Abstract
Mitomycin C (MMC) is a widely employed chemotherapeutic agent, particularly in non-muscle invasive bladder cancer (NMIBC), where it functions by inducing DNA cross-linking and promoting tumor cell apoptosis. However, the tumor microenvironment (TME) significantly influences the therapeutic efficacy of MMC. Among the key regulators within the TME, the complement system and the coagulation pathway play a crucial role in modulating immune responses to cancer therapies, including MMC. This article explores the interaction between platinum nanoparticles (PtNPs) with human serum (HS) of NMIBC patients (T1 and Ta subtypes) at three different points: before the chemotherapy instillation of MMC (t0) and three (t3) and six months (t6) after the treatment with MMC. This novel nanoproteomic strategy allowed the identification of a TME proteomic signature associated with the response to MMC treatment. Importantly, two proteins involved in the immune response were found to be deregulated across all patients (T1 and Ta subtypes) during MMC treatment: prothrombin (F2) downregulated and complement component C7 (C7) upregulated. By understanding how these biomarker proteins interact with MMC treatment, novel therapeutic strategies can be developed to enhance treatment outcomes and overcome resistance in NMIBC.
1. Introduction
Bladder cancer (BC) ranks among the top ten most frequently diagnosed cancers on a global scale, with approximately 573,000 new cases identified each year and an annual mortality of around 213,000 []. NMIBC comprises the majority of cases and is categorized into Ta, T1, and carcinoma in situ (CIS) stages based on the tumor’s depth of invasion []. Although NMIBC has a lower mortality risk compared to muscle-invasive bladder cancer (MIBC), its high recurrence and progression rates necessitate rigorous surveillance and intervention strategies [,].
MMC is a widely used chemotherapeutic agent, particularly in the treatment of NMIBC, where it is typically administered intravesically []. MMC functions by inducing DNA cross-linking, resulting in cell cycle arrest and tumor cell apoptosis []. Despite its effectiveness, the response to MMC can be variable, and the TME plays a critical role in determining the therapeutic outcomes.
Among the various components of the TME, the complement system has emerged as an influential factor in regulating the immune response to cancer therapies, modulating tumor cell sensitivity to DNA-damaging agents like mitomycin C (MMC) []. Importantly, recent studies suggest that the complement system’s activation can modulate the efficacy of MMC in BC [].
Proteomics approaches have significantly advanced our understanding of the TME by enabling the comprehensive characterization of proteins involved in tumor progression, immune evasion, and therapy resistance [,,,,]. Particularly, sequential window acquisition of all theoretical fragment ion mass spectra (SWATH-MS) has emerged as a powerful mass spectrometry (MS) technique for the comprehensive and reproducible analysis of circulating proteins derived from the TME. Unlike traditional data-dependent acquisition (DDA) methods, sequential window acquisition of all theoretical mass spectra (SWATH-MS) enables data-independent acquisition (DIA), allowing for the quantification of thousands of proteins across multiple samples with high reproducibility and depth []. This approach is particularly advantageous for analyzing circulating tumor-derived proteins, such as cytokines, growth factors, and extracellular vesicle-associated proteins, which play crucial roles in tumor progression, immune modulation, and metastasis []. Recent studies have demonstrated that SWATH-MS can accurately profile TME-associated circulating proteins in plasma and serum, facilitating the identification of non-invasive biomarkers for cancer diagnosis and treatment response monitoring [,,,]. As advancements in DIA-based proteomics continue, SWATH-MS is expected to further revolutionize the study of the TME and its systemic effects on the host.
Biological samples exhibit a high level of complexity, with protein concentrations spanning a broad dynamic range, thereby posing significant challenges to proteomic analysis []. To reduce sample complexity, fractionation or enrichment of specific cellular compartments can be performed prior to MS analysis.
The integration of nanomaterials into proteomics has led to the emergence of nanoproteomics, a rapidly growing research field []. It is well established that when nanomaterials are dispersed in physiological fluids, they spontaneously interact with proteins, forming a dynamic layer known as the PC. Notably, disease-related biomarkers constitute less than 1% of the total serum protein content, making their detection particularly challenging. Due to their high surface area and tunable physicochemical properties, nanoparticles (NPs) can function as selective sorbents, preferentially binding low-abundance proteins in serum samples, thereby facilitating biomarker enrichment prior to MS-based analysis [,,,,,,]. The characterization of the PC surrounding NPs offers significant advantages over conventional proteomic approaches, enhancing the likelihood of discovering novel molecular biomarkers [].
Particularly, the unique physicochemical properties of PtNPs [] render them effective sorbent nanomaterials with significant potential for biomedical applications. Our previous studies provide robust protocols for the in vitro formation of the PC around PtNPs (2.40 ± 0.30 nm) following their interaction with HS []. The combined approach using PtNPs, electrophoretic separation (SDS-PAGE), and liquid chromatography–tandem mass spectrometry (LC-MS/MS) has proven effective in discovering novel circulating proteins linked to therapeutic response and resistance in HER2-positive breast cancer undergoing neoadjuvant chemotherapy []. In a comparable approach, our research group utilized silver nanoparticles (AgNPs; 9.73 ± 1.70 nm) as nanoscale scavenging platforms in conjunction with SWATH-MS to perform a comprehensive quantitative analysis of serum proteome alterations in two NMIBC subtypes: T1 and Ta. The ex vivo characterization of the protein coronas (PCs) formed on AgNPs facilitated the discrimination of two principal classes of serum proteins exhibiting differential abundance in NMIBC patients relative to healthy controls: components of the complement and coagulation cascades and various apolipoproteins [].
Due to colloidal stability, their high surface-area-to-volume ratio, and their ability to conjugate with biomolecules [], in the present work, PtNPs (2.40 ± 0.30 nm) were incubated with serum samples of NMIBC patients (n = 42) and healthy controls (n = 42) before the chemotherapy instillation of MMC (t0) and three (t3) and six months (t6) after treatment. Subsequently, an in-depth quantitative analysis of the PCs was performed using SWATH-MS to identify novel molecular targets associated with the distinct intrinsic subtypes of NMIBC, as well as for the identification of a TME proteomic signature associated with the MMC response (see Figure 1).
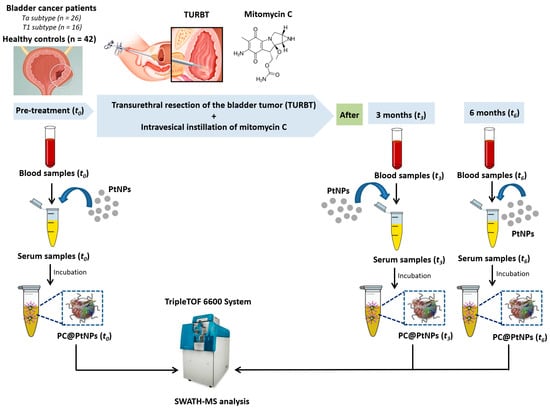
Figure 1.
Diagrammatic overview of the protocol for PC assembly on PtNPs (2.40 ± 0.30 nm), following ex vivo incubation with HS from n = 42 healthy controls (HCs) and n = 42 NMIBC patients. Samples were collected at baseline (t0, pre-treatment) and three (t3) and six months (t6) post-mitomycin C (MMC) instillation.
2. Results
2.1. Incubation of PtNPs (2.40 ± 0.30 nm) with HS Samples: Ex Vivo Formation and Comprehensive Characterization of the PC
HS samples from n = 42 HC and n = 42 NMIBC patients (n = 26 with the Ta subtype and n = 16 with the T1 subtype) were collected at times t0, t3, and t6 and processed and analyzed in the same manner (see Section 4.1).
PtNPs with a size of 2.40 ± 0.30 nm were prepared by a chemical reduction method [,,] (see Section 4.3, Table S1). Proteins present in triplicate serum samples were first reduced using dithiothreitol (DTT) and subsequently alkylated with iodoacetamide (IAA). Following these modifications, the proteins were incubated with PtNPs averaging 2.40 ± 0.30 nm in diameter to facilitate the formation of PCs (see Figure 2) [,].
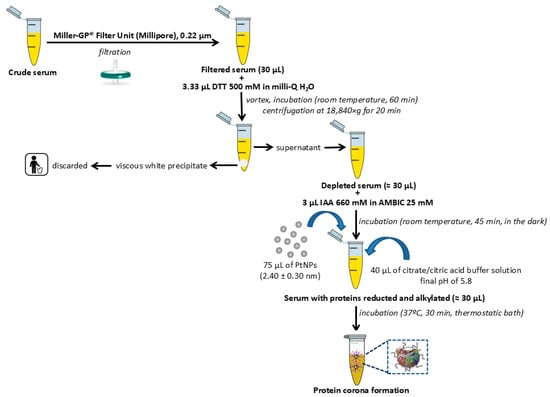
Figure 2.
Flowchart depicting the general pretreatment, depletion with DDT, and alkylation with IAA of human serum samples before the incubation with PtNPs (2.40 ± 0.30 nm) for the PC formation.
PtNPs were incubated separately with pooled HS samples obtained from n = 42 HC and n = 42 patients diagnosed with NMIBC. Following incubation, the PC-coated PtNPs were isolated by centrifugation and subsequently analyzed using transmission electron microscopy (TEM) and dynamic light scattering (DLS). Adsorption of serum proteins onto the nanoparticle surfaces induced a measurable increase in hydrodynamic diameter, with average sizes shifting from 2.40 ± 0.30 nm (bare PtNPs) to 2.55 ± 0.17 nm in the HC group and 2.65 ± 0.18 nm in the NMIBC group.
The association of cationic proteins with the surface of PtNPs is likely responsible for the observed shift in zeta potential, with the initially highly negative charge of −38.7 mV for unmodified PtNPs becoming less negative upon protein adsorption, reaching −25.3 mV in the HC condition and −27.2 mV in the NMIBC context [,].
2.2. Quantitative Analysis of the Protein Corona-Coated PtNPs by SWATH-MS Before the Chemotherapy Instillation of MMC (t0) and Three (t3) and Six Months (t6) After Treatment
Proteins bound to PtNPs were isolated using Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis (SDS-PAGE) and subsequently enzymatically digested according to established protocols [,]. The generated peptides were then subjected to quantitative analysis employing the advanced label-free proteomics technique, SWATH-MS.
The comparison of the ex vivo PC profiles enabled the identification of proteins exhibiting differential expression between HC and the two NMIBC subtypes (Ta and T1) across multiple time points (t0, t3, and t6). All analyses were subjected to statistical filtering, retaining only those proteins with a significance threshold of p-value ≤ 0.05.
2.3. Differentially Expressed Proteins in the Blood Serum of Control Patients and NMIBC Patients Before the Chemotherapy Instillation of MMC (t0)
At point t0, a total of 39 proteins were identified as differentially expressed in NMIBC patients with the T1 subtype, including 25 upregulated and 14 downregulated proteins. In contrast, 62 proteins were differentially expressed in patients with the Ta subtype, comprising 53 upregulated and 9 downregulated proteins (see Table 1). Comprehensive lists of candidate protein biomarkers exhibiting upregulation or downregulation in both NMIBC subtypes relative to healthy controls, along with the corresponding fold-change values, are provided in Tables S2 and S5.

Table 1.
Total number of differentially expressed proteins (upregulated and downregulated; p-value ≤ 0.05) specific to the NMIBC subtypes (T1 and Ta) identified by SWATH-MS analysis of protein corona-coated PtNPs (2.40 ± 0.30 nm) at time point t0 (data for the T1 subtype are highlighted in blue color and for the Ta subtype in orange).
The Venn diagram depicting statistically significant upregulated and downregulated proteins reveals that 26 proteins are commonly altered in both the T1 and Ta NMIBC subtypes (see Figure 3). Notably, 21 of these 26 proteins were consistently upregulated across both subtypes: alpha-1-antichymotrypsin (SERPINA3), alpha-1-antitrypsin (SERPINA1), alpha-1B-glycoprotein (A1BG), biotinidase (BTD), coagulation factor IX (F9), coagulation factor XII (F12), complement C4-B (C4B), complement component C7 (C7), complement component C8 alpha chain (C8A), complement component C9 (C9), galectin-3-binding protein (LGALS3BP), immunoglobulin alpha-2 heavy chain, immunoglobulin delta heavy chain, immunoglobulin heavy constant gamma 3 (IGHG3), immunoglobulin kappa variable 1-33 (IGKV1-33), lumican (LUM), monocyte differentiation antigen CD14 (CD14), plasminogen (PLG), platelet glycoprotein Ib alpha chain (GP1BA), serum amyloid A-4 protein (SAA4), and testis-expressed protein 33 (CIMIP4). On the other hand, 5 of these 26 proteins were found to be downregulated in the T1 and Ta NMIBC subtypes: alpha-2-HS-glycoprotein (AHSG), apolipoprotein M (APOM), carboxypeptidase N subunit 2 (CPN2), prothrombin (F2), and serum paraoxonase/arylesterase 1 (PON1) (see Tables S2 and S5).
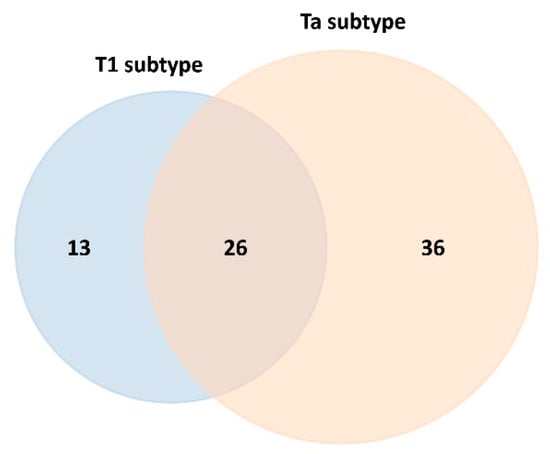
Figure 3.
Venn diagram depicting the overlap and subtype-specific differentially expressed serum proteins adsorbed onto PtNPs (2.40 ± 0.30 nm) after 30 min ex vivo incubation with HS from NMIBC patients of subtypes T1 and Ta (data for the T1 subtype are highlighted in blue color and for the Ta subtype in orange).
SWATH-MS analysis further enabled the identification of subtype-specific proteins (see Figure 3), with 13 proteins uniquely associated with the T1 subtype (4 upregulated and 9 downregulated) and 36 proteins specific to the Ta subtype (32 upregulated and 4 downregulated), as detailed in Table 2 and Table 3, respectively.

Table 2.
Differentially expressed proteins identified in NMIBC patients with the T1 subtype (t0) compared to HC following SWATH-MS analysis of PC-coated PtNPs (2.40 ± 0.30 nm). Proteins were classified as potential biomarkers if they exhibited a statistically significant difference (p-value ≤ 0.05) and a fold change (FC) greater than 1.1 (downregulated in T1 subtype: dark blue color) or less than 0.8 (upregulated in T1 subtype: light blue).

Table 3.
Differentially expressed proteins identified in NMIBC patients with the Ta subtype (t0) compared to HC following SWATH-MS analysis of PC-coated PtNPs (2.40 ± 0.30 nm). Proteins were classified as potential biomarkers if they exhibited a statistically significant difference (p-value ≤ 0.05) and an FC greater than 1.1 (downregulated in Ta subtype: dark orange color) or less than 0.8 (upregulated in Ta subtype: light orange).
2.4. The Biological Role of the NMIBC-Related Proteins Identified in the PtNP–Protein Corona Before the Chemotherapy Instillation of MMC (t0)
To analyze the global alterations in the serum proteome associated with NMIBC at baseline (t0), the STRING database was used to perform protein–protein interaction (PPI) network analysis. This approach enabled the identification of 26 commonly dysregulated proteins shared between the T1 and Ta subtypes, which exhibited significant differential expression between HC and NMIBC patients. Additionally, subtype-specific analyses revealed 13 proteins uniquely altered in the T1 subtype and 36 in the Ta subtype.
The analysis revealed that the dysregulated serum proteins are mainly involved in the immune response pathway. From the 26 proteins found to be commonly deregulated in both NMIBC subtypes, 8 proteins were implicated in the immune response pathway: coagulation factor XII (F12), complement C4-B (C4B), complement component C7 (C7), complement component C8 alpha chain (C8A), complement component C9 (C9), immunoglobulin kappa variable 1-33 (IGKV1-33), monocyte differentiation antigen CD14 (CD14), and prothrombin (F2). From these, C4B, C7, C8A, C9, F2, and F12 also participate in the complement and coagulation cascades pathways.
Particularly, 5 of the 13 biomarker proteins from the T1 subtype are involved in the immune response pathway, such as complement C4-A (C4A), C4b-binding protein beta chain (C4BPB), complement factor D (CFD), kininogen-1 (KNG1), and platelet basic protein (PPBP). From these, C4B, C4BPB, CFC, and KNG1 also participate in the complement and coagulation cascade pathways.
In the case of the Ta subtype, 15 from the 36 biomarker proteins are vinculated with the immune response pathway: N-acetylmuramoyl-L-alanine amidasa (PGLYRP2), complement C1q subcomponent subunit A (C1QA), complement C1r subcomponent (C1R), complement C1s subcomponent (C1S), C4b-binding protein alpha chain (C4BPA), complement component C6 (C6), complement component C8 beta chain (C8B), complement component C8 gamma chain (C8G), complement factor H (CFH), complement factor I (CFI), clusterin (CLU), ficolin-2 (FCN2), fibrinogen alpha chain (FGA), immunoglobulin heavy variable 4-38-2 (IGHV4-38-2 or LOC102723407), and plasma protease C1 inhibitor (SERPING1). From these, C1QA, C1R, C1S, C4BPA, C6, C8B, C8G, CFH, CFI, CLU, FGA, and SERPING1 are also involved in the complement and coagulation cascade pathways.
2.5. Comparison of the Differentially Expressed Proteins in the Blood Serum of Control and NMIBC Patients Before Treatment (t0) and Three (t1) and Six Months (t2) After the Chemotherapy Instillation of MMC
As presented in Table 4, a total of 39, 49, and 62 proteins were found to be differentially expressed by liquid chromatography–tandem mass spectrometry (LC-MS/MS) in all serum samples from NMIBC patients with the T1 subtype, whereas 62, 43, and 60 proteins were identified in all serum samples from patients with the Ta subtype at time points t0, t3, and t6, respectively (see Table 4 and Tables S2–S7). A comparative analysis of the results obtained across the three points revealed that 9 and 13 proteins were commonly identified in T1 and Ta cases, respectively (see Table 4 and Figure 4).

Table 4.
The total amount of differently expressed proteins (upregulated and downregulated, p-value ≤ 0.05) and those specific to different NMIBC subtypes (T1 and Ta) found after the SWATH-MS analysis of PC-coated PtNPs (2.40 ± 0.30 nm) at different times (t0, t3, and t6).
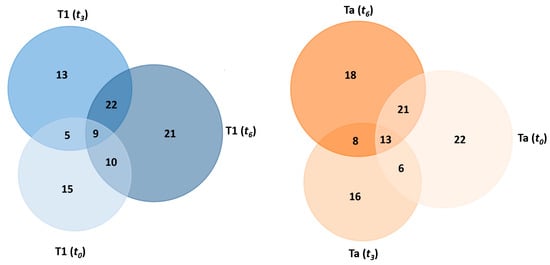
Figure 4.
Venn diagram showing the number of shared and specific deregulated proteins identified in the PC-coated PtNPs (2.40 ± 0.30 nm) after their incubation (30 min) with HS samples from NMIBC patients of the T1 subtype (left) and Ta subtype (right) at different times (t0, t3, and t6).
Among the nine proteins commonly identified in the T1 subtype across all time points, four were consistently downregulated in all cases: insulin-like growth factor-binding protein complex acid-labile subunit (IGFALS), carboxypeptidase N subunit 2 (CPN2), prothrombin (F2), and apolipoprotein M (APOM). Three proteins were consistently upregulated: complement component C7 (C7), complement component C9 (C9), and immunoglobulin kappa variable 1-33 (IGKV1-33). In contrast, complement component C4-B (C4B) and apolipoprotein F (APOF) exhibited either upregulation or downregulation depending on the time point.
Among the 13 proteins commonly identified in the Ta subtype across all time points, four were consistently downregulated: N-acetylmuramoyl-L-alanine amidase (PGLYRP2), carboxypeptidase N subunit 2 (CPN2), prothrombin (F2), and apolipoprotein M (APOM). In contrast, nine proteins were consistently upregulated: monocyte differentiation antigen CD14 (CD14), complement component C7 (C7), fibulin-1 (FBLN1), immunoglobulin lambda variable 3-21 (IGLV3-21), immunoglobulin heavy variable 3-49 (IGHV3-49), immunoglobulin lambda-1 light chain (X), immunoglobulin lambda variable 3-25 (IGLV3-25), apolipoprotein A-II (APOA2), and hemoglobin subunit beta (HBB).
Notably, apolipoprotein M (APOM), carboxypeptidase N subunit 2 (CPN2), and prothrombin (F2) were consistently downregulated across all patients (T1 and Ta subtypes) at all time points (t0, t3, and t6), whereas complement component C7 (C7) was consistently upregulated.
2.6. The Biological Role of the NMIBC-Related Proteins Identified in the PtNP–Protein Corona Before (t0) and After (t3 and t6) the Chemotherapy Instillation of MMC
To analyze global changes in the serum proteome associated with the response to chemotherapy instillation of MMC in NMIBC patients with the T1 and Ta subtypes, proteins identified at the three time points were examined using the STRING software version 10.0 (see Figure 5). Consistent with the findings at time point t0, the analysis indicated that dysregulated serum proteins at t3 and t6 are primarily involved in the immune response pathway.
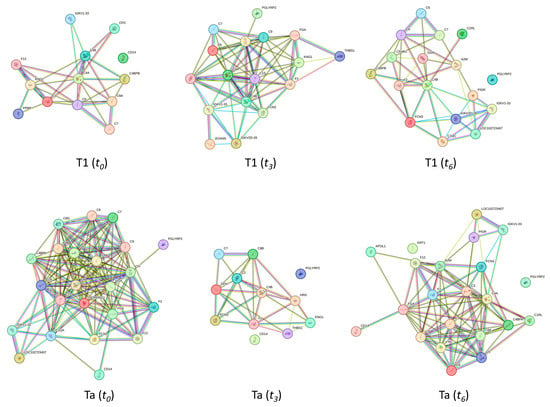
Figure 5.
Clusters found in the protein–protein interaction network map based on the STRING database, highlighting proteins associated with the immune response pathway. At the top, clusters with 13, 17, and 17 deregulated proteins related to the immune response pathway found in the T1 subtype at t0, t3, and t6, respectively, are shown. A total of 23, 12, and 22 proteins related to the immune response pathway were deregulated in the Ta subtype at times t0, t3, and t6, respectively (down).
Figure 5 presents the clusters identified in the protein–protein interaction network map based on the STRING database, highlighting proteins associated with the immune response pathway. A total of 13, 17, and 17 proteins related to the immune response pathway were deregulated in the T1 subtype at t0, t3, and t6, respectively. However, as Figure 4 shows, 23, 12, and 22 proteins related to the immune response pathway were deregulated in the Ta subtype at times t0, t3, and t6, respectively.
Table 5 summarizes the variation in a total of 45 immune-response-related proteins during treatment for both subtypes T1 and Ta. From them, while 22 of these proteins were commonly found to be deregulated in the T1 and the Ta subtypes (C4A, C4B, C7, C8A, C9, CD14, F12, F2, IGKV1-33, KNG1, C1S, C2, C6, CFI, FCN3, FGA, PGLYRP2, THBS1, A2M, C1RL, LOC102723407, and PIGR), 8 proteins were specific to the T1 subtype (C4BPB, CFD, PPBP, IGKV2D-28, JCHAIN, C1QC, CFHR1, and GSN), and 15 proteins were specific to the Ta subtype (C1QA, C1R, C4BPA, C8B, C8G, CFH, CLU, FCN2, SERPING1, HRG, APOL1, C3, C5, KRT1, and VTN (see Figure 5 and Table 5)).

Table 5.
Deregulated immune-response-related proteins in NMIBC patients with subtypes T1 and Ta at different time points (t0, t3, and t6). The upward-pointing arrow indicates upregulated, while the downward-pointing arrow indicates downregulated.
Importantly, as previously mentioned, among all proteins involved in the immune response, F2 was consistently downregulated across all patients (T1 and Ta subtypes) at all time points (t0, t3, and t6), whereas C7 was consistently upregulated.
3. Discussion
MMC functions by inducing DNA cross-linking, resulting in cell cycle arrest and tumor cell apoptosis []. The response to MMC in BC is influenced by various factors, including the TME, which plays a key role in modulating the effects of chemotherapy [,,]. Among the various components of the TME, the complement system has emerged as an influential factor in regulating the immune response to cancer therapies, modulating tumor cell sensitivity to DNA-damaging agents like MMC [].
The complement system consists of over 30 proteins that mediate a cascade of immune responses, including opsonization, inflammation, and cell lysis. These proteins are tightly regulated, and their activation can have both pro- and anti-tumor effects depending on the context [,,]. Complement activation generates anaphylatoxins such as C3a and C5a, which act as chemoattractants for myeloid-derived suppressor cells (MDSCs) and tumor-associated macrophages (TAMs) [,]. Binding of C5a to C5aR1 on these immune cells promotes their migration into the tumor microenvironment, where they secrete immunosuppressive molecules, including reactive oxygen and nitrogen species (ROS/RNS) and interleukin-10 (IL-10). These factors attenuate CD8+ T cell-mediated clearance of MMC-damaged tumor cells, thereby undermining chemotherapy efficacy [,,,]. Furthermore, complement activation can influence tumor cell resistance to MMC through several mechanisms. Tumor cells often overexpress membrane complement regulatory proteins (mCRPs) such as CD55, CD46, and CD59, which inhibit complement activation and protect against complement-mediated cytotoxicity [,]. For instance, CD59 prevents the formation of the membrane attack complex (MAC), thereby shielding tumor cells from lysis [,]. Additionally, sublytic levels of MAC deposition can activate intracellular signaling pathways, including PI3K/Akt, MAPK/Erk, and NF-κB/STAT3, leading to enhanced tumor cell proliferation, survival, and upregulation of drug efflux transporters [,,].
In the present work, among all deregulated complement proteins, only C7 was consistently upregulated across all patients (T1 and Ta subtypes) at all time points (t0, t3, and t6).
C7 is a critical component of the membrane attack complex (MAC). While the full MAC assembly can cause cell lysis, sub-lytic MAC deposition—facilitated by elevated C7—activates intracellular pathways in tumor cells, including Ca2+- and G-protein-mediated signals, and transcription factors such as EGR1, IRF1, AREG, and CXCL1. This activation enhances tumor proliferation, survival, and adaptive resistance mechanisms []. Furthermore, in hepatocellular carcinoma models, higher nuclear C7 expression upregulates stemness-associated genes (OCT4, SOX2, and MYC) through LSF-1 activation. These changes support tumor-initiating cell expansion, survival, and treatment resistance—including likely resistance to DNA-damaging agents like MMC []. Studies have also found that elevated tumor C7 expression correlates with distinct immune phenotypes and can stratify prognosis. Although more research is needed across cancer types, data indicate that higher C7 may reflect a microenvironment that favors immune suppression and resistance to therapy [].
On the other hand, tumor-associated coagulation proteins significantly influence the response to MMC therapy by creating a protective microenvironment around cancer cells. Specifically, fibrin deposition, initiated through tumor-expressed tissue factor (TF), forms dense “fibrin clot shields” that physically impede cytotoxic immune cell binding and hinder drug penetration, thereby reducing MMC-induced tumor cell clearance []. Additionally, in vitro models show that these fibrin-based shields confer resistance to chemotherapeutic agents by creating a scaffold that isolates tumor cells and limits effector function of NK and LAK cells []. Beyond the physical barrier, coagulation proteases such as thrombin and factor Xa activate protease-activated receptors (PARs) and integrin-mediated survival signaling pathways in tumor cells, promoting PI3K/Akt and PTEN/AKT axis activation. This signaling enhances tumor proliferation, survival, and chemoresistance, and it is potentiated by fibrin–integrin β1 interactions that reinforce drug resistance phenotypes []. Furthermore, the cross-talk between coagulation and inflammation fosters an immunosuppressive tumor microenvironment that limits immune-mediated clearance of MMC-damaged cells and may worsen therapeutic outcome []. These findings suggest that elevated coagulation activity may serve as indicators of reduced MMC efficacy, while targeting coagulation pathways could enhance chemotherapy response.
In the present work, among all deregulated coagulation proteins, only F2 was consistently downregulated across all patients (T1 and Ta subtypes) at all time points (t0, t3, and t6). Tumor-associated coagulation activation drives prothrombin conversion to thrombin, which affects MMC therapy efficacy through both biological and physical mechanisms. Thrombin signals via protease-activated receptors (PAR-1 and PAR-2) on tumor, stromal, and endothelial cells, inducing oncogenic pathways such as PI3K/Akt, MAPK/Erk, and NF-κB and thereby promoting cell survival, angiogenesis, and metastasis [,]. Simultaneously, thrombin enhances fibrin deposition and platelet activation, contributing to a fibrin matrix “shield” around tumor cells that impedes MMC penetration and protects against immune-mediated clearance []. Moreover, thrombin-induced PAR activation drives an immunosuppressive microenvironment by recruiting immunosuppressive populations such as M2-like macrophages, T regulatory cells, and myeloid-derived suppressor cells via release of cytokines like IL-6, TNF-α, and MCP-1 [,]. This milieu diminishes immune-mediated eradication of MMC-compromised tumor cells and fosters tumor resilience.
In contrast, reduced F2 levels and, consequently, diminished thrombin activity have been shown in preclinical models to significantly impair tumor growth and survival signaling. In genetically modified mice expressing approximately 10% of normal prothrombin, subcutaneous colon adenocarcinoma growth was reduced nearly threefold compared to wild-type mice []. Similarly, in pancreatic ductal adenocarcinoma models (KPC2 cells), both genetic reduction and pharmacologic inhibition of prothrombin dramatically suppressed tumor mass in vivo []. These findings highlight that thrombin is a potent driver of tumor progression via mechanisms involving PAR-mediated signaling, angiogenesis, cellular proliferation, and metastasis [].
Consequently, low prothrombin levels may weaken protective tumor microenvironment features such as fibrin-based shielding, thrombin-triggered survival signaling (via PAR-1 and related pathways), and immunosuppressive cell recruitment. These conditions are expected to enhance MMC delivery, attenuate DNA repair and survival pathways in tumor cells, and improve therapeutic response. Although direct clinical studies in the context of MMC are limited, the mechanistic rationale strongly suggests that patients with low circulating prothrombin levels may experience better responses to MMC by lacking the thrombin-fueled protection that typically impairs drug efficacy.
Although a direct clinical correlation between C7 levels and MMC response has not yet been well-established, measuring tumor and/or circulating C7 may help identify tumors with high sub-lytic MAC activity and stemness-associated resistance. Higher C7 levels may reflect the presence of these resistance mechanisms, predicting a reduced response to MMC therapy. Conversely, patients with low circulating F2 may experience better responses to MMC by lacking the thrombin-fueled protection that typically impairs drug efficacy. Clinically, this suggests that low F2 plus high C7 levels may inversely influence MMC response: reduced coagulation enhances MMC effectiveness, whereas elevated C7 may undermine it by activating pro-survival and stemness pathways.
Understanding the interplay between complement activation, prothrombin, and MMC offers potential therapeutic strategies to overcome resistance mechanisms in NMIBC. C7 could potentially complement other biomarkers, such as F2, to stratify patients more likely to benefit from MMC chemotherapy versus those needing alternative or combinational treatments.
Complement modulation, particularly inhibiting C7, could enhance MMC’s efficacy. Therapies that target C7’s immunosuppressive effects might reduce tumor cell survival and improve immune cell activation, thereby improving MMC’s therapeutic outcome. Combining MMC with strategies that disrupt both complement activation and coagulation may represent a promising approach to overcoming resistance to MMC and enhancing tumor cell clearance.
4. Materials and Methods
4.1. Patient Study Group and Biological Samples
Between January 2018 and June 2019, a total of 42 individuals diagnosed with NMIBC, ranging in age from 31 to 86 years, were enrolled at the University Hospital Lucus Augusti (HULA) in Lugo, Spain. An equal number of healthy individuals, matched by age, were included as controls. The NMIBC cohort was further stratified into two pathological categories: 26 patients presented with Ta stage tumors, while 16 were classified as having T1-stage disease [,].
In accordance with current guidelines, a single immediate postoperative intravesical instillation of a chemotherapeutic agent—specifically 40 mg MMC—after transurethral resection of the bladder tumor (TURBT) is recommended for all NMIBC patients, as numerous randomized trials and meta-analyses have shown that this strategy significantly reduces tumor recurrence (risk reduction ~25–38%) without affecting progression or survival []. This effect appears most pronounced in patients with solitary, small, and low-grade tumors, and benefits persist even when subsequent adjuvant intravesical therapies are administered. To maximize the efficacy of the 40 mg MMC instillation, it should be administered as promptly as possible—preferably within two hours post-TURBT, either in the recovery room or the operating theatre [,]. In the present study, all NMIBC patients received a single immediate intravesical instillation of 40 mg MMC following TURBT [,,,,].
Cystoscopy at 3 months following TURBT was carried out for all NMIC patients with the Ta subtype. Patients with the T1 subtype underwent cystoscopy and urinary cytology at 3 months [,,].
Peripheral venous blood samples were collected at three times: before NMIBC patients underwent surgery and/or before receiving any treatment (t0) and three months (t3) and six months (t6) after TURBT. Blood samples from the 42 age-matched HC were collected in parallel at the same timepoints.
Peripheral blood was drawn using 9 mL VACUETTE® Serum Clot Activator Tubes (Greiner Bio-One, Kremsmünster, Austria). Following a 15 min clotting period at room temperature, samples were centrifuged at 1800× g for 5 min at 4 °C. The resulting serum was carefully aliquoted into sterile cryogenic vials, immediately frozen, and stored at −80 °C until analysis.
Ethical approval for the study was granted by the Clinical Research Ethics Committee (CEIC) of Galicia, Spain (reference number: 2017/419). All procedures were carried out in accordance with the principles outlined in the Declaration of Helsinki. Written informed consent was obtained from all participants prior to their inclusion in the study.
4.2. Chemicals and Reagents
All chemicals and solvents employed were of HPLC/LC-MS or electrophoresis grade. The following reagents were obtained from Merck (Barcelona, Spain): ammonium bicarbonate (AMBIC, ≥99.5%), ammonium persulfate (APS, ≥98%), β-mercaptoethanol (≥99%), glycerol (86–88%), chloroplatinic acid (H2PtCl6, ≥99%), sodium carbonate (≥99%), sodium citrate tribasic dihydrate (≥99%), tannic acid, trifluoroacetic acid (TFA, ≥99%), tris (hydroxymethyl)aminomethane (Tris-base), trizma base (≥99.9%), trypsin (from bovine pancreas), and urea. The acrylamide/bis-acrylamide solution (30%, 37.5:1) was sourced from Serva (Heidelberg, Germany). Reagents purchased from Bio-Rad (Madrid, Spain) included bromophenol blue, a Coomassie Brilliant Blue R-250 (CCB) staining solution, DL-dithiothreitol (DTT), iodoacetamide (IAA, ≥99%), sodium dodecyl sulfate (SDS), N,N,N′,N′-tetramethylethylenediamine (TEMED), and a molecular weight marker for sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE; range: 6.5–200 kDa). All solvents were provided by Panreac Química SLU (Barcelona, Spain).
4.3. Synthesis of Citrate-Coated PtNPs (2.40 ± 0.30 nm) and Ex Vivo Protein Corona Formation
PtNPs were synthesized using a modified version of a previously described chemical reduction protocol [,] (see Figure 6). Briefly, 200 µL of freshly prepared sodium borohydride (NaBH4, 50 mM) was added dropwise to an aqueous mixture containing 1 mL of chloroplatinic acid (H2PtCl6, 16 mM), 1 mL of trisodium citrate (40 mM), and 38 mL of deionized water. The reaction was maintained under vigorous stirring at room temperature, during which a color change to brownish-yellow signaled the formation of well-dispersed PtNPs. Stirring continued for an additional 1 h to ensure complete nanoparticle formation.
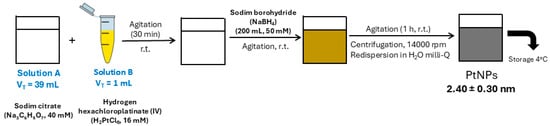
Figure 6.
Synthesis of citrate-coated PtNPs (2.40 ± 0.30 nm).
To eliminate excess trisodium citrate (SC) and sodium borohydride (NaBH4), the PtNPs suspension was subjected to three cycles of centrifugation at 24,610× g for 30 min. The purified nanoparticles were subsequently redispersed in Milli-Q water. The ex vivo formation of the PC was carried out according to the procedure outlined in Figure 2, as described in previous studies [,]. Characterization of the colloidal PtNPs and confirmation of PC formation were performed using TEM with a JEOL JEM-1011 instrument (Santiago de Compostela, Spain). ζ-potential measurements of the PtNPs were carried out in triplicate at 25 °C using a Malvern Zetasizer Nano ZS (Santiago de Compostela, Spain).
4.4. Depletion of Multiple Highly Abundant Proteins in Serum Samples
Three aliquots of human serum (30 µL each) were individually filtered through 0.22 µm Millipore Miller-GP® filter units (Merck Millipore, Burlington, MA, USA) to remove particulate contaminants. Each filtered sample was then treated with 3.3 µL of freshly prepared DTT (500 mM), briefly vortexed, and incubated for 60 min to induce protein precipitation, which was evidenced by the formation of a viscous white pellet. Samples were subsequently centrifuged at 18,840× g for 20 min, and the resulting supernatants were collected for downstream protein alkylation and nanoparticle-assisted fractionation.
4.5. Isolation of Low-Abundance Proteins: Ex Vivo Protein Corona Formation
Following the depletion of high-abundance serum proteins using DTT, individual serum aliquots from each patient were processed for the enrichment and analysis of low-abundance proteins, with potential biomarker relevance. Protein alkylation was performed by incubating each aliquot with IAA at room temperature for 45 min in the absence of light to prevent degradation. Subsequently, 75 µL of PtNPs (2.40 ± 0.30 nm) was added to each sample, along with 40 µL of citrate/citric acid buffer to adjust the pH to 5.8. The mixtures were incubated at 37 °C for 30 min under continuous shaking in a thermostatic water bath to facilitate PC formation. Following incubation, nanoparticle–protein complexes were isolated via centrifugation at 24,610× g for 30 min. The resulting pellets were washed three times with 25 µL of citrate/citric acid buffer and centrifuged under identical conditions to remove non-specifically bound proteins. The final PC complexes associated with the PtNPs were reconstituted and separated by SDS-PAGE using PowerPac™ Basic Power Supply (Bio-Rad, Madrid, Spain). Gels were stained, and specific protein bands of interest were excised and subjected to in-gel tryptic digestion, following previously published protocols established by our group [,].
4.6. Quantification of the Proteins Presented in the Corona-Coated PtNPs by SWATH-MS
SWATH-MS acquisition and data processing were performed following a previously established protocol, with slight modifications []. Briefly, a spectral library was constructed using pooled serum samples representative of each study group—HC, NMIBC patients with the Ta subtype, and those with the T1 subtype—via DDA on a micro-LC system. Peptide peak extraction was carried out using the MS/MSALL add-in within PeakView Software (version 2.2, Sciex, Foster City, CA, USA), in combination with the SWATH Acquisition MicroApp (version 2.0, Sciex). Only peptides with a confidence score exceeding 99% were retained for inclusion in the spectral library.
SWATH-MS analyses were conducted on a TripleTOF® 6600 LC-MS/MS instrument (Sciex) using a DIA workflow. Peak extraction and relative quantification were performed using PeakView (v. 2.2), and the resulting data were exported as mrkvw files into MarkerView software (Sciex) version 1.4 for statistical analysis. For each identified protein, the mean MS peak area across biological replicates was used for quantification. Group comparisons were conducted using a Student’s t-test within MarkerView based on the cumulative peak areas of all quantified transitions per protein. For protein quantitation, only peptides with a False Discovery Rate (FDR) below 1% were considered. The mean area sums of all the transitions derived for each protein in each sample will be used in a Student’s t-test to determine how well each variable distinguishes the two groups, which will be presented as a p-value. The fold change (FC) was derived from the ratio of the geometric means of the two groups, equivalently obtained by transforming data via logarithm, computing the arithmetic ratio, and then applying the inverse transformation. For each library, its set of differentially expressed proteins (p-value < 0.05) with an FC > 1.1 or <0.8 was selected based on previously reported methods [,,,].
4.7. Protein Functional Interaction Network Analysis
Analyses of functional protein interaction networks, incorporating both direct (physical) and indirect protein–protein interactions (PPIs), were performed using the STRING database (version 10.0; http://string-db.org, accessed on 15 January 2021) [].
5. Conclusions
The clinical objective of this study is to identify a tumor microenvironment (TME)-associated proteomic signature that is associated with the response to mitomycin C (MMC) treatment in patients with NMIBC, specifically the Ta and T1 subtypes. To achieve this, a comparative quantitative proteomic analysis was performed on the PCs formed following the incubation of PtNPs with HS samples obtained from NMIBC patients at three distinct time points: prior to MMC instillation (t0) and at three (t3) and six (t6) months post-treatment.
This analysis revealed that many of the dysregulated serum proteins across these time points were enriched in immune-response-related pathways, underscoring the influence of systemic immune changes on therapeutic response. Notably, two proteins showed consistent differential expression across all patients and time points: F2 was consistently downregulated, while C7 was consistently upregulated in both Ta and T1 subtypes.
These findings suggest that C7 and F2 act as functionally divergent markers within the TME: Elevated C7 may drive MMC resistance through immune evasion and survival signaling, whereas reduced F2 may enhance MMC efficacy by weakening coagulation-driven tumor protection. These two proteins may therefore serve as complementary biomarkers for stratifying NMIBC patients based on likely therapeutic response. Furthermore, targeting these pathways—either by inhibiting C7-mediated complement activation or modulating thrombin-driven coagulation signaling—may provide a rational basis for combinatorial strategies aimed at overcoming resistance and improving MMC-based treatment outcomes.
Understanding this interplay between immune and coagulation systems provides valuable insight into the mechanisms underpinning MMC sensitivity and resistance in NMIBC and lays the groundwork for future therapeutic interventions aimed at reprogramming the TME to enhance clinical efficacy.
Supplementary Materials
The supporting information can be downloaded at https://www.mdpi.com/article/10.3390/ijms26157413/s1.
Author Contributions
Conceptualization, C.N.; methodology, B.B.G., F.J.C.-N., D.P.-F., L.R.-S., S.B.B. and C.N.; software, S.B.B. and C.N.; formal analysis, S.B.B. and C.N.; investigation, S.B.B. and C.N.; resources, B.B.G. and C.N.; writing—original draft preparation, C.N.; writing—review and editing, C.N.; visualization, C.N.; supervision, C.N.; project administration, C.N.; funding acquisition, C.N. All authors have read and agreed to the published version of the manuscript.
Funding
This research has been funded by the “Instituto de Salud Carlos III (ISCIII)” through grant PI22/00025 and also by the Spanish Ministry of Science, Innovation, and Universities through the program “Consolidación Investigadora 2024” (CNS2024-154855).
Institutional Review Board Statement
The research was carried out in full compliance with the ethical standards set forth in the Declaration of Helsinki, and it received formal approval from the Clinical Research Ethics Committee (CEIC) of Galicia, Spain, under protocol number 2017/419.
Informed Consent Statement
Informed consent was obtained from all subjects involved in this study.
Data Availability Statement
The original contributions presented in this study are included in the article/Supplementary Materials. Further inquiries can be directed to the corresponding author.
Acknowledgments
The authors thank all participating patients.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
| CIS | Carcinoma in situ |
| DDA | Data-dependent acquisition |
| DIA | Data-independent acquisition |
| DLS | Dynamic light scattering |
| DTT | Dithiothreitol |
| HC | Healthy controls |
| HS | Human serum |
| IAA | Iodoacetic acid |
| LC-MS/MS | Liquid chromatography–tandem mass spectrometry |
| MDSCs | Myeloid-derived suppressor cells |
| MIBC | Muscle-invasive bladder cancer |
| MS | Mass spectrometry |
| NMIBC | Non-muscle invasive bladder cancer |
| PC | Protein corona |
| PPI | Protein–protein interaction |
| PtNPs | Platinum nanoparticles |
| SDS-PAGE | Sodium dodecyl sulfate–polyacrylamide gel electrophoresis |
| SWATH-MS | Sequential window acquisition of all theoretical mass spectra |
| TAMs | Tumor-associated macrophages |
| TEM | Transmission electron microscopy |
| TURBT | Transurethral resection of the bladder tumor |
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Babjuk, M.; Burger, M.; Capoun, O.; Cohen, D.; Compérat, E.M.; Dominguez Escrig, J.L.; Gontero, P.; Liedberg, F.; Masson-Lecomte, A.; Mostafid, A.H.; et al. European Association of Urology Guidelines on Non-muscle-invasive Bladder Cancer (Ta, T1, and Carcinoma in Situ). Eur. Urol. 2022, 81, 75–94. [Google Scholar] [CrossRef]
- Jiang, L.-J.; Guo, S.-B.; Huang, Z.-Y.; Li, X.-L.; Jin, X.-H.; Huang, W.-J.; Tian, X.-P. PHB promotes bladder cancer cell epithelial-mesenchymal transition via the Wnt/β-catenin signaling pathway. Pathol. Res. Pract. 2023, 247, 154536. [Google Scholar] [CrossRef]
- Jiang, L.-J.; Guo, S.-B.; Zhou, Z.-H.; Li, Z.-Y.; Zhou, F.-J.; Yu, C.-P.; Li, M.; Huang, W.-J.; Liu, Z.-W.; Tian, X.-P. Snai2-mediated upregulation of NADSYN1 promotes bladder cancer progression by interacting with PHB. Clin. Transl. Med. 2024, 14, e1555. [Google Scholar] [CrossRef]
- Scilipoti, P.; Ślusarczyk, A.; Angelis, M.; Soria, F.; Pradere, B.; Krajewski, W.; D’Andrea, D.; Mari, A.; Giudice, F.; Pichler, R.; et al. European Association of Urology Young Academic Urologists Urothelial Carcinoma Working Group. The Role of Mitomycin C in Intermediate-risk Non-muscle-invasive Bladder Cancer: A Systematic Review and Meta-analysis. Eur. Urol. Oncol. 2024, 7, 1293–1302. [Google Scholar] [CrossRef]
- Volpato, M.; Seargent, J.; Loadman, P.M.; Phillips, R.M. Formation of DNA interstrand cross-links as a marker of Mitomycin C bioreductive activation and chemosensitivity. Eur. J. Cancer 2005, 41, 1331–1338. [Google Scholar] [CrossRef]
- O’Brien, R.M.; Cannon, A.; Reynolds, J.V.; Lysaght, J.; Lynam-Lennon, N.; Lynam-Lennon, N. Complement in Tumourigenesis and the Response to Cancer Therapy. Cancers 2021, 13, 1209. [Google Scholar] [CrossRef] [PubMed]
- Oresta, B.; Pozzi, C.; Hurle, R.; Lazzeri, M.; Faccani, C.; Colombo, P.; Elefante, G.; Casale, P.; Guazzoni, G.; Rescigno, M. MP63-17 Mitomycin C triggers immunogenic cell death in bladder cancer cells. J. Urol. 2019, 101, e903. [Google Scholar] [CrossRef]
- Wolters, D.A.; Washburn, M.P.; Yates, J.R. An automated multidimensional protein identification technology for shotgun proteomics. Anal. Chem. 2001, 73, 5683–5690. [Google Scholar] [CrossRef]
- Capello, M.; Katayama, H.; Hanash, S.M. Proteomic Profiling of the Tumor Microenvironment. Methods Mol. Biol. 2022, 2435, 157–167. [Google Scholar] [PubMed]
- Urbiola-Salvador, V.; Miroszewska, D.; Jabłońska, A.; Qureshi, T.; Chen, Z. Proteomics approaches to characterize the immune responses in cancer. BBA-Mol. Cell Res. 2022, 1869, 11926. [Google Scholar] [CrossRef]
- Beckabir, W.; Wobker, S.E.; Damrauer, J.S.; Midkiff, B.; De la Cruz, G.; Makarov, V.; Flick, L.; Woodcock, M.G.; Grivas, P.; Bjurlin, M.A.; et al. Spatial Relationships in the Tumor Microenvironment Demonstrate Association with Pathologic Response to Neoadjuvant Chemoimmunotherapy in Muscle-invasive Bladder Cancer. Eur. Urol. 2024, 85, 242–253. [Google Scholar] [CrossRef]
- Feng, C.; Wang, X.; Tao, Y.; Xie, Y.; Lai, Z.; Li, Z.; Hu, J.; Tang, S.; Pan, L.; He, L.; et al. Single-Cell Proteomic Analysis Dissects the Complexity of Tumor Microenvironment in Muscle Invasive Bladder Cancer. Cancers 2021, 13, 5440. [Google Scholar] [CrossRef] [PubMed]
- Gillet, L.C.; Navarro, P.; Tate, S.; Röst, H.; Selevsek, N.; Reiter, L.; Bonner, R.; Aebersold, R. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: A new concept for consistent and accurate proteome analysis. Mol. Cell. Proteom. 2012, 11, O111.016717. [Google Scholar] [CrossRef] [PubMed]
- Anjo, S.I.; Santa, C.; Manadas, B. SWATH-MS as a tool for biomarker discovery: From basic research to clinical applications. Proteomics 2017, 17, 1600278. [Google Scholar] [CrossRef] [PubMed]
- Min, L.; Zhu, S.; Wei, R.; Zhao, Y.; Liu, S.; Li, P.; Zhang, S. Integrating SWATH-MS Proteomics and Transcriptome Analysis Identifies CHI3L1 as a Plasma Biomarker for Early Gastric Cancer. Mol. Ther.-Oncolytics 2020, 17, 257–266. [Google Scholar] [CrossRef]
- Chang, Q.; Chen, Y.; Yin, J.; Wang, T.; Dai, Y.; Wu, Z.; Guo, Y.; Wang, L.; Zhao, Y.; Yuan, H.; et al. Comprehensive Urinary Proteome Profiling Analysis Identifies Diagnosis and Relapse Surveillance Biomarkers for Bladder Cancer. J. Proteome Res. 2024, 23, 2241–2252. [Google Scholar] [CrossRef]
- Blanco-Pintos, T.; Regueira-Iglesias, A.; Relvas, M.; Alonso-Sampedro, M.; Chantada-Vázquez, M.P.; Balsa-Castro, C.; Tomás, I. Using SWATH-MS to identify new molecular biomarkers in gingival crevicular fluid for detecting periodontitis and its response to treatment. J. Clin. Periodontol. 2024, 51, 1342–1358. [Google Scholar] [CrossRef]
- Kim, J.; Jin, P.; Yang, W.; Kim, W.J. Proteomic profiling of bladder cancer for precision medicine in the clinical setting: A review for the busy urologist. Investig. Clin. Urol. 2020, 61, 539–554. [Google Scholar] [CrossRef]
- Zhang, Q.; Faca, V.; Hanash, S. Mining the plasma proteome for disease applications across seven logs of protein abundance. J. Proteome Res. 2011, 10, 46–50. [Google Scholar] [CrossRef]
- Jia, L.; Lu, Y.; Shao, J.; Liang, X.J.; Xu, Y. Nanoproteomics: A new sprout from emerging links between nanotechnology and proteomics. Trends Biotechnol. 2013, 31, 99–107. [Google Scholar] [CrossRef]
- Chantada-Vázquez, M.P.; Castro López, A.; Bravo, S.B.; Vázquez-Estévez, S.; Acea-Nebril, B.; Núñez, C. Proteomic analysis of the bio-corona formed on the surface of (Au, Ag, Pt)-nanoparticles in human serum. Colloids Surf. B Biointerfaces 2019, 177, 141–148. [Google Scholar] [CrossRef]
- Chantada-Vázquez, M.D.P.; Castro López, A.; García Vence, M.; Vázquez-Estévez, S.; Acea-Nebril, B.; Calatayud, D.G.; Jardiel, T.; Bravo, S.B.; Núñez, C. Proteomic investigation on bio-corona of Au, Ag and Fe nanoparticles for the discovery of triple negative breast cancer serum protein biomarkers. J. Proteom. 2020, 212, 103581. [Google Scholar] [CrossRef]
- Chantada-Vázquez, M.D.P.; García-Vence, M.; Vázquez-Estévez, S.; Bravo, S.B.; Núñez, C. Identification of a Profile of Neutrophil-Derived Granule Proteins in the Surface of Gold Nanoparticles after Their Interaction with Human Breast Cancer Sera. Nanomaterials 2020, 10, 1223. [Google Scholar] [CrossRef] [PubMed]
- Chantada-Vázquez, M.D.P.; López, A.C.; García-Vence, M.; Acea-Nebril, B.; Bravo, S.B.; Núñez, C. Protein corona gold nanoparticles fingerprinting reveals a profile of blood coagulation proteins in the serum of Her2-overexpressing breast cancer patients. Int. J. Mol. Sci. 2020, 21, 8449. [Google Scholar] [CrossRef] [PubMed]
- García-Vence, M.; Chantada-Vázquez, M.D.P.; Cameselle-Teijeiro, J.M.; Bravo, S.B.; Núñez, C. A Novel nanoproteomic approach for the identification of molecular targets associated with thyroid tumors. Nanomaterials 2020, 10, 2370. [Google Scholar] [CrossRef]
- Blanco-Gómez, B.; López-Cortés, R.; Casas-Nebra, F.J.; Vázquez-Estévez, S.; Pérez-Fentes, D.; Chantada-Vázquez, M.D.P.; Bravo, S.B.; Núñez, C. Detection of Circulating Serum Protein Biomarkers of Non-Muscle Invasive Bladder Cancer after Protein Corona-Silver Nanoparticles Analysis by SWATH-MS. Nanomaterials 2021, 11, 2384. [Google Scholar] [CrossRef]
- Chantada-Vázquez, M.D.P.; Conde-Amboage, M.; Graña-López, L.; Vázquez-Estévez, S.; Bravo, S.B.; Núñez, C. Circulating Proteins Associated with Response and Resistance to Neoadjuvant Chemotherapy in HER2-Positive Breast Cancer. Cancers 2022, 14, 1087. [Google Scholar] [CrossRef]
- Lai, Z.W.; Yan, Y.; Caruso, F.; Nice, E.C. Emerging techniques in proteomics for probing nano-bio interactions. ACS Nano 2012, 6, 10438–10448. [Google Scholar] [CrossRef] [PubMed]
- Jeyaraj, M.; Gurunathan, S.; Qasim, M.; Kang, M.-H.; Kim, J.-H. A Comprehensive Review on the Synthesis, Characterization, and Biomedical Application of Platinum Nanoparticles. Nanomaterials 2019, 9, 1719. [Google Scholar] [CrossRef]
- Wu, G.-W.; He, S.-B.; Peng, H.; Deng, H.-H.; Liu, A.-L.; Lin, X.-H.; Xia, X.-H.; Chen, W. Citrate-capped platinum nanoparticle as a smart probe for ultrasensitive mercury sensing. Anal. Chem. 2014, 86, 10955–10960. [Google Scholar] [CrossRef] [PubMed]
- Dobrovolskaia, M.A.; Patri, A.K.; Zheng, J.; Clogston, J.D.; Ayub, N.; Aggarwal, P.; Neun, B.W.; Hall, J.B.; McNeil, S.E. Interaction of colloidal gold nanoparticles with human blood: Effects on particle size and analysis of plasma protein binding profiles. Nanomedicine 2009, 5, 106–117. [Google Scholar] [CrossRef] [PubMed]
- García-Álvarez, R.; Hadjidemetriou, M.; Sánchez-Iglesias, A.; Liz-Marzán, L.M.; Kostarelos, K. In vivo formation of protein corona on gold nanoparticles: The effect of size and shape. Nanoscale 2018, 10, 1256–1264. [Google Scholar] [CrossRef]
- Wang, C.; Li, A.; Yang, S.; Qiao, R.; Zhu, X.; Zhang, J. CXCL5 promotes mitomycin C resistance in non-muscle invasive bladder cancer by activating EMT and NF-κB pathway. Biochem. Biophys. Res. Commun. 2018, 498, 862–868. [Google Scholar] [CrossRef]
- Zhang, C.-J.; Shen, Z.-J.; Pan, C.-W.; Zhong, S.; Li, T.; Zhang, M.-G. Engagement of integrinβ1 induces resistance of bladder cancer cells to mitomycin-C. Urology 2012, 79, 638–643. [Google Scholar] [CrossRef]
- Vallo, S.; Rutz, J.; Kautsch, M.; Winkelmann, R.; Michaelis, M.; Wezel, F.; Bartsch, G.; Haferkamp, A.; Rothweiler, F.; Blaheta, R.A.; et al. Blocking integrin β1 decreases adhesion in chemoresistant urothelial cancer cell lines. Oncol. Lett. 2017, 14, 5513–5518. [Google Scholar] [CrossRef]
- Sarma, J.V.; Ward, P.A. The complement system. Cell Tissue Res. 2011, 343, 227–235. [Google Scholar] [CrossRef]
- Markiewski, M.M.; DeAngelis, R.A.; Benencia, F.; Ricklin-Lichtsteiner, S.K.; Koutoulaki, A.; Gerard, C.; Coukos, G.; Lambris, J.D. Modulation of the antitumor immune response by complement. Nat. Immunol. 2008, 9, 1225–1235. [Google Scholar] [CrossRef]
- Merle, N.S.; Roumenina, L.T. The complement system as a target in cancer immunotherapy. Eur. J. Immunol. 2024, 54, e2350820. [Google Scholar] [CrossRef]
- Lee, K.S.W.; Zhang, Q.; Suwa, T.; Clark, H.; Olcina, M.M. The role of the complement system in the response to cytotoxic therapy. Semin. Immunol. 2025, 77, 101927. [Google Scholar] [CrossRef] [PubMed]
- Kolev, M.; Das, M.; Gerber, M.; Baver, S.; Deschatelets, P.; Markiewski, M.M. Inside-Out of Complement in Cancer. Front. Immunol. 2022, 13, 931273. [Google Scholar] [CrossRef]
- Revel, M.; Merle, N.S. Local and Cell-intrinsic complement: The new player in cancer progression. Semin. Immunol. 2025, 79, 101976. [Google Scholar] [CrossRef] [PubMed]
- Ajona, D.; Ortiz-Espinosa, S.; Pio, R.; Lecanda, F. Complement in Metastasis: A Comp in the Camp. Front. Immunol. 2019, 10, 669. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Dubinett, S. Inhibiting C5a/C5aR axis reduces myeloid-derived suppressor cells and enhances PD-1 blockade therapy in lung cancer. Transl. Cancer Res. 2017, 6, S944–S948. [Google Scholar] [CrossRef]
- Liu, H.; Wang, Z.; Zhou, Y.; Yang, Y. MDSCs in breast cancer: An important enabler of tumor progression and an emerging therapeutic target. Front. Immunol. 2023, 14, 1199273. [Google Scholar] [CrossRef]
- Kourtzelis, I.; Rafail, S. The dual role of complement in cancer and its implication in anti-tumor therapy. Ann. Transl. Med. 2016, 4, 265. [Google Scholar] [CrossRef] [PubMed]
- Macor, P.; Capolla, S.; Tedesco, F. Complement as a Biological Tool to Control Tumor Growth. Front. Immunol. 2018, 9, 2203. [Google Scholar] [CrossRef] [PubMed]
- Patel, B.; Silwal, A.; Eltokhy, M.A.; Gaikwad, S.; Curcic, M.; Patel, J.; Prasad, S. Deciphering CD59: Unveiling Its Role in Immune Microenvironment and Prognostic Significance. Cancers 2024, 16, 3699. [Google Scholar] [CrossRef]
- Couves, E.C.; Gardner, S.; Voisin, T.B.; Bickel, J.K.; Stansfeld, P.J.; Tate, E.W.; Bubeck, D. Structural basis for membrane attack complex inhibition by CD59. Nat. Commun. 2023, 14, 890. [Google Scholar] [CrossRef]
- Rutkowski, M.J.; Sughrue, M.E.; Kane, A.J.; Mills, S.A.; Parsa, A.T. Cancer and the Complement Cascade. Mol. Cancer Res. 2010, 8, 1453–1465. [Google Scholar] [CrossRef]
- Towner, L.D.; Wheat, R.A.; Hughes, T.R.; Morgan, B.P. Complement Membrane Attack and Tumorigenesis. J. Biol. Chem. 2016, 291, 14927–14938. [Google Scholar] [CrossRef]
- Vlaicu, S.I.; Tatomir, A.; Rus, V.; Rus, H. Role of C5b-9 and RGC-32 in Cancer. Front. Immunol. 2019, 10, 1054. [Google Scholar] [CrossRef] [PubMed]
- Oliveira-Tore, C.F.; Moraes, A.G.; Plácido, H.M.B.S.; Signorini, N.M.D.L.; Fontana, P.D.; Godoy, T.P.B.; Boldt, A.B.W.; Messias, I. Non-canonical extracellular complement pathways and the complosome paradigm in cancer: A scoping review. Front. Immunol. 2025, 16, 1519465. [Google Scholar] [CrossRef]
- Gunji, Y.; Gorelik, E. Role of fibrin coagulation in protection of murine tumor cells from destruction by cytotoxic cells. Cancer Res. 1988, 48, 5216–5221. [Google Scholar]
- Moreno, P. Exploring the Complex Interactions of Immunity, Coagulation, and Tumor Cells. J. Thromb. Circ. 2024, 10, 265. [Google Scholar]
- Wahab, R.; Hasan, M.M.; Azam, Z.; Grippo, P.J.; Al-Hilal, T.A. The role of coagulome in the tumor immune microenvironment. Adv. Drug. Deliv. Rev. 2023, 200, 115027. [Google Scholar] [CrossRef]
- Beitia, M.; Romano, P.; Larrinaga, G.; Solano-Iturri, J.D.; Salis, A.; Damonte, G.; Bruzzone, M.; Ceppi, M.; Profumo, A. The Activation of Prothrombin Seems to Play an Earlier Role than the Complement System in the Progression of Colorectal Cancer: A Mass Spectrometry Evaluation. Diagnostics 2020, 10, 1077. [Google Scholar] [CrossRef]
- Alexander, E.T.; Gilmour, S.K. Immunomodulatory role of thrombin in cancer progression. Mol. Carcinog. 2022, 61, 527–536. [Google Scholar] [CrossRef]
- Cantrell, R.; Palumbo, J.S. The thrombin-inflammation axis in cancer progression. Thromb. Res. 2020, 191 (Suppl. S1), S117–S122. [Google Scholar] [CrossRef] [PubMed]
- Sinitsky, M.Y.; Kutikhin, A.G.; Tsepokina, A.V.; Shishkova, D.K.; Asanov, M.A.; Yuzhalin, A.E.; Minina, V.I.; Ponasenko, A.V. Mitomycin C induced genotoxic stress in endothelial cells is associated with differential expression of proinflammatory cytokines. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2020, 858–860, 503252. [Google Scholar] [CrossRef] [PubMed]
- Adams, G.N.; Rosenfeldt, L.; Frederick, M.; Miller, W.; Waltz, D.; Kombrinck, K.; McElhinney, K.E.; Flick, M.J.; Monia, B.P.; Revenko, A.S.; et al. Colon Cancer Growth and Dissemination Relies upon Thrombin, Stromal PAR-1, and Fibrinogen. Cancer Res. 2015, 75, 4235–4243. [Google Scholar] [CrossRef]
- Yang, Y.; Stang, A.; Schweickert, P.G.; Lanman, N.A.; Paul, E.N.; Monia, B.P.; Revenko, A.S.; Palumbo, J.S.; Mullins, E.S.; Elzey, B.D.; et al. Thrombin Signaling Promotes Pancreatic Adenocarcinoma through PAR-1–Dependent Immune Evasion. Cancer Res. 2019, 79, 3417–3430. [Google Scholar] [CrossRef]
- Reddel, C.J.; Tan, C.W.; Chen, V.M. Thrombin Generation and Cancer: Contributors and Consequences. Cancers 2019, 11, 100. [Google Scholar] [CrossRef]
- Brierley, J.D.; Gospodarowicz, M.K.; Wittekind, C. TNM Classification of Malignant Tumors, 8th ed.; UICC International Union Against Cancer; Wiley-Blackwell and UICC: New York, NY, USA, 2017. [Google Scholar]
- Soukup, V.; Capoun, O.; Cohen, D.; Hernández, V.; Babjuk, M.; Burger, M.; Compérat, E.; Gontero, P.; Lam, T.; MacLennan, S.; et al. Prognostic performance and reproducibility of the 1973 and 2004/2016 World Health Organization grading classification systems in non-muscle-invasive bladder cancer: A European association of urology non-muscle-invasive bladder cancer guidelines panel systematic review. Eur. Urol. 2017, 72, 801–813. [Google Scholar] [PubMed]
- Kang, M.; Jeong, C.W.; Kwak, C.; Kim, H.H.; Ku, J.H. Single, immediate postoperative instillation of chemotherapy in non-muscle invasive bladder cancer: A systematic review and network meta-analysis of randomized clinical trials using different drugs. Oncotarget 2016, 7, 45479. [Google Scholar] [CrossRef]
- Sylvester, R.J.; Oosterlinck, W.; Witjes, J.A. The schedule and duration of intravesical chemotherapy in patients with non-muscle-invasive bladder cancer: A systematic review of the published results of randomized clinical trials. Eur. Urol. 2008, 53, 709. [Google Scholar] [CrossRef] [PubMed]
- Sylvester, R.J.; Oosterlinck, W.; Holmang, S.; Sydes, M.R.; Birtle, A.; Gudjonsson, S.; De Nunzio, C.; Okamura, K.; Kaasinen, E.; Solsona, E.; et al. Systematic Review and Individual Patient Data Meta-analysis of Randomized Trials Comparing a Single Immediate Instillation of Chemotherapy After Transurethral Resection with Transurethral Resection Alone in Patients with Stage pTa-pT1 Urothelial Carcinoma of the Bladder: Which Patients Benefit from the Instillation? Eur. Urol. 2016, 69, 231. [Google Scholar]
- Tolley, D.A.; Parmar, M.K.; Grigor, K.M.; Lallemand, G.; Benyon, L.L.; Fellows, J.; Freedman, L.S.; Grigor, K.M.; Hall, R.R.; Hargreave, T.B.; et al. The effect of intravesical mitomycin C on recurrence of newly diagnosed superficial bladder cancer: A further report with 7 years of follow up. J. Urol. 1996, 155, 1233–1238. [Google Scholar] [CrossRef] [PubMed]
- Solsona, E.; Iborra, I.; Ricos, J.V.; Monrós, J.L.; Casanova, J.; Dumont, R. Effectiveness of a single immediate mitomycin C instillation in patients with low-risk superficial bladder cancer: Short and long-term followup. J. Urol. 1999, 161, 1120–1123. [Google Scholar] [CrossRef]
- Barghi, M.R.; Rahmani, M.R.; Moghaddam, S.M.H.; Jahanbin, M. Immediate intravesical instillation of mitomycin C after transurethral resection of bladder tumor in patients with low-risk superficial transitional cell carcinoma of bladder. Urol. Oncol. 2006, 3, 220–224. [Google Scholar]
- Oddens, J.R.; van der Meijden, A.P.; Sylvester, R. One immediate postoperative instillation of chemotherapy in low risk Ta, T1 bladder cancer patients. Is it always safe? Eur. Urol. 2004, 46, 336–338. [Google Scholar] [CrossRef]
- Racioppi, M.; Porreca, A.; Foschi, N.; Delicato, G.; Destito, A.; D’Addessi, A. Bladder perforation: A potential risk of early endovesical chemotherapy with mitomycin C. Urol. Int. 2005, 75, 373–375. [Google Scholar] [CrossRef]
- Holmang, S.; Johansson, S.L. Stage Ta-T1 bladder cancer: The relationship between findings at first followup cystoscopy and subsequent recurrence and progression. J. Urol. 2002, 167, 1634. [Google Scholar] [CrossRef] [PubMed]
- Soukup, V.; Babjuk, M.; Bellmunt, J.; Dalbagni, G.; Giannarini, G.; Hakenberg, O.W.; Herr, H.; Lechevallier, E.; Ribal, M.J. Follow-up after surgical treatment of bladder cancer: A critical analysis of the literature. Eur. Urol. 2012, 62, 290–302. [Google Scholar] [CrossRef] [PubMed]
- Solsona, E.; Iborra, I.; Dumont, R.; Rubio-Briones, J.; Casanova, J.; Almenar, S. The 3-month clinical response to intravesical therapy as a predictive factor for progression in patients with high risk superficial bladder cancer. J. Urol. 2000, 164, 685–689. [Google Scholar] [CrossRef]
- Pursiheimo, A.; Vehmas, A.P.; Afzal, S.; Suomi, T.; Chand, T.; Strauss, L.; Poutanen, M.; Rokka, A.; Corthals, G.L.; Elo, L.L. Optimization of Statistical Methods Impact on Quantitative Proteomics Data. J. Proteome Res. 2015, 14, 4118–4126. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, D.J.; Smyth, G.K. Testing significance relative to a fold-change threshold is a TREAT. Bioinformatics 2009, 25, 765–771. [Google Scholar] [CrossRef]
- Gao, Y.; Wang, X.; Sang, Z.; Li, Z.; Liu, F.; Mao, J.; Yan, D.; Zhao, Y.; Wang, H.; Li, P.; et al. Quantitative proteomics by SWATH-MS reveals sophisticated metabolic reprogramming in hepatocellular carcinoma tissues. Sci. Rep. 2017, 7, 45913. [Google Scholar] [CrossRef]
- Wu, J.X.; Song, X.; Pascovici, D.; Zaw, T.; Care, N.; Krisp, C.; Molloy, M.P. SWATH Mass Spectrometry Performance Using Extended Peptide MS/MS Assay Libraries. Mol. Cell. Proteom. 2016, 15, 2501–2514. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).