology 2023, 55, 397–399. [Google Scholar] [CrossRef] [PubMed]
- Kerschner, H.; Cabal, A.; Hartl, R.; Machherndl-Spandl, S.; Allerberger, F.; Ruppitsch, W.; Apfalter, P. Hospital Outbreak Caused by Linezolid Resistant Enterococcus faecium in Upper Austria. Antimicrob. Resist. Infect. Control 2019, 8, 150. [Google Scholar] [CrossRef]
- Kerschner, H.; Rosel, A.C.; Hartl, R.; Hyden, P.; Stoeger, A.; Ruppitsch, W.; Allerberger, F.; Apfalter, P. Oxazolidinone Resistance Mediated by optrA in Clinical Enterococcus faecalis Isolates in Upper Austria: First Report and Characterization by Whole Genome Sequencing. Microb. Drug Resist. 2021, 27, 685–690. [Google Scholar] [CrossRef]
- Do Prado, G.V.B.; Marchi, A.P.; Moreno, L.Z.; Rizek, C.; Amigo, U.; Moreno, A.M.; Rossi, F.; Guimaraes, T.; Levin, A.S.; Costa, S.F. Virulence and Resistance Pattern of a Novel Sequence Type of Linezolid-Resistant Enterococcus faecium Identified by Whole-Genome Sequencing. J. Glob. Antimicrob. Resist. 2016, 6, 27–31. [Google Scholar] [CrossRef] [PubMed]
- Strateva, T.V.; Hristova, P.; Stoeva, T.J.; Hitkova, H.; Peykov, S. First Detection and Genomic Characterization of Linezolid-Resistant Enterococcus faecalis Clinical Isolates in Bulgaria. Microorganisms 2025, 13, 195. [Google Scholar] [CrossRef]
- Yu, Z.; Chen, Z.; Cheng, H.; Zheng, J.; Li, D.; Deng, X.; Pan, W.; Yang, W.; Deng, Q. Complete Genome Sequencing and Comparative Analysis of the Linezolid-Resistant Enterococcus faecalis Strain DENG1. Arch. Microbiol. 2014, 196, 513–516. [Google Scholar] [CrossRef]
- Chen, M.; Pan, H.; Lou, Y.; Wu, Z.; Zhang, J.; Huang, Y.; Yu, W.; Qiu, Y. Epidemiological Characteristics and Genetic Structure of Linezolid-Resistant Enterococcus faecalis. Infect. Drug Resist. 2018, 11, 2397–2409. [Google Scholar] [CrossRef] [PubMed]
- Bao, P.; Zhang, Z.; Zhao, W.; Jiang, Y.; Wang, D. Isolation and Whole-Genome Sequencing of a New Type of Linezolid-Resistant Enterococcus faecalis from Two Cases of Infective Endocarditis Following Renal Transplantation. J. Glob. Antimicrob. Resist. 2020, 20, 346–347. [Google Scholar] [CrossRef]
- Brajerova, M.; Nyc, O.; Drevinek, P.; Krutova, M. Genomic Insights into the Spread of Vancomycin- and Tigecycline-Resistant Enterococcus faecium ST117. Ann. Clin. Microbiol. Antimicrob. 2025, 24, 36. [Google Scholar] [CrossRef] [PubMed]
- Misiakou, M.-A.; Hertz, F.B.; Schønning, K.; Häussler, S.; Nielsen, K.L. Emergence of Linezolid-Resistant Enterococcus faecium in a Tertiary Hospital in Copenhagen. Microb. Genom. 2023, 9, mgen001055. [Google Scholar] [CrossRef] [PubMed]
- Hammerum, A.M.; Karstensen, K.T.; Roer, L.; Kaya, H.; Lindegaard, M.; Porsbo, L.J.; Kjerulf, A.; Pinholt, M.; Holzknecht, B.J.; Worning, P.; et al. Surveillance of Vancomycin-Resistant Enterococci Reveals Shift in Dominating Clusters from vanA to vanB Enterococcus faecium Clusters, Denmark, 2015 to 2022. Eurosurveillance 2024, 29, 2300633. [Google Scholar] [CrossRef]
- Sassi, M.; Guérin, F.; Zouari, A.; Beyrouthy, R.; Auzou, M.; Fines-Guyon, M.; Potrel, S.; Dejoies, L.; Collet, A.; Boukthir, S.; et al. Emergence of optrA-Mediated Linezolid Resistance in Enterococci from France, 2006–2016. J. Antimicrob. Chemother. 2019, 74, 1469–1472. [Google Scholar] [CrossRef]
- Olearo, F.; Both, A.; Belmar Campos, C.; Hilgarth, H.; Klupp, E.-M.; Hansen, J.L.; Maurer, F.P.; Christner, M.; Aepfelbacher, M.; Rohde, H. Emergence of Linezolid-Resistance in Vancomycin-Resistant Enterococcus faecium ST117 Associated with Increased Linezolid-Consumption. Int. J. Med. Microbiol. 2021, 311, 151477. [Google Scholar] [CrossRef]
- Bender, J.K.; Fleige, C.; Lange, D.; Klare, I.; Werner, G. Rapid Emergence of Highly Variable and Transferable Oxazolidinone and Phenicol Resistance Gene optrA in German Enterococcus spp. Clinical Isolates. Int. J. Antimicrob. Agents 2018, 52, 819–827. [Google Scholar] [CrossRef]
- Bender, J.K.; Fleige, C.; Klare, I.; Fiedler, S.; Mischnik, A.; Mutters, N.T.; Dingle, K.E.; Werner, G. Detection of a cfr(B) Variant in German Enterococcus faecium Clinical Isolates and the Impact on Linezolid Resistance in Enterococcus spp. PLoS ONE 2016, 11, e0167042. [Google Scholar] [CrossRef]
- Bakthavatchalam, Y.D.; Vasudevan, K.; Babu, P.; Neeravi, A.R.; Narasiman, V.; Veeraraghavan, B. Genomic Insights of optrA-Carrying Linezolid-Resistant Enterococcus faecium Using Hybrid Assembly: First Report from India. J. Glob. Antimicrob. Resist. 2021, 25, 331–336. [Google Scholar] [CrossRef]
- Egan, S.A.; Shore, A.C.; O’Connell, B.; Brennan, G.I.; Coleman, D.C. Linezolid Resistance in Enterococcus faecium and Enterococcus faecalis from Hospitalized Patients in Ireland: High Prevalence of the MDR Genes optrA and poxtA in Isolates with Diverse Genetic Backgrounds. J. Antimicrob. Chemother. 2020, 75, 1704–1711. [Google Scholar] [CrossRef]
- Lazaris, A.; Coleman, D.C.; Kearns, A.M.; Pichon, B.; Kinnevey, P.M.; Earls, M.R.; Boyle, B.; O’Connell, B.; Brennan, G.I.; Shore, A.C. Novel Multiresistance cfr Plasmids in Linezolid-Resistant Methicillin-Resistant Staphylococcus epidermidis and Vancomycin-Resistant Enterococcus faecium (VRE) from a Hospital Outbreak: Co-Location of cfr and optrA in VRE. J. Antimicrob. Chemother. 2017, 72, 3252–3257. [Google Scholar] [CrossRef] [PubMed]
- Labecka, L.; Ķibilds, J.; Cīrulis, A.; Čeirāne, E.; Zeltiņa, I.; Reinis, A.; Vilima, B.; Rudzīte, D.; Erts, R.; Mauliņa, I.; et al. Evaluation of Antimicrobial Resistance in Clinical Isolates of Enterococcus spp. Obtained from Hospital Patients in Latvia. Medicina 2024, 60, 850. [Google Scholar] [CrossRef]
- Hu, Y.; Won, D.; Nguyen, L.P.; Osei, K.M.; Seo, Y.; Kim, J.; Lee, Y.; Lee, H.; Yong, D.; Choi, J.R.; et al. Prevalence and Genetic Analysis of Resistance Mechanisms of Linezolid-Nonsusceptible Enterococci in a Tertiary Care Hospital Examined via Whole-Genome Sequencing. Antibiotics 2022, 11, 1624. [Google Scholar] [CrossRef]
- Ruiz-Ripa, L.; Feßler, A.T.; Hanke, D.; Eichhorn, I.; Azcona-Gutiérrez, J.M.; Pérez-Moreno, M.O.; Seral, C.; Aspiroz, C.; Alonso, C.A.; Torres, L.; et al. Mechanisms of Linezolid Resistance Among Enterococci of Clinical Origin in Spain—Detection of optrA- and cfr(D)-Carrying Enterococcus faecalis. Microorganisms 2020, 8, 1155. [Google Scholar] [CrossRef] [PubMed]
- Pincus, N.B.; Joshi, T.; Gatesy, S.W.M.; Al-Heeti, O.; Moore, W.J.; Bachta, K.E.R. Breakthrough Daptomycin-, Linezolid-, Vancomycin-Resistant Enterococcus faecium Bacteremia during Protracted Daptomycin Therapy: A Case Report. IDCases 2022, 29, e01593. [Google Scholar] [CrossRef] [PubMed]
- Deshpande, L.M.; Ashcraft, D.S.; Kahn, H.P.; Pankey, G.; Jones, R.N.; Farrell, D.J.; Mendes, R.E. Detection of a New cfr-Like Gene, cfr (B), in Enterococcus faecium Isolates Recovered from Human Specimens in the United States as Part of the SENTRY Antimicrobial Surveillance Program. Antimicrob. Agents Chemother. 2015, 59, 6256–6261. [Google Scholar] [CrossRef]
- Chilambi, G.S.; Nordstrom, H.R.; Evans, D.R.; Ferrolino, J.A.; Hayden, R.T.; Marón, G.M.; Vo, A.N.; Gilmore, M.S.; Wolf, J.; Rosch, J.W.; et al. Evolution of Vancomycin-Resistant Enterococcus faecium during Colonization and Infection in Immunocompromised Pediatric Patients. Proc. Natl. Acad. Sci. USA 2020, 117, 11703–11714. [Google Scholar] [CrossRef]
- Chacko, K.I.; Sullivan, M.J.; Beckford, C.; Altman, D.R.; Ciferri, B.; Pak, T.R.; Sebra, R.; Kasarskis, A.; Hamula, C.L.; Van Bakel, H. Genetic Basis of Emerging Vancomycin, Linezolid, and Daptomycin Heteroresistance in a Case of Persistent Enterococcus faecium Bacteremia. Antimicrob. Agents Chemother. 2018, 62, e02007-17. [Google Scholar] [CrossRef]
- Gargis, A.S.; Spicer, L.M.; Kent, A.G.; Zhu, W.; Campbell, D.; McAllister, G.; Ewing, T.O.; Albrecht, V.; Stevens, V.A.; Sheth, M.; et al. Sentinel Surveillance Reveals Emerging Daptomycin-Resistant ST736 Enterococcus faecium and Multiple Mechanisms of Linezolid Resistance in Enterococci in the United States. Front. Microbiol. 2022, 12, 807398. [Google Scholar] [CrossRef]
- Wardenburg, K.E.; Potter, R.F.; D’Souza, A.W.; Hussain, T.; Wallace, M.A.; Andleeb, S.; Burnham, C.-A.D.; Dantas, G. Phenotypic and Genotypic Characterization of Linezolid-Resistant Enterococcus faecium from the USA and Pakistan. J. Antimicrob. Chemother. 2019, 74, 3445–3452. [Google Scholar] [CrossRef]
- Mendes, R.E.; Deshpande, L.; Streit, J.M.; Sader, H.S.; Castanheira, M.; Hogan, P.A.; Flamm, R.K. ZAAPS Programme Results for 2016: An Activity and Spectrum Analysis of Linezolid Using Clinical Isolates from Medical Centres in 42 Countries. J. Antimicrob. Chemother. 2018, 73, 1880–1887. [Google Scholar] [CrossRef]
- Marshall, S.H.; Donskey, C.J.; Hutton-Thomas, R.; Salata, R.A.; Rice, L.B. Gene Dosage and Linezolid Resistance in Enterococcus faecium and Enterococcus faecalis. Antimicrob. Agents Chemother. 2002, 46, 3334–3336. [Google Scholar] [CrossRef]
- Boumghar-Bourtchaï, L.; Dhalluin, A.; Malbruny, B.; Galopin, S.; Leclercq, R. Influence of Recombination on Development of Mutational Resistance to Linezolid in Enterococcus faecalis JH2-2. Antimicrob. Agents Chemother. 2009, 53, 4007–4009. [Google Scholar] [CrossRef]
- Miller, W.R.; Munita, J.M.; Arias, C.A. Mechanisms of Antibiotic Resistance in Enterococci. Expert. Rev. Anti-Infect. Ther. 2014, 12, 1221–1236. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Wu, W.; Ni, M.; Liu, Y.; Zhang, J.; Xia, F.; He, W.; Wang, Q.; Wang, Z.; Cao, B.; et al. Linezolid-Resistant Clinical Isolates of Enterococci and Staphylococcus cohnii from a Multicentre Study in China: Molecular Epidemiology and Resistance Mechanisms. Int. J. Antimicrob. Agents 2013, 42, 317–321. [Google Scholar] [CrossRef]
- Yi, M.; Zou, J.; Zhao, J.; Tang, Y.; Yuan, Y.; Yang, B.; Huang, J.; Xia, P.; Xia, Y. Emergence of optrA-Mediated Linezolid Resistance in Enterococcus faecium: A Molecular Investigation in a Tertiary Hospital of Southwest China from 2014–2018. Infect. Drug Resist. 2022, 15, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Baccani, I.; Antonelli, A.; Galano, A.; Bartalesi, F.; Bartoloni, A.; Rossolini, G.M. Linezolid-Resistant Enterococcus faecalis Infection Following Prolonged Low-Dosage Linezolid Treatment for Multidrug-Resistant Tuberculosis. Clin. Infect. Dis. 2017, 65, 2159–2160. [Google Scholar] [CrossRef] [PubMed]
- McHugh, M.P.; Parcell, B.J.; Pettigrew, K.A.; Toner, G.; Khatamzas, E.; El Sakka, N.; Karcher, A.M.; Walker, J.; Weir, R.; Meunier, D.; et al. Presence of optrA-Mediated Linezolid Resistance in Multiple Lineages and Plasmids of Enterococcus faecalis Revealed by Long Read Sequencing. Microbiology 2022, 168, 1–7. [Google Scholar] [CrossRef]
- Lázaro-Perona, F.; Navarro-Carrera, P.; Bloise, I.; Prieto-Casado, P.; García-Pérez, I.; Paradela, A.; Corrales, F.; Cacho-Calvo, J.; Mingorance, J. Multiple Mechanisms Drive Linezolid Resistance in Clinical Enterococcus faecium Isolates by Increasing poxtA Gene Expression. J. Glob. Antimicrob. Resist. 2025, 42, 113–119. [Google Scholar] [CrossRef]
- Deshpande, L.M.; Castanheira, M.; Flamm, R.K.; Mendes, R.E. Evolving Oxazolidinone Resistance Mechanisms in a Worldwide Collection of Enterococcal Clinical Isolates: Results from the SENTRY Antimicrobial Surveillance Program. J. Antimicrob. Chemother. 2018, 73, 2314–2322. [Google Scholar] [CrossRef]
- Wilson, D.N.; Hauryliuk, V.; Atkinson, G.C.; O’Neill, A.J. Target Protection as a Key Antibiotic Resistance Mechanism. Nat. Rev. Microbiol. 2020, 18, 637–648. [Google Scholar] [CrossRef]
- Brenciani, A.; Morroni, G.; Schwarz, S.; Giovanetti, E. Oxazolidinones: Mechanisms of Resistance and Mobile Genetic Elements Involved. J. Antimicrob. Chemother. 2022, 77, 2596–2621. [Google Scholar] [CrossRef]
- Bender, J.K.; Fleige, C.; Klare, I.; Werner, G. Development of a Multiplex-PCR to Simultaneously Detect Acquired Linezolid Resistance Genes cfr, optrA and poxtA in Enterococci of Clinical Origin. J. Microbiol. Methods 2019, 160, 101–103. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, Y.; Bollas, A.; Wang, Y.; Au, K.F. Nanopore Sequencing Technology, Bioinformatics and Applications. Nat. Biotechnol. 2021, 39, 1348–1365. [Google Scholar] [CrossRef] [PubMed]
- Kehrenberg, C.; Schwarz, S.; Jacobsen, L.; Hansen, L.H.; Vester, B. A New Mechanism for Chloramphenicol, Florfenicol and Clindamycin Resistance: Methylation of 23S Ribosomal RNA at A2503. Mol. Microbiol. 2005, 57, 1064–1073. [Google Scholar] [CrossRef]
- Giessing, A.M.B.; Jensen, S.S.; Rasmussen, A.; Hansen, L.H.; Gondela, A.; Long, K.; Vester, B.; Kirpekar, F. Identification of 8-Methyladenosine as the Modification Catalyzed by the Radical SAM Methyltransferase cfr That Confers Antibiotic Resistance in Bacteria. RNA 2009, 15, 327–336. [Google Scholar] [CrossRef] [PubMed]
- Guerin, F.; Sassi, M.; Dejoies, L.; Zouari, A.; Schutz, S.; Potrel, S.; Auzou, M.; Collet, A.; Lecointe, D.; Auger, G.; et al. Molecular and Functional Analysis of the Novel cfr(D) Linezolid Resistance Gene Identified in Enterococcus faecium. J. Antimicrob. Chemother. 2020, 75, 1699–1703. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Deng, Z.; Shen, Y.; Wei, W.; Xiang, Q.; Liu, Z.; Hanf, K.; Huang, S.; Lv, Z.; Cao, T.; et al. High Prevalence and Plasmidome Diversity of optrA-Positive Enterococci in a Shenzhen Community, China. Front. Microbiol. 2024, 15, 1505107. [Google Scholar] [CrossRef]
- Schwarz, S.; Zhang, W.; Du, X.-D.; Krüger, H.; Feßler, A.T.; Ma, S.; Zhu, Y.; Wu, C.; Shen, J.; Wang, Y. Mobile Oxazolidinone Resistance Genes in Gram-Positive and Gram-Negative Bacteria. Clin. Microbiol. Rev. 2021, 34, 1–63. [Google Scholar] [CrossRef]
- Diaz, L.; Kiratisin, P.; Mendes, R.E.; Panesso, D.; Singh, K.V.; Arias, C.A. Transferable Plasmid-Mediated Resistance to Linezolid Due to cfr in a Human Clinical Isolate of Enterococcus faecalis. Antimicrob. Agents Chemother. 2012, 56, 3917–3922. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, D.; Zhang, J.; Liu, L.; Zhang, Z.; Liu, C.; Hu, S.; Wu, L.; He, Z.; Sun, H. Genomic Epidemiology Reveals Multiple Mechanisms of Linezolid Resistance in Clinical Enterococci in China. Ann. Clin. Microbiol. Antimicrob. 2024, 23, 41. [Google Scholar] [CrossRef]
- Kuroda, M.; Sekizuka, T.; Matsui, H.; Suzuki, K.; Seki, H.; Saito, M.; Hanaki, H. Complete Genome Sequence and Characterization of Linezolid-Resistant Enterococcus faecalis Clinical Isolate KUB3006 Carrying a cfr(B)-Transposon on Its Chromosome and optrA-Plasmid. Front. Microbiol. 2018, 9, 2576. [Google Scholar] [CrossRef]
- Martínez-Ayala, P.; Perales-Guerrero, L.; Gómez-Quiroz, A.; Avila-Cardenas, B.B.; Gómez-Portilla, K.; Rea-Márquez, E.A.; Vera-Cuevas, V.C.; Gómez-Quiroz, C.A.; Briseno-Ramírez, J.; De Arcos-Jiménez, J.C. Whole-Genome Sequencing of Linezolid-Resistant and Linezolid-Intermediate-Susceptibility Enterococcus faecalis Clinical Isolates in a Mexican Tertiary Care University Hospital. Microorganisms 2025, 13, 684. [Google Scholar] [CrossRef]
- Sharkey, L.K.R.; Edwards, T.A.; O’Neill, A.J. ABC-F Proteins Mediate Antibiotic Resistance through Ribosomal Protection. MBio 2016, 7, e01975-15. [Google Scholar] [CrossRef] [PubMed]
- Sharkey, L.K.R.; O’Neill, A.J. Antibiotic Resistance ABC-F Proteins: Bringing Target Protection into the Limelight. ACS Infect. Dis. 2018, 4, 239–246. [Google Scholar] [CrossRef]
- Crowe-McAuliffe, C.; Graf, M.; Huter, P.; Takada, H.; Abdelshahid, M.; Nováček, J.; Murina, V.; Atkinson, G.C.; Hauryliuk, V.; Wilson, D.N. Structural Basis for Antibiotic Resistance Mediated by the Bacillus subtilis ABCF ATPase VmlR. Proc. Natl. Acad. Sci. USA 2018, 115, 8978–8983. [Google Scholar] [CrossRef] [PubMed]
- Murina, V.; Kasari, M.; Takada, H.; Hinnu, M.; Saha, C.K.; Grimshaw, J.W.; Seki, T.; Reith, M.; Putrinš, M.; Tenson, T.; et al. ABCF ATPases Involved in Protein Synthesis, Ribosome Assembly and Antibiotic Resistance: Structural and Functional Diversification across the Tree of Life. J. Mol. Biol. 2019, 431, 3568–3590. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lv, Y.; Cai, J.; Schwarz, S.; Cui, L.; Hu, Z.; Zhang, R.; Li, J.; Zhao, Q.; He, T.; et al. A Novel Gene, optrA, That Confers Transferable Resistance to Oxazolidinones and Phenicols and Its Presence in Enterococcus faecalis and Enterococcus faecium of Human and Animal Origin. J. Antimicrob. Chemother. 2015, 70, 2182–2190. [Google Scholar] [CrossRef]
- Freitas, A.R.; Tedim, A.P.; Novais, C.; Lanza, V.F.; Peixe, L. Comparative Genomics of Global optrA-Carrying Enterococcus faecalis Uncovers a Common Chromosomal Hotspot for optrA Acquisition within a Diversity of Core and Accessory Genomes. Microb. Genom. 2020, 6, e000350. [Google Scholar] [CrossRef]
- Elghaieb, H.; Freitas, A.R.; Abbassi, M.S.; Novais, C.; Zouari, M.; Hassen, A.; Peixe, L. Dispersal of Linezolid-Resistant Enterococci Carrying poxtA or optrA in Retail Meat and Food-Producing Animals from Tunisia. J. Antimicrob. Chemother. 2019, 74, 2865–2869. [Google Scholar] [CrossRef]
- Li, Y.; Jiang, T.; Mao, J.; Xu, F.; Zhang, R.; Yan, J.; Cai, J.; Xie, Y. Prevalence and Genetic Diversity of optrA-Positive Enterococci Isolated from Patients in an Anorectal Surgery Ward of a Chinese Hospital. Front. Microbiol. 2024, 15, 1481162. [Google Scholar] [CrossRef]
- Antonelli, A.; D’Andrea, M.M.; Brenciani, A.; Galeotti, C.L.; Morroni, G.; Pollini, S.; Varaldo, P.E.; Rossolini, G.M. Characterization of poxtA, a Novel Phenicol–Oxazolidinone–Tetracycline Resistance Gene from an MRSA of Clinical Origin. J. Antimicrob. Chemother. 2018, 73, 1763–1769. [Google Scholar] [CrossRef] [PubMed]
- Dejoies, L.; Sassi, M.; Schutz, S.; Moreaux, J.; Zouari, A.; Potrel, S.; Collet, A.; Lecourt, M.; Auger, G.; Cattoir, V. Genetic Features of the poxtA Linezolid Resistance Gene in Human Enterococci from France. J. Antimicrob. Chemother. 2021, 76, 1978–1985. [Google Scholar] [CrossRef] [PubMed]
- Baccani, I.; Antonelli, A.; Di Pilato, V.; Coppi, M.; Di Maggio, T.; Spinicci, M.; Villagran, A.L.; Revollo, C.; Bartoloni, A.; Rossolini, G.M. Detection of poxtA2, a Presumptive poxtA Ancestor, in a Plasmid from a Linezolid-Resistant Enterococcus gallinarum Isolate. Antimicrob. Agents Chemother. 2021, 65, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Gagetti, P.; Faccone, D.; Ceriana, P.; Lucero, C.; Menocal, A.; Argentina, G.L.; Corso, A. Emergence of optrA-Mediated Linezolid Resistance in Clinical Isolates of Enterococcus faecalis from Argentina. J. Glob. Antimicrob. Resist. 2023, 35, 335–341. [Google Scholar] [CrossRef]
- Pan, P.; Sun, L.; Shi, X.; Huang, X.; Yin, Y.; Pan, B.; Hu, L.; Shen, Q. Analysis of Molecular Epidemiological Characteristics and Antimicrobial Susceptibility of Vancomycin-Resistant and Linezolid-Resistant Enterococcus in China. BMC Med. Genom. 2024, 17, 174. [Google Scholar] [CrossRef]
- Chen, H.; Wang, X.; Yin, Y.; Li, S.; Zhang, Y.; Wang, Q.; Wang, H. Molecular Characteristics of Oxazolidinone Resistance in Enterococci from a Multicenter Study in China. BMC Microbiol. 2019, 19, 162. [Google Scholar] [CrossRef]
- Wu, W.; Xiao, S.; Han, L.; Wu, Q. Antimicrobial Resistance, Virulence Gene Profiles, and Molecular Epidemiology of Enterococcal Isolates from Patients with Urinary Tract Infections in Shanghai, China. Microbiol. Spectr. 2025, 13, e01217-24. [Google Scholar] [CrossRef]
- Yang, P.; Li, J.; Lv, M.; He, P.; Song, G.; Shan, B.; Yang, X. Molecular Epidemiology and Horizontal Transfer Mechanism of optrA -Carrying Linezolid-Resistant Enterococcus faecalis. Pol. J. Microbiol. 2024, 73, 349–362. [Google Scholar] [CrossRef]
- Saavedra, S.Y.; Bernal, J.F.; Montilla-Escudero, E.; Torres, G.; Rodríguez, M.K.; Hidalgo, A.M.; Ovalle, M.V.; Rivera, S.; Perez-Gutierrez, E.; Duarte, C. Vigilancia nacional de aislamientos clínicos de Enterococcus faecalis resistentes al linezolid portadores del gen optrA en Colombia, 2014–2019. Rev. Panam. Salud Pública 2020, 44, 1. [Google Scholar] [CrossRef]
- Tsilipounidaki, K.; Gerontopoulos, A.; Papagiannitsis, C.; Petinaki, E. First Detection of an optrA-Positive, Linezolid-Resistant ST16 Enterococcus faecalis from Human in Greece. New Microbes New Infect. 2019, 29, 100515. [Google Scholar] [CrossRef]
- Egan, S.A.; Corcoran, S.; McDermott, H.; Fitzpatrick, M.; Hoyne, A.; McCormack, O.; Cullen, A.; Brennan, G.I.; O’Connell, B.; Coleman, D.C. Hospital Outbreak of Linezolid-Resistant and Vancomycin-Resistant ST80 Enterococcus faecium Harbouring an optrA-Encoding Conjugative Plasmid Investigated by Whole-Genome Sequencing. J. Hosp. Infect. 2020, 105, 726–735. [Google Scholar] [CrossRef]
- Segawa, T.; Hisatsune, J.; Ishida-Kuroki, K.; Sugawara, Y.; Masuda, K.; Tadera, K.; Kashiyama, S.; Yokozaki, M.; Le, M.N.-T.; Kawada-Matsuo, M.; et al. Complete Genome Sequence of optrA-Carrying Enterococcus faecalis Isolated from Open Pus in a Japanese Patient. J. Glob. Antimicrob. Resist. 2023, 33, 276–278. [Google Scholar] [CrossRef]
- Wardal, E.; Żabicka, D.; Skalski, T.; Kubiak-Pulkowska, J.; Hryniewicz, W.; Sadowy, E. Characterization of a Tigecycline-, Linezolid- and Vancomycin-Resistant Clinical Enteroccoccus faecium Isolate, Carrying vanA and vanB Genes. Infect. Dis. Ther. 2023, 12, 2545–2565. [Google Scholar] [CrossRef]
- Kamus, L.; Auger, G.; Gambarotto, K.; Houivet, J.; Ramiandrisoa, M.; Picot, S.; Lugagne-Delpon, N.; Jaffar-Bandjee, M.-C.; Zouari, A.; Birer, A.; et al. Investigation of a vanA Linezolid- and Vancomycin-Resistant Enterococcus faecium Outbreak in the Southwest Indian Ocean (Reunion Island). Int. J. Antimicrob. Agents 2022, 60, 106686. [Google Scholar] [CrossRef]
- Farman, M.; Yasir, M.; Al-Hindi, R.R.; Farraj, S.A.; Jiman-Fatani, A.A.; Alawi, M.; Azhar, E.I. Genomic Analysis of Multidrug-Resistant Clinical Enterococcus faecalis Isolates for Antimicrobial Resistance Genes and Virulence Factors from the Western Region of Saudi Arabia. Antimicrob. Resist. Infect. Control 2019, 8, 55. [Google Scholar] [CrossRef] [PubMed]
- On, Y.; Lee, S.Y.; Yoo, J.S.; Kim, J.W. Molecular Epidemiology and Genetic Context of optrA-Carrying Linezolid-Resistant Enterococci from Humans and Animals in South Korea. Antibiotics 2025, 14, 571. [Google Scholar] [CrossRef] [PubMed]
- Càmara, J.; Camoez, M.; Tubau, F.; Pujol, M.; Ayats, J.; Ardanuy, C.; Domínguez, M.Á. Detection of the Novel optrA Gene Among Linezolid-Resistant Enterococci in Barcelona, Spain. Microb. Drug Resist. 2019, 25, 87–93. [Google Scholar] [CrossRef]
- Rodríguez-Lucas, C.; Fernández, J.; Vázquez, X.; De Toro, M.; Ladero, V.; Fuster, C.; Rodicio, R.; Rodicio, M.R. Detection of the optrA Gene Among Polyclonal Linezolid-Susceptible Isolates of Enterococcus faecalis Recovered from Community Patients. Microb. Drug Resist. 2022, 28, 773–779. [Google Scholar] [CrossRef] [PubMed]
- Fang, H.; Fröding, I.; Ullberg, M.; Giske, C.G. Genomic Analysis Revealed Distinct Transmission Clusters of Vancomycin-Resistant Enterococcus faecium ST80 in Stockholm, Sweden. J. Hosp. Infect. 2021, 107, 12–15. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.; Guo, Y.; Chen, C.; Fuzhu, Y.; Xiao, S.; Zhu, D.; Wang, M.; Xu, X. Draft Genome Sequence of Linezolid-Resistant Enterococcus faecalis Clinical Isolate HS0914. Genome Announc. 2014, 2, 1. [Google Scholar] [CrossRef]
- More Information for Sivextro (Tedizolid). FDA 2019. Available online: https://www.fda.gov/drugs/drug-approvals-and-databases/more-information-sivextro-tedizolid (accessed on 20 August 2025).
- Sivextro European Medicines Agency (EMA). Available online: https://www.ema.europa.eu/en/medicines/human/EPAR/sivextro (accessed on 20 August 2025).
- Rybak, J.M.; Roberts, K. Tedizolid Phosphate: A Next-Generation Oxazolidinone. Infect. Dis. Ther. 2015, 4, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Im, W.B.; Choi, S.H.; Park, J.-Y.; Choi, S.H.; Finn, J.; Yoon, S.-H. Discovery of Torezolid as a Novel 5-Hydroxymethyl-Oxazolidinone Antibacterial Agent. Eur. J. Med. Chem. 2011, 46, 1027–1039. [Google Scholar] [CrossRef]
- Carvalhaes, C.G.; Sader, H.S.; Flamm, R.K.; Streit, J.M.; Mendes, R.E. Assessment of Tedizolid In Vitro Activity and Resistance Mechanisms against a Collection of Enterococcus spp. Causing Invasive Infections, Including Isolates Requiring an Optimized Dosing Strategy for Daptomycin from U.S. and European Medical Centers, 2016 to 2018. Antimicrob. Agents Chemother. 2020, 64, e00175-20. [Google Scholar] [CrossRef]
- Carvalhaes, C.G.; Sader, H.S.; Streit, J.M.; Mendes, R.E. Five-Year Analysis of the In Vitro Activity of Tedizolid against a Worldwide Collection of Indicated Species Causing Clinical Infections: Results from the Surveillance of Tedizolid Activity and Resistance (STAR) Programme. JAC-Antimicrob. Resist. 2022, 4, dlac088. [Google Scholar] [CrossRef]
- Shoji, R.; Maeda, M.; Yamaguchi, K.; Takuma, T.; On, R.; Ugajin, K.; Okatomi, D.; Fukuchi, K.; Tokimatsu, I.; Ishino, K. Resistance Mechanisms and Tedizolid Susceptibility in Clinical Isolates of Linezolid-Resistant Bacteria in Japan. JAC-Antimicrob. Resist. 2025, 7, dlaf097. [Google Scholar] [CrossRef]
- Goda, N.B.; El-Ganiny, A.M.; El-khamissy, T.R.; Najar, F.Z.; Kadry, A.A. Identification of Genetic Mutations Conferring Tedizolid Resistance in MRSA Mutants. Eur. J. Clin. Microbiol. Infect. Dis. 2025, 44, 1917–1924. [Google Scholar] [CrossRef]
- Coll, F.; Gouliouris, T.; Blane, B.; Yeats, C.A.; Raven, K.E.; Ludden, C.; Khokhar, F.A.; Wilson, H.J.; Roberts, L.W.; Harrison, E.M.; et al. Antibiotic Resistance Determination Using Enterococcus faecium Whole-Genome Sequences: A Diagnostic Accuracy Study Using Genotypic and Phenotypic Data. Lancet Microbe 2024, 5, e151–e163. [Google Scholar] [CrossRef]
- Wenzler, E.; Santarossa, M.; Meyer, K.A.; Harrington, A.T.; Reid, G.E.; Clark, N.M.; Albarillo, F.S.; Bulman, Z.P. In Vitro Pharmacodynamic Analyses Help Guide the Treatment of Multidrug-Resistant Enterococcus faecium and Carbapenem-Resistant Enterobacter cloacae Bacteremia in a Liver Transplant Patient. Open Forum Infect. Dis. 2020, 7, ofz545. [Google Scholar] [CrossRef]
- Beh, J.Q.; Daniel, D.S.; Judd, L.M.; Wick, R.R.; Kelley, P.; Cronin, K.M.; Sherry, N.L.; Howden, B.P.; Connor, C.H.; Webb, J.R. Genomics to Understand the Global Landscape of Linezolid Resistance in Enterococcus faecium and Enterococcus faecalis. Microb. Genom. 2025, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Yang, F.; Lu, Y.; Huang, W. Identification of Enterococcus faecalis in a Patient with Urinary-Tract Infection Based on Metagenomic Next-Generation Sequencing: A Case Report. BMC Infect. Dis. 2020, 20, 467. [Google Scholar] [CrossRef] [PubMed]
- Paudel, R.; Nepal, H.P. Linezolid Resistance in Vancomycin Resistant Enterococci: A Worrisome Situation. Int. J. Basic. Clin. Pharmacol. 2021, 10, 464. [Google Scholar] [CrossRef]
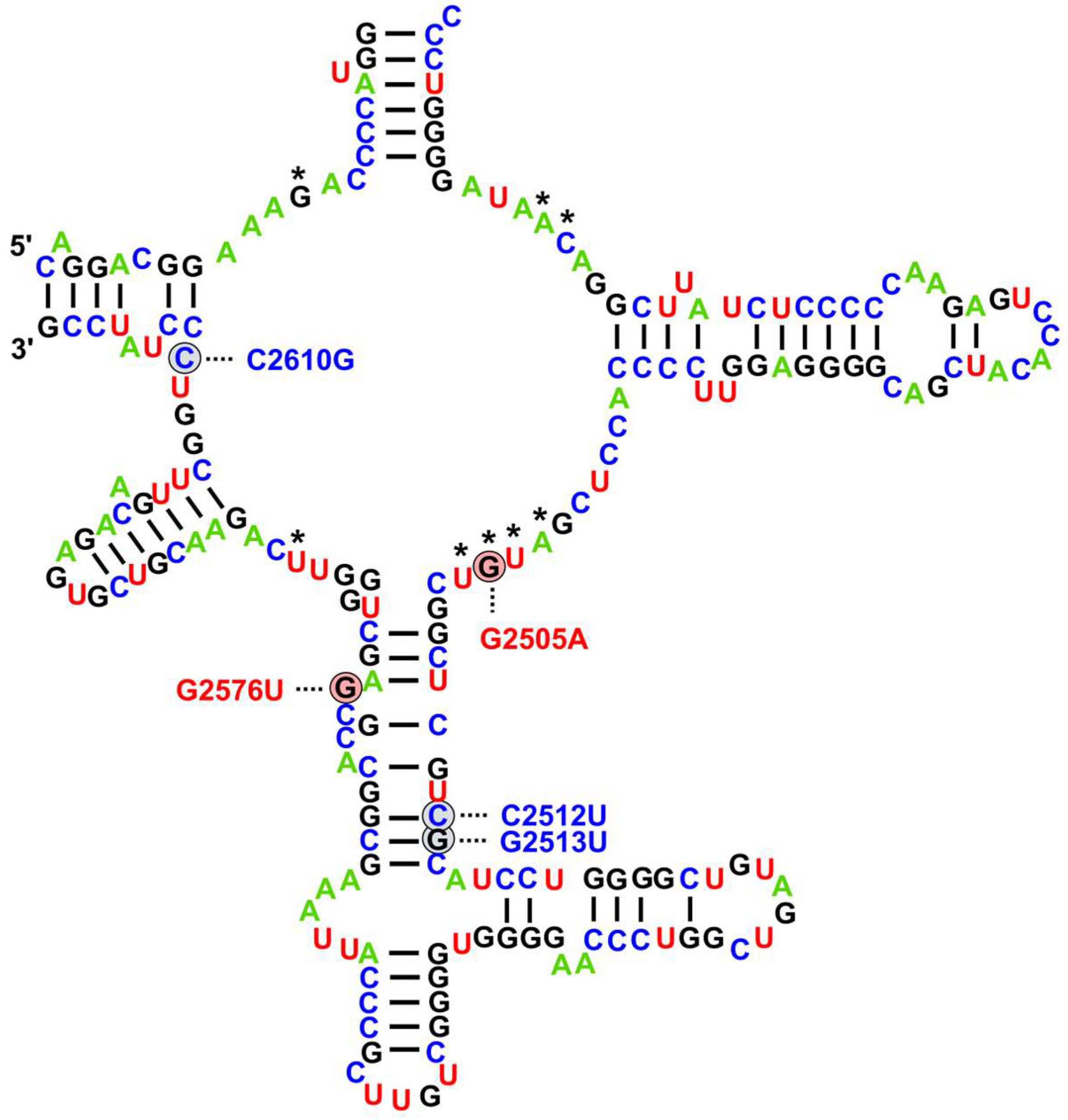
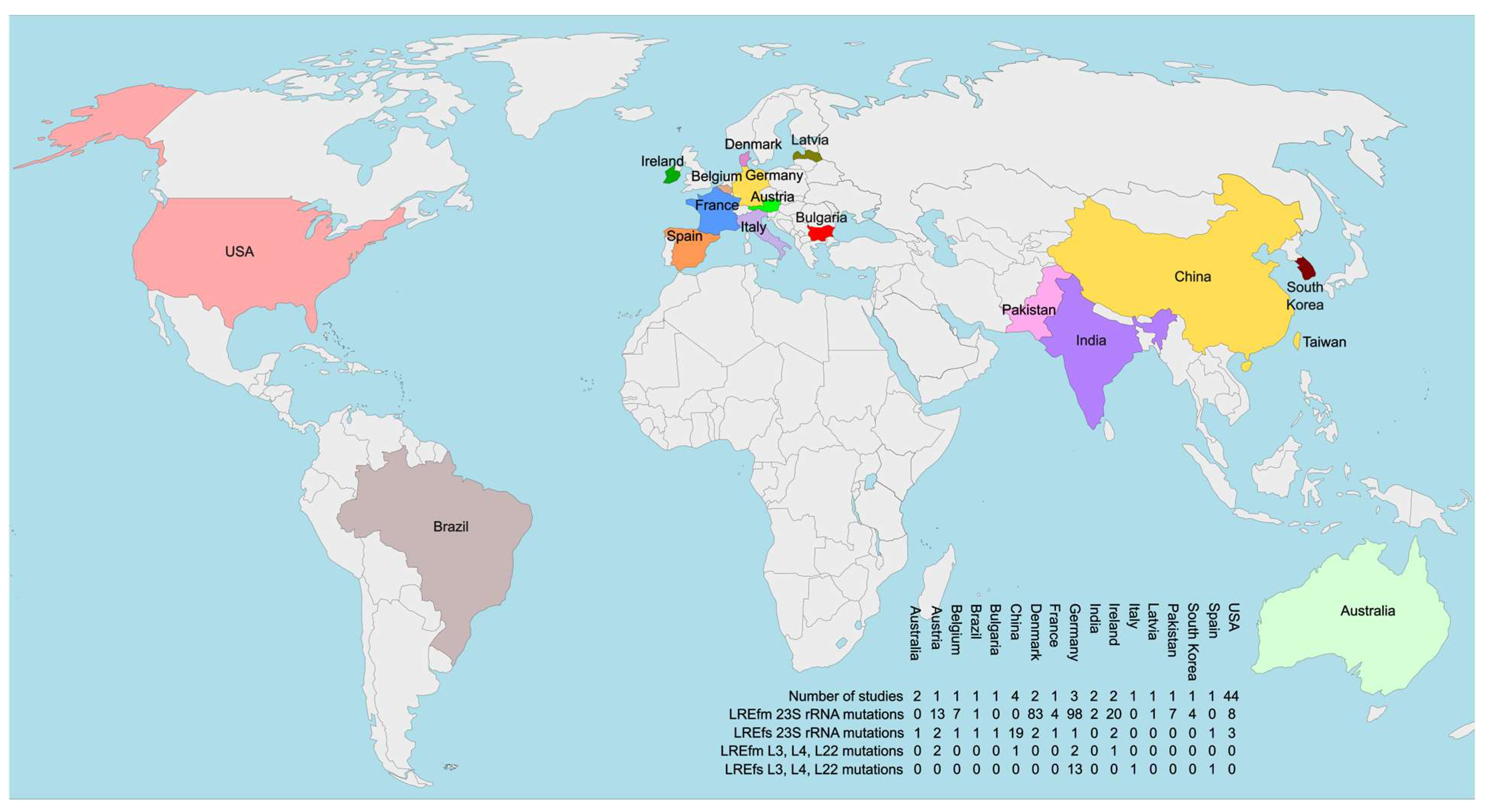
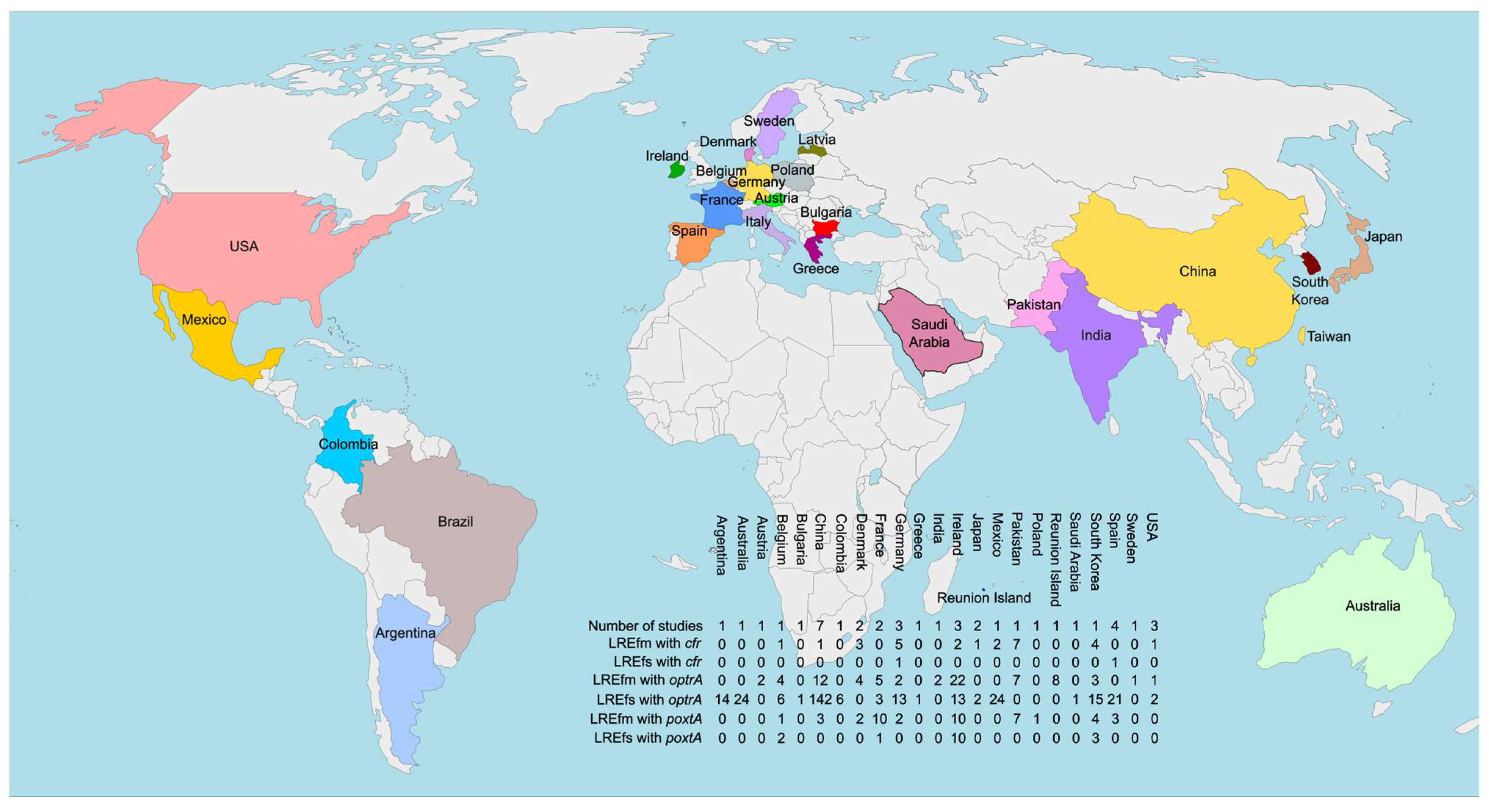
| Country | Year | Genomes with 23S rRNA Mutations | Additional Resistance Mechanisms | Source |
|---|---|---|---|---|
| Australia | 2016–2021 | 1 LREfs with G2576U (3 loci affected) | No Data | [69] |
| Austria | 2014–2017 | 13 LREfm with G2576U and 3 of them also harbor A2598G variant | No Data | [70] |
| Austria | 2017 | 1 LREfs with G388A/D130N (1 locus), T2802 C/I934M (1 locus) and T2838C/- (2 loci); 1 LREfs with G388A/D130N (3 loci), T2802 C/I934M (1 locus) and T2838C/- (2 loci) | optrA_4, T301C/F101L in L4; optrA_3, T301C/F101L in L4 | [71] |
| Belgium | 2013–2025 | 7 LREfm with G2576U | 1 LREFm with cfr | [49] |
| Brazil | 2013 | 1 LREfm with G2576U | No Data | [72] |
| Bulgaria | 2018 | 1 LREfs with G2576U (3 loci) 1 LREfs with C2163T (1 locus) | No Data optrA | [73] |
| China | - | 1 LREfs with G2576U (2 loci) | No Data | [74] |
| China | 2011–2015 | 16 LREfs with G2576U | 10 LREfs with optrA | [75] |
| China | 2018 | 2 LREfs with novel mutations | No Data | [76] |
| Czech Republic | 2021 | 1 LREfm with G2576U | No Data | [77] |
| Denmark | 2014–2023 | 52 LREfm with G2576U | 1 LREfm with cfr + poxtA | [78] |
| Denmark | 2015–2022 | 31 LREfm with G2576U | 1 LREfm with optrA | [79] |
| France | 2006–2016 | 4 LREfm with G2576U | 3 LREfm with optrA | [80] |
| Germany | 2014–2018 | 96 LREfm with G2576U | 1 LREfm with poxtA | [81] |
| Germany | 2007–2017 | 1 LREfs with G2576U | optrA | [82] |
| Germany | 2013–2015 | 1 LREfm with G2576U (4 affected loci) 1 LREfm with G2576U (2 affected loci) | cfr(B) cfr(B) | [83] |
| India | 2019 | 2 LREfm with G2592T | 2 LREfm with optrA | [84] |
| Ireland | 2016–2019 | 19 LREfm with G2576U (1–5 affected loci) 2 LREfs with G2576U | 1 LREfm with poxtA No Data | [85] |
| Ireland | 2013–2014 | 1 LREfm with G2576U (4 loci affected) | optrA, cfr, T150A in L3 | [86] |
| Latvia | 2021–2022 | 1 LREfm with G2576U | No Data | [87] |
| Pakistan | 2021–2023 | 7 LREfm with G2576U | 6 LREfm with optrA | [46] |
| South Korea | 2019–2020 | 4 LREfm with G2576U | No Data | [88] |
| Spain | 2017–2018 | 1 LREfs with G2576U | optrA | [89] |
| USA | - | 3 LREfm with G2576U (2 × 3 loci affected and 1 × 2 loci affected) | No Data | [90] |
| USA | 2012–2013 | 1 LREfm with G2576U (1 locus affected) | cfr(B) | [91] |
| USA | 2009–2019 | 4 LREfm with G2576U | No Data | [92] |
| USA | 2015 | 1 LREfm with G2576U (2–3 loci affected) | No Data | [93] |
| USA | 2017 | 4 LREfm with G2576U | No Data | [47] |
| USA | 2013–2016 | 2 LREfm with G2576U | No Data | [94] |
| USA | 2018–2019 | 3 LREfs with G2576U | No Data | [51] |
| USA | 2015–2016 | 29 LREfm with G2576U | No Data | [95] |
| ZAAPS programme | - | 2 LREfm with G2576U | No Data | [96] |
| Isolate | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sequence type | 80 | 203 | 787 | 203 | 203 | 203 | 17 | 17 | 787 | 787 | 789 | 787 | 789 | 117 | 80 | 80 | 80 | 80 |
| G2576 copy number | 2 | 3 | 3 | 2 | 2 | 2 | 4 | 5 | 1 | 3 | 3 | 2 | 4 | 2 | 3 | 2 | 3 | 3 |
| LNZ MIC (mg/L) | ≥ 256 | ≥ 256 | ≥ 256 | ≥ 256 | 16 | 48 | 48 | 64 | 32 | 32 | ≥ 256 | ≥ 256 | ≥ 256 | 32 | ≥ 256 | 64 | 16 | 64 |
| Country | Year | Genomes with L3 and/or L4 Mutations | Additional Resistance Mechanisms | Source |
|---|---|---|---|---|
| Austria | 2017 | 1 LREfm with T301C/F101L in L4 1 LREfm with T301C/F101L in L4 + C120T/- in L22 | optrA + 23S mutations (see Table 1) optrA + 23S mutations (see Table 1) | [71] |
| China | 2014–2018 | 1 LREfm with Ser77Thr in L22 | No Data | [101] |
| Germany | 2007–2017 | 13 LREfs with F101L in L4 | optrA | [82] |
| Germany | 2013–2015 | 2 LREfm with 211GGT213 (glycine insertion) in L4 | cfr(B) | [83] |
| Ireland | 2013–2014 | 1 LREfm with T150A in L3 | G2576U (4 loci affected), optrA, cfr | [86] |
| Italy | 2016 | 1 LREfs with 71Gly72 in L4 | No Data | [102] |
| Scotland | 2014–2017 | 5 LREfs with T150A in L3 and F101L in L4 1 LREfs with T150A in L3 and F101L in L4 | optrA optrA + cfr(D) | [103] |
| Spain | 2023 | 1 LREfm with Lys68Glu in L4 | poxtA | [104] |
| SENTRY Program | 2008–2016 | 4 LREfs with F101L in L4 1 LREfs with F101L in L4 | optrA optrA + cfr | [105] |
| Country | Year | Genomes Harboring cfr | Additional Resistance Mechanisms | Source |
|---|---|---|---|---|
| Belgium | 2013–2021 | 1 LREfm with cfr(B) | G2576U | [49] |
| China | 2011–2022 | 1 LREfm with cfr(D) | optrA | [116] |
| Denmark | 2014–2023 | 1 LREfm with cfr 1 LREfm with cfr | poxtA + G2576U poxtA | [78] |
| Denmark | 2015–2022 | 1 LREfm with cfrB | No Data | [79] |
| Germany | 2007–2017 | 1 LREfs with cfr | No Data | [82] |
| Germany | 2013–2015 | 1 LREfm with cfr(B) 2 LREfm with cfr(B) 2 LREfm with cfr(B) | No Data G2576U 211GGT213 insertion in the gene for L4 | [83] |
| Ireland | 2016–2019 | 1 LREfm with cfr(D) | optrA | [85] |
| Ireland | 2013–2014 | 1 LREfm wit cfr | G2576U (4 copies), T150A in L3, optrA | [86] |
| Italy | 2 LREfm with cfr(D) 1 LREfs with cfr(D) | optrA optrA | [52] | |
| Japan | 2017 | 1 LREfs with cfr(B) | optrA | [117] |
| Mexico | 2023–2024 | 2 LREfs with cfr(A) | optrA | [118] |
| Pakistan | 2021–2023 | 6 LREfm with cfr(D) 1 LREfm with cfr(D) | poxtA optrA + poxtA | [46] |
| Scotland | 2014–2017 | 1 LREfs with cfr(D) | optr(A) + T150A in L3 + F101L in L4 | [103] |
| South Korea | 2019–2020 | 3 LREfm with cfr(D) 1 LREfm with cfr(D) | poxtA optrA + poxtA | [88] |
| Spain | 2017–2018 | 1 LREfs with cfr(D) | optrA | [89] |
| USA | 2012–2013 | 1 LREfm with cfr(B) | No data | [91] |
| SENTRY Program | 2008–2016 | 1 LREfs with cfr 1 LREfs with cfr | optrA optrA + F101L in L4 | [105] |
| Country | Year | Genomes Harboring optrA/poxtA | Additional Resistance Mechanisms | Source |
|---|---|---|---|---|
| Argentina | 2016–2021 | 14 LREfs with optrA | No Data | [130] |
| Austarlia | 2016–2021 | 24 LREfs with optrA | No Data | [69] |
| Austria | 2017 | 2 LREfm with optrA | 23S rRNA mutations (Table 1) and L4 mutations (Table 3) | [71] |
| Belgium | 2013–2021 | 3 LREfm with optrA 1 LREfm with optrA and poxtA 61 LREfs with optrA 2 LREfs with poxtA | No Data No Data No Data No Data | [49] |
| Bulgaria | 2023 | 1 LEEfs with optrA | 23S rRNA mutation C2163T | [73] |
| China | 2012–2021 | 11 LREfs with optrA | No Data | [131] |
| China | 2014–2017 | 6 LREfm with optrA 1 LREfm with optrA + poxtA 1 LREfm with poxtA | No Data No Data No Data | [101] |
| China | 2011–2022 | 2 LREfm with optrA 1 LREfm with optrA 1 LREfm with poxtA 61 LREfs with optrA | No Data cfr(D) No Data No Data | [116] |
| China | 2009–2013 | 1 LREfm with optrA 12 LREfs with optrA | No Data No Data | [132] |
| China | 2022–2023 | 5 LREfs with optrA | No Data | [133] |
| China | 2011–2015 | 10 LREfs with optrA 13 LREfs with optrA | G2576U No Data | [75] |
| China | 2018–2022 | 30 LREfs with optrA | No Data | [134] |
| Colombia | 2014–2019 | 6 LREFs with optrA | No Data | [135] |
| Denmark | 2014–2023 | 1 LREFm with poxtA 1 LREFm with poxtA | cfr cfr + G3576U | [78] |
| Denmark | 2015–2022 | 3 LREfm with optrA 1 LREfm with optrA | No Data G2576U | [79] |
| France | 2006–2016 | 3 LREfm with optrA 2 LREfm with optrA 3 LREfs with optrA | G2576U No Data No Data | [80] |
| France | 2016–2020 | 10 LREfm with poxtA 1 LREfs with poxtA | No Data No Data | [128] |
| Germany | 2014–2018 | 1 LREfm with poxtA 1 LREfm with poxtA | G2567U No Data | [81] |
| Germany | 2007–2017 | 1 LREfm with optrA 1 LREfm with optrA 13 LREfs with optrA | No Data G2576U No Data | [82] |
| Greece | 2018 | 1 LREfs with optrA | No Data | [136] |
| India | 2019 | 1 LREfm with optrA 1LREfm with optrA 2 copies | G2592T G2592T | [84] |
| Ireland | 2019 | 19 LREfm with optrA | No Data | [137] |
| Ireland | 2016–2019 | 1 LREfm with optrA + poxtA 1 LREfm with optrA 1 LREfm with optrA 9LREfm with poxtA 13 LREfs with optrA 10 LREfs with poxtA | No Data cfr(D) No Data No Data No Data No Data | [85] |
| Ireland | 2013–2014 | 1 LREfm with optrA | G2576U (4 copies), T150A in L3, cfr | [86] |
| Italy | 2 LREfm with optrA 1 LREfs with optrA | cfr(D) cfr(D) | [52] | |
| Japan | 2017 | 1 LREfs with optrA | cfr(B) | [117] |
| Japan | 2021 | 1 LREfs with optrA | No Data | [138] |
| Mexico | 2023–2024 | 22 LREfs with optrA 2 LREfs with optrA | No Data cfr(A) | [118] |
| Pakistan | 2021–2023 | 6 LREfm with optrA 1 LREfm with optrA + poxtA 6 LREfm with poxtA | G2576U cfr(D) cfr(D) | [46] |
| Poland | 2020 | 1 LREfm with poxtA | No Data | [139] |
| Reunion Island | 2015–2019 | 8LREfm with optrA | No data | [140] |
| Saudi Arabia | 2014–2015 | 1 LREfs with optrA | No Data | [141] |
| Scotland | 2014–2017 | 5 LREfs with optrA 1 LREfs with optrA | T150A in L3 + F101L in L4 cfr(D) + T150A in L3 + F101L in L4 | [103] |
| South Korea | 2017–2019 | 2 LREfs with optrA | No data | [142] |
| South Korea | 2019–2020 | 2 LREfm with optrA 1 LREfm with optrA + poxtA 3 LREfm with poxtA 15 LREfs with optrA 3 LREfs with poxtA | No Data cfr(D) cfr(D) No Data No Data | [88] |
| Spain | 2016–2017 | 5 LREfs with optrA | No Data | [143] |
| Spain | 2017–2018 | 11 LREfs with optrA 1 LREfs with optrA 1 LREfs with optrA | No Data cfr(D) G2576U | [89] |
| Spain | 2023 | 2 LREfm with poxtA 1 LREfm with poxtA | No Data Lys68Glu in L4 | [104] |
| Spain | 2018 | 3 LREfs with optrA | No Data | [144] |
| Sweden | 2017–2020 | 1 LREfm with optrA | No Data | [145] |
| USA | 2013–2016 | 1 LREfs with optrA | No Data | [94] |
| USA | 2018–2019 | 1 LREfm with optrA 1 LREfs with optrA | No Data No Data | [51] |
| SENTRY Program | 2008–2016 | 4 LREfs with optrA 1 LREfs with optrA 1 LREfs with optrA | F101L in L4 cfr + F101L cfr | [105] |
| ZAAPS Program | - | 8 LREfs with optrA | No Data | [96] |
| Mechanisms | Involved Mutations/Determinants | Associated Phenotypes | Geographic Distribution of WGS Studies and Clinical Significance |
|---|---|---|---|
| Modifications of LNZ binding pocket | 23S rRNA mutations; G2576U most frequent | LNZ MICs: 8– ≥ 256, generally higher with more 23S rRNA loci affected, but many exceptions; TDZ MICs: 4–8× lower, except T2504A (MIC 32) | Asia, Australia, Europe, North America, and South America; mainly LREfm; non-transferable |
| Modifications of LNZ binding pocket | L3, L4 and L22 ribosomal proteins | LNZ MICs ≤8 in most cases; TDZ—important in staphylococci | Asia, Europe; mainly LREfs; non-transferable |
| Target modification | cfr genes | Unclear significance in LRE; low LNZ MICs; TDZ unaffected, clinically important in staphylococci | Asia, Europe, North America; mainly LREfm; transferrable—reservoir for staphylococci |
| Ribosome protection | optrA and poxtA variants | Varying MICs dependant on the variant; TDZ MICs 4–8× lower | Global dissemination; mainly LREfs; easily transferrable (especially optrA); optrA is the most widespread LNZ resistance mechanism in clinical LREfs |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peykov, S.; Kirov, B.; Strateva, T. Linezolid in the Focus of Antimicrobial Resistance of Enterococcus Species: A Global Overview of Genomic Studies. Int. J. Mol. Sci. 2025, 26, 8207. https://doi.org/10.3390/ijms26178207
Peykov S, Kirov B, Strateva T. Linezolid in the Focus of Antimicrobial Resistance of Enterococcus Species: A Global Overview of Genomic Studies. International Journal of Molecular Sciences. 2025; 26(17):8207. https://doi.org/10.3390/ijms26178207
Chicago/Turabian StylePeykov, Slavil, Boris Kirov, and Tanya Strateva. 2025. "Linezolid in the Focus of Antimicrobial Resistance of Enterococcus Species: A Global Overview of Genomic Studies" International Journal of Molecular Sciences 26, no. 17: 8207. https://doi.org/10.3390/ijms26178207
APA StylePeykov, S., Kirov, B., & Strateva, T. (2025). Linezolid in the Focus of Antimicrobial Resistance of Enterococcus Species: A Global Overview of Genomic Studies. International Journal of Molecular Sciences, 26(17), 8207. https://doi.org/10.3390/ijms26178207







