Abstract
Molecular methods have become integral to microbiological research for microbial identification. This literature review focuses on the application of molecular methods in examining airborne bacteria and fungi in healthcare facilities. In January 2024, a comprehensive electronic search was carried out in esteemed databases including PubMed, Web of Science, and Scopus, employing carefully selected keywords such as ((bacteria) OR (virus) OR (fungi)) AND (aerosol) AND ((hospital) OR (healthcare) OR (dental office)) AND ((molecular) OR (PCR) OR (NGS) OR (RNA) OR (DNA) OR (metagenomic) OR (microarray)), following the PRISMA protocol. The review specifically targets healthcare environments with elevated concentrations of pathogenic bacteria. A total of 487 articles were initially identified, but only 13 met the inclusion criteria and were included in the review. The study disclosed that the prevalent molecular methodology for appraising aerosol quality encompassed the utilization of the PCR method, incorporating either 16S rRNA (bacteria) or 18S rRNA (fungi) amplification techniques. Notably, five diverse molecular techniques, specifically PFGE, DGGE, SBT, LAMP, and DNA hybridization methods, were implemented in five distinct studies. These molecular tests exhibited superior capabilities compared to traditional bacterial and fungal cultures, providing precise strain identification. Additionally, the molecular methods allowed the detection of gene sequences associated with antibiotic resistance. In conclusion, molecular testing offers significant advantages over classical microbiological culture, providing more comprehensive information.
1. Introduction
An aerosol is characterized as a mixture of solid or liquid particles suspended in a gas, such as air, with varying dimensions and origins [1]. Notably, a significant proportion of particles in aerosols consists of bacteria and has drawn attention from researchers due to their potential to transmit infections. Healthcare facilities in particular merit special consideration due to heightened exposure to bacteria-contaminated aerosols compared to other environments [2,3,4,5,6]. Concerns about nosocomial infections, including the airborne transmission of pathogenic bacterial strains, have been highlighted in healthcare settings [7]. Scientists have predominantly used culturing techniques to analyze the microbiological content of aerosols, involving the cultivation of bacteria from a sample on specific media (both solid and liquid) to form countable colonies [8,9,10]. However, this method has limitations, including low sensitivity, as not all bacteria can be cultured in the laboratory, leading to an underestimation of overall bacterial diversity. Furthermore, culture techniques have low specificity as they do not provide detailed information on the exact taxonomic origin, genus, or species of bacteria present in the sample [8].
In recent years, advanced molecular methods have been employed for the molecular-level study of bacterial communities [11,12]. These techniques focus on the extraction and analysis of bacterial genetic material, eliminating the need for culturing. This approach enhances our understanding of the oral microbiota, encompassing both cultivable and non-cultivable bacteria [13]. Notable methods in this category are polymerase chain reaction (PCR), reverse transcription polymerase chain reaction (RT-PCR), quantitative polymerase chain reaction (qPCR), denaturing gradient gel electrophoresis (DGGE), DNA microarrays, fluorescence in situ hybridization (FISH) and DNA hybridization [14,15,16,17,18,19,20]. The polymerase chain reaction (PCR) is a widely used molecular biology technique that focuses on amplifying specific DNA regions. Its product can then be sequenced and used for bacterial identification by targeting specific bacterial genes or regions and detecting them in the sample if present [14,17]. Among PCR methods, we can distinguish variations in the conventional PCR method, such as asymmetric PCR, nested PCR, multiplex PCR, competitive PCR, qPCR, and RT-PCR [21]. The advanced version, qPCR, quantifies the amount of DNA in a sample, offering insights into bacterial DNA quantity and contamination extent [15,17,22]. Isothermal PCR methods deserve additional attention. Unlike classic PCR methods, they do not require thermocycling but can be performed at constant temperature and simple conditions [23]. In turn, Next-Generation Sequencing (NGS) allows for the sequencing of large numbers of DNA fragments simultaneously. In the context of aerosol samples, NGS can provide a comprehensive overview of all bacterial species present, allowing for detailed analysis of microbial diversity [16]. Another method used to identify bacterial genetic material is the DNA microarray technique, which consists of many nucleic acid sequences on small surfaces that can be used to locate and measure the gene expression [24]. DGGE is a technique used to separate DNA fragments based on their melting behavior. It is often employed to analyze the diversity of microbial communities in aerosol samples by separating PCR-amplified 16S rRNA gene fragments [20]. FISH involves the use of fluorescently labeled nucleic acid probes to specifically bind to target DNA or RNA sequences in microbial cells. It is a direct detection method that can be applied to assess the abundance and spatial distribution of specific microorganisms in aerosol samples [18,25]. Furthermore, DNA hybridization involves the hybridization of DNA probes with complementary sequences in the target microorganisms. It can be used to detect specific microbial species in aerosols [26].
Various nucleic acid amplification techniques are used as molecular methods to evaluate microbial quality in aerosols. One of the amplification techniques applied in aerosol studies focuses on the 16S ribosomal RNA gene, a ubiquitous component in all bacteria. Sequencing this gene enables the identification of various bacterial species within the sample [27]. This method allows for a high-resolution analysis of the bacterial community, aiding in understanding the diversity and composition of aerosolized bacteria. Furthermore, metagenomic analysis entails sequencing all genetic material in a sample, offering insights not only into bacterial species but also into other microorganisms like viruses and fungi that might be present in the aerosols [22]. Advanced molecular techniques, including metagenomic sequencing, are increasingly employed to overcome limitations, providing a more comprehensive understanding of the microbial composition in aerosols, encompassing unculturable species and potential pathogens [28]. Moreover, loop-mediated isothermal amplification is a method for isothermally amplifying DNA, known for its simplicity and rapid detection of specific bacterial DNA [29]. This technique is particularly useful in quickly identifying and quantifying specific bacterial strains in aerosol samples. Additionally, microarray technology enables the simultaneous detection of multiple bacterial species. DNA probes specific to different bacteria are immobilized on a solid surface, and the presence of complementary DNA from the aerosol sample indicates the bacterial species [30]. The use of microarrays enhances the ability to detect and differentiate a broad range of bacterial species in aerosol samples.
Molecular techniques employed for microbial identification in healthcare unit aerosols offer notable advantages, including high precision, sensitivity, and rapid detection of microbial presence, enabling swift responses to potential infections [27,28,29]. The specificity of these methods allows for the accurate differentiation of microbial species, and their multiplexing capabilities enable the simultaneous detection of multiple pathogens [30]. However, the adoption of molecular techniques in healthcare settings faces challenges such as cost constraints, as the equipment and reagents can be expensive. Technical expertise is essential for successful implementation, and the reliance on sophisticated equipment may be a limitation in resource-limited settings [14]. Additionally, the potential for false positives or negatives, influenced by contamination and sample quality, poses considerations. Moreover, the accessibility of molecular diagnostic technologies may be limited in certain healthcare units, particularly in less developed regions, thereby affecting their widespread use for microbial identification in aerosols [14].
The aim of this systematic review was to assess the molecular tests employed for the analysis of aerosols in diverse healthcare settings. The second objective was to investigate whether the implementation of molecular methodologies contributes to enhanced precision in the assessment of bacterial quality within aerosols in healthcare settings. Conducting such a literature review is intended to inspire researchers to pursue additional studies utilizing highly accurate tests to evaluate microbiological hazards in medical facilities. Moreover, a systematic review on this specific topic has not been published to date. This endeavor sought to comprehend the effectiveness, accuracy, and intended applications of selected molecular methods in the examination of bioaerosols.
2. Materials and Methods
2.1. Focused Question
This systematic review followed the PICO framework as follows: PICO question: In the case of bacteria and fungi included in aerosols (population), what would be the best method to identify and examine (outcome) these microorganisms from a molecular aspect (investigated condition)?
2.2. Protocol
A detailed presentation and description of selected articles used in this systematic review were outlined in accordance with the PRISMA statement [31]. (Figure 1) The systematic review was registered on the Open Science Framework under the following link: https://osf.io/ysk4t/ (accessed on 2 April 2024).
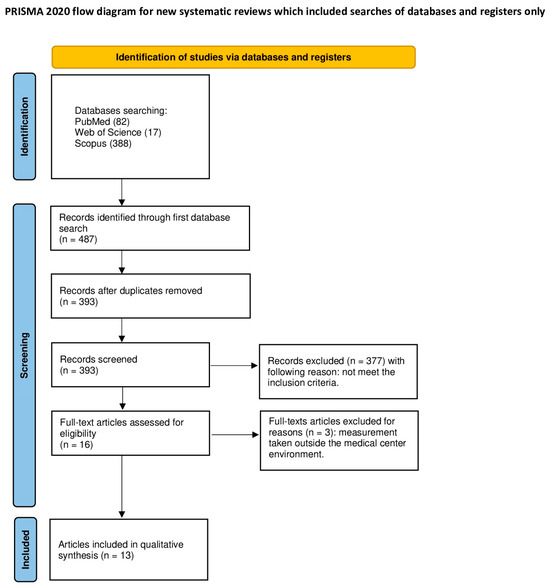
Figure 1.
PRISMA 2020 flow diagram.
2.3. Eligibility Criteria
The reviewers agreed to include only the articles which met the following criteria listed below:
- Studies that focused on aerosols in healthcare environments (dental clinics, rehabilitation centers, nursing centers, sanatoriums, different hospital areas);
- Presence of bacteria and fungi;
- Application of molecular methods to recognize the bacteria;
- In vivo studies;
- In vitro studies;
- Full-text articles;
- Studies in English.
The exclusion criteria the reviewers agreed upon were as follows:
- Aerosols not focused on aerosols in healthcare environments;
- Studies not including the presence of bacteria and fungi;
- Studies without molecular analysis;
- Non-English studies;
- Systematic review articles;
- Reviews;
- Meta-analysis.
No restrictions were imposed regarding the year of publication.
2.4. Information Sources, Search Strategy, and Study Selection
In January 2024, a comprehensive electronic search was carried out in esteemed databases including PubMed, Web of Science (WoS), and Scopus, employing carefully selected keywords such as ((bacteria) OR (virus) OR (fungi)) AND (aerosol) AND ((hospital) OR (healthcare) OR (dental office)) AND ((molecular) OR (PCR) OR (NGS) OR (RNA) OR (DNA) OR (metagenomic) OR (microarray)), following the PRISMA protocol. In PubMed and WoS, the results were refined to titles, authors, and abstracts, while in the Scopus database, the results were narrowed down to titles, authors, and keywords. The search was restricted to studies investigating bacteria and fungi in aerosols using molecular methods. The inclusion criteria encompassed both in vivo and in vitro studies published in English. Studies that did not meet the predefined criteria were excluded. Non-English papers, meta-analyses, and other reviews or systematic reviews were not considered.
2.5. Data Collection Process, Data Items
The articles that met the inclusion criteria were extracted by two independent reviewers (J.M., J.K.). The following data were used: first author, year of publication, study design, article title, methods used to examine bacteria and fungi contained in aerosols, and their effectiveness and results. The extracted information was then entered into a standardized Excel file.
2.6. Risk of Bias in Individual Studies
At the initial stage of study selection, each reviewer individually checked the titles and abstracts to minimize potential reviewer bias. Cohen’s k test was used as a tool to determine the level of agreement among reviewers [32]. Any disagreement about the inclusion or exclusion of an article in the review was resolved by discussion between the authors.
2.7. Quality Assessment
Two reviewers (J.M. and J.K.) independently conducted separate screenings of the included studies to assess the quality of each study. The criteria for evaluating study design, implementation, and analysis included information such as the origin of the aerosol, method of aerosol collection, type of molecular test performed, presence of a control group, sample size calculation, and the number of samples in a group exceeding 10. The studies were assigned scores ranging from 0 to 6 points, with assessments as follows: 0–2 points indicated a high risk, 3–4 points denoted a moderate risk, and 5–6 points indicated a low risk. Any discrepancies in scoring were resolved via discussion until a consensus was reached.
2.8. Risk of Bias across Studies
The scores of each study were calculated, and an overall estimated risk of bias (low, moderate, high) was made for each included study, as recommended in the Cochrane Handbook for Systematic Reviews of Interventions [33].
3. Results
3.1. Study Selection
The PubMed, Scopus, and WoS (Web of Science) database search identified 487 articles potentially applicable for the analysis. After eliminating all duplications, 393 articles were screened. The first screening, which included titles and abstracts, allowed us to exclude 377 articles as they did not focus on the topic of this review. In total, 16 articles were subjected to a full-text analysis, from which 3 were excluded due to not meeting the inclusion criteria (measurements of the exhaled air or non-English papers). In total, 13 articles were qualified for analysis in this systematic review [3,26,34,35,36,37,38,39,40,41,42,43,44].
3.2. General Characteristics of the Included Studies
In this systematic review, a total of thirteen articles were included. Most of the studies examined the aerosols present in different hospital areas such as wards, operating rooms, nursing stations, waiting rooms, textile rooms, corridors, etc. [3,26,34,35,36,37,38,39,40,41,42,43]. Among all included studies, one focused on aerosols produced during dental prophylaxis with an ultrasonic scaler [44]. A general characteristic of the included studies has been demonstrated in Table 1.

Table 1.
General characteristics of studies.
Most papers included in this review focus on bacteria detection [3,36,37,39,40,41,42]. In addition to examining bacteria, five studies also consider the content of fungi in the air [26,34,35,38,43]. To collect samples for testing, in most cases, specially prepared air samplers were used [3,26,34,35,36,37,38,39,40,42,43]. There were also exceptions where a Petri dish [44] or a specially created microchip [41] was used to collect the aerosol sample.
The predominant molecular technique in the reviewed studies was the polymerase chain reaction (PCR) method, which was applied in eight included studies [3,26,35,37,38,39,42,43]. Chen, P-S et al., in their study, used a modified method of conventional PCR, which was RT-qPCR, which allows direct analysis of viral RNA and enables quantitative analysis of gene expression and RNA quantity [42]. This method focuses on amplifying the 16S rRNA region, selected for its notable conservation [26,34,35,37,38,39,42,43]. Additionally, Habibi et al. employed the 18S rRNA region sequencing in case of fungal identification [38]. Another method of sequencing the genetic material of mushrooms was used by Núñez et al., using ITS sequencing [34]. These methods involve amplifying the DNA chain of bacteria or fungi extracted from samples, enabling their precise identification. The identification process involves comparing the obtained sequences with a comprehensive public database to assess the similarity with sequences present in the gene bank [39]. Notably, this method offers a meticulous approach to determining the taxonomic origin of bacteria or fungi, specifically their classification within a distinct genus and species [3,26,35,37,38,39,42,43].
Retamal-Valdes B et al. [44] employed an alternative molecular methodology, opting for DNA hybridization instead of PCR. This method diverges significantly in its procedural aspects from PCR. The obtained DNA underwent meticulous preparation under specific thermal conditions for an appropriate duration. Subsequently, digoxigenin-labeled whole genomic DNA probes targeting 40 bacterial species were utilized for hybridization. The detection process involved the utilization of anti-digoxigenin antibodies conjugated with alkaline phosphatase, and chemiluminescence detection using a specialized scanner facilitated the identification of specific bacterial species. Analogous to the PCR method, this technique effectively detected the targeted bacterial species [44].
Molecular techniques, such as PCR, offer versatile applications for conducting various molecular tests, as exemplified in a study conducted by Gilbert et al. [43]. In this investigation, PCR was utilized to amplify the extracted DNA, which was subsequently applied to an agarose gel for Denaturing gradient gel electrophoresis (DGGE). This approach uses the PCR, which is loaded in a gel and subjected to a process that allows you to distinguish strips corresponding to different bacterial strains [43]. A different method that involves electrophoresis was used by Wu B et al. [36], where Pulsed Gel Electrophoresis (PFGE) was applied. This method differs from the others in that it is based only on DNA denaturation and not on its amplification. However, this method, despite its different nature, allowed to show a wide genetic polymorphism among examined E. coli samples [36]. Other molecular methods used in the qualified studies were SBT [40] and LAMP [41].
Molecular methods are an instrument in detecting genes associated with antibiotic resistance. Wu B. et al. [36], in their study targeting Extended-Spectrum Beta-Lactamase (ESBL)-producing Escherichia coli, employed both the conventional culture method for identifying antibiotic resistance and the PCR method for testing the presence of TEM, SHV, and CTX m genes. Unique primers and specific parameters for annealing, extension, and the number of cycles were utilized for each gene. The investigation successfully identified four strains positive for the SHV gene, eight strains positive for the TEM gene, and five strains positive for the CTX m gene. Gene identification proved unattainable via classical methods, which only facilitated Minimum Inhibitory Concentration (MIC) testing to assess resistance to individual antibiotics [36].
Other studies conducted in healthcare facilities such as dental clinics have shown that prevalent bioaerosol pathogens include periopathogens and saliva-borne bacteria, such as S. mutans. The molecular genetic profiling of bacteria in generated bioaerosols emphasizes the imperative need for individual protection measures among employees to avoid potential infections [44]. A parallel study by Handorean A. et al. examined airborne bacteria associated with linens used by patients, subsequently cleaned and stored by medical staff. The identified bacteria, including Staphylococcus, Propionibacteria, Corynebacteria, Lactobacillus, and Streptococcus spp., pose a significant pathogenic risk to employees [37]. A detailed characteristic of the included is presented in Table 2.

Table 2.
Microbiological culture and molecular methods in the evaluation of aerosol quality.
3.3. Main Study Outcomes
In all thirteen of the studies included, a consistent application of molecular methodologies was observed for the precise identification of bacterial or fungal genera or exact strains [3,26,34,35,36,37,38,39,40,41,42,43,44]. Additionally, eight investigations integrated traditional microbiological techniques, such as traditional culturing, to allow for comparative analyses with the employed molecular methods [3,26,34,35,36,40,43,44]. In all instances, the classic microbiological culture was not employed with the primary goal of identifying specific bacterial strains. Instead, in most of the presented studies, microbiological culture served the purpose of quantifying the bacterial load. This quantification involved assessing the quantity of bacteria in the compared samples using the unit CFU/m3 [3,26,34,35,36,40,43]. This methodology provided a means to evaluate bacterial concentrations across diverse contexts, encompassing various phases of dental procedures, distinct hospital zones, and comparisons between indoor and outdoor settings [3,26,34,35,36,40,43]. While conventional assessments of bacterial diversity relied on culture-based morphology or histopathology, molecular testing emerged as indispensable for precise strain identification and determining the exact species within cultured bacteria [3,34,35,36,37,38,39,40,42,43,44]. The identification of fungal strains was slightly different. In most studies also focusing on fungi, culture was the only form of strain identification [26,34,35,38,43]. Only Núñez et al. and Habibi et al. in their research they used molecular methods to identify fungal strains [34,38].
Furthermore, molecular testing exhibited superiority over classical methods by uncovering a broader spectrum of bacterial species. In the studies by Wu B et al. [36] and Yousefzadeh A et al. [26], bacterial culture served to evaluate antibiotic resistance via the assessment of the inhibition zone and examination of the Minimum Inhibitory Concentration (MIC) values. However, molecular techniques such as PCR offered heightened precision by identifying specific genes responsible for antibiotic resistance, elevating the accuracy of such assessments [26,36]. In addition, bacterial culture was also conducted to prepare material for extracting DNA, which was then subjected to further molecular tests [3,26,34,35,36,40,43,44]. However, in the research conducted by Handorean et al., Habibi et al., Perkins et al., Jiang et al., and Chen et al., the molecular test was performed directly from the collected air samples without the need to perform bacterial culture. Thanks to this, researchers immediately obtained accurate results in the time needed only to perform the molecular method, omitting the time needed for incubation [37,38,39,41,42].
In seven investigations, a predominant molecular methodology was employed, focusing on the PCR amplification of the 16S rRNA gene region [26,34,35,37,38,39,42,43]. In two studies focusing on fungi identification, different kinds of amplification were used: 18S rRNA sequencing and ITS rRNA sequencing [34,38]. The use of this method made it possible to recognize the phylogenetic origin of the studied bacteria contained in the collected aerosol samples. In studies conducted by Wu b et al. and Gilbert Y et al., the 16S rRNA amplification facilitated the electrophoresis analysis of DNA material, providing an alternative means of identifying bacterial species [36,43]. The LAMP method used in the study by Jiang X et al. deserves particular attention. This method, compared to other popular methods, is characterized by a short implementation time, which makes the identification of a given bacterium in a hospital ward much faster than would be possible using another method [41].
Another molecular technique employed in the conducted research within this review was Checkerboard DNA-DNA hybridization [44]. This method, akin to other molecular approaches, was utilized for the identification of bacterial strains present in collected samples. However, the procedure necessitated meticulous preparation, involving the addition of a specific buffer to previously cultivated colonies on plates. Subsequent to this preparatory step, the samples were transferred to Eppendorf tubes before undergoing molecular testing. In contrast to the PCR method, this approach was characterized by increased time requirements and complexity due to the specific demands of sample preparation [44].
3.4. Quality Assessment of the Included Studies
Among the articles included in the review, four were appraised as high-quality studies, achieving scores of 5/6 [26,34] or 6/6 [38,40] points. Furthermore, five studies were characterized by a moderate risk of bias, with scores of 4/6 points [35,36,37] or 3/6 points [3,39]. No studies were excluded due to low quality (high risk of bias), as the missing information was deemed non-essential for the thoroughness of the review. The precise risk of bias for each included study is outlined in Table 3.

Table 3.
The results of the quality assessment and risk of bias across the studies.
4. Discussion
The aim of this systematic review was to assess the molecular tests employed for the analysis of aerosols in diverse healthcare settings. The second objective was to investigate whether the implementation of molecular methods contributes to enhanced precision in the assessment of bacterial and fungal quality within aerosols in healthcare settings. The investigation delineated that the prevailing molecular method in aerosol quality evaluations was the PCR method [3,26,34,35,36,37,38,39,42,43] using 16S rRNA [3,26,34,35,37,38,39,42,43] or using 18S rRNA sequencing [38] or ITS sequencing [34] in case of fungal analysis. Nevertheless, five of the included studies utilized PFGE [36], DGGE [43], DNA hybridization [44], LAMP [41], or sequence-based typing SBT [40]. As the 16S rRNA sequencing was the most applied type of genetic material analysis, it can be said that it is a powerful and widely used tool for studying bacterial diversity and taxonomy in various environments [26]. Furthermore, 16S rRNA together with 18S rRNA sequencing is advocated as the optimal tool for elucidating the phylogenetic origin of a given bacterium and fungi, respectively, given its conserved nature and minimal susceptibility to alterations [38,45].
In the conducted investigations, molecular methods served as instrumental tools in delineating the precise composition of bacterial and fungal entities within the sampled aerosols [3,26,34,35,36,37,38,39,40,41,42,43,44]. The markedly abbreviated timeframe of these molecular assays (several hours instead of a few days) stands as a notable advantage over conventional techniques, where the cultivation of bacterial cultures necessitates prolonged incubation periods [29,46]. This advantage was strongly emphasized in a study performed by Jiang et al., which shows the advantage of the LAMP method over other molecular methods, indicating a significantly shorter waiting time for the result [41]. Noteworthy is the capacity of these assays to yield comprehensive insights immediately post-sample collection, obviating the requirement for bacterial culture [37,38,39,41,42,47,48]. Such molecular analyses furnish exhaustive details concerning species, genera, and taxonomic diversity, both in the case of bacteria and fungi [37,38,39]. Beyond its efficacy in elucidating bacterial origin and strain-specific identification, molecular testing emerges as a potent tool for the detection of antibiotic resistance—a pivotal facet in comprehending the potential dissemination of antibiotic resistance within a particular strain across the population. The molecular method involves capturing the appropriate sequence in the genetic material that encodes the appropriate protein that confers antibiotic resistance to a given strain [49].
It is essential to acknowledge the ubiquitous presence of aerosols, especially within healthcare environments, where the concentration of pathogenic microorganisms may exceed permissible standards [26,37]. This situation poses a potential risk of exposing healthcare workers and patients to infections associated with airborne bacteria and fungi [37,42]. Dental offices warrant special attention due to the generation of aerosols during various procedures [50]. Ultrasound scaling, a widely employed dental procedure, generates substantial aerosol quantities containing bacteria from the patient’s saliva, gingival sulcus, or pocket, where the most pathogenic oral bacteria reside [51]. The research presented in this article revealed the methods of reducing and eliminating the number of bacteria in aerosols generated during dental procedures. Retamal-Valdes B et al. [44] observed that the use of chlorhexidine or a cetylpyridinium chloride+zinc lactate+sodium fluoride mixture rinse reduces the number of orange complex bacteria, presenting promising results. Ensuring proper disinfection and ventilation in dental rooms is crucial, as aerosols settle in various office areas, facilitating the spread of microorganisms and thereby posing a threat to employees and subsequent patients [44].
In numerous studies, the initial step prior to molecular analysis included culturing bacteria on various agar media [3,26,34,35,36,37,38,40,41,42,43,44,52]. The culture procedure was aimed at quantifying the cultured bacteria in terms of the number of colonies [3,26,34,35,36,40,43] and preliminary identification [40]. Wu B et al. also used bacterial culture to examine antibiotic resistance [36]. Additionally, the culture method was mainly used to determine colony-forming units (CFU), which allowed for the assessment of the total bacterial load in the studies performed [26,34,35,36,40,43,44]. While all studies incorporating bacterial culture later used it for molecular testing, some studies decided to skip the culture step and move directly to molecular testing to increase accuracy and information [37,38,39,41,42]. In the case of identifying fungal strains, the identification was slightly different. In most cases, culture was sufficient to assess the exact species [26,35]. Only in two cases, fungi samples were subjected to a more thorough molecular analysis to determine their species, which precisely described the fungal species [34,38].
This systematic review emphasized the precision achieved in the analysis of bioaerosols via molecular studies. However, it is crucial to acknowledge certain limitations. The features of molecular methods used in research highlight their advantages over classical methods. However, it is important to acknowledge that molecular methods also face certain challenges. One significant challenge is the cost. Wang Y et al. reported that the cost of examining one species using molecular methods can be up to 10 times more expensive than the classic seeding method [53]. Due to the high expenses involved, these methods may not be feasible in economically slower developing countries [46]. Furthermore, some molecular methods still necessitate a previous bacterial culture, thereby extending the time required to obtain results by the duration of the culture time [54]. The advantage here is the identification of ridges, in which molecular identification is not necessary, and therefore performing such a test is cheaper and more accessible [26,35]. Another issue with molecular methods is the presence of mutations. The reliability of these methods is particularly questioned in the context of sequencing the 16S rRNA, which is traditionally considered a conservative part of the genetic material. Recent research indicates that horizontal gene transfer may occur, potentially influencing the 16S rRNA region. This discovery implies that the 16S rRNA’s conservative nature may no longer hold, rendering its use in bacterial identification potentially ineffective [55].
Last but not least, it is worth highlighting that the molecular techniques employed in the included papers in the review also allow for identifying antibiotic resistance genes, focusing on those directly associated with resistance or previously linked to such processes in research or available in databases. By targeting specific genes or genomic regions known to confer resistance, researchers can elucidate the underlying mechanisms driving resistance development in pathogens [56]. Notably, many pathogens regulate their pathogenicity and virulence via a resistance phenotype, sometimes covertly. This phenotype, often governed by complex regulatory networks, can modulate the expression of various genes involved in antibiotic resistance pathways. Researchers can decipher the intricate interplay between resistance mechanisms and microbial fitness via gene expression profiling and functional genomics studies [57]. This phenotype interacts within the genomic network with other genes, leading to bacteriostasis, enhanced fitness, and eventual resistance via a gradual mechanism influenced by time intervals under selective pressure. These processes often entail intricate genetic circuits and transduction chains involving activating or repressing specific genes in response to external stimuli [58]. This interpretation underscores the significance of integrating identification efforts with mechanisms to prevent health risks, including identifying virulent factors. Understanding the complex interplay between antibiotic resistance mechanisms and microbial pathogenesis is essential for developing effective strategies to mitigate the spread of resistant pathogens and safeguard public health [59].
Given that PCR is the predominant research method, there is a lack or scarcity of data obtained from alternative genetic material examination methods, such as FISH, PFGE, DGGE, and DNA hybridization. It should be noted that the above-mentioned molecular methods are being improved and replaced by new, improved methods. A good example is isothermal PCR methods, which do not require large temperature changes during the cycle and can be conducted in constant conditions. Modern non-PCR methods such as Transcription-Based Amplification (TBA) or Strand Displacement Amplification (SDA) also deserve attention. The first of them uses reverse transcriptase to amplify the genetic material, while the second uses DNA polymerase. Their advantage, similar to isothermal processes, is that they carry out processes under constant temperature conditions without the need to use thermocyclers [60]. Many of the modern methods allow you to overcome the limitations associated with conventional methods. An example is Next-Generation Sequencing (NGS), which allows for the analysis of genetic material without the need to perform culture and allows us to focus on longer DNA fragments [61]. To enhance the comprehensive assessment of accuracy and methodological superiority, further investigations are warranted, incorporating a diverse range of modern molecular techniques. These approaches may offer distinct characteristics that can significantly contribute to the nuanced examination of bioaerosols.
5. Conclusions
In conclusion, most of the included studies predominantly utilized molecular methods, specifically 16S rRNA gene amplification via PCR for bacterial identification. In the case of fungi identification, in most cases, traditional culture was sufficient to provide information on the identification of a given strain. However, the molecular test also proves to be accurate in this case to confirm the initial identification. This approach consistently identified bacterial and fungal strains, providing precise insights into their origin, genera, and species. Molecular analyses were effective in detecting antibiotic resistance genes. Molecular methods were found to be superior to classical bacterial cultures. They offer more information for strain identification, while the culture mainly provides information on the total bacteria load expressed in CFU/m3. However, the culture method seems to be sufficient in the case of fungal identification. Additionally, molecular methods enable the detection of specific gene sequences in the bacterial genetic material, aiding in the identification of genes responsible for antibiotic resistance. Moreover, they require less time to obtain the result—in the case of a culture, it takes several days, and in the case of a molecular test, only a few hours. While molecular testing presents significant advantages over classical microbiological culture, further research is needed to compare various molecular tests and determine the most optimal approach. Overall, this review emphasizes the pivotal role of molecular methods in comprehensively characterizing bioaerosols, understanding their pathogenicity, and assessing health risks for healthcare professionals and patients.
Author Contributions
Conceptualization, J.M. and J.K.; methodology, J.M.; software, J.M.; formal analysis, J.M. and I.Z.; data curation, J.M. and J.K.; writing—original draft preparation, J.M. and J.K.; writing—review and editing, J.M., I.Z., K.G.-L. and M.D.; supervision, J.M.; project administration, J.M., T.G. and M.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Availability of supporting data—the datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Jones, R.M.; Brosseau, L.M. Aerosol Transmission of Infectious Disease. J. Occup. Environ. Med. 2015, 57, 501–508. [Google Scholar] [CrossRef] [PubMed]
- Pathak, B.; Khataniar, A.; Das, B.; Upadhyaya, S.; Medhi, A.; Bhuyan, P.K.; Buragohain, A.K.; Borah, D. Spatio-Temporal Diversity of Biological Aerosols over Northeast India: A Metagenomic Approach. Environ. Sci. Pollut. Res. 2022, 29, 64096–64111. [Google Scholar] [CrossRef] [PubMed]
- Angenent, L.T.; Kelley, S.T.; Amand, A.S.; Pace, N.R.; Hernandez, M.T. Molecular Identification of Potential Pathogens in Water and Air of a Hospital Therapy Pool. Proc. Natl. Acad. Sci. USA 2005, 29, 4860–4865. [Google Scholar] [CrossRef] [PubMed]
- Grydaki, N.; Colbeck, I.; Mendes, L.; Eleftheriadis, K.; Whitby, C. Bioaerosols in the Athens Metro: Metagenetic Insights into the PM10 Microbiome in a Naturally Ventilated Subway Station. Environ. Int. 2021, 146, 106186. [Google Scholar] [CrossRef] [PubMed]
- Matys, J.; Grzech-Leśniak, K. Dental Aerosol as a Hazard Risk for Dental Workers. Materials 2020, 13, 5109. [Google Scholar] [CrossRef] [PubMed]
- Grzech-Leśniak, K.; Matys, J. The Effect of Er:Yag Lasers on the Reduction of Aerosol Formation for Dental Workers. Materials 2021, 14, 2857. [Google Scholar] [CrossRef] [PubMed]
- Santarpia, J.L.; Rivera, D.N.; Herrera, V.L.; Morwitzer, M.J.; Creager, H.M.; Santarpia, G.W.; Crown, K.K.; Brett-Major, D.M.; Schnaubelt, E.R.; Broadhurst, M.J.; et al. Aerosol and Surface Contamination of SARS-CoV-2 Observed in Quarantine and Isolation Care. Sci. Rep. 2020, 10, 12732. [Google Scholar] [CrossRef] [PubMed]
- Austin, B. The Value of Cultures to Modern Microbiology. Antonie Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2017, 110, 1247–1256. [Google Scholar] [CrossRef]
- Perkins, S.D.; Angenent, L.T. Potential Pathogenic Bacteria in Metalworking Fluids and Aerosols from a Machining Facility. FEMS Microbiol. Ecol. 2010, 74, 643–654. [Google Scholar] [CrossRef]
- Matys, J.; Gedrange, T.; Dominiak, M.; Grzech-Leśniak, K. Quantitative Evaluation of Aerosols Produced in the Dental Office during Caries Treatment: A Randomized Clinical Trial. J. Clin. Med. 2023, 12, 4597. [Google Scholar] [CrossRef]
- Lagier, J.C.; Edouard, S.; Pagnier, I.; Mediannikov, O.; Drancourt, M.; Raoult, D. Current and Past Strategies for Bacterial Culture in Clinical Microbiology. Clin. Microbiol. Rev. 2015, 28, 208–236. [Google Scholar] [CrossRef]
- Nammour, S.; El Mobadder, M.; Maalouf, E.; Namour, M.; Namour, A.; Rey, G.; Matamba, P.; Matys, J.; Zeinoun, T.; Grzech-Leśniak, K. Clinical Evaluation of Diode (980 Nm) Laser-Assisted Nonsurgical Periodontal Pocket Therapy: A Randomized Comparative Clinical Trial and Bacteriological Study. Photobiomodulation Photomed. Laser Surg. 2021, 39, 10–22. [Google Scholar] [CrossRef]
- Mbareche, H.; Brisebois, E.; Veillette, M.; Duchaine, C. Bioaerosol Sampling and Detection Methods Based on Molecular Approaches: No Pain No Gain. Sci. Total Environ. 2017, 599–600, 2095–2104. [Google Scholar] [CrossRef] [PubMed]
- Riley, L.W. Laboratory Methods in Molecular Epidemiology: Bacterial Infections. Microbiol. Spectr. 2018, 6, 10-1128. [Google Scholar] [CrossRef] [PubMed]
- Thornton, B.; Basu, C. Rapid and Simple Method of QPCR Primer Design. Methods Mol. Biol. 2015, 1275, 173–179. [Google Scholar] [CrossRef]
- Aly, S.M.; Sabri, D.M. Next Generation Sequencing (NGS): A Golden Tool in Forensic Toolkit. Arch. Med. Sądowej Kryminol. 2015, 65, 260–271. [Google Scholar] [CrossRef]
- Heid, C.A.; Stevens, J.; Livak, K.J.; Williams, P.M. Real Time Quantitative PCR. Genome Res. 1996, 6, 986–994. [Google Scholar] [CrossRef] [PubMed]
- Chrzanowska, N.M.; Kowalewski, J.; Lewandowska, M.A. Use of Fluorescence in Situ Hybridization (FISH) in Diagnosis and Tailored Therapies in Solid Tumors. Molecules 2020, 25, 1864. [Google Scholar] [CrossRef]
- Campos, R.; Borme, J.; Guerreiro, J.R.; Machado, G.; Cerqueira, M.F.; Petrovykh, D.Y.; Alpuim, P. Attomolar Label-Free Detection of Dna Hybridization with Electrolyte-Gated Graphene Field-Effect Transistors. ACS Sens. 2019, 4, 286–293. [Google Scholar] [CrossRef]
- Oueslati, A.; Hassen, W.; Ellafi, A.; Alibi, S.; Jaziri, A.; Bachkouel, S.; Oueslati, I.; Snoussi, M.; Adnan, M.; Patel, M.; et al. Assessment of Bacterial Diversity of Industrial Poultry Wastewater by Denaturing Gradient Gel Electrophoresis (DGGE) and the Cultivation Method in Order to Inform Its Reuse in Agriculture. BioMed Res. Int. 2022, 2022, 12. [Google Scholar] [CrossRef]
- Malhotra, S.; Sharma, S.; Bhatia, N.; Kumar, P.; Hans, C. Molecular Methods in Microbiology and Their Clinical Application. J. Mol. Genet. Med. 2014, 8. [Google Scholar] [CrossRef]
- Sleator, R.D.; Shortall, C.; Hill, C. Metagenomics. Lett. Appl. Microbiol. 2008, 47, 361–366. [Google Scholar] [CrossRef]
- Zhao, Y.; Chen, F.; Li, Q.; Wang, L.; Fan, C. Isothermal Amplification of Nucleic Acids. Chem. Rev. 2015, 115, 12491–12545. [Google Scholar] [CrossRef] [PubMed]
- Bumgarner, R. Overview of DNA microarrays: Types, applications, and their future. Curr. Protoc. Mol. Biol. 2013, 101, 22.1.1–22.1.11. [Google Scholar] [CrossRef] [PubMed]
- Tamminga, G.G.; Jansen, G.J.; Wiersma, M. Evaluation of a Fluorescence in Situ Hybridization (FISH)-Based Method for Detection of SARS-CoV-2 in Saliva. PLoS ONE 2022, 17, e0277367. [Google Scholar] [CrossRef]
- Yousefzadeh, A.; Maleki, A.; Athar, S.D.; Darvishi, E.; Ahmadi, M.; Mohammadi, E.; Tang, V.T.; Kalmarzi, R.N.; Kashefi, H. Evaluation of bio-aerosols type, density, and modeling of dispersion in inside and outside of different wards of educational hospital. Environ. Sci. Pollut. Res. 2022, 29, 14143–14157. [Google Scholar] [CrossRef]
- Johnson, J.S.; Spakowicz, D.J.; Hong, B.Y.; Petersen, L.M.; Demkowicz, P.; Chen, L.; Leopold, S.R.; Hanson, B.M.; Agresta, H.O.; Gerstein, M.; et al. Evaluation of 16S RRNA Gene Sequencing for Species and Strain-Level Microbiome Analysis. Nat. Commun. 2019, 10, 5029. [Google Scholar] [CrossRef] [PubMed]
- Núñez, A.; de Paz, G.A.; Rastrojo, A.; García, A.M.; Alcamí, A.; Montserrat Gutiérrez-Bustillo, A.; Moreno, D.A. Monitoring of Airborne Biological Particles in Outdoor Atmosphere. Part 2: Metagenomics Applied to Urban Environments. Int. Microbiol. 2016, 19, 69–80. [Google Scholar] [CrossRef]
- Soroka, M.; Wasowicz, B.; Rymaszewska, A. Loop-Mediated Isothermal Amplification (Lamp): The Better Sibling of PCR? Cells 2021, 10, 1931. [Google Scholar] [CrossRef]
- Cordero, F.; Botta, M.; Calogero, R.A. Microarray Data Analysis and Mining Approaches. Brief. Funct. Genom. Proteom. 2007, 6, 265–281. [Google Scholar] [CrossRef]
- Page, M.J.; Moher, D.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. PRISMA 2020 Explanation and Elaboration: Updated Guidance and Exemplars for Reporting Systematic Reviews. BMJ 2021, 372, n160. [Google Scholar] [CrossRef] [PubMed]
- Watson, P.F.; Petrie, A. Method Agreement Analysis: A Review of Correct Methodology. Theriogenology 2010, 73, 1167–1179. [Google Scholar] [CrossRef] [PubMed]
- Higgins, J.P.T.; Thomas, J.; Chandler, J.; Cumpston, M.; Li, T.; Page, M.J. Cochrane Handbook for Systematic Reviews of Interventions; Cochrane Training; Cochrane: London, UK, 2021. [Google Scholar]
- Núñez, A.; García, A.M. The aerobiome in a hospital environment: Characterization, seasonal tendencies and the effect of window opening ventilation. Build. Environ. 2023, 230, 110024. [Google Scholar] [CrossRef]
- Nimra, A.; Ali, Z.; Sultan, S.; Nasir, Z.A.; Sidra, S.; Hussain, A. Molecular sequencing and phylogenetic analysis of bioaerosols in hospital wards with different ventilation conditions. J. Infect. Dev. Ctries. 2022, 16, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Qi, C.; Wang, L.; Yang, W.; Zhou, D.; Wang, M.; Dong, Y.; Weng, H.; Li, C.; Hou, X.; et al. Detection of Microbial Aerosols in Hospital Wards and Molecular Identification and Dissemination of Drug Resistance of Escherichia coli. Environ. Int. 2020, 137, 105479. [Google Scholar] [CrossRef] [PubMed]
- Handorean, A.; Robertson, C.E.; Harris, J.K.; Frank, D.; Hull, N.; Kotter, C.; Stevens, M.J.; Baumgardner, D.; Pace, N.R.; Hernandez, M. Microbial Aerosol Liberation from Soiled Textiles Isolated during Routine Residuals Handling in a Modern Health Care Setting. Microbiome 2015, 3, 72. [Google Scholar] [CrossRef] [PubMed]
- Habibi, N.; Uddin, S.; Behbehani, M.; Al Salameen, F.; Razzack, N.A.; Zakir, F.; Shajan, A.; Alam, F. Bacterial and fungal communities in indoor aerosols from two Kuwaiti hospitals. Front. Microbiol. 2022, 13, 955913. [Google Scholar] [CrossRef]
- Perkins, S.D.; Mayfield, J.; Fraser, V.; Angenent, L.T. Potentially Pathogenic Bacteria in Shower Water and Air of a Stem Cell Transplant Unit. Appl. Environ. Microbiol. 2009, 75, 5363–5372. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Montagna, M.T.; Cristina, M.L.; De Giglio, O.; Spagnolo, A.M.; Napoli, C.; Cannova, L.; Deriu, M.G.; Delia, S.A.; Giuliano, A.; Guida, M.; et al. Serological and molecular identification of Legionella spp. in water and surrounding air samples in Italian healthcare facilities. Environ. Res. 2016, 146, 47–50. [Google Scholar] [CrossRef]
- Jiang, X.; Liu, Y.; Liu, Q.; Jing, W.; Qin, K.; Sui, G. Rapid Capture and Analysis of Airborne Staphylococcus aureus in the Hospital Using a Microfluidic Chip. Micromachines 2016, 7, 169. [Google Scholar] [CrossRef]
- Chen, P.-S.; Lin, C.K.; Tsai, F.T.; Yang, C.-Y.; Lee, C.-H.; Liao, Y.-S.; Yeh, C.-Y.; King, C.-C.; Wu, J.-L.; Wang, Y.-C.; et al. Quantification of Airborne Influenza and Avian Influenza Virus in a Wet Poultry Market using a Filter/Real-time qPCR Method. Aerosol Sci. Technol. 2009, 43, 290–297. [Google Scholar] [CrossRef]
- Gilbert, Y.; Veillette, M.; Duchaine, C. Airborne bacteria and antibiotic resistance genes in hospital rooms. Aerobiologia 2010, 26, 185–194. [Google Scholar] [CrossRef]
- Retamal-Valdes, B.; Soares, G.M.; Stewart, B.; Figueiredo, L.C.; Faveri, M.; Miller, S.; Zhang, Y.P.; Feres, M. Effectiveness of a Pre-Procedural Mouthwash in Reducing Bacteria in Dental Aerosols: Randomized Clinical Trial. Braz. Oral Res. 2017, 31, e21. [Google Scholar] [CrossRef] [PubMed]
- Clarridge, J.E. Impact of 16S RRNA Gene Sequence Analysis for Identification of Bacteria on Clinical Microbiology and Infectious Diseases. Clin. Microbiol. Rev. 2004, 17, 840–862. [Google Scholar] [CrossRef]
- Başpınar, E.Ö.; Dayan, S.; Bekçibaşı, M.; Tekin, R.; Ayaz, C.; Deveci, Ö.; Hoşoğlu, S. Comparison of Culture and PCR Methods in the Diagnosis of Bacterial Meningitis. Braz. J. Microbiol. 2017, 48, 232–236. [Google Scholar] [CrossRef]
- Ku, D.N.; Ku, S.K.; Helfman, B.; McCarty, N.A.; Wolff, B.J.; Winchell, J.M.; Anderson, L.J. Ability of Device to Collect Bacteria from Cough Aerosols Generated by Adults with Cystic Fibrosis [Version 1; Referees: 2 Approved]. F1000Research 2016, 5, 1920. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, M.; Ramsheh, M.Y.; Williams, C.M.L.; Auty, J.; Haldar, K.; Abdulwhhab, M.; Brightling, C.E.; Barer, M.R. Face Mask Sampling Reveals Antimicrobial Resistance Genes in Exhaled Aerosols from Patients with Chronic Obstructive Pulmonary Disease and Healthy Volunteers. BMJ Open Respir. Res. 2018, 5, e000321. [Google Scholar] [CrossRef]
- Shu, Q.; Rajagopal, M.; Fan, J.; Zhan, L.; Kong, X.; He, Y.; Rotcheewaphan, S.; Lyon, C.J.; Sha, W.; Zelazny, A.M.; et al. Peptidomic analysis of mycobacterial secreted proteins enables species identification. View 2022, 3, 20210019. [Google Scholar] [CrossRef]
- Innes, N.; Johnson, I.G.; Al-Yaseen, W.; Harris, R.; Jones, R.; KC, S.; McGregor, S.; Robertson, M.; Wade, W.G.; Gallagher, J.E. A Systematic Review of Droplet and Aerosol Generation in Dentistry. J. Dent. 2021, 105, 103556. [Google Scholar] [CrossRef]
- Lahdentausta, L.; Sanmark, E.; Lauretsalo, S.; Korkee, V.; Nyman, S.; Atanasova, N.; Oksanen, L.; Zhao, J.; Hussein, T.; Hyvärinen, A.; et al. Aerosol Concentrations and Size Distributions during Clinical Dental Procedures. Heliyon 2022, 8, e11074. [Google Scholar] [CrossRef]
- Stockwell, R.E.; Chin, M.; Johnson, G.R.; Wood, M.E.; Sherrard, L.J.; Ballard, E.; O’Rourke, P.; Ramsay, K.A.; Kidd, T.J.; Jabbour, N.; et al. Transmission of Bacteria in Bronchiectasis and Chronic Obstructive Pulmonary Disease: Low Burden of Cough Aerosols. Respirology 2019, 24, 980–987. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Guo, G.; Wang, H.; Yang, X.; Shao, F.; Yang, C.; Gao, W.; Shao, Z.; Zhang, J.; Luo, J.; et al. Comparative study of bacteriological culture and real-time fluorescence quantitative PCR (RT-PCR) and multiplex PCR-based reverse line blot (mPCR/RLB) hybridization assay in the diagnosis of bacterial neonatal meningitis. BMC Pediatr. 2014, 14, 224. [Google Scholar] [CrossRef] [PubMed]
- Pandya, S.; Ravi, K.; Srinivas, V.; Jadhav, S.; Khan, A.; Arun, A.; Riley, L.W.; Madhivanan, P. Comparison of culture-dependent and culture-independent molecular methods for characterization of vaginal microflora. J Med. Microbiol. 2017, 66, 149–153. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Dash, H.R.; Mangwani, N.; Chakraborty, J.; Kumari, S. Understanding molecular identification and polyphasic taxonomic approaches for genetic relatedness and phylogenetic relationships of microorganisms. J. Microbiol. Methods 2014, 103, 80–100. [Google Scholar] [CrossRef] [PubMed]
- López-Pérez, M.; Aguirre-Garrido, F.; Herrera-Zúñiga, L.; Fernández, F.J. Gene as a Dynamical Notion: An Extensive and Integrative Vision. Redefining the Gene Concept, from Traditional to Genic-Interaction, as a New Dynamical Version. BioSystems 2023, 234, 105060. [Google Scholar] [CrossRef] [PubMed]
- Antunes, L.C.S.; Imperi, F.; Carattoli, A.; Visca, P. Deciphering the Multifactorial Nature of Acinetobacter Baumannii Pathogenicity. PLoS ONE 2011, 6, e22674. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Fussenegger, M. Designing Cell Function: Assembly of Synthetic Gene Circuits for Cell Biology Applications. Nat. Rev. Mol. Cell Biol. 2018, 19, 507–525. [Google Scholar] [CrossRef] [PubMed]
- Martínez, J.L.; Baquero, F. Interactions among Strategies Associated with Bacterial Infection: Pathogenicity, Epidemicity, and Antibiotic Resistance. Clin. Microbiol. Rev. 2002, 15, 647–679. [Google Scholar] [CrossRef]
- Kurkela, S.; Brown, D.W.G. Molecular diagnostic techniques. Medicine 2009, 37, 535–540. [Google Scholar] [CrossRef]
- Ari, Ş.; Arikan, M. Next-generation sequencing: Advantages, disadvantages, and future. In Plant Omics: Trends and Applications; Springer: Berlin/Heidelberg, Germany, 2016; pp. 109–135. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).