ArfGAP with Dual Pleckstrin Homology Domains 2 Promotes Hypertrophy of Cultured Neonatal Cardiomyocytes
Abstract
1. Introduction
2. Results
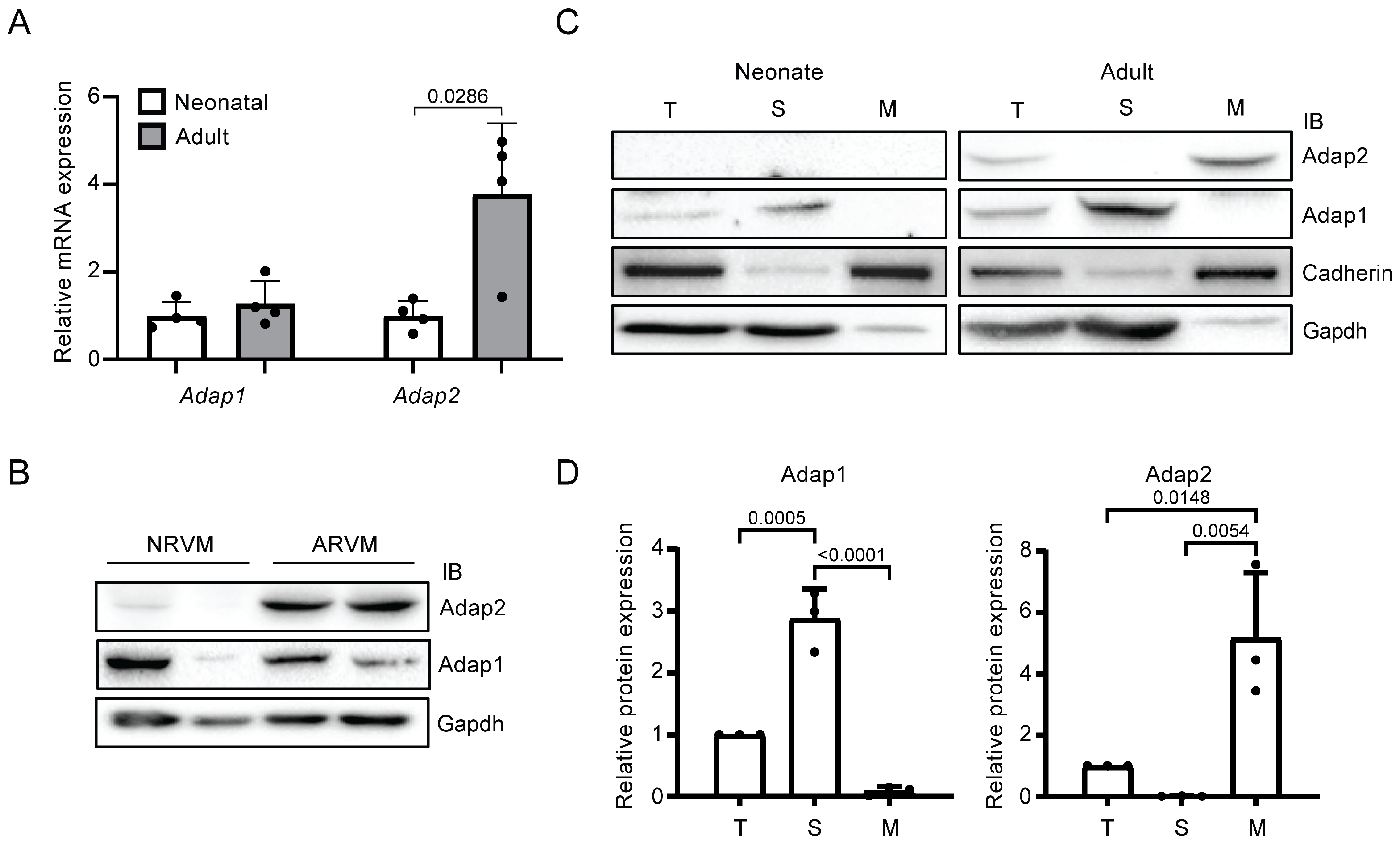
2.1. Adap2 Is Predominantly Localized, Unlike Adap1, at the Membranes of Adult Rat Hearts
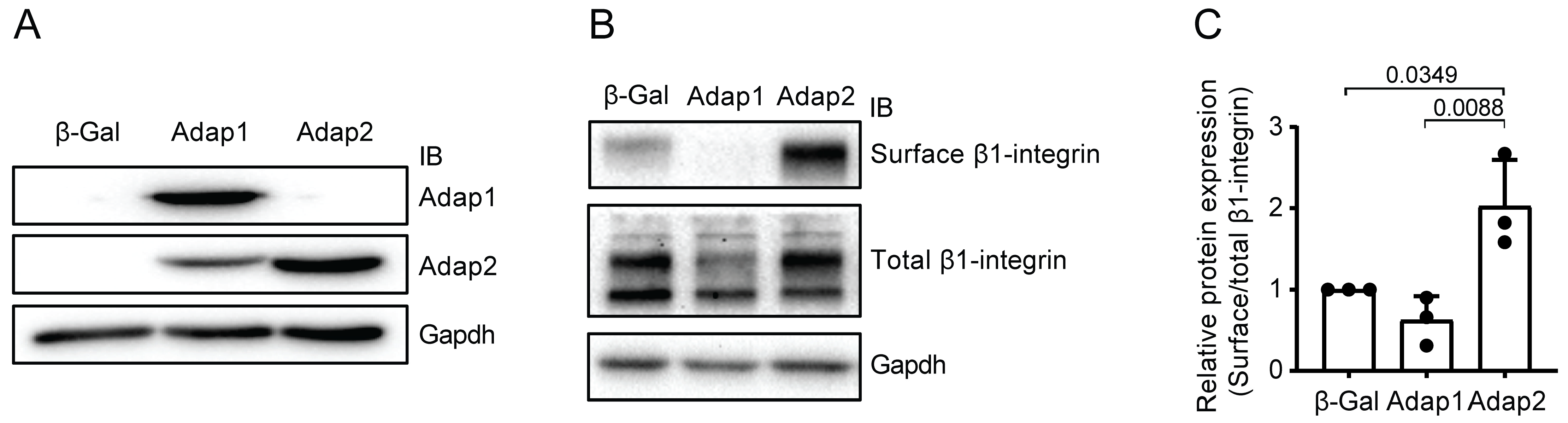
2.2. Adap2 Increases β1-Integrin Cell Surface Localization
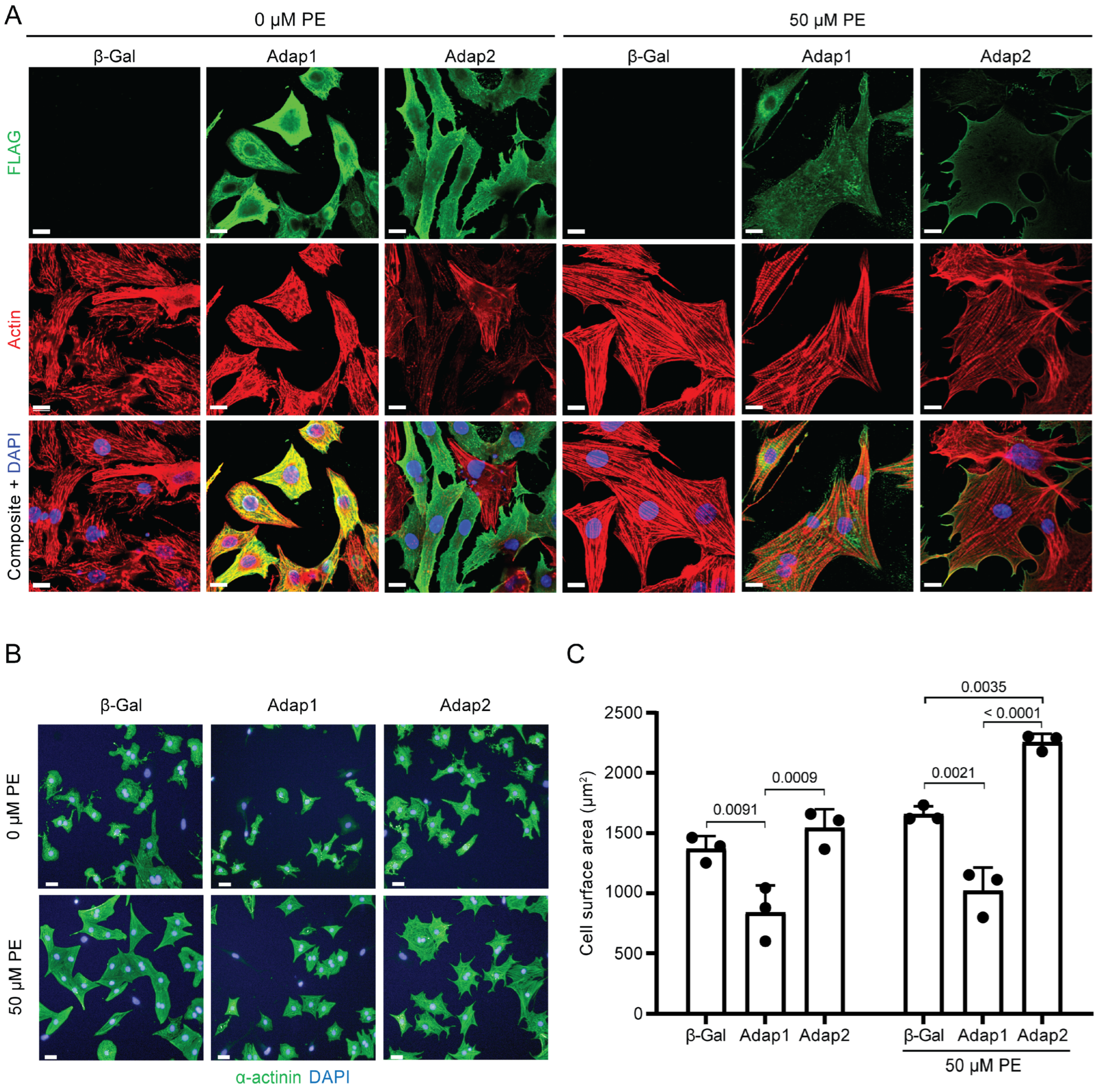
2.3. Phenylephrine Stimulation Enhances Adap2 Localization at the Sarcolemma and Promotes Cell Spreading of Cultured Cardiomyocytes
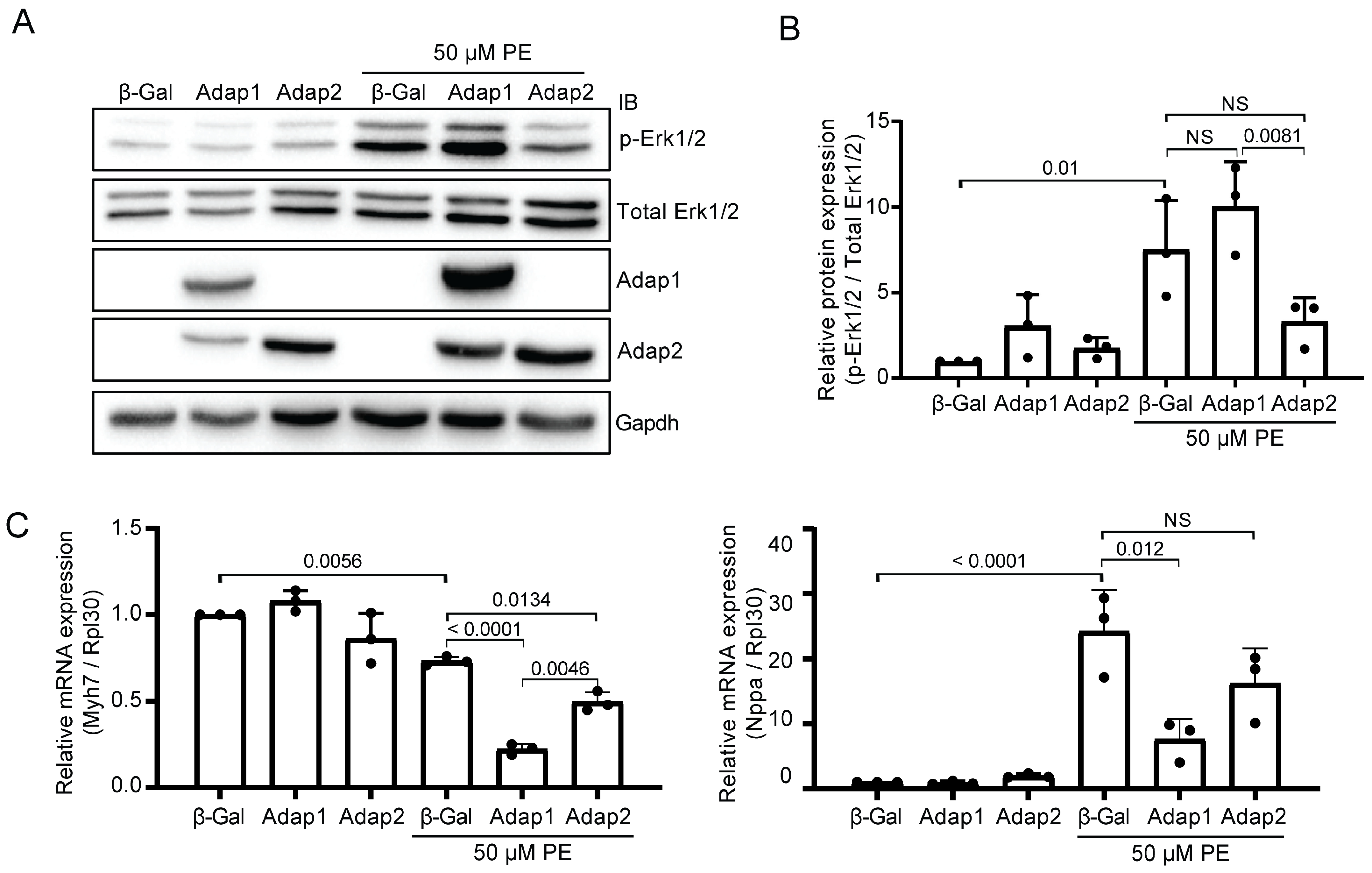
2.4. Adap2 Attenuates Part of the Phenylephrine-Induced Hypertrophic Signaling
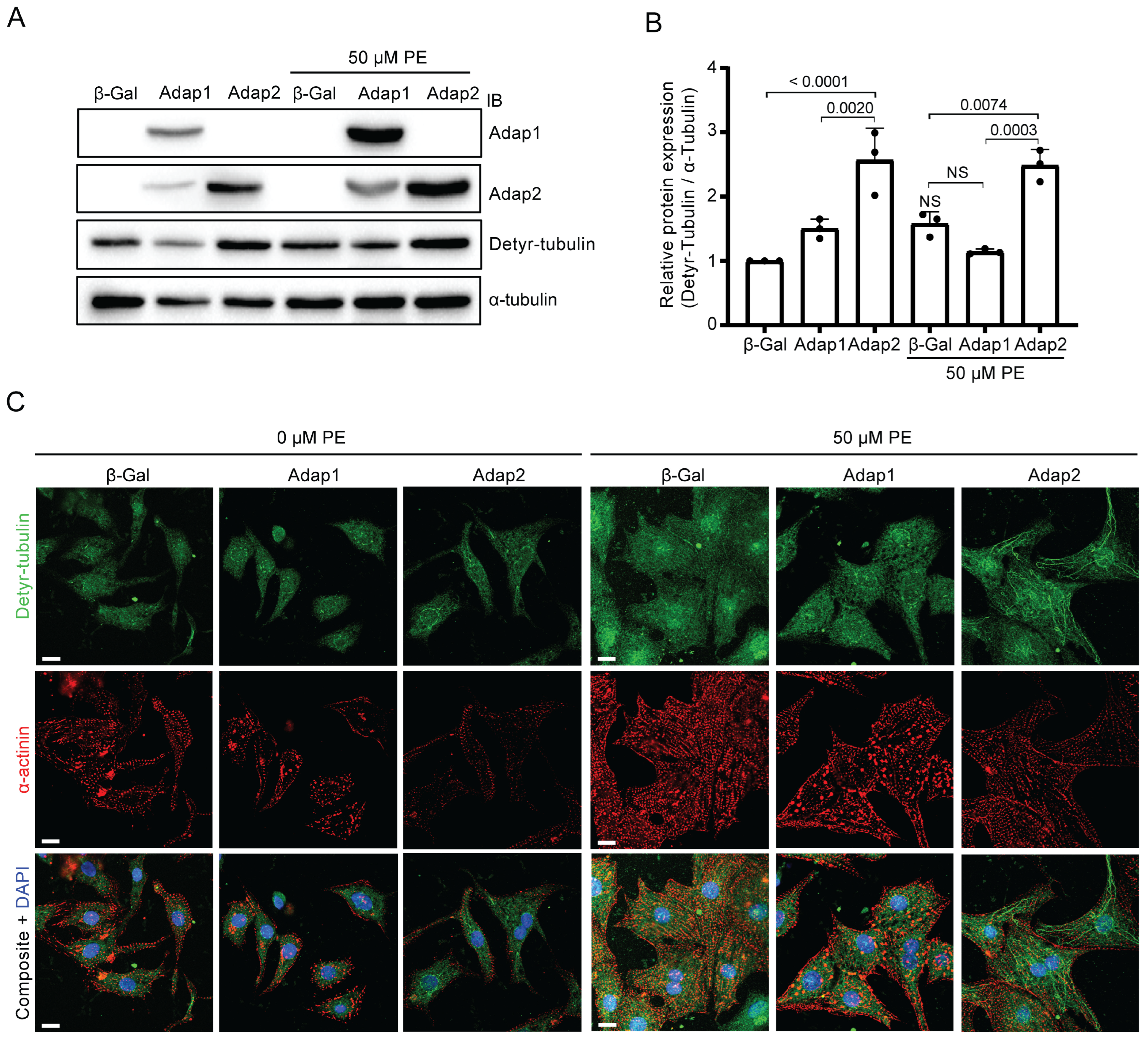
2.5. Adap2 Increases the Detyrosinated Tubulin Levels in Cardiomyocytes
3. Discussion
4. Materials and Methods
4.1. Animals and Experimental Protocols
4.2. Rat Heart Fractionation and Subcellular Enrichment
4.3. Adenovirus Generation
4.4. Adult Rat Ventricular Myocytes Isolation
4.5. Neonatal Rat Ventricular Myocytes Culture
4.6. Quantitative PCR and mRNA Expression Analysis
4.7. Protein Expression Analysis
4.8. Confocal Microscopy and Immunofluorescence Analysis
4.9. Surface β1-Integrin Pull-Down Assay
4.10. Statistical Analysis
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Giguere, H.; Dumont, A.A.; Berthiaume, J.; Oliveira, V.; Laberge, G.; Auger-Messier, M. ADAP1 limits neonatal cardiomyocyte hypertrophy by reducing integrin cell surface expression. Sci. Rep. 2018, 8, 13605. [Google Scholar] [CrossRef] [PubMed]
- Whitley, P.; Gibbard, A.M.; Koumanov, F.; Oldfield, S.; Kilgour, E.E.; Prestwich, G.D.; Holman, G.D. Identification of centaurin-alpha2: A phosphatidylinositide-binding protein present in fat, heart and skeletal muscle. Eur. J. Cell Biol. 2002, 81, 222–230. [Google Scholar] [CrossRef] [PubMed]
- Venkateswarlu, K.; Oatey, P.B.; Tavare, J.M.; Jackson, T.R.; Cullen, P.J. Identification of centaurin-alpha1 as a potential in vivo phosphatidylinositol 3,4,5-trisphosphate-binding protein that is functionally homologous to the yeast ADP-ribosylation factor (ARF) GTPase-activating protein, Gcs1. Biochem. J. 1999, 340, 359–363. [Google Scholar] [CrossRef]
- Hammonds-Odie, L.P.; Jackson, T.R.; Profit, A.A.; Blader, I.J.; Turck, C.W.; Prestwich, G.D.; Theibert, A.B. Identification and cloning of centaurin-alpha. A novel phosphatidylinositol 3,4,5-trisphosphate-binding protein from rat brain. J. Biol. Chem. 1996, 271, 18859–18868. [Google Scholar] [CrossRef]
- Venkateswarlu, K.; Cullen, P.J. Molecular cloning and functional characterization of a human homologue of centaurin-alpha. Biochem. Biophys. Res. Commun. 1999, 262, 237–244. [Google Scholar] [CrossRef]
- Aggensteiner, M.; Reiser, G. Expression of the brain-specific membrane adapter protein p42IP4/centaurin alpha, a Ins(1,3,4,5)P4/PtdIns(3,4,5)P3 binding protein, in developing rat brain. Brain Res. Dev. Brain Res. 2003, 142, 77–87. [Google Scholar] [CrossRef]
- Venturin, M.; Carra, S.; Gaudenzi, G.; Brunelli, S.; Gallo, G.R.; Moncini, S.; Cotelli, F.; Riva, P. ADAP2 in heart development: A candidate gene for the occurrence of cardiovascular malformations in NF1 microdeletion syndrome. J. Med. Genet. 2014, 51, 436–443. [Google Scholar] [CrossRef]
- Venturin, M.; Bentivegna, A.; Moroni, R.; Larizza, L.; Riva, P. Evidence by expression analysis of candidate genes for congenital heart defects in the NF1 microdeletion interval. Ann. Hum. Genet. 2005, 69, 508–516. [Google Scholar] [CrossRef]
- Venkateswarlu, K.; Brandom, K.G.; Yun, H. PI-3-kinase-dependent membrane recruitment of centaurin-alpha2 is essential for its effect on ARF6-mediated actin cytoskeleton reorganisation. J. Cell Sci. 2007, 120, 792–801. [Google Scholar] [CrossRef][Green Version]
- Venkateswarlu, K.; Brandom, K.G.; Lawrence, J.L. Centaurin-alpha1 is an in vivo phosphatidylinositol 3,4,5-trisphosphate-dependent GTPase-activating protein for ARF6 that is involved in actin cytoskeleton organization. J. Biol. Chem. 2004, 279, 6205–6208. [Google Scholar] [CrossRef]
- Li, T.; Guo, Y. ADP-Ribosylation Factor Family of Small GTP-Binding Proteins: Their Membrane Recruitment, Activation, Crosstalk and Functions. Front. Cell Dev. Biol. 2022, 10, 813353. [Google Scholar] [CrossRef]
- Casalou, C.; Ferreira, A.; Barral, D.C. The Role of ARF Family Proteins and Their Regulators and Effectors in Cancer Progression: A Therapeutic Perspective. Front. Cell Dev. Biol. 2020, 8, 217. [Google Scholar] [CrossRef] [PubMed]
- Heasman, S.J.; Ridley, A.J. Mammalian Rho GTPases: New insights into their functions from in vivo studies. Nat. Rev. Mol. Cell Biol. 2008, 9, 690–701. [Google Scholar] [CrossRef] [PubMed]
- Radhakrishna, H.; Al-Awar, O.; Khachikian, Z.; Donaldson, J.G. ARF6 requirement for Rac ruffling suggests a role for membrane trafficking in cortical actin rearrangements. J. Cell Sci. 1999, 112, 855–866. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, N.; Scott, D.W.; Castle, J.D.; Casanova, J.E.; Schwartz, M.A. Arf6 and microtubules in adhesion-dependent trafficking of lipid rafts. Nat. Cell Biol. 2007, 9, 1381–1391. [Google Scholar] [CrossRef]
- Etienne-Manneville, S. Microtubules in cell migration. Annu. Rev. Cell Dev. Biol. 2013, 29, 471–499. [Google Scholar] [CrossRef]
- D’Souza-Schorey, C.; Chavrier, P. ARF proteins: Roles in membrane traffic and beyond. Nat. Rev. Mol. Cell Biol. 2006, 7, 347–358. [Google Scholar] [CrossRef]
- Ross, R.S.; Pham, C.; Shai, S.Y.; Goldhaber, J.I.; Fenczik, C.; Glembotski, C.C.; Ginsberg, M.H.; Loftus, J.C. Beta1 integrins participate in the hypertrophic response of rat ventricular myocytes. Circ. Res. 1998, 82, 1160–1172. [Google Scholar] [CrossRef]
- Maitra, N.; Flink, I.L.; Bahl, J.J.; Morkin, E. Expression of alpha and beta integrins during terminal differentiation of cardiomyocytes. Cardiovasc. Res. 2000, 47, 715–725. [Google Scholar] [CrossRef]
- Keller, R.S.; Shai, S.Y.; Babbitt, C.J.; Pham, C.G.; Solaro, R.J.; Valencik, M.L.; Loftus, J.C.; Ross, R.S. Disruption of integrin function in the murine myocardium leads to perinatal lethality, fibrosis, and abnormal cardiac performance. Am. J. Pathol. 2001, 158, 1079–1090. [Google Scholar] [CrossRef]
- Kuwahara, K.; Nishikimi, T.; Nakao, K. Transcriptional regulation of the fetal cardiac gene program. J. Pharmacol. Sci. 2012, 119, 198–203. [Google Scholar] [CrossRef]
- Mutlak, M.; Kehat, I. Extracellular signal-regulated kinases 1/2 as regulators of cardiac hypertrophy. Front. Pharmacol. 2015, 6, 149. [Google Scholar] [CrossRef]
- Vigil-Garcia, M.; Demkes, C.J.; Eding, J.E.C.; Versteeg, D.; de Ruiter, H.; Perini, I.; Kooijman, L.; Gladka, M.M.; Asselbergs, F.W.; Vink, A.; et al. Gene expression profiling of hypertrophic cardiomyocytes identifies new players in pathological remodelling. Cardiovasc. Res. 2021, 117, 1532–1545. [Google Scholar] [CrossRef] [PubMed]
- Zuccotti, P.; Cartelli, D.; Stroppi, M.; Pandini, V.; Venturin, M.; Aliverti, A.; Battaglioli, E.; Cappelletti, G.; Riva, P. Centaurin-alpha(2) interacts with beta-tubulin and stabilizes microtubules. PLoS ONE 2012, 7, e52867. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.; Shan, X.L.; Wang, S.N.; Chen, H.H.; Zhao, P.; Qian, D.D.; Xu, M.; Guo, W.; Zhang, C.; Lu, R. Trans-cinnamaldehyde suppresses microtubule detyrosination and alleviates cardiac hypertrophy. Eur. J. Pharmacol. 2022, 914, 174687. [Google Scholar] [CrossRef] [PubMed]
- Fassett, J.T.; Hu, X.; Xu, X.; Lu, Z.; Zhang, P.; Chen, Y.; Bache, R.J. AMPK attenuates microtubule proliferation in cardiac hypertrophy. Am. J. Physiol. Heart Circ. Physiol. 2013, 304, H749–H758. [Google Scholar] [CrossRef]
- Akhmanova, A.; Maiato, H. Closing the tubulin detyrosination cycle. Science 2017, 358, 1381–1382. [Google Scholar] [CrossRef]
- Kerr, J.P.; Robison, P.; Shi, G.; Bogush, A.I.; Kempema, A.M.; Hexum, J.K.; Becerra, N.; Harki, D.A.; Martin, S.S.; Raiteri, R.; et al. Detyrosinated microtubules modulate mechanotransduction in heart and skeletal muscle. Nat. Commun. 2015, 6, 8526. [Google Scholar] [CrossRef]
- Sanyal, C.; Pietsch, N.; Ramirez Rios, S.; Peris, L.; Carrier, L.; Moutin, M.J. The detyrosination/re-tyrosination cycle of tubulin and its role and dysfunction in neurons and cardiomyocytes. Semin. Cell Dev. Biol. 2023, 137, 46–62. [Google Scholar] [CrossRef]
- Chen, C.Y.; Caporizzo, M.A.; Bedi, K.; Vite, A.; Bogush, A.I.; Robison, P.; Heffler, J.G.; Salomon, A.K.; Kelly, N.A.; Babu, A.; et al. Suppression of detyrosinated microtubules improves cardiomyocyte function in human heart failure. Nat. Med. 2018, 24, 1225–1233. [Google Scholar] [CrossRef]
- Piquereau, J.; Novotova, M.; Fortin, D.; Garnier, A.; Ventura-Clapier, R.; Veksler, V.; Joubert, F. Postnatal development of mouse heart: Formation of energetic microdomains. J. Physiol. 2010, 588, 2443–2454. [Google Scholar] [CrossRef]
- Salameh, S.; Ogueri, V.; Posnack, N.G. Adapting to a new environment: Postnatal maturation of the human cardiomyocyte. J. Physiol. 2023, 601, 2593–2619. [Google Scholar] [CrossRef] [PubMed]
- Talman, V.; Teppo, J.; Poho, P.; Movahedi, P.; Vaikkinen, A.; Karhu, S.T.; Trost, K.; Suvitaival, T.; Heikkonen, J.; Pahikkala, T.; et al. Molecular Atlas of Postnatal Mouse Heart Development. J. Am. Heart Assoc. 2018, 7, e010378. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, B.; Li, J.; Coba, M.P. Synaptic GAP and GEF Complexes Cluster Proteins Essential for GTP Signaling. Sci. Rep. 2017, 7, 5272. [Google Scholar] [CrossRef] [PubMed]
- Yucel, D.; Solinsky, J.; van Berlo, J.H. Isolation of Cardiomyocytes from Fixed Hearts for Immunocytochemistry and Ploidy Analysis. J. Vis. Exp. 2020, 164, e60938. [Google Scholar] [CrossRef]
- Randazzo, P.A.; Andrade, J.; Miura, K.; Brown, M.T.; Long, Y.Q.; Stauffer, S.; Roller, P.; Cooper, J.A. The Arf GTPase-activating protein ASAP1 regulates the actin cytoskeleton. Proc. Natl. Acad. Sci. USA 2000, 97, 4011–4016. [Google Scholar] [CrossRef]
- Gasilina, A.; Vitali, T.; Luo, R.; Jian, X.; Randazzo, P.A. The ArfGAP ASAP1 Controls Actin Stress Fiber Organization via Its N-BAR Domain. iScience 2019, 22, 166–180. [Google Scholar] [CrossRef]
- Miura, K.; Jacques, K.M.; Stauffer, S.; Kubosaki, A.; Zhu, K.; Hirsch, D.S.; Resau, J.; Zheng, Y.; Randazzo, P.A. ARAP1: A point of convergence for Arf and Rho signaling. Mol. Cell 2002, 9, 109–119. [Google Scholar] [CrossRef]
- Davidson, A.C.; Humphreys, D.; Brooks, A.B.; Hume, P.J.; Koronakis, V. The Arf GTPase-activating protein family is exploited by Salmonella enterica serovar Typhimurium to invade nonphagocytic host cells. mBio 2015, 6, e02253-14. [Google Scholar] [CrossRef]
- Tanna, C.E.; Goss, L.B.; Ludwig, C.G.; Chen, P.W. Arf GAPs as Regulators of the Actin Cytoskeleton-An Update. Int. J. Mol. Sci. 2019, 20, 442. [Google Scholar] [CrossRef]
- Song, J.; Khachikian, Z.; Radhakrishna, H.; Donaldson, J.G. Localization of endogenous ARF6 to sites of cortical actin rearrangement and involvement of ARF6 in cell spreading. J. Cell Sci. 1998, 111, 2257–2267. [Google Scholar] [CrossRef] [PubMed]
- Pasqualato, S.; Menetrey, J.; Franco, M.; Cherfils, J. The structural GDP/GTP cycle of human Arf6. EMBO Rep. 2001, 2, 234–238. [Google Scholar] [CrossRef] [PubMed]
- Hongu, T.; Funakoshi, Y.; Fukuhara, S.; Suzuki, T.; Sakimoto, S.; Takakura, N.; Ema, M.; Takahashi, S.; Itoh, S.; Kato, M.; et al. Arf6 regulates tumour angiogenesis and growth through HGF-induced endothelial beta1 integrin recycling. Nat. Commun. 2015, 6, 7925. [Google Scholar] [CrossRef] [PubMed]
- Robison, P.; Caporizzo, M.A.; Ahmadzadeh, H.; Bogush, A.I.; Chen, C.Y.; Margulies, K.B.; Shenoy, V.B.; Prosser, B.L. Detyrosinated microtubules buckle and bear load in contracting cardiomyocytes. Science 2016, 352, aaf0659. [Google Scholar] [CrossRef]
- Swiatlowska, P.; Sanchez-Alonso, J.L.; Wright, P.T.; Novak, P.; Gorelik, J. Microtubules regulate cardiomyocyte transversal Young’s modulus. Proc. Natl. Acad. Sci. USA 2020, 117, 2764–2766. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
| Gene Name | Gene Description | Sequences (5′ → 3′) |
|---|---|---|
| Adap1 | ArfGAP with dual PH domains 1 | Fwd: CAAAGACCCTCTGGATGCCTT |
| Rev: GGTGACTCTGGGTTGACGG | ||
| Adap2 | ArfGAP with dual PH domains 2 | Fwd: CATCACTCCGGAGCGGAAAT |
| Rev: CAGCCAGATCCTCCGAGATG | ||
| Myh7 | Myosin heavy chain 7 | Fwd: CAACCTGTCCAAGTTCCGCA |
| Rev: GGCATCCTTAGGGTTGGGTAG | ||
| Nppa | Natriuretic peptide A | Fwd: CGGCACTTAGCTCCCTCTCT |
| Rev: GTTGCAGCCTAGTCCGCTCT | ||
| Rpl30 | Ribosomal protein L30 | Fwd: TCTTGGCGTCTGATCTTGGT |
| Rev: AAGTTGGAGCCGAGAGTTGA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Berthiaume, J.; Dumont, A.-A.; Dumont, L.; Roy, M.-F.; Giguère, H.; Auger-Messier, M. ArfGAP with Dual Pleckstrin Homology Domains 2 Promotes Hypertrophy of Cultured Neonatal Cardiomyocytes. Int. J. Mol. Sci. 2025, 26, 7588. https://doi.org/10.3390/ijms26157588
Berthiaume J, Dumont A-A, Dumont L, Roy M-F, Giguère H, Auger-Messier M. ArfGAP with Dual Pleckstrin Homology Domains 2 Promotes Hypertrophy of Cultured Neonatal Cardiomyocytes. International Journal of Molecular Sciences. 2025; 26(15):7588. https://doi.org/10.3390/ijms26157588
Chicago/Turabian StyleBerthiaume, Jonathan, Audrey-Ann Dumont, Lauralyne Dumont, Marie-Frédérique Roy, Hugo Giguère, and Mannix Auger-Messier. 2025. "ArfGAP with Dual Pleckstrin Homology Domains 2 Promotes Hypertrophy of Cultured Neonatal Cardiomyocytes" International Journal of Molecular Sciences 26, no. 15: 7588. https://doi.org/10.3390/ijms26157588
APA StyleBerthiaume, J., Dumont, A.-A., Dumont, L., Roy, M.-F., Giguère, H., & Auger-Messier, M. (2025). ArfGAP with Dual Pleckstrin Homology Domains 2 Promotes Hypertrophy of Cultured Neonatal Cardiomyocytes. International Journal of Molecular Sciences, 26(15), 7588. https://doi.org/10.3390/ijms26157588





