Genome-Wide Identification, Phylogenetic Analysis, and Expression Pattern of Polyamine Biosynthesis Gene Family in Pepper
Abstract
1. Introduction
2. Results
2.1. Identification of PA Biosynthesis Genes in Pepper
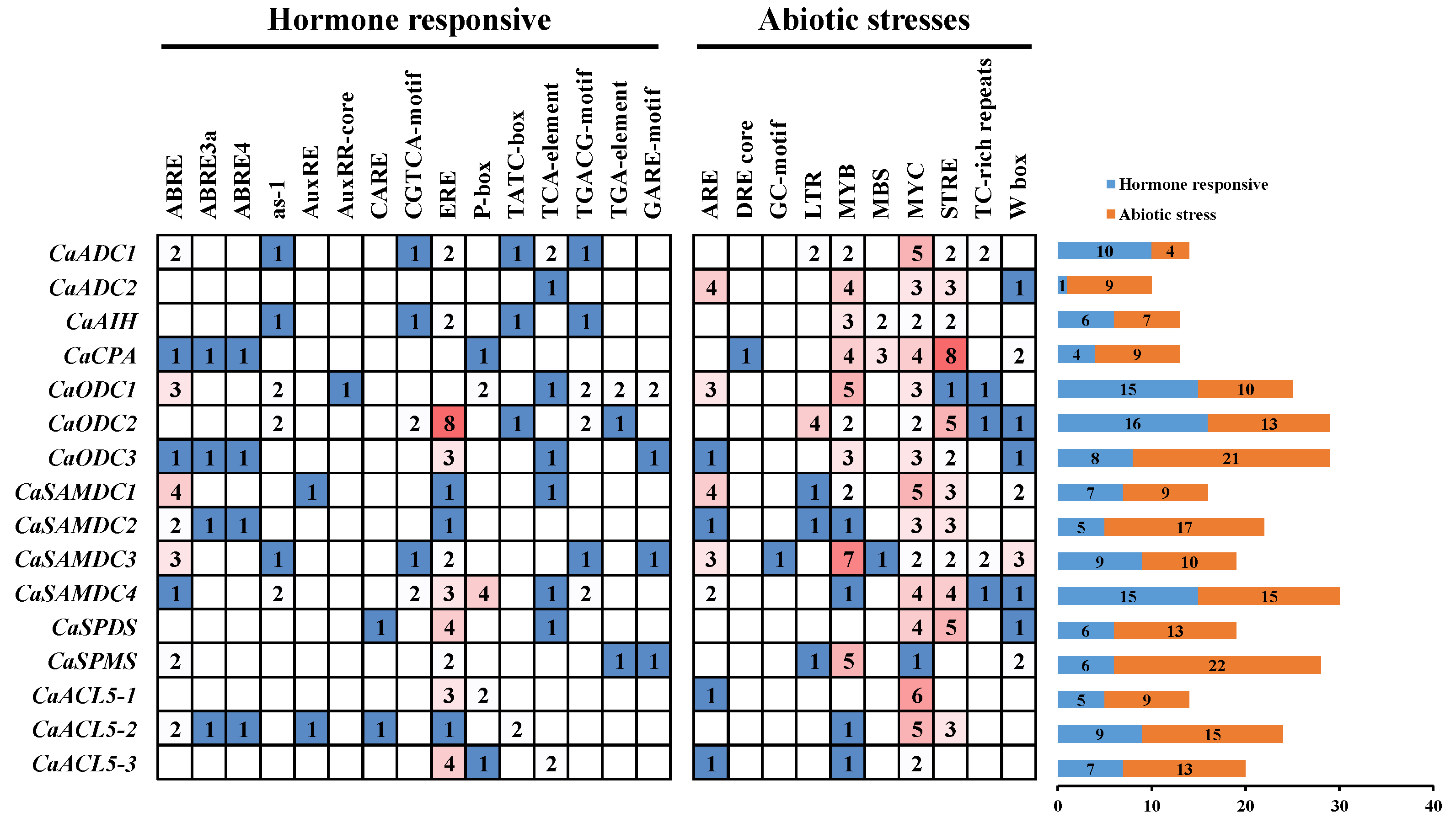
2.2. Cis-Acting Elements in the Promoters of PA Biosynthesis Genes in Pepper
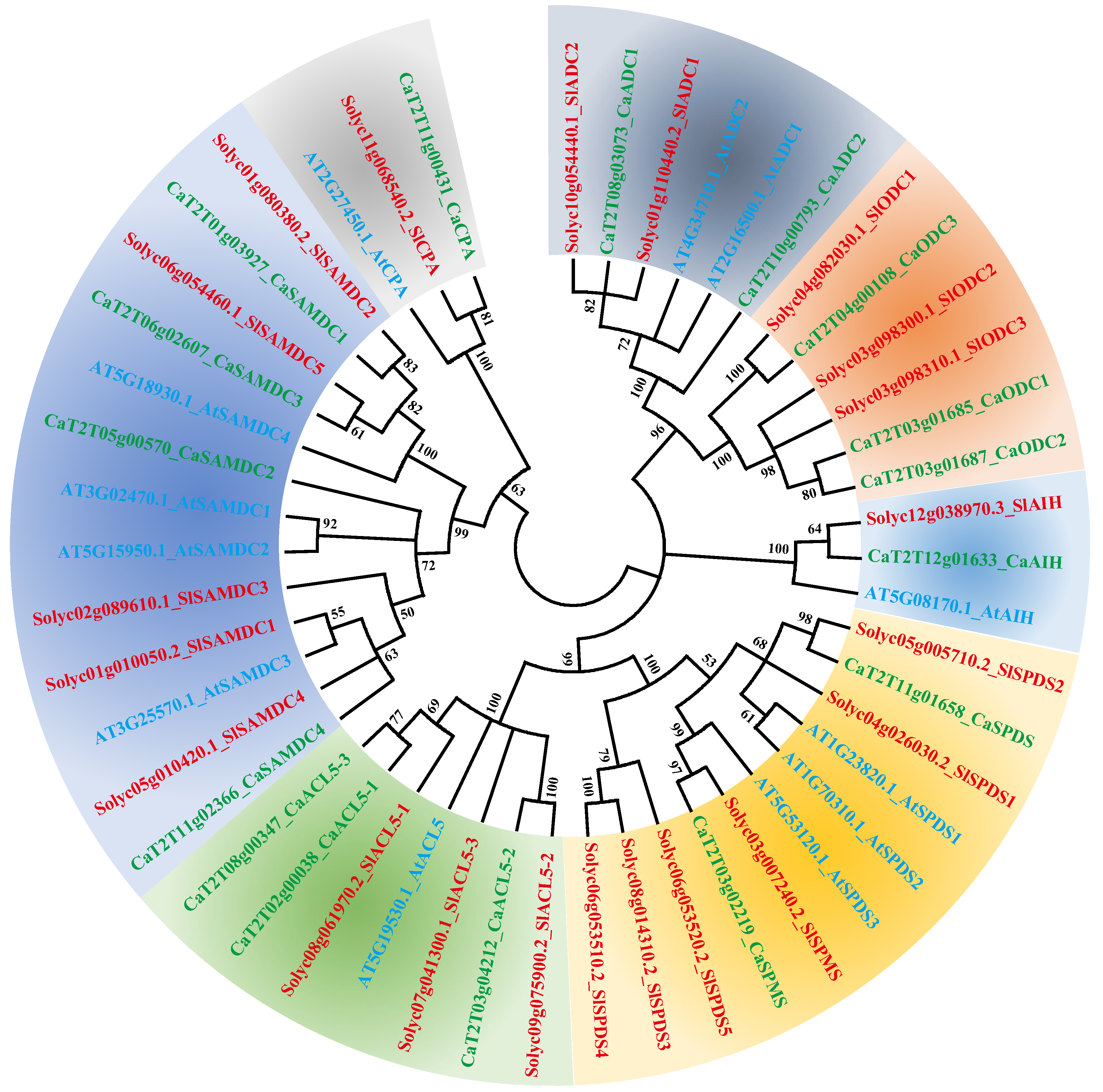
2.3. Phylogenetic Analysis of PA Biosynthesis Genes
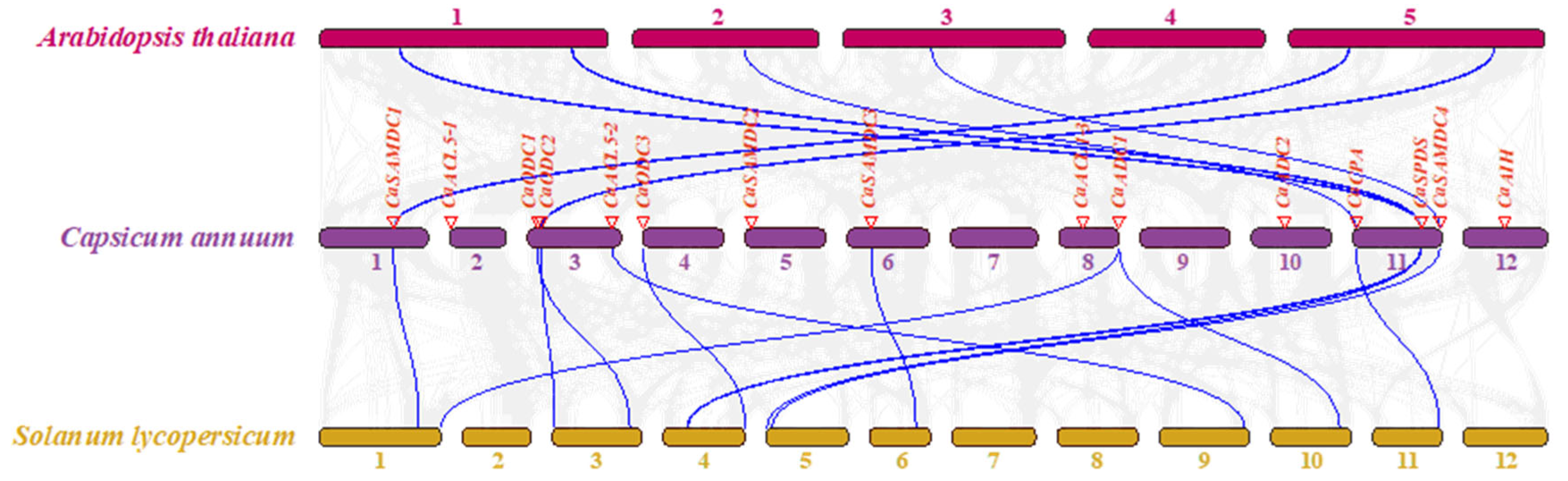
2.4. Chromosomal Localization and Duplication of PA Biosynthesis Genes in Pepper
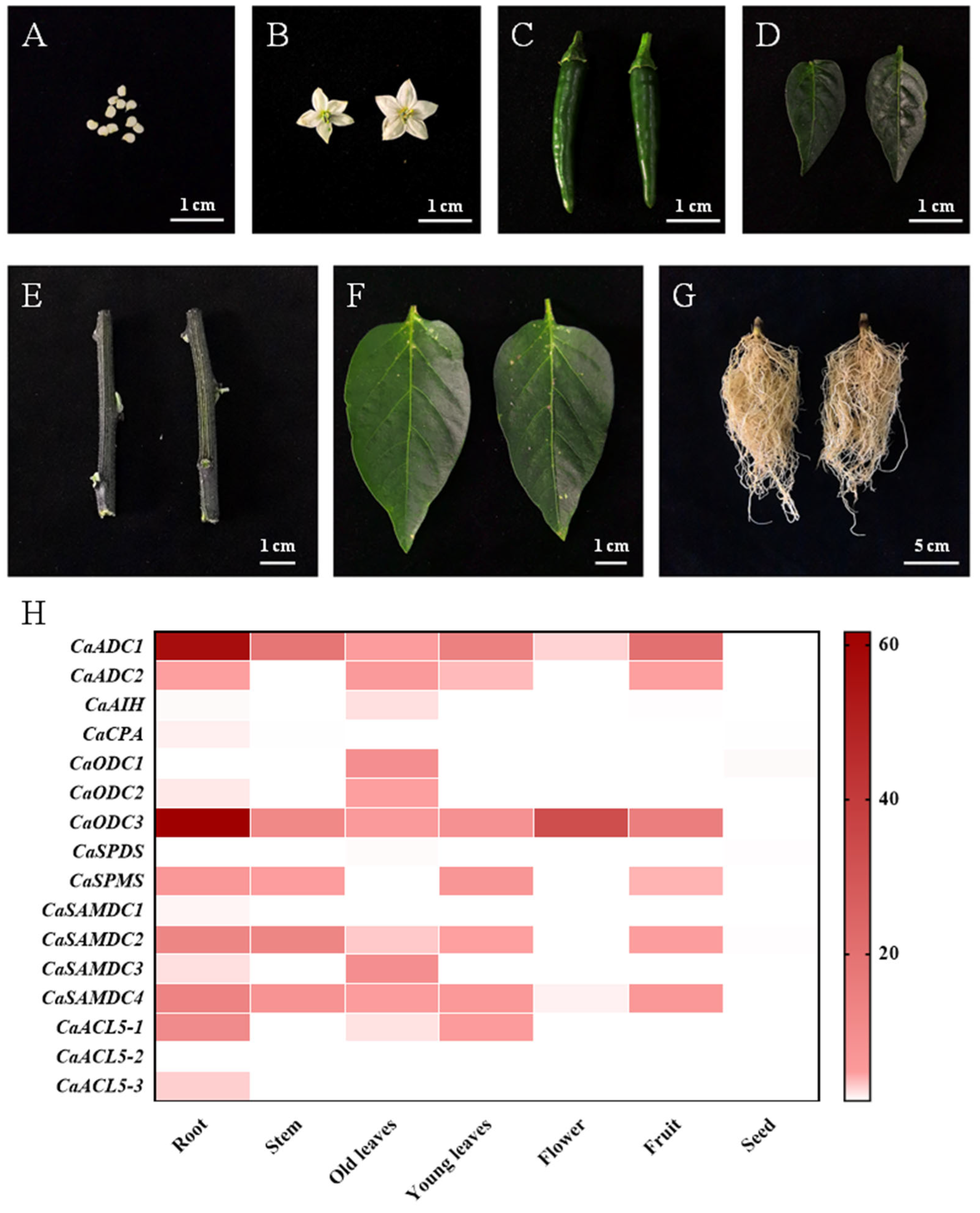
2.5. Organ-Specific Expression Patterns of Pepper PA Biosynthesis Genes
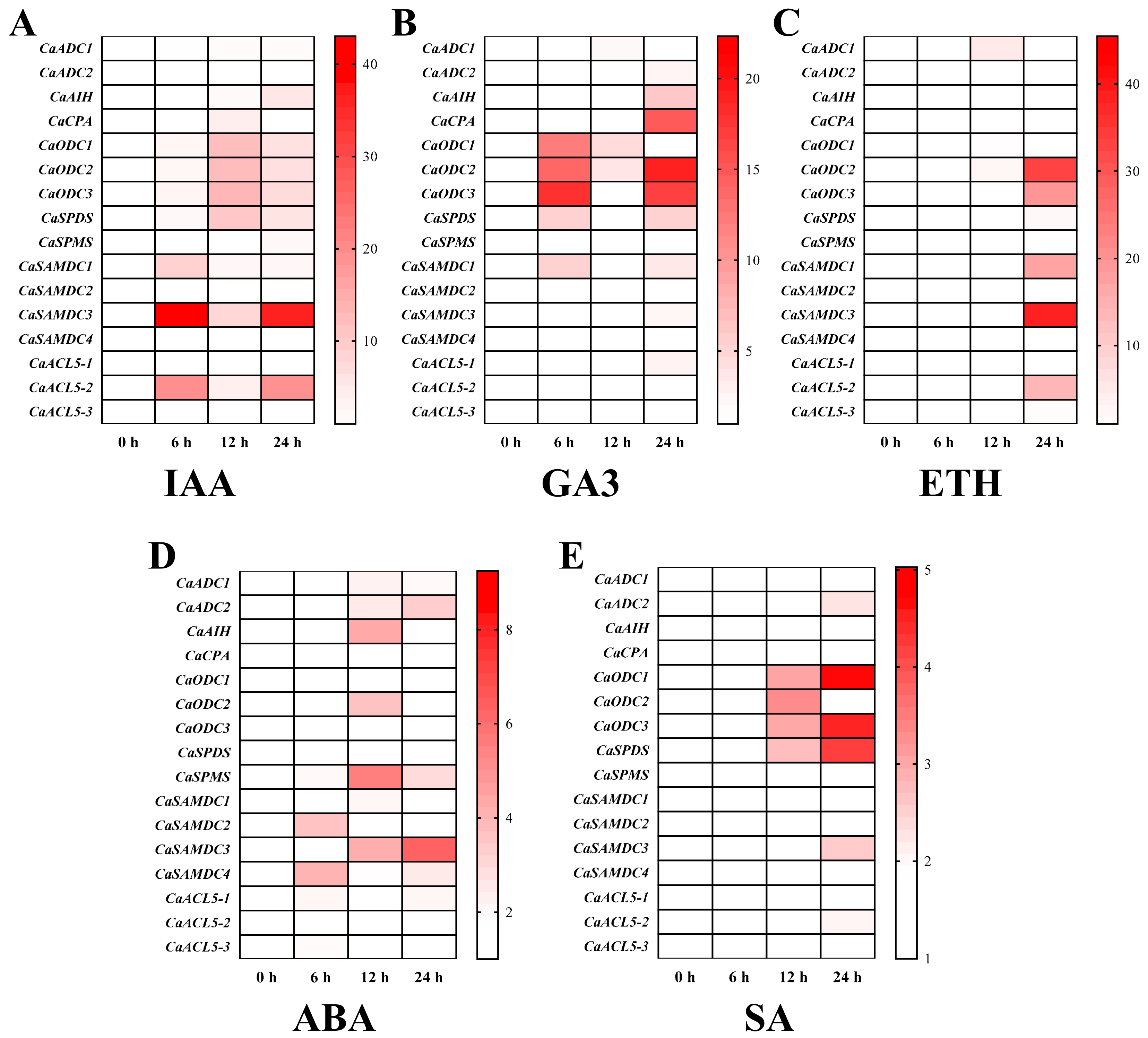
2.6. Effect of Exogenous Phytohormones on PA Biosynthesis Gene Expression
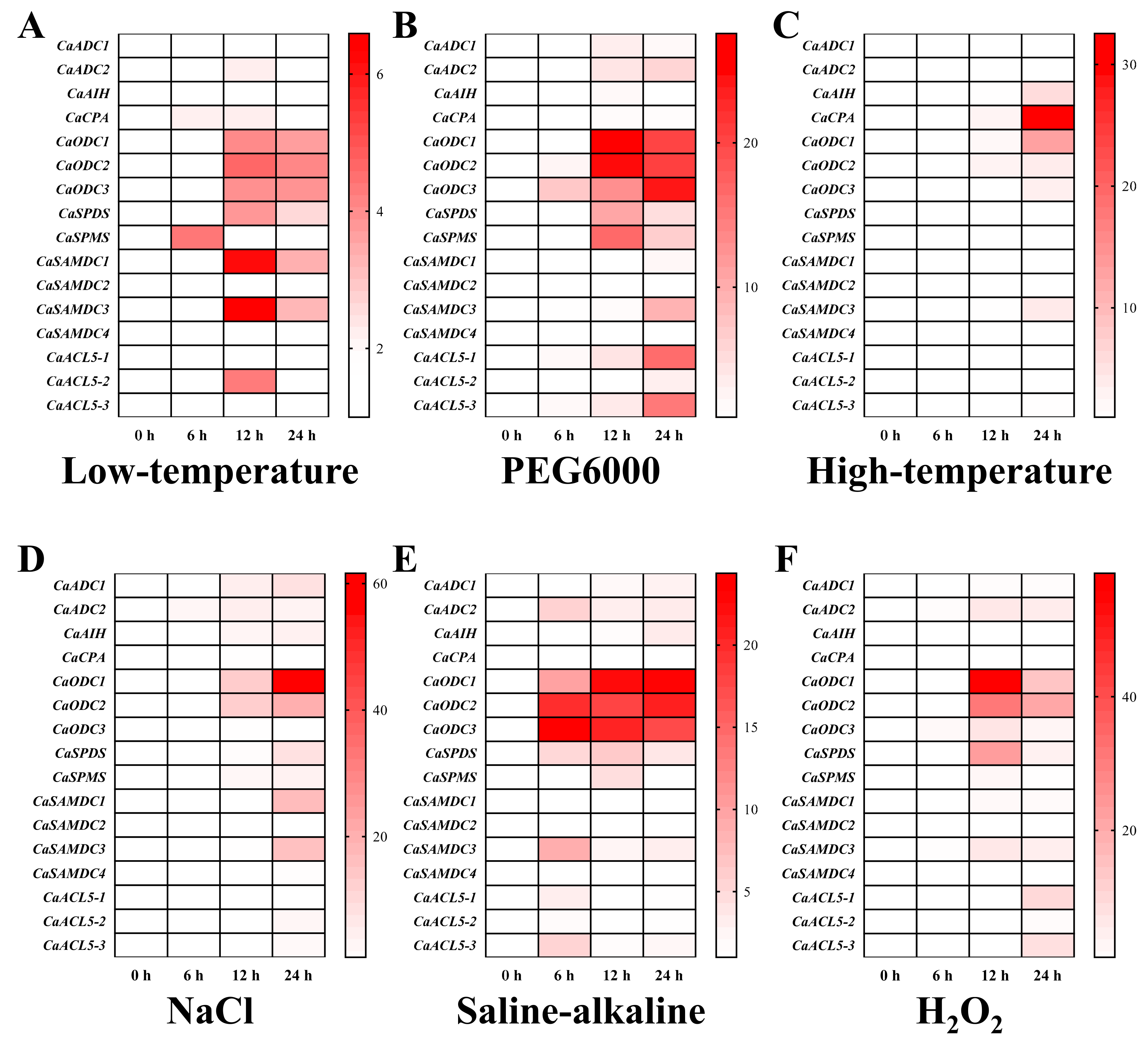
2.7. Differential Expression of PA Biosynthesis Genes During Abiotic Stress
3. Discussion
4. Materials and Methods
4.1. Plant Growth Conditions and Treatments
4.2. Identification of PA Biosynthesis Genes
4.3. Gene Structure, Subcellular Localization and Protein Physicochemical Properties Analysis
4.4. Cis-Element Prediction for PA Biosynthesis Gene Promoters
4.5. Sequence Alignment and Phylogenetic Analysis
4.6. Chromosomal Location and Duplication Analysis
4.7. Total RNA Extraction and Gene Expression Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Correction Statement
References
- Tabor, C.W.; Tabor, H. Polyamines. Annu. Rev. Biochem. 1984, 53, 747–790. [Google Scholar] [CrossRef]
- Takahashi, T.; Kakehi, J. Polyamines: Ubiquitous polycations with unique roles in growth and stress responses. Ann. Bot. 2010, 105, 1–6. [Google Scholar] [CrossRef]
- Xuan, M.; Gu, X.; Li, J.; Huang, D.; Xue, C.; He, Y. Polyamines: Their significance for maintaining health and contributing to diseases. Cell Commun. Signal. 2023, 21, 348. [Google Scholar] [CrossRef]
- Alcázar, R.; Altabella, T.; Marco, F.; Bortolotti, C.; Reymond, M.; Koncz, C.; Carrasco, P.; Tiburcio, A.F. Polyamines: Molecules with regulatory functions in plant abiotic stress tolerance. Planta 2010, 231, 1237–1249. [Google Scholar] [CrossRef] [PubMed]
- Fuell, C.; Elliott, K.A.; Hanfrey, C.C.; Franceschetti, M.; Michael, A.J. Polyamine biosynthetic diversity in plants and algae. Plant Physiol. Biochem. 2010, 48, 513–520. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.X.; Li, Y.B.; Liu, H.P.; Kurtenbach, R. Putrescine transformation to other forms of polyamines in filling grain embryos functioned in enhancing the resistance of maize plants to drought stress. Plant Physiol. Biochem. 2023, 197, 107654. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Chan, Z. Improvement of plant abiotic stress tolerance through modulation of the polyamine pathway. J. Integr. Plant Biol. 2014, 56, 114–121. [Google Scholar] [CrossRef]
- Takahashi, Y. ACL5 acquired strict thermospermine synthesis activity during the emergence of vascular plants. New Phytol. 2024, 242, 2669–2681. [Google Scholar] [CrossRef]
- Planas-Portell, J.; Gallart, M.; Tiburcio, A.F.; Altabella, T. Copper-containing amine oxidases contribute to terminal polyamine oxidation in peroxisomes and apoplast of Arabidopsis thaliana. BMC Plant Biol. 2013, 13, 109. [Google Scholar] [CrossRef]
- Tiburcio, A.F.; Alcázar, R. Potential Applications of Polyamines in Agriculture and Plant Biotechnology. In Polyamines: Methods in Molecular Biology; Humana Press: New York, NY, USA, 2018; pp. 489–508. [Google Scholar]
- Richards, F.J.; Coleman, R.G. Occurrence of Putrescine in Potassium-deficient Barley. Nature 1952, 170, 460. [Google Scholar] [CrossRef]
- Pál, M.; Szalai, G.; Janda, T. Speculation: Polyamines are important in abiotic stress signaling. Plant Sci. 2015, 237, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.S.; Tuteja, N. Polyamines and abiotic stress tolerance in plants. Plant Signal. Behav. 2010, 5, 26–33. [Google Scholar] [CrossRef]
- Minocha, R.; Majumdar, R.; Minocha, S.C. Polyamines and abiotic stress in plants: A complex relationship. Front. Plant Sci. 2014, 5, 175. [Google Scholar] [CrossRef]
- Urano, K.; Yoshiba, Y.; Nanjo, T.; Ito, T.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Arabidopsis stress-inducible gene for arginine decarboxylase AtADC2 is required for accumulation of putrescine in salt tolerance. Biochem. Biophys. Res. Commun. 2004, 313, 369–375. [Google Scholar] [CrossRef]
- Do, P.T.; Degenkolbe, T.; Erban, A.; Heyer, A.G.; Kopka, J.; Köhl, K.I.; Hincha, D.K.; Zuther, E. Dissecting Rice Polyamine Metabolism under Controlled Long-Term Drought Stress. PLoS ONE 2013, 8, e60325. [Google Scholar] [CrossRef]
- Li, Z.Y.; Chen, S.Y. Differential accumulation of the S-adenosyl-methionine decarboxylase transcript in rice seedlings in response to salt and drought stresses. Theor. Appl. Genet. 1999, 100, 782–788. [Google Scholar] [CrossRef]
- Zhu, X.; Li, Q.; Hu, J.; Wang, M.; Li, X. Molecular Cloning and Characterization of Spermine Synthesis Gene Associated with Cold Tolerance in Tea Plant (Camellia sinensis). Appl. Biochem. Biotechnol. 2015, 177, 1055–1068. [Google Scholar] [CrossRef]
- Kusbah, M.M.; Yelenosky, G. Evaluation of Polyamine and Proline Levels during Low Temperature Acclimation of Citrus. Plant Physiol. 1987, 1987, 692–695. [Google Scholar] [CrossRef] [PubMed]
- Zapata, P.J.; Serrano, M.; Pretel, M.T.; Amorós, A.; Botella, M.Á. Polyamines and ethylene changes during germination of different plant species under salinity. Plant Sci. 2004, 167, 781–788. [Google Scholar] [CrossRef]
- Zhang, J.; Liang, L.; Xiao, J.; Xie, Y.; Zhu, L.; Xue, X.; Xu, L.; Zhou, P.; Ran, J.; Huang, Z.; et al. Genome-Wide Identification of Polyamine Oxidase (PAO) Family Genes: Roles of CaPAO2 and CaPAO4 in the Cold Tolerance of Pepper (Capsicum annuum L.). Int. J. Mol. Sci. 2022, 23, 9999. [Google Scholar] [CrossRef]
- Zhang, J.; Xie, M.; Yu, G.; Wang, D.; Xu, Z.; Liang, L.; Xiao, J.; Xie, Y.; Tang, Y.; Sun, G.; et al. CaSPDS, a Spermidine Synthase Gene from Pepper (Capsicum annuum L.), Plays an Important Role in Response to Cold Stress. Int. J. Mol. Sci. 2023, 24, 5013. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Sun, M.; Jiang, X.; Sun, H.; Dang, X.; Cong, H.; Qiao, F. Glycinebetaine Biosynthesis in Response to Osmotic Stress Depends on Jasmonate Signaling in Watermelon Suspension Cells. Front. Plant Sci. 2018, 9, 1469. [Google Scholar] [CrossRef]
- Yang, F.; Lu, C.; Wei, Y.; Wu, J.; Ren, R.; Gao, J.; Ahmad, S.; Jin, J.; Xv, Y.; Liang, G.; et al. Organ-Specific Gene Expression Reveals the Role of the Cymbidium ensifolium-miR396/Growth-Regulating Factors Module in Flower Development of the Orchid Plant Cymbidium ensifolium. Front. Plant Sci. 2022, 12, 799778. [Google Scholar] [CrossRef]
- Napieraj, N.; Janicka, M.; Reda, M. Interactions of Polyamines and Phytohormones in Plant Response to Abiotic Stress. Plants 2023, 12, 1159. [Google Scholar] [CrossRef]
- Ebeed, H.T. Genome-wide analysis of polyamine biosynthesis genes in wheat reveals gene expression specificity and involvement of STRE and MYB-elements in regulating polyamines under drought. BMC Genom. 2022, 23, 734. [Google Scholar] [CrossRef]
- Alhag, A.; Song, J.; Dahro, B.; Wu, H.; Khan, M.; Salih, H.; Liu, J.H. Genome-wide identification and expression analysis of Polyamine Uptake Transporter gene family in sweet orange (Citrus sinensis). Plant Biol. 2021, 23, 1157–1166. [Google Scholar] [CrossRef]
- Liu, T.; Huang, B.; Chen, L.; Xian, Z.; Song, S.; Chen, R.; Hao, Y. Genome-wide identification, phylogenetic analysis, and expression profiling of polyamine synthesis gene family members in tomato. Gene 2018, 661, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Sen, S.; Ghosh, D.; Mohapatra, S. Modulation of polyamine biosynthesis in Arabidopsis thaliana by a drought mitigating Pseudomonas putida strain. Plant Physiol. Biochem. 2018, 129, 180–188. [Google Scholar] [CrossRef]
- Scott, J.D.; Pawson, T. Cell Signaling in Space and Time: Where Proteins Come Together and When They’re Apart. Science 2009, 326, 1220–1224. [Google Scholar] [CrossRef] [PubMed]
- Hillis, D.M. Phylogenetic analysis. Curr. Biol. 1997, 7, 129–131. [Google Scholar] [CrossRef]
- Killiny, N.; Nehela, Y. Citrus Polyamines: Structure, Biosynthesis, and Physiological Functions. Plants 2020, 9, 426. [Google Scholar] [CrossRef]
- Chen, D.; Shao, Q.; Yin, L.; Younis, A.; Zheng, B. Polyamine Function in Plants: Metabolism, Regulation on Development, and Roles in Abiotic Stress Responses. Front. Plant Sci. 2019, 9, 1945. [Google Scholar] [CrossRef]
- Sémon, M.; Wolfe, K.H. Consequences of genome duplication. Curr. Opin. Genet. Dev. 2007, 17, 505–512. [Google Scholar] [CrossRef]
- Moore, R.C.; Purugganan, M.D. The evolutionary dynamics of plant duplicate genes. Curr. Opin. Plant Biol. 2005, 8, 122–128. [Google Scholar] [CrossRef]
- Xie, T.; Zeng, L.; Chen, X.; Rong, H.; Wu, J.; Batley, J.; Jiang, J.; Wang, Y. Genome-Wide Analysis of the Lateral Organ Boundaries Domain Gene Family in Brassica Napus. Genes 2020, 11, 280. [Google Scholar] [CrossRef]
- Hernandez-Garcia, C.M.; Finer, J.J. Identification and validation of promoters and cis-acting regulatory elements. Plant Sci. 2014, 217–218, 109–119. [Google Scholar] [CrossRef]
- Li, Z.; Huang, Y.; Shen, Z.; Wu, M.; Huang, M.; Hong, S.B.; Xu, L.; Zang, Y. Advances in functional studies of plant MYC transcription factors. Theor. Appl. Genet. 2024, 137, 195. [Google Scholar] [CrossRef] [PubMed]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; Zhu, X.; Huang, X.; Liu, J.H. Overexpression of a stress-responsive MYB transcription factor of Poncirus trifoliata confers enhanced dehydration tolerance and increases polyamine biosynthesis. Plant Physiol. Biochem. 2014, 78, 71–79. [Google Scholar] [CrossRef]
- Martínez-Pastor, M.T.; Marchler, G.; Schüller, C.; Marchler-Bauer, A.; Ruis, H.; Estruch, F. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE). EMBO J. 1996, 15, 2227–2235. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Fujita, Y.; Sayama, H.; Kidokoro, S.; Maruyama, K.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 2010, 61, 672–685. [Google Scholar] [CrossRef]
- Song, J.; Sun, P.; Kong, W.; Xie, Z.; Li, C.; Liu, J.H. SnRK2.4-mediated phosphorylation of ABF2 regulates ARGININE DECARBOXYLASE expression and putrescine accumulation under drought stress. New Phytol. 2022, 238, 216–236. [Google Scholar] [CrossRef]
- Khajuria, A.; Sharma, N.; Bhardwaj, R.; Ohri, P. Emerging Role of Polyamines in Plant Stress Tolerance. Curr. Protein Pept. Sci. 2018, 19, 1114–1123. [Google Scholar] [CrossRef]
- Ohri, P.; Bhardwaj, R.; Bali, S.; Kaur, R.; Jasrotia, S.; Khajuria, A.; Parihar, R.D. The Common Molecular Players in Plant Hormone Crosstalk and Signaling. Curr. Protein Pept. Sci. 2015, 16, 369–388. [Google Scholar] [CrossRef]
- Bagni, N.; Ruiz-Carrasco, K.; Franceschetti, M.; Fornalè, S.; Fornasiero, R.B.; Tassoni, A. Polyamine metabolism and biosynthetic gene expression in Arabidopsis thaliana under salt stress. Plant Physiol. Biochem. 2006, 44, 776–786. [Google Scholar] [CrossRef]
- Alcázar, R.; Bitrián, M.; Bartels, D.; Koncz, C.; Altabella, T.; Tiburcio, A.F. Polyamine metabolic canalization in response to drought stress in Arabidopsis and the resurrection plant Craterostigma plantagineum. Plant Signal. Behav. 2014, 6, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Cuevas, J.C.; López-Cobollo, R.; Alcázar, R.; Zarza, X.; Koncz, C.; Altabella, T.; Salinas, J.; Tiburcio, A.F.; Ferrando, A. Putrescine Is Involved in Arabidopsis Freezing Tolerance and Cold Acclimation by Regulating Abscisic Acid Levels in Response to Low Temperature. Plant Physiol. 2008, 148, 1094–1105. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.J.; Zhang, Z.; Kitashiba, H.; Honda, C.; Ubi, B.; Kita, M.; Moriguchi, T. Molecular cloning and functional characterization of two apple S-adenosylmethionine decarboxylase genes and their different expression in fruit development, cell growth and stress responses. Gene 2005, 350, 41–50. [Google Scholar] [CrossRef]
- Zhu, M.; Chen, G.; Wu, J.; Wang, J.; Wang, Y.; Guo, S.; Shu, S. Identification of cucumber S-adenosylmethionine decarboxylase genes and functional analysis of CsSAMDC3 in salt tolerance. Front. Plant Sci. 2023, 14, 1076153. [Google Scholar] [CrossRef]
- Yariuchi, Y.; Okamoto, T.; Noutoshi, Y.; Takahashi, T. Responses of Polyamine-Metabolic Genes to Polyamines and Plant Stress Hormones in Arabidopsis Seedlings. Cells 2021, 10, 3283. [Google Scholar] [CrossRef]
- Banerjee, A.; Roychoudhury, A. The Regulatory Signaling of Gibberellin Metabolism and Its Crosstalk with Phytohormones in Response to Plant Abiotic Stresses. In Plant Signaling Molecules; Elsevier: Amsterdam, The Netherlands, 2019; pp. 333–339. [Google Scholar]
- Chen, W.; Wang, X.; Sun, J.; Wang, X.; Zhu, Z.; Ayhan, D.H.; Yi, S.; Yan, M.; Zhang, L.; Meng, T.; et al. Two telomere-to-telomere gapless genomes reveal insights into Capsicum evolution and capsaicinoid biosynthesis. Nat. Commun. 2024, 15, 4295. [Google Scholar] [CrossRef]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Geourjon, C.; Deleage, G. SOPMA: Significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Bioinformatics 1995, 11, 681–684. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.P.; Chou, K.C.; Shen, H.B. A New Method for Predicting the Subcellular Localization of Eukaryotic Proteins with Both Single and Multiple Sites: Euk-mPLoc 2.0. PLoS ONE 2010, 5, e9931. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]


| Gene ID from PGDB | Name | Position | Coding Sequences (bp) | Intron | Subcellular Localization |
|---|---|---|---|---|---|
| CaT2T08g03073 | CaADC1 | Chr08: 184132402~184134576 | 2175 | 0 | Mitochondrion. |
| CaT2T10g00793 | CaADC2 | Chr10: 100641424~100645196 | 1296 | 4 | Mitochondrion. |
| CaT2T12g01633 | CaAIH | Chr12: 128776924~128789031 | 1557 | 10 | Extracell. Mitochondrion. |
| CaT2T11g00431 | CaCPA | Chr11: 10439223~10444141 | 846 | 7 | Cytoplasm. |
| CaT2T03g01685 | CaODC1 | Chr03: 30721970~30723123 | 1086 | 0 | Mitochondrion. |
| CaT2T03g01687 | CaODC2 | Chr03: 30758389~30759591 | 1203 | 0 | Mitochondrion. |
| CaT2T04g00108 | CaODC3 | Chr04: 1757160~1758467 | 1308 | 0 | Mitochondrion. |
| CaT2T01g03927 | CaSAMDC1 | Chr01: 227578235~227579245 | 1011 | 0 | Cytoplasm. |
| CaT2T05g00570 | CaSAMDC2 | Chr05: 19555986~19564951 | 2469 | 7 | Cytoplasm. |
| CaT2T06g02607 | CaSAMDC3 | Chr06: 73468151~73469155 | 1005 | 0 | Cytoplasm. Extracell. |
| CaT2T11g02366 | CaSAMDC4 | Chr11: 268736228~268737310 | 1083 | 0 | Cytoplasm. Extracell. Nucleus. |
| CaT2T11g01658 | CaSPDS | Chr11: 210985086~210989836 | 1104 | 7 | Cytoplasm. Nucleus. |
| CaT2T03g02219 | CaSPMS | Chr03: 42757075~42764512 | 1044 | 9 | Cytoplasm. |
| CaT2T02g00038 | CaACL5-1 | Chr02: 4353693~4360371 | 1032 | 9 | Cytoplasm. Nucleus. |
| CaT2T03g04212 | CaACL5-2 | Chr03: 259804076~259808320 | 756 | 6 | Cytoplasm. |
| CaT2T08g00347 | CaACL5-3 | Chr08: 74094704~74100057 | 654 | 6 | Cytoplasm. |
| Name | Number of Amino Acid (aa) | Molecular Weight (kDa) | Theoretical pI | Instability Index | Aliphatic Index | Grand Average of Hydropathicity |
|---|---|---|---|---|---|---|
| CaADC1 | 724 | 78.15 | 5.08 | 43.64 | 88.23 | −0.053 |
| CaADC2 | 431 | 48.02 | 6.32 | 48.50 | 97.59 | −0.058 |
| CaAIH | 518 | 56.44 | 5.54 | 30.86 | 90.62 | −0.117 |
| CaCPA | 281 | 31.49 | 5.78 | 32.50 | 78.15 | −0.334 |
| CaODC1 | 361 | 40.00 | 7.65 | 40.41 | 95.93 | −0.060 |
| CaODC2 | 400 | 43.89 | 5.84 | 41.00 | 94.57 | 0.025 |
| CaODC3 | 435 | 46.94 | 5.59 | 40.71 | 85.29 | 0.007 |
| CaSAMDC1 | 336 | 37.93 | 5.40 | 44.60 | 79.14 | −0.061 |
| CaSAMDC2 | 822 | 92.00 | 7.79 | 42.51 | 77.53 | −0.360 |
| CaSAMDC3 | 334 | 37.51 | 5.73 | 44.60 | 77.90 | −0.082 |
| CaSAMDC4 | 360 | 39.38 | 5.28 | 39.59 | 82.89 | −0.099 |
| CaSPDS | 367 | 40.34 | 5.09 | 51.20 | 87.85 | −0.142 |
| CaSPMS | 347 | 38.66 | 5.17 | 43.92 | 87.09 | −0.165 |
| CaACL5-1 | 343 | 39.08 | 5.31 | 34.00 | 84.14 | −0.278 |
| CaACL5-2 | 251 | 28.44 | 5.25 | 33.27 | 81.16 | −0.414 |
| CaACL5-3 | 217 | 24.63 | 5.40 | 29.10 | 77.24 | −0.327 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, D.; Zhao, X.; Hu, Q.; Wang, S.; Zhang, Y.; Xu, Z. Genome-Wide Identification, Phylogenetic Analysis, and Expression Pattern of Polyamine Biosynthesis Gene Family in Pepper. Int. J. Mol. Sci. 2025, 26, 8208. https://doi.org/10.3390/ijms26178208
Lin D, Zhao X, Hu Q, Wang S, Zhang Y, Xu Z. Genome-Wide Identification, Phylogenetic Analysis, and Expression Pattern of Polyamine Biosynthesis Gene Family in Pepper. International Journal of Molecular Sciences. 2025; 26(17):8208. https://doi.org/10.3390/ijms26178208
Chicago/Turabian StyleLin, Duo, Xianqi Zhao, Qingshan Hu, Su Wang, Yan Zhang, and Zijian Xu. 2025. "Genome-Wide Identification, Phylogenetic Analysis, and Expression Pattern of Polyamine Biosynthesis Gene Family in Pepper" International Journal of Molecular Sciences 26, no. 17: 8208. https://doi.org/10.3390/ijms26178208
APA StyleLin, D., Zhao, X., Hu, Q., Wang, S., Zhang, Y., & Xu, Z. (2025). Genome-Wide Identification, Phylogenetic Analysis, and Expression Pattern of Polyamine Biosynthesis Gene Family in Pepper. International Journal of Molecular Sciences, 26(17), 8208. https://doi.org/10.3390/ijms26178208





