Abstract
The APETALA2/ethylene response factor (AP2/ERF) family of transcription factors (TFs) is one of the largest and most important TF families in plants. This family plays a crucial role in regulating growth, development, and responses to both biotic and abiotic stresses. This study provides a comprehensive overview of the structure, classification, and distribution of AP2/ERF TFs in various plant species, with particular emphasis on their roles in responses to biotic stress. These findings provide valuable insights for future research on AP2/ERF TFs and their potential applications in crop improvement through molecular breeding.
1. Introduction
Transcription factors (TFs) are defined as DNA-binding proteins that specifically bind to cis-acting elements within promoter regions. The expression of target genes is regulated by TFs at specific times and in specific locations with a certain intensity, resulting in the activation or repression of gene transcription [1]. To date, approximately 60 TF families have been identified in plants, including AP2/ERF, bZIP, C2H2, MYB, MADS, NAC, and WRKY [2].
The APETALA2/ethylene response factor (AP2/ERF) family of TFs is one of the largest and most significant important TF families in plants and is characterized by at least one AP2 domain of approximately 60 amino acids. This family is extensively involved in plant growth and development and in responses to biotic and abiotic stresses [3,4,5]. The AP2/ERF TFs include five subfamilies. The APETALA2 (AP2) subfamily plays crucial roles in regulating various aspects of plant growth and development, such as flower development [6,7,8], leaf morphology [9], epidermal-cell identity in leaves [10], spikelet-meristem determinacy [11], embryonic development [12], and seed growth [13]. The ethylene response factor (ERF) subfamily is essential for hormone signal transduction [14,15], responses to biotic and abiotic stresses [16,17], plant development [18], and metabolic regulation [19,20,21]. The dehydration-responsive-element-binding (DREB) subfamily is primarily associated with supporting the plant’s responses to abiotic stresses, specifically drought and cold stresses [22,23]. The related to ABI3/VP1 (RAV) subfamily plays important roles in regulating leaf senescence [24], ethylene (ET) [25] and brassinosteroid (BR) signaling [26], and responses to biotic and abiotic stresses [27]. However, the functional characterization of the soloist subfamily remains limited, and its regulatory mechanisms and biological significance are not well understood. Although numerous reviews on AP2/ERF have been published, few studies have focused on its role in plant responses to biotic stress. Recent research findings together have clarified the regulatory network of the AP2/ERF family in response to biotic stress and have shown that it involves hormone signaling pathways, MAPK cascades, cell-wall reinforcement, and epigenetic regulation. These efforts provide a theoretical basis and technical support for the development of stress-resistant and high-yield crop varieties, thereby contributing to the advancement of sustainable agriculture.
2. The Structure and Classification of AP2/ERF TFs
The AP2/ERF TFs are characterized by the AP2 domain, a DNA-binding motif of approximately 60–70 amino acid residues [28]. Sakuma et al. classified the AP2/ERF TFs into five subfamilies (AP2, ERF, DREB, RAV, and Soloist) based on the number of AP2 domains and other structural features [3]. All AP2/ERF-domain sequences contain a single α-helix and three β-sheets [29]. The earliest identified subfamily, AP2, contains two AP2 domains [30]. The ERF subfamily was first isolated from tobacco by Ohme-Takagi et al. in 1995 and contains a single AP2 domain [31]. The DREB subfamily was initially identified in Arabidopsis thaliana by Stockinger et al. [32]. Like the ERF subfamily, the DREB subfamily contains only a single AP2 domain. However, the AP2-domain amino acid residues at positions 14 and 19 differ between DREB and ERF TFs. The RAV subfamily, first reported by Kagaya et al. in 1999, contains both an AP2 domain and a B3-like domain [33]. The Soloist subfamily also contains an AP2 domain, but its amino acid motif and conformation are significantly distinct from those of other subfamily members. Nakano et al. proposed an alternative classification by merging the DREB and ERF subfamilies into a single group, categorizing AP2/ERF TFs into four classes [34]. Both classification methods are widely used in the literature as primary approaches to AP2/ERF TF family classification (Figure 1).
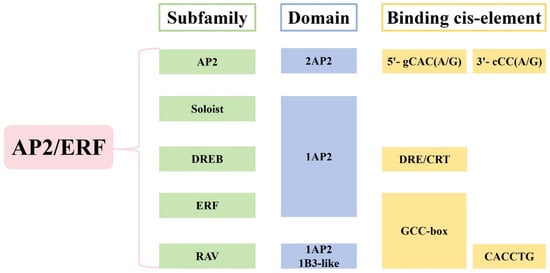
Figure 1.
The structure and classification of AP2/ERF TFs.
Due to differences in the amino acid sequences and nuclear-localization signals of its two AP2 domains, the AP2 subfamily can be further divided into the AP2 and ANT groups [35]. Nole-Wilson et al. proposed that the first AP2 domain of ANT binds to the 5′-gCAC(A/G) sequence, whereas the second domain binds to the 3′-cCC(A/G) sequence [36]. Members of the AP2 subfamily are primarily involved in regulating processes such as flowering and lateral root formation in plants. Previous studies collectively referred to the ERF and DREB subfamilies as the ERF subfamily. Later, based on the cis-acting elements bound by ERF transcription factors, researchers divided this group into the ERF and DREB subfamilies [37]. Members of the ERF subfamily bind the ethylene-responsive GCC-box element and regulate ethylene signaling and responses to abiotic stresses [38]. The DREB subfamily binds to drought- and cold-stress-responsive elements (DRE/CRT), thereby inducing expression of related genes and enhancing plant tolerance to abiotic stresses [22,23]. The key distinction between the ERF and DREB subfamilies lies in the amino acid residues at the 14th and 19th positions within their AP2/ERF domains. Within the ERF subfamily, the 14th and 19th residues are alanine and aspartic acid, respectively, whereas in the DREB subfamily, they are valine and glutamic acid [37]. This difference in residues determines each transcription factor’s specific binding to distinct cis-acting elements. In the RAV subfamily, an AP2/ERF domain at the N-terminus binds the GCC-box element, whereas the B3 domain at the C-terminus specifically binds the CACCTG sequence [39,40].
3. The Distribution and Quantity of AP2/ERF TFs Involved in Responses to Biotic Stresses
Advances in technologies such as whole-genome sequencing have led to the identification of an increasing number of AP2/ERF TFs in plants. Within the AP2/ERF TF family, the ERF and DREB subfamilies have the most members, followed by the AP2 subfamily, while the RAV and Soloist subfamilies contain the fewest [41,42,43]. The AP2/ERF TF family has now been identified in various plant species. For example, Arabidopsis thaliana has 122 ERF genes, 18 AP2 genes, 6 RAV genes, and 1 Soloist gene [34]. In sorghum, 105 ERF genes, 16 AP2 genes, 4 RAV genes, and 1 Soloist gene have been reported [44]. Similarly, buckwheat contains 116 ERF genes, 15 AP2 genes, and 3 RAV genes [45].
AP2/ERF TFs typically activate the expression of downstream genes responsive to biotic stress, thereby enhancing plant resistance to various pathogens and insect pests (Figure 2 and Table 1) [46,47]. Biotic stresses, such as bacterial, fungal, and viral infections, as well as insect pests, can lead to crop-yield reduction or even plant death [48,49].
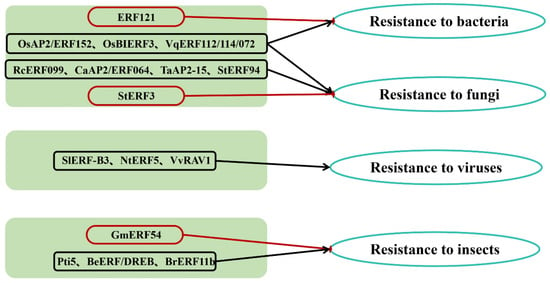
Figure 2.
The role of AP2/ERF in biotic stress.

Table 1.
AP2/ERF TFs involved in different responses to biotic stresses.
3.1. Bacterial Diseases
Bacterial diseases affect a wide range of plants, causing various symptoms such as leaf spots, blights, wilts, and cankers. Pillai et al. demonstrated that overexpression of the OsAP2/ERF152 gene in Arabidopsis thaliana induced callose deposition and other immune responses, thereby enhancing resistance to bacterial and fungal infections [50]. Overexpression of the rice OsBIERF3 gene enhances resistance to both fungal and bacterial pathogens [52]. Additionally, VqERF112, VqERF114 and VqERF072 from Chinese wild grape (Vitis quinquangularis) positively regulate resistance to Pseudomonas syringae pv. tomato DC3000 and Botrytis cinerea [53]. Conversely, in Brassica oleracea, the ERF121 TF increases susceptibility to Xanthomonas infections by disrupting plant defense pathways [51].
3.2. Fungal Diseases
Fungal diseases, caused by pathogenic fungi, are the most prevalent and destructive plant diseases, comprising over 80% of all known plant diseases. There are two main types of fungal pathogens: biotrophs and nectrotrophs. Together with bacteria, they cause vascular wilts, leaf spots, cankers, and other symptoms, infecting various parts of the plant. Biotrophic pathogens first penetrate epidermal cells and multiply in the intercellular spaces by feeding on living host tissue. Most biotrophs, including Pseudomonas syringae, are host-specific. Necrotrophic pathogens kill host-plant cells using toxic metabolites and then feed on the remains. Most necrotrophs infect a wide range of hosts. RcERF099 is an important positive regulator of resistance to Botrytis cinerea infections in rose petals [54]. Overexpression of CaAP2/ERF064 enhances resistance to Phytophthora blight in Capsicums (peppers) [55]. Silencing TaAP2-15 increases susceptibility to Puccinia striiformis f. sp. tritici in wheat, promoting pathogen growth [56]. In potato, overexpression of StERF94 inhibits fungal proliferation in cellular tissues, thereby enhancing disease resistance [57]. Conversely, StERF3 negatively regulates resistance to Phytophthora infestans, the causal agent of late blight [58].
3.3. Viral Diseases
Viral diseases cause not only local lesions but also systemic damage that leads to stunting, chlorosis and malformations affecting different parts of the plant, although they rarely kill their hosts. Tomato yellow leaf curly virus (TYLCV) causes leaf curling and yellowing, plant dwarfism, and growth inhibition in tomato. A comprehensive analysis revealed that five SlERF-B3 TFs are differentially expressed in resistant and susceptible tomato during TYLCV infection [59]. In tobacco, overexpression of NtERF5 enhances resistance to tobacco mosaic virus [60]. Grapevine leafroll-associated virus 2 (GLRaV-2) is a common virus associated with grapevine leafroll disease. The GLRaV-2 p24 protein interacts with VvRAV1, thereby suppressing host defense responses and facilitating accumulation and infection of GLRaV-2 in grape [61].
3.4. Insect Pests
Insect pests often damage plants through feeding or by transmitting plant pathogens, reducing growth and yield. Ulti-host common cutworm (CCW) is a major herbivore in low latitudes of China which feeds almost all soybean tissues [78]. Overexpression of GmERF54 decreases resistance to CCW in transgenic soybean [62]. In herbivore-induced defense responses, expression of Pti5 is induced by potato aphid feeding, and the ERF Pti5 gene contributes to resistance to potato aphids in tomato [63]. Bambusa emeiensis is one of the most economically important bamboos, and its BeERF/DREB subfamily plays a critical role inresponding to herbivore attack [79]. BrERF11b enhances plant resistance to both chewing and sap-sucking insects [65].
4. Regulation of Hormones in Responses to Biotic Stress
The AP2/ERF TF family serves as a pivotal regulatory hub connecting plant hormone signaling pathways (Figure 3 and Table 1). They induce plant hormone responses by activating target genes or acting as response factors. Current research indicates that AP2/ERF TFs primarily regulate responses to biotic stress via plant hormone signaling pathways such as those mediated by ethylene (ET), jasmonic acid (JA), and salicylic acid (SA) [80,81].
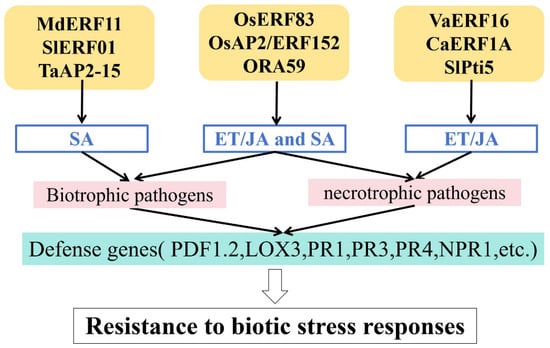
Figure 3.
Regulation of hormones in responses to biotic stress.
4.1. SA-Mediated Regulation
SA is a plant hormone involved in defense signaling; it activates pathogenesis-related (PR) genes, thereby modulating responses to biotic stress. The SA signaling pathway confers resistance to biotrophic pathogens [82,83].
Wang et al. showed that MdERF11 enhances resistance to gray mold disease in apple trees by promoting the expression of genes involved in SA synthesis [66]. Yang et al. reported that expression of SlERF01 is strongly induced by Stemphylium lycopersici and exogenous applications of SA, thereby enhancing resistance to Stemphylium lycopersici in tomato [67]. TaAP2-15 acts as a positive regulator of resistance to Puccinia striiformis f. sp. tritici (Pst) in wheat via an SA-induced mechanism [56].
4.2. ET/JA-Mediated Regulation
In addition to SA, AP2/ERF TFs regulate disease resistance through the ET/JA signaling pathway. The ET/JA signaling pathways are particularly effective against necrotrophic pathogens such as Botrytis cinerea and Fusarium oxysporum [84,85]. VaERF16 acts as a positive regulatory factor by enhancing the ET/JA-signaling-dependent response to hormone signals, thereby improving resistance to Botrytis cinerea in grape [68]. In hot pepper, overexpression of CaERF1A enhances resistance to necrotrophic fungal pathogens by upregulating an ET/JA-synthesis-related gene [69]. Silencing of the SlPti5 gene in tomato weakens the ET/JA signaling pathway, impairing immune responses and reducing resistance to Botrytis cinerea [64].
4.3. ET/JA- and SA-Mediated Regulation
More importantly, AP2/ERF TFs mediate crosstalk between the SA and JA/ET signaling pathways. Tezuka et al. demonstrated that treatment with ET, JA, and SA induces OsERF83 expression, thereby enhancing the expression of PR genes in transgenic rice and significantly improving resistance to rice blast disease [70]. Pillai et al. found that expression of OsAP2/ERF152 upregulates SA- and JA/ET-responsive defense genes, enhancing resistance to bacterial and fungal pathogens in Arabidopsis thaliana [50]. In Arabidopsis thaliana, ORA59 is a key positive regulator of the ET/JA-mediated defense pathway that protects against necrotrophic pathogens, but it is repressed by SA signaling [71].
5. Regulation of MAPK in Responses to Biotic Stress
In plants, MAPKs generally act downstream of sensors/receptors that recognize endogenous stimuli or exogenous stimuli, coordinating plant growth, development, and immunity. Activated MAPKs mediate the phosphorylation of various downstream substrates, such as protein kinases, TFs, structural proteins, and other enzymes, to activate cellular responses [86,87].
Multiple AP2/ERF TFs have been shown to be substrates of MAPKs, linking them to plant defense responses. MAPK cascades are important signaling modules in responses to biotic stress (Figure 4 and Table 1). In Arabidopsis thaliana, the partially redundant MAPKs MPK3/MPK6 phosphorylate AtERF6 and AtERF72, enhancing resistance to Botrytis cinerea via different pathways [72,73]. In rice, MAP kinase BWMK1 phosphorylates OsEREBP1, promoting its binding to the GCC-box in PR gene promoters and thus enhancing PR gene expression and disease resistance [74]. Moreover, the soybean GmMKK4-GmMPK6 module phosphorylates GmERF113, promoting the stability of its protein product and enhancing transcriptional activity; it thereby increases the expression of defensive genes and enhances immune responses to Phytophthora sojae [75].
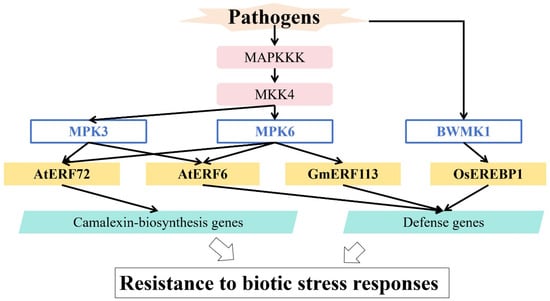
Figure 4.
Regulation of MAPK in responses to biotic stress.
6. Regulation of Cell Wall in Responses to Biotic Stress
In plants, the cell wall is a physical barrier against adverse stress and pathogen invasion, and it plays a crucial role in signaling under conditions of biotic stress [88,89]. Lignin, a key component of the cell wall, enhances the wall’s defensive capabilities [90,91].
Multiple investigations have reported that AP2/ERF TFs critically regulate plant defenses by altering lignin biosynthesis (Figure 5 and Table 1). For example, ERF139 in Populus plays a role in the lignin-synthesis pathway by inducing the expression of genes related to secondary cell-wall synthesis, thereby enhancing resistance to biotic stress [76]. In Arabidopsis thaliana, AtERF114 activates the lignin biosynthetic gene AtPAL1 by directly binding to its promoter, increasing lignin accumulation and enhancing immunity [77]. Similarly, MdERF114 promotes lignin accumulation by regulating the expression or transcriptional activity of MdPRX63, resulting in enhanced lignin deposition and disease resistance [48].
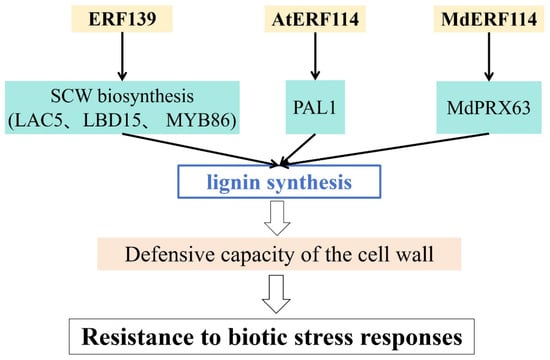
Figure 5.
Regulation of the cell wall in responses to biotic stress.
7. Epigenetic Regulation of the AP2/ERF TF Family in Plant Growth and Development
Epigenetic regulation plays a significant role in plant growth and development [92]. It primarily affects DNA methylation and histone modifications (including methylation, acetylation, phosphorylation, and ubiquitination) [93,94]. Current research on AP2/ERF TFs has mainly focused on histone modifications (Figure 6).
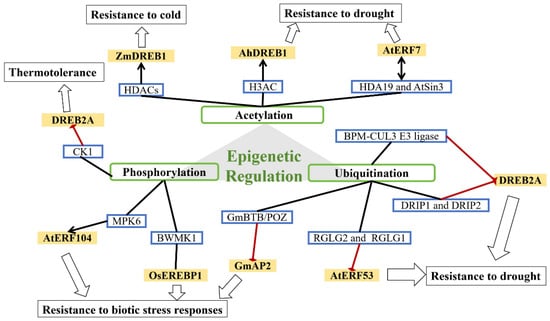
Figure 6.
Epigenetic regulation of the AP2/ERF TF family in plant growth and development.
The level of histone acetylation is controlled by the dynamic balance between histone acetyltransferases (HATs) and histone deacetylases (HDACs) [95]. In maize, HDACs cause global histone deacetylation, promoting the expression of ZmDREB1 under cold stress [96]. In peanut, histone acetylation regulates AhDREB1 under conditions of osmotic stress by increasing H3ac levels, thereby improving drought resistance [97]. Song et al. showed that in Arabidopsis thaliana, AtERF7 interacts with the transcriptional corepressor AtSin3, which is associated with histone deacetylase 19 (HDA19). HDA19 and AtSin3 enhance the transcriptional-repression activity of AtERF7. Overexpression of AtERF7 in transgenic Arabidopsis thalian decreases drought tolerance [98].
Phosphorylation modulates AP2/ERF protein transactivity and stability. Phosphorylation by different kinases usually results in a functional change in the target protein by changing activity, cellular location, or association with other proteins. For example, the rice MAP kinase BWMK1 phosphorylates OsEREBP1, enhancing its DNA-binding activity to the GCC box element and activating expression of pathogenesis-related genes [74]. Phosphorylation by MPK6 releases AtERF104, which in turn activates defense genes such as PDF1.1 and PDF1.2 [99]. Under nonstress conditions, the negative-regulatory domain (NRD) of DREB2A is phosphorylated primarily by casein kinase 1 (CK1), which facilitates DREB2A degradation and thereby negatively regulates its stability [100].
Ubiquitination may affect proteins in various ways, including targeting them for proteasome-mediated degradation, altering cellular localization, modifying their activity, and enhancing or inhibiting protein–protein interactions. In Arabidopsis, BTB/POZ–MATH (BPM) adaptor proteins of the CUL3 E3 ligase complex mediate DREB2A degradation to coordinate drought-stress responses [101,102]. Similarly, DRIP1 and DRIP2 function as E3 ubiquitin ligases that ubiquitinate DREB2A, targeting it for degradation via the 26S proteasome and thus negatively regulating the drought response [103]. RGLG2 and its closest homolog, RGLG1, have E3-ligase activity, mediating AtERF53 ubiquitination and degradation via the 26S proteasome pathway and thus negatively regulating drought-stress responses [104]. In soybean, the GmBTB/POZ protein promotes ubiquitination and degradation of the AP2/ERF-like TF GmAP2, thereby regulating the defense response against Phytophthora sojae [105].
8. Challenges and Future Directions for AP2/ERF TFs
AP2/ERF TFs are ubiquitous in plants and play critical roles in mediating resistance to diverse biotic stresses, including bacterial, fungal, viral, and insect challenges. Since the cloning of the first AP2/ERF TFs in Arabidopsis thaliana, substantial progress has been achieved in elucidating their functions; however, many questions remain to be resolved. To date, research on the roles of AP2/ERF TFs in responses to biotic stress has largely focused on plants such as Oryza sativa and Arabidopsis thaliana, whereas studies in other plants have concentrated on responses to abiotic stresses. AP2/ERF TFs are predominantly characterized as positive regulators, whereas their negative regulatory functions remain underexplored. Therefore, novel AP2/ERF family members that regulate multiple facets of plant responses to biotic stress, as well as their underlying mechanisms, warrant exploration in future research.
Additionally, with the advancement of molecular biology and high-throughput sequencing technologies, genetic engineering, such as the use of CRISPR/Cas9 to breed resistant varieties, has arisen as a new method for improving some crops. Therefore, future research can build on existing methodologies by using molecular-biology techniques to investigate AP2/ERF TFs and their roles in responses to biotic stress in a wider range of plants. Key areas of focus should include the similarities and differences in the regulatory mechanisms of AP2/ERF TFs in various responses to biotic stress, their roles in coordinating multiple signaling pathways, and the mechanisms though which they exert their effects. With the identification of novel AP2/ERF TFs and the elucidation of the underlying mechanisms, AP2/ERFs may serve as promising targets in breeding for disease resistance in many crops.
Author Contributions
A.-M.L., W.-G.C. and D.-L.H. designed the manuscript; Z.-L.S.,with the assistance from A.-M.L., M.W., Z.-L.C., C.-X.Q., F.L., B.-Q.Z., Y.-Q.P., W.-G.C. and D.-L.H. drafted the manuscript. All authors contributed to this work and approved the submitted version. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Science and Technology Major Project of Guangxi (AA23073001), National Natural Science Foundation of China (32360247), the Natural Science Foundation of Guangxi (2023GXNSFAA026453), the Science and Technology Major Project of Guangxi (AA22036003 and AA22117002), the Fundamental Research Fund of Guangxi Academy of Agriculture Sciences (2021YT010).
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Badis, G.; Berger, M.F.; Philippakis, A.A.; Talukder, S.; Gehrke, A.R.; Jaeger, S.A.; Chan, E.T.; Metzler, G.; Vedenko, A.; Chen, X.; et al. Diversity and complexity in DNA recognition by transcription factors. Science 2009, 324, 1720–1723. [Google Scholar] [CrossRef]
- Xie, W.; Ding, C.; Hu, H.; Dong, G.; Zhang, G.; Qian, Q.; Ren, D. Molecular Events of Rice AP2/ERF Transcription Factors. Int. J. Mol. Sci. 2022, 23, 12013. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, Y.; Liu, Q.; Dubouzet, J.G.; Abe, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 2002, 290, 998–1009. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Hu, L.; Jiang, W. Understanding AP2/ERF transcription factor responses and tolerance to various abiotic stresses in plants: A comprehensive review. Int. J. Mol. Sci. 2024, 25, 893. [Google Scholar] [CrossRef] [PubMed]
- Feng, K.; Hou, X.-L.; Xing, G.-M.; Liu, J.-X.; Duan, A.-Q.; Xu, Z.-S.; Li, M.-Y.; Zhuang, J.; Xiong, A.-S. Advances in AP2/ERF super-family transcription factors in plant. Crit. Rev. Biotechnol. 2020, 40, 750–776. [Google Scholar] [CrossRef]
- Elliott, R.C.; Betzner, A.S.; Huttner, E.; Oakes, M.P.; Tucker, W.Q.; Gerentes, D.; Perez, P.; Smyth, D.R. AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell 1996, 8, 155–168. [Google Scholar] [CrossRef]
- Aukerman, M.J.; Sakai, H. Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell 2003, 15, 2730–2741. [Google Scholar] [CrossRef]
- Krizek, B.A.; Eaddy, M. AINTEGUMENTA-LIKE6 regulates cellular differentiation in flowers. Plant Mol. Biol. 2012, 78, 199–209. [Google Scholar] [CrossRef]
- Jiang, F.; Guo, M.; Yang, F.; Duncan, K.; Jackson, D.; Rafalski, A.; Wang, S.; Li, B. Mutations in an AP2 transcription factor-like gene affect internode length and leaf shape in maize. PLoS ONE 2012, 7, e37040. [Google Scholar] [CrossRef]
- Moose, S.P.; Sisco, P.H. Glossy15, an APETALA2-like gene from maize that regulates leaf epidermal cell identity. Genes Dev. 1996, 10, 3018–3027. [Google Scholar] [CrossRef]
- Chuck, G.; Meeley, R.B.; Hake, S. The control of maize spikelet meristem fate by the APETALA2-like gene indeterminate spikelet1. Genes Dev. 1998, 12, 1145–1154. [Google Scholar] [CrossRef] [PubMed]
- Boutilier, K.; Offringa, R.; Sharma, V.K.; Kieft, H.; Ouellet, T.; Zhang, L.; Hattori, J.; Liu, C.M.; van Lammeren, A.A.; Miki, B.L.; et al. Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 2002, 14, 1737–1749. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Yu, X.; Cao, B.; Peng, L.; Gao, Y.; Feng, T.; Li, H.; Ren, Z. LkAP2L2, an AP2/ERF transcription factor gene of Larix kaempferi, with pleiotropic roles in plant branch and seed development. Russ. J. Genet. 2017, 53, 1335–1342. [Google Scholar] [CrossRef]
- Shinshi, H.; Usami, S.; Ohme-Takagi, M. Identification of an ethylene-responsive region in the promoter of a tobacco class I chitinase gene. Plant Mol. Biol. 1995, 27, 923–932. [Google Scholar] [CrossRef]
- Müller, M.; Munné-Bosch, S. Ethylene Response Factors: A Key Regulatory Hub in Hormone and Stress Signaling. Plant Physiol. 2015, 169, 32–41. [Google Scholar] [CrossRef]
- Dubouzet, J.G.; Sakuma, Y.; Ito, Y.; Kasuga, M.; Dubouzet, E.G.; Miura, S.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J. 2003, 33, 751–763. [Google Scholar] [CrossRef]
- Yao, Y.; He, R.J.; Xie, Q.L.; Zhao, X.H.; Deng, X.M.; He, J.B.; Song, L.; He, J.; Marchant, A.; Chen, X.Y.; et al. ETHYLENE RESPONSE FACTOR 74 (ERF74) plays an essential role in controlling a respiratory burst oxidase homolog D (RbohD)-dependent mechanism in response to different stresses in Arabidopsis. New Phytol. 2017, 213, 1667–1681. [Google Scholar] [CrossRef]
- Banno, H.; Ikeda, Y.; Niu, Q.W.; Chua, N.H. Overexpression of Arabidopsis ESR1 induces initiation of shoot regeneration. Plant Cell 2001, 13, 2609–2618. [Google Scholar] [CrossRef]
- van der Fits, L.; Memelink, J. ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism. Science 2000, 289, 295–297. [Google Scholar] [CrossRef]
- Deng, B.; Huang, Z.; Ge, F.; Liu, D.; Lu, R.; Chen, C. An AP2/ERF family transcription factor PnERF1 raised the biosynthesis of saponins in Panax notoginseng. J. Plant Growth Regul. 2017, 36, 691–701. [Google Scholar] [CrossRef]
- Yu, Z.X.; Li, J.X.; Yang, C.Q.; Hu, W.L.; Wang, L.J.; Chen, X.Y. The jasmonate-responsive AP2/ERF transcription factors AaERF1 and AaERF2 positively regulate artemisinin biosynthesis in Artemisia annua L. Mol. Plant 2012, 5, 353–365. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 1994, 6, 251–264. [Google Scholar] [CrossRef]
- Thomashow, M.F. Plant Cold Acclimation: Freezing Tolerance Genes and Regulatory Mechanisms. Annu. Rev. Plant Biol. 1999, 50, 571–599. [Google Scholar] [CrossRef] [PubMed]
- Woo, H.R.; Kim, J.H.; Kim, J.; Kim, J.; Lee, U.; Song, I.J.; Kim, J.H.; Lee, H.Y.; Nam, H.G.; Lim, P.O. The RAV1 transcription factor positively regulates leaf senescence in Arabidopsis. J. Exp. Bot. 2010, 61, 3947–3957. [Google Scholar] [CrossRef] [PubMed]
- Sohn, K.H.; Lee, S.C.; Jung, H.W.; Hong, J.K.; Hwang, B.K. Expression and functional roles of the pepper pathogen-induced transcription factor RAV1 in bacterial disease resistance, and drought and salt stress tolerance. Plant Mol. Biol. 2006, 61, 897–915. [Google Scholar] [CrossRef]
- Alonso, J.M.; Stepanova, A.N.; Solano, R.; Wisman, E.; Ferrari, S.; Ausubel, F.M.; Ecker, J.R. Five components of the ethylene-response pathway identified in a screen for weak ethylene-insensitive mutants in Arabidopsis. Proc. Natl. Acad. Sci. USA 2003, 100, 2992–2997. [Google Scholar] [CrossRef]
- Hu, X.; Cheng, X.; Jiang, H.; Zhu, S.; Cheng, B.; Xiang, Y. Genome-wide analysis of cyclins in maize (Zea mays). Genet. Mol. Res. 2010, 9, 1490–1503. [Google Scholar] [CrossRef]
- Owji, H.; Hajiebrahimi, A.; Seradj, H.; Hemmati, S. Identification and functional prediction of stress responsive AP2/ERF transcription factors in Brassica napus by genome-wide analysis. Comput. Biol. Chem. 2017, 71, 32–56. [Google Scholar] [CrossRef]
- Allen, M.D.; Yamasaki, K.; Ohme-Takagi, M.; Tateno, M.; Suzuki, M. A novel mode of DNA recognition by a β-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA. EMBO J. 1998, 17, 5484–5496. [Google Scholar] [CrossRef]
- Jofuku, K.D.; den Boer, B.G.; Van Montagu, M.; Okamuro, J.K. Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 1994, 6, 1211–1225. [Google Scholar] [CrossRef]
- Ohme-Takagi, M.; Shinshi, H. Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 1995, 7, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Stockinger, E.J.; Gilmour, S.J.; Thomashow, M.F. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA 1997, 94, 1035–1040. [Google Scholar] [CrossRef] [PubMed]
- Kagaya, Y.; Ohmiya, K.; Hattori, T. RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucleic Acids Res. 1999, 27, 470–478. [Google Scholar] [CrossRef]
- Nakano, T.; Suzuki, K.; Fujimura, T.; Shinshi, H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 2006, 140, 411–432. [Google Scholar] [CrossRef]
- Shigyo, M.; Ito, M. Analysis of gymnosperm two-AP2-domain-containing genes. Dev. Genes Evol. 2004, 214, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Nole-Wilson, S.; Krizek, B.A. DNA binding properties of the Arabidopsis floral development protein AINTEGUMENTA. Nucleic Acids Res. 2000, 28, 4076–4082. [Google Scholar] [CrossRef]
- Zhang, J.; Liao, J.; Ling, Q.; Xi, Y.; Qian, Y. Genome-wide identification and expression profiling analysis of maize AP2/ERF superfamily genes reveal essential roles in abiotic stress tolerance. BMC Genom. 2022, 23, 125. [Google Scholar] [CrossRef]
- Hao, D.; Ohme-Takagi, M.; Sarai, A. Unique mode of GCC box recognition by the DNA-binding domain of ethylene-responsive element-binding factor (ERF domain) in plant. J. Biol. Chem. 1998, 273, 26857–26861. [Google Scholar] [CrossRef]
- Giraudat, J.; Hauge, B.M.; Valon, C.; Smalle, J.; Parcy, F.; Goodman, H.M. Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 1992, 4, 1251–1261. [Google Scholar] [CrossRef]
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef]
- Zhang, G.; Chen, M.; Chen, X.; Xu, Z.; Guan, S.; Li, L.C.; Li, A.; Guo, J.; Mao, L.; Ma, Y. Phylogeny, gene structures, and expression patterns of the ERF gene family in soybean (Glycine max L.). J. Exp. Bot. 2008, 59, 4095–4107. [Google Scholar] [CrossRef]
- Rashid, M.; Guangyuan, H.; Guangxiao, Y.; Hussain, J.; Xu, Y. AP2/ERF Transcription Factor in Rice: Genome-Wide Canvas and Syntenic Relationships between Monocots and Eudicots. Evol. Bioinform. 2012, 8, 321–355. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Ma, R.; Xu, D.; Bi, H.; Xia, Z.; Peng, H. Genome-Wide Identification and Analysis of the AP2 Transcription Factor Gene Family in Wheat (Triticum aestivum L.). Front. Plant Sci. 2019, 10, 1286. [Google Scholar] [CrossRef]
- Yan, H.W.; Hong, L.; Zhou, Y.Q.; Jiang, H.Y.; Zhu, S.W.; Fan, J.; Cheng, B.J. A genome-wide analysis of the ERF gene family in sorghum. Genet. Mol. Res. 2013, 12, 2038–2055. [Google Scholar] [CrossRef]
- Liu, M.; Sun, W.; Ma, Z.; Zheng, T.; Huang, L.; Wu, Q.; Zhao, G.; Tang, Z.; Bu, T.; Li, C.; et al. Genome-wide investigation of the AP2/ERF gene family in tartary buckwheat (Fagopyum Tataricum). BMC Plant Biol. 2019, 19, 84. [Google Scholar] [CrossRef] [PubMed]
- Zang, Z.; Wang, Z.; Zhao, F.; Yang, W.; Ci, J.; Ren, X.; Jiang, L.; Yang, W. Maize ethylene response factor ZmERF061 is required for resistance to Exserohilum turcicum. Front. Plant Sci. 2021, 12, 630413. [Google Scholar] [CrossRef]
- Reboledo, G.; Agorio, A.; Vignale, L.; Alvarez, A.; Ponce De León, I. The moss-specific transcription factor PpERF24 positively modulates immunity against fungal pathogens in Physcomitrium patens. Front. Plant Sci. 2022, 13, 908682. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, Q.; Li, X.; Zhang, Z.; Ai, S.; Liu, C.; Ma, F.; Li, C. MdERF114 enhances the resistance of apple roots to Fusarium solani by regulating the transcription of MdPRX63. Plant Physiol. 2023, 192, 2015–2029. [Google Scholar] [CrossRef]
- Xie, Z.; Nolan, T.M.; Jiang, H.; Yin, Y. AP2/ERF transcription factor regulatory networks in hormone and abiotic stress responses in Arabidopsis. Front. Plant Sci. 2019, 10, 228. [Google Scholar] [CrossRef] [PubMed]
- Pillai, S.E.; Kumar, C.; Dasgupta, M.; Kumar, B.K.; Vungarala, S.; Patel, H.K.; Sonti, R.V. Ectopic expression of a cell-wall-degrading enzyme-induced OsAP2/ERF152 leads to resistance against bacterial and fungal infection in Arabidopsis. Phytopathology 2020, 110, 726–733. [Google Scholar] [CrossRef]
- Zlobin, N.; Lebedeva, M.; Monakhova, Y.; Ustinova, V.; Taranov, V. An ERF121 transcription factor from Brassica oleracea is a target for the conserved TAL-effectors from different Xanthomonas campestris pv. campestris strains. Mol. Plant Pathol. 2021, 22, 618–624. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.; Wang, H.; Gao, Y.; Bi, Y.; Xiong, X.; Yan, Y.; Wang, J.; Li, D.; Song, F. ERF transcription factor OsBIERF3 positively contributes to immunity against fungal and bacterial diseases but negatively regulates cold tolerance in rice. Int. J. Mol. Sci. 2022, 23, 606. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, W.; Wang, Y. Heterologous expression of Chinese wild grapevine VqERFs in Arabidopsis thaliana enhance resistance to Pseudomonas syringae pv. tomato DC3000 and to Botrytis cinerea. Plant Sci. 2020, 293, 110421. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Liu, X.; Shu, L.; Zhang, H.; Zhang, S.; Song, Y.; Zhang, Z. Global analysis of the AP2/ERF gene family in rose (Rosa chinensis) genome unveils the role of RcERF099 in Botrytis resistance. BMC Plant Biol. 2020, 20, 533. [Google Scholar] [CrossRef]
- Jin, J.-H.; Zhang, H.-X.; Ali, M.; Wei, A.-M.; Luo, D.-X.; Gong, Z.-H. The CaAP2/ERF064 regulates dual functions in pepper: Plant cell death and resistance to Phytophthora capsici. Genes 2019, 10, 541. [Google Scholar] [CrossRef] [PubMed]
- Hawku, M.D.; Goher, F.; Islam, M.A.; Guo, J.; He, F.; Bai, X.; Yuan, P.; Kang, Z.; Guo, J. TaAP2-15, an AP2/ERF transcription factor, is positively involved in wheat resistance to Puccinia striiformis f. sp. tritici. Int. J. Mol. Sci. 2021, 22, 2080. [Google Scholar] [CrossRef]
- Charfeddine, M.; Samet, M.; Charfeddine, S.; Bouaziz, D.; Gargouri Bouzid, R. Ectopic expression of StERF94 transcription factor in potato plants improved resistance to Fusarium solani infection. Plant Mol. Biol. Rep. 2019, 37, 450–463. [Google Scholar] [CrossRef]
- Tian, Z.; He, Q.; Wang, H.; Liu, Y.; Zhang, Y.; Shao, F.; Xie, C. The potato ERF transcription factor StERF3 negatively regulates resistance to Phytophthora infestans and salt tolerance in potato. Plant Cell Physiol. 2015, 56, 992–1005. [Google Scholar] [CrossRef]
- Huang, Y.; Zhang, B.L.; Sun, S.; Xing, G.M.; Wang, F.; Li, M.Y.; Tian, Y.S.; Xiong, A.S. AP2/ERF transcription factors involved in response to tomato yellow leaf curly virus in tomato. Plant Genome 2016, 9, plantgenome2015.2009.0082. [Google Scholar] [CrossRef]
- Fischer, U.; Dröge-Laser, W. Overexpression of NtERF5, a new member of the tobacco ethylene response transcription factor family enhances resistance to tobacco mosaic virus. Mol. Plant-Microbe Interact. 2004, 17, 1162–1171. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, X.; Li, H.; Wang, J.; Zeng, Q.; Huang, W.; Huang, H.; Xie, Y.; Yu, S.; Kan, Q. GLRaV-2 protein p24 suppresses host defenses by interaction with a RAV transcription factor from grapevine. Plant Physiol. 2022, 189, 1848–1865. [Google Scholar] [CrossRef] [PubMed]
- Cai, L.; Qin, R.; Li, X.; Liu, X.; Yu, D.; Wang, H. GmERF54, an ERF Transcription Factor, Negatively Regulates the Resistance of Soybean to the Common Cutworm (Spodoptera litura Fabricius). Agronomy 2023, 13, 596. [Google Scholar] [CrossRef]
- Wu, C.; Avila, C.A.; Goggin, F.L. The ethylene response factor Pti5 contributes to potato aphid resistance in tomato independent of ethylene signalling. J. Exp. Bot. 2015, 66, 559–570. [Google Scholar] [CrossRef]
- Qiong, T.; Zheng, X.-D.; Jun, G.; Ting, Y. Tomato SlPti5 plays a regulative role in the plant immune response against Botrytis cinerea through modulation of ROS system and hormone pathways. J. Integr. Agric. 2022, 21, 697–709. [Google Scholar] [CrossRef]
- Wu, J.; Gao, H.; Zhu, X.; Li, D. An ERF transcription factor enhances plant resistance to Myzus persicae and Spodoptera litura. Biotechnol. Biotechnol. Equip. 2020, 34, 946–954. [Google Scholar] [CrossRef]
- Wang, J.H.; Gu, K.D.; Han, P.L.; Yu, J.Q.; Wang, C.K.; Zhang, Q.Y.; You, C.X.; Hu, D.G.; Hao, Y.J. Apple ethylene response factor MdERF11 confers resistance to fungal pathogen Botryosphaeria dothidea. Plant Sci. 2020, 291, 110351. [Google Scholar] [CrossRef]
- Yang, H.; Shen, F.; Wang, H.; Zhao, T.; Zhang, H.; Jiang, J.; Xu, X.; Li, J. Functional analysis of the SlERF01 gene in disease resistance to S. lycopersici. BMC Plant Biol. 2020, 20, 376. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Zhang, X.; Zhang, Q.; Chai, S.; Yin, W.; Gao, M.; Li, Z.; Wang, X. The transcription factors VaERF16 and VaMYB306 interact to enhance resistance of grapevine to Botrytis cinerea infection. Mol. Plant Pathol. 2022, 23, 1415–1432. [Google Scholar] [CrossRef]
- Huh, S.U. Functional analysis of hot pepper ethylene responsive factor 1A in plant defense. Plant Signal. Behav. 2022, 17, 2027137. [Google Scholar] [CrossRef]
- Tezuka, D.; Kawamata, A.; Kato, H.; Saburi, W.; Mori, H.; Imai, R. The rice ethylene response factor OsERF83 positively regulates disease resistance to Magnaporthe oryzae. Plant Physiol. Biochem. 2019, 135, 263–271. [Google Scholar] [CrossRef]
- Huang, L.J.; Zhang, J.; Lin, Z.; Yu, P.; Lu, M.; Li, N. The AP2/ERF transcription factor ORA59 regulates ethylene-induced phytoalexin synthesis through modulation of an acyltransferase gene expression. J. Cell. Physiol. 2024, 239, e30935. [Google Scholar] [CrossRef]
- Meng, X.; Xu, J.; He, Y.; Yang, K.-Y.; Mordorski, B.; Liu, Y.; Zhang, S. Phosphorylation of an ERF transcription factor by Arabidopsis MPK3/MPK6 regulates plant defense gene induction and fungal resistance. Plant Cell 2013, 25, 1126–1142. [Google Scholar] [CrossRef]
- Li, Y.; Liu, K.; Tong, G.; Xi, C.; Liu, J.; Zhao, H.; Wang, Y.; Ren, D.; Han, S. MPK3/MPK6-mediated phosphorylation of ERF72 positively regulates resistance to Botrytis cinerea through directly and indirectly activating the transcription of camalexin biosynthesis enzymes. J. Exp. Bot. 2022, 73, 413–428. [Google Scholar] [CrossRef]
- Cheong, Y.H.; Moon, B.C.; Kim, J.K.; Kim, C.Y.; Kim, M.C.; Kim, I.H.; Park, C.Y.; Kim, J.C.; Park, B.O.; Koo, S.C. BWMK1, a rice mitogen-activated protein kinase, locates in the nucleus and mediates pathogenesis-related gene expression by activation of a transcription factor. Plant Physiol. 2003, 132, 1961–1972. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Jiang, L.; Du, B.; Ning, B.; Ding, X.; Zhang, C.; Song, B.; Liu, S.; Zhao, M.; Zhao, Y. GmMKK4-activated GmMPK6 stimulates GmERF113 to trigger resistance to Phytophthora sojae in soybean. Plant J. 2022, 111, 473–495. [Google Scholar] [CrossRef] [PubMed]
- Wessels, B.; Seyfferth, C.; Escamez, S.; Vain, T.; Antos, K.; Vahala, J.; Delhomme, N.; Kangasjärvi, J.; Eder, M.; Felten, J. An AP 2/ERF transcription factor ERF 139 coordinates xylem cell expansion and secondary cell wall deposition. New Phytol. 2019, 224, 1585–1599. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, Y.; Ren, J.; Jia, F.; Zeng, H.; Li, G.; Yang, X. Ethylene-responsive factor ERF114 mediates fungal pathogen effector PevD1-induced disease resistance in Arabidopsis thaliana. Mol. Plant Pathol. 2022, 23, 819–831. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.; Gai, J.; Ji, D.; Ren, Z. A study on leaf-feeding insect species on soyabeans in the Nanjing area. Soybean Sci. 1997, 16, 12–20. [Google Scholar]
- Li, Y.; Luo, C.; Chen, Y.; Xiao, X.; Fu, C.; Yang, Y. Transcriptome-based discovery of AP2/ERF transcription factors and expression profiles under herbivore stress conditions in bamboo (Bambusa emeiensis). J. Plant Biol. 2019, 62, 297–306. [Google Scholar] [CrossRef]
- Phukan, U.J.; Jeena, G.S.; Tripathi, V.; Shukla, R.K. Regulation of Apetala2/Ethylene Response Factors in Plants. Front. Plant Sci. 2017, 8, 150. [Google Scholar] [CrossRef]
- Amorim, L.L.; da Fonseca Dos Santos, R.; Neto, J.P.B.; Guida-Santos, M.; Crovella, S.; Benko-Iseppon, A.M. Transcription factors involved in plant resistance to pathogens. Curr. Protein Pept. Sci. 2017, 18, 335–351. [Google Scholar] [CrossRef]
- Yang, L.; Li, B.; Zheng, X.-y.; Li, J.; Yang, M.; Dong, X.; He, G.; An, C.; Deng, X.W. Salicylic acid biosynthesis is enhanced and contributes to increased biotrophic pathogen resistance in Arabidopsis hybrids. Nat. Commun. 2015, 6, 7309. [Google Scholar] [CrossRef]
- Kou, M.-Z.; Bastías, D.A.; Christensen, M.J.; Zhong, R.; Nan, Z.-B.; Zhang, X.-X. The plant salicylic acid signalling pathway regulates the infection of a biotrophic pathogen in grasses associated with an Epichloë endophyte. J. Fungi 2021, 7, 633. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Han, X.; Feng, D.; Yuan, D.; Huang, L.-J. Signaling crosstalk between salicylic acid and ethylene/jasmonate in plant defense: Do we understand what they are whispering? Int. J. Mol. Sci. 2019, 20, 671. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhang, L.; Xiang, S.; Chen, Y.; Zhang, H.; Yu, D. The transcription factor WRKY75 positively regulates jasmonate-mediated plant defense to necrotrophic fungal pathogens. J. Exp. Bot. 2021, 72, 1473–1489. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Su, J.; Zhang, Y.; Xu, J.; Zhang, S. Conveying endogenous and exogenous signals: MAPK cascades in plant growth and defense. Curr. Opin. Plant Biol. 2018, 45, 1–10. [Google Scholar] [CrossRef]
- Zhang, M.; Zhang, S. Mitogen-activated protein kinase cascades in plant signaling. J. Integr. Plant Biol. 2022, 64, 301–341. [Google Scholar] [CrossRef]
- Miedes, E.; Vanholme, R.; Boerjan, W.; Molina, A. The role of the secondary cell wall in plant resistance to pathogens. Front. Plant Sci. 2014, 5, 358. [Google Scholar] [CrossRef]
- Yang, C.; Liu, R.; Pang, J.; Ren, B.; Zhou, H.; Wang, G.; Wang, E.; Liu, J. Poaceae-specific cell wall-derived oligosaccharides activate plant immunity via OsCERK1 during Magnaporthe oryzae infection in rice. Nat. Commun. 2021, 12, 2178. [Google Scholar] [CrossRef]
- Cesarino, I. Structural features and regulation of lignin deposited upon biotic and abiotic stresses. Curr. Opin. Biotechnol. 2019, 56, 209–214. [Google Scholar] [CrossRef]
- Vanholme, R.; De Meester, B.; Ralph, J.; Boerjan, W. Lignin biosynthesis and its integration into metabolism. Curr. Opin. Biotechnol. 2019, 56, 230–239. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Ding, Y.; Sun, X.; Xie, S.; Wang, D.; Liu, X.; Su, L.; Wei, W.; Pan, L.; Zhou, D.-X. Histone deacetylase HDA9 negatively regulates salt and drought stress responsiveness in Arabidopsis. J. Exp. Bot. 2016, 67, 1703–1713. [Google Scholar] [CrossRef]
- Xie, S.-S.; Duan, C.-G. Epigenetic regulation of plant immunity: From chromatin codes to plant disease resistance. Abiotech 2023, 4, 124–139. [Google Scholar] [CrossRef] [PubMed]
- Giovannoni, J.; Nguyen, C.; Ampofo, B.; Zhong, S.; Fei, Z. The epigenome and transcriptional dynamics of fruit ripening. Annu. Rev. Plant Biol. 2017, 68, 61–84. [Google Scholar] [CrossRef]
- Liu, X.; Yang, S.; Zhao, M.; Luo, M.; Yu, C.-W.; Chen, C.-Y.; Tai, R.; Wu, K. Transcriptional repression by histone deacetylases in plants. Mol. Plant 2014, 7, 764–772. [Google Scholar] [CrossRef]
- Hu, Y.; Zhang, L.; Zhao, L.; Li, J.; He, S.; Zhou, K.; Yang, F.; Huang, M.; Jiang, L.; Li, L. Trichostatin A selectively suppresses the cold-induced transcription of the ZmDREB1 gene in maize. PLoS ONE 2011, 6, e22132. [Google Scholar] [CrossRef]
- Zhang, B.; Su, L.; Hu, B.; Li, L. Expression of AhDREB1, an AP2/ERF transcription factor gene from peanut, is affected by histone acetylation and increases abscisic acid sensitivity and tolerance to osmotic stress in Arabidopsis. Int. J. Mol. Sci. 2018, 19, 1441. [Google Scholar] [CrossRef] [PubMed]
- Song, C.-P.; Agarwal, M.; Ohta, M.; Guo, Y.; Halfter, U.; Wang, P.; Zhu, J.-K. Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. The Plant Cell 2005, 17, 2384–2396. [Google Scholar] [CrossRef]
- Bethke, G.; Unthan, T.; Uhrig, J.F.; Pöschl, Y.; Gust, A.A.; Scheel, D.; Lee, J. Flg22 regulates the release of an ethylene response factor substrate from MAP kinase 6 in Arabidopsis thaliana via ethylene signaling. Proc. Natl. Acad. Sci. USA 2009, 106, 8067–8072. [Google Scholar] [CrossRef]
- Mizoi, J.; Kanazawa, N.; Kidokoro, S.; Takahashi, F.; Qin, F.; Morimoto, K.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Heat-induced inhibition of phosphorylation of the stress-protective transcription factor DREB2A promotes thermotolerance of Arabidopsis thaliana. J. Biol. Chem. 2019, 294, 902–917. [Google Scholar] [CrossRef]
- Weber, H.; Hellmann, H. Arabidopsis thaliana BTB/POZ-MATH proteins interact with members of the ERF/AP2 transcription factor family. FEBS J. 2009, 276, 6624–6635. [Google Scholar] [CrossRef] [PubMed]
- Morimoto, K.; Ohama, N.; Kidokoro, S.; Mizoi, J.; Takahashi, F.; Todaka, D.; Mogami, J.; Sato, H.; Qin, F.; Kim, J.-S. BPM-CUL3 E3 ligase modulates thermotolerance by facilitating negative regulatory domain-mediated degradation of DREB2A in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, E8528–E8536. [Google Scholar] [CrossRef] [PubMed]
- Qin, F.; Sakuma, Y.; Tran, L.-S.P.; Maruyama, K.; Kidokoro, S.; Fujita, Y.; Fujita, M.; Umezawa, T.; Sawano, Y.; Miyazono, K.-i. Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress–responsive gene expression. The Plant Cell 2008, 20, 1693–1707. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.-C.; Hsieh, E.-J.; Chen, J.-H.; Chen, H.-Y.; Lin, T.-P. Arabidopsis RGLG2, functioning as a RING E3 ligase, interacts with AtERF53 and negatively regulates the plant drought stress response. Plant Physiol. 2012, 158, 363–375. [Google Scholar] [CrossRef]
- Zhang, C.; Gao, H.; Sun, Y.; Jiang, L.; He, S.; Song, B.; Liu, S.; Zhao, M.; Wang, L.; Liu, Y. The BTB/POZ domain protein GmBTB/POZ promotes the ubiquitination and degradation of the soybean AP2/ERF-like transcription factor GmAP2 to regulate the defense response to Phytophthora sojae. J. Exp. Bot. 2021, 72, 7891–7908. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).