Abstract
We previously demonstrated that patients with metastatic unresectable stage IIIb–IV melanoma receiving cetirizine (a second-generation H1 antagonist antihistamine) premedication with immunotherapy had better outcomes than those not receiving cetirizine. In this retrospective study, we searched for a gene signature potentially predictive of the response to the addition of cetirizine to checkpoint inhibition (nivolumab or pembrolizumab with or without previous ipilimumab). Transcriptomic analysis showed that inducible T cell costimulator ligand (ICOSLG) expression directly correlated with the disease control rate (DCR) when detected with a loading value > 0.3. A multivariable logistic regression model showed a positive association between the DCR and ICOSLG expression for progression-free survival and overall survival. ICOSLG expression was associated with CD64, a specific marker of M1 macrophages, at baseline in the patient samples who received cetirizine concomitantly with checkpoint inhibitors, but this association was not present in subjects who had not received cetirizine. In conclusion, our results show that the clinical advantage of concomitant treatment with cetirizine during checkpoint inhibition in patients with malignant melanoma is associated with high ICOSLG expression, which could predict the response to immune checkpoint inhibitor blockade.
1. Introduction
Checkpoint inhibition has become a mainstay of treatment for advanced cutaneous melanoma, providing improved survival compared with previous therapies [1,2]. However, a proportion of patients still experience poor outcomes after immunotherapy, as confirmed by several studies [3,4,5,6,7]. As a result, identifying predictive biomarkers of the response to immunotherapy is an important area of research, aiming to prevent overtreatment, unnecessary risk of toxicity, and wasteful resource usage. Since gene expression signatures in liquid biopsies are non-invasive repeatable tests that may provide high specificity and sensitivity [8], they are widely investigated as potential predictive or prognostic markers in cancer patients, specifically in subjects with cutaneous melanoma [9,10,11].
Patients with unresectable cutaneous melanoma were observed to have improved overall survival (OS) and progression-free survival (PFS) when premedicated with cetirizine (a second-generation H1 antagonist antihistamine) at the beginning of checkpoint-inhibiting immunotherapy compared with non-premedicated subjects [12]. Nonetheless, some patients did not show clinical benefits from adding cetirizine [12]. The effect of cetirizine administration was associated with a shift in circulating macrophages from the M2 to the M1 phenotype. This observation suggests that the antihistamine may promote the antitumor activity of macrophages through an interferon-gamma pathway that drives macrophages from the immunosuppressive phenotype to the M1 phenotype, which has antitumoral activities based on phagocytosis and antigen presentation [13]. Building on these results and taking advantage of our experience with gene expression profiling techniques [14,15], we aimed to identify a gene expression signature linked to the immune state of patients and associated with the clinical response to the addition of cetirizine to checkpoint inhibition.
In a retrospective study, we compared clinical outcomes and blood sample transcriptomic analysis in patients with unresectable cutaneous malignant melanoma treated with checkpoint inhibitors (the anti-PD1 nivolumab or pembrolizumab, with or without previous anti-CTLA4 ipilimumab), either with or without cetirizine premedication.
2. Results
2.1. Patient Characteristics and Outcomes
Blood and tissue samples were collected between April 2016 and June 2018. Patient characteristics at baseline are reported in Table 1. Overall, 116 patients were included in the study; 51 (44.0%) were females, 22 (20.4%) had a BRAF mutation, 86 (79.6%) had a wild-type BRAF (BRAF state was missing for eight patients), and 84 (75.7%) had brain metastases. Sixty-seven (57.7%) patients had received cetirizine before immunotherapy and 49 (42.3%) had not. These two groups of patients had similar characteristics. However, those treated with cetirizine had a higher frequency of mutated BRAF and central nervous system (CNS) metastases, along with more elevated levels of LDH. Table 1 reports anti-PD-1 agents used in the included patients. Nivolumab was administered at a dose of 3 mg/kg every 2 weeks and pembrolizumab at a dose of 2 mg/kg mg every 3 weeks. Ipilimumab was administrated at a dose of 3 mg/kg IV every 3 weeks for a maximum of 4 doses.

Table 1.
Demographic and clinical data at baseline.
At the first disease response assessment (3 months), patients treated with cetirizine had significantly better response to therapy; the DCR was recorded in 55 (DCR: 47.4%) patients. Respondents were 38/67 (DCR: 56.7%) among the patients who had received cetirizine and 17/49 (DCR: 34.7%) among those who had not. The objective response rate (ORR) for the whole cohort was 29.3% of patients but was higher in the cetirizine group than in the group without cetirizine (ORR: 22.4%).
2.2. Gene Expression Signatures
The analysis of blood samples collected at baseline showed that a gene expression signature was associated (the cut-off point was 1.76 at the 26th percentile) with response to anti-PD-1 and cetirizine treatment. It included 15 genes (TRAF1, S100A9, S100A8, S100A12, OLR1, NF1, MMP9, LDHA, ITGAM, ICOSLG, HLA-DPA1, FSTL3, CST2, CLEC5A, and ANLN). Only the increased expression of inducible T cell costimulator ligand (ICOSLG) with a loading value > 0.3 directly correlated with a higher probability of response to treatment (Supplementary Figure S1A). The other genes of the signature were negatively correlated with the probability of response. The accuracy of the association between signature expression and the DCR is shown in Supplementary Figure S1B. The expression of the signature was significantly more frequent in responders than in non-responders (p < 0.01) (Figure 1).
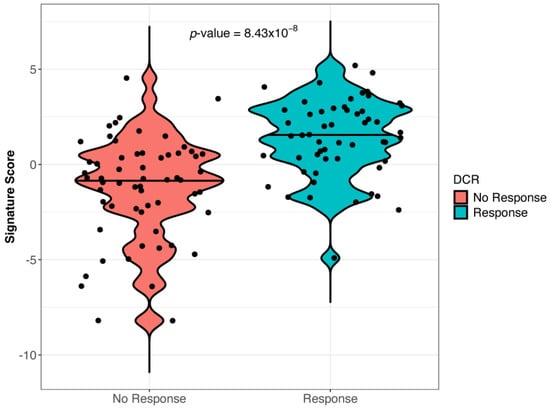
Figure 1.
Gene signature expression in responder and non-responder patients (Wilcoxon test, p = 8.3 × 10−8). The signature included 15 genes, namely TRAF1, S100A9, S100A8, S100A12, OLR1, NF1, MMP9, LDHA, ITGAM, ICOSLG, HLA-DPA1, FSTL3, CST2, CLEC5A, and ANLN.
To investigate the effect of high versus low expression of the ICOSLG signature on oncological outcomes, the best cut-off point of signature expression was determined by the Simon permutation approach, and a low-expression group with 30 patients and a high-expression group with 86 patients were identified (Supplementary Figure S2).
Among patients with low ICOSLG expression, PFS was not different between the two groups of patients with or without cetirizine (Figure 2A). In contrast, among patients with high ICOSLG expression, PFS was significantly better in those who had received cetirizine (Figure 2B).

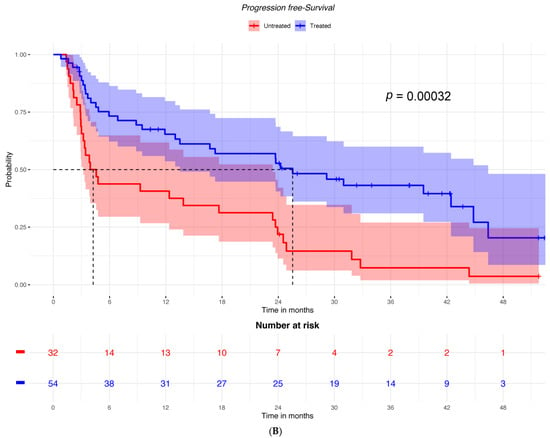
Figure 2.
PFS according to having received or not received cetirizine in (A) patients with low ICOSLG expression (n = 30) and (B) patients with high ICOSLG expression (N = 86).
Accordingly, OS was not significantly different between groups treated with cetirizine versus untreated patients with low ICOSLG expression (HR = 0.789; 95% CI: 0.36–1.73) (Figure 3A). Conversely, among patients with high ICOSLG expression, it was higher in those who had received cetirizine (HR = 0.449; 95% CI: 0.25–0.82) (Figure 3B).

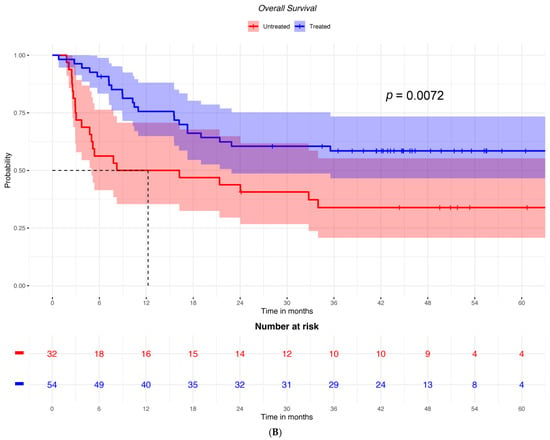
Figure 3.
OS in patients treated or not treated with cetirizine and (A) with low ICOSLG expression (n = 30) or (B) with high ICOSLG expression (N = 86).
A multivariable analysis with a logistic regression model showed that patients treated with cetirizine had a better PFS than untreated ones (Figure 4A). Additionally, these results were confirmed by dichotomizing data, finding an improved DCR with cetirizine treatment in patients with high ICOSLG expression (Supplementary Figure S3A).

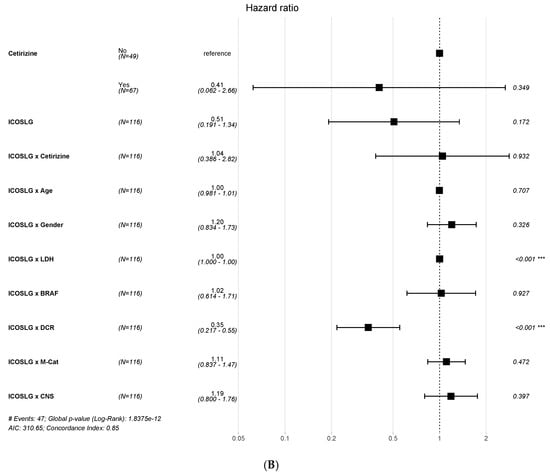
Figure 4.
Multivariable survival analysis according to PFS (A) and OS (B) for ICOSLG expression. * p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001.
A similar association of ICOSLG expression was found for OS (Figure 4B and Supplementary Figure S3B). ICOSLG was associated with LDH expression, but the association was irrelevant as the HR was =1 for both PFS and OS.
Patients treated with cetirizine had a higher risk of colitis than the overall population (25.5% vs. 5.1%, p = 0.02), independent of ICOSLG expression. In comparison, patients with high ICOSLG expression had an increased risk of arthralgia (33.9% vs. 8.3%, p = 0.02), colitis (22.6% vs. 0, p = 0.01), pancreatitis (17.7% vs. 0, p = 0.03), and skin reactions (50% vs. 16.7%, p = 0.01).
The ICOSLG expression at baseline was associated with the blood expression of high-affinity IgG Fc receptor 1 (FCGR1A), coding for CD64, in the overall population (Rho = −0.219; p = 0.018) and patients treated with cetirizine (Rho = −0.287; p = 0.018) but not in the untreated ones. After 3 months, this association was not present in any subgroup.
3. Discussion
A previous study [12] demonstrated that patients with metastatic unresectable melanoma receiving cetirizine premedication with immunotherapy had better outcomes than those not receiving cetirizine. Since a proportion of patients had poorer results after immune checkpoint inhibitor (ICI) blockade despite cetirizine premedication, we conducted this retrospective study with the aim of identifying a predictive biomarker of the response to ICI blockade immunotherapy in patients with advanced unresectable stage IIIb–IV cutaneous melanoma with concomitant administration of cetirizine.
Circulating biomarkers and tumor molecular signatures have been previously investigated in advanced melanoma to identify patients with resistance hallmarks and a reduced probability of response to checkpoint inhibition. A tumor inflammation signature including 16 genes was associated with response to anti-PD-1 [16]; tumor mutational burden and microsatellite instability are considered predictive tissue markers of metastasis, based on results from KEYNOTE-158 [17,18,19]. Our transcriptomic analysis showed a gene expression signature associated with response to anti-PD-1 treatment in this patient population. The signature included 15 genes, but only ICOSLG was directly correlated to the DCR when detected with a loading value > 0.3 (Supplementary Figure S1). ICOSLG has a relevant role in immune antibody response modulation [20] and was a suitable candidate as a possible predictive biomarker for response to treatment. Therefore, we decided to select it for further analysis. The clinical relevance of ICOSLG expression was evaluated by comparing PFS and OS Kaplan–Meier curves of patients with high versus low gene expression; the findings showed that only when the ICOSLG expression is high, patients receiving cetirizine have better outcomes than those not receiving it. This result was confirmed by a multivariable model of logistic regression showing a positive association of the DCR with ICOSLG expression for PFS and OS in patients receiving cetirizine and anti-PD-1 versus those receiving only anti-PD-1. The advantage of receiving cetirizine in those with ICOSLG expression was present even though the cetirizine subgroup had a higher frequency of negative predictive factors such as BRAF mutation, an elevated LDH level, and CNS metastases. On the contrary, the association of ICOSLG with LDH had no association with survival.
We observed an association between ICOSLG expression and CD64 expression at baseline in the overall patient sample and those who received cetirizine concomitantly with ICI. However, this association was absent in subjects who had not received cetirizine, nor was it detected after 3 months of treatment in any subgroup.
ICOSLG is an immune checkpoint expressed by antigen-presenting cells and other cell types, including mesenchymal stem cells, endothelial cells, fibroblasts, and tumor cells [21,22]. It is a ligand for the T cell-specific cell surface receptor ICOS, and their interaction promotes T cell responses. ICOS is found in T regulatory cells in melanoma patients, and cells with high expression can induce diverse immune responses in responder cells depending on activation signals and cytokines in the microenvironment [23]. ICOSLG is more highly expressed by M2-type than M1-type macrophages, and the activity of M1 and M2 is modulated by ICOSLG activation [24].
CD64 is a functional high-affinity IgG Fc receptor coded by the FCGR1A gene in humans [25]. It is a specific marker of M1 macrophages, playing a role in immune functions, including phagocytosis, antigen presentation, degranulation, cytokine production, immune complex clearance, and inflammation (UniProt https://www.uniprot.org/uniprot/P12314, accessed on 7 February 2024) [26,27]. Its expression was increased in cetirizine-treated patient blood macrophages after 3 months of checkpoint inhibition compared with baseline, and CD64 expression correlated with genes linked to the interferon pathway [12].
We speculate that the high expression of both ICOSLG and CD64 before ICI treatment in a subset of patients may be linked to a baseline immune activation condition [28,29]. These subjects benefit from concomitant cetirizine treatment, which may promote M1 polarization [12,30] and reduce ICOSLG expression in immune cells, resulting in the loss of the ICOSLG/CD64 association after 3 months of treatment.
In conclusion, our results show that the clinical advantage of concomitant treatment with cetirizine during checkpoint inhibition in patients with malignant melanoma is associated with high expression of ICOSLG. This gene signature could predict the response to ICI blockade, and a prospective study is necessary to confirm this hypothesis. Additionally, treatment with cetirizine is associated with an increased risk of colitis, while high ICOSLG expression predicts a higher risk of colitis, arthralgia, pancreatitis, and skin reactions.
4. Materials and Methods
4.1. Study Design
A retrospective cohort study was carried out at the Istituto Nazionale Tumori–IRCCS–Fondazione “G. Pascale”, Naples, Italy, upon communication with the local Ethics Committee [protocol no. 32/22 oss]. The study was performed in accordance with the revised version of the Declaration of Helsinki (52nd WMA General Assembly, Edinburgh, UK, October 2000).
Data were retrieved from clinical records. Consecutive adult patients with histologically confirmed unresectable or metastatic melanoma (stage IIIb–IV), treated with an anti-PD-1 agent (nivolumab or pembrolizumab), either first-line or pretreated with ipilimumab, and aged over 18 years, were enrolled between April 2014 and June 2018. The study compared the group of patients exposed to cetirizine to the not exposed group. The concurrent use of cetirizine at each immunotherapy administration was retrospectively ascertained. Cetirizine was used as a premedication on the day of immune therapy (10 mg, once). B-RAF mutation status was assessed at baseline in tumor tissue samples. All patients provided their written informed consent. Raw data generated in this study have been deposited in Zenodo: https://doi.org/10.5281/zenodo.13838853.
4.2. Evaluation of Outcomes
RECIST 1.1 criteria were used to evaluate the tumor response as complete response (CR), partial response (PR), stable disease (SD), or progressive disease. The following parameters were collected at baseline: demographic data, concurrent use of cetirizine, history of allergy, BRAF mutation status, American Joint Committee on Cancer (AJCC) distant metastases category, lactate dehydrogenase (LDH) serum level, and the presence of brain metastases.
The treatment response was evaluated during the first tumor response assessment, conducted 3 months after starting immunotherapy. During the follow-up, the following data were collected: PFS (defined as the time from the administration of the first dose of an anti-PD-1 agent to documented radiological progression, death, or loss at follow-up, whichever occurred first), OS (defined as the time from the administration of the first dose of an anti-PD-1 agent to death or loss at follow-up, whichever occurred first), the disease control rate (DCR; defined as the sum of CR, PR, and SD > 1 year), and the objective response (OR; defined as the sum of CR and PR).
4.3. Transcriptomic Analysis
To conduct a gene expression profile analysis, baseline peripheral blood samples were collected from enrolled patients treated with an anti-PD-1 agent. RNA from whole blood was extracted using the QIAamp RNA Blood Mini Kit (Qiagen, Hilden, Germany). A quantification and purity assessment were performed with a NanoDrop spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA, USA). The purified RNA of PBMCs was used for hybridization and subjected to gene profiling analysis on NanoString nCounter through the PanCancer IO 360 panel [31], characterized by 770 human genes involved in the interplay between the tumor microenvironment and immune response. Gene expression data were normalized using nSolver Version 4.0 Software; NanoString counts were normalized to External RNA Controls Consortium (ERCC; https://www.nist.gov/programs-projects/external-rna-controls-consortium, accessed on 30 November 2023) technical controls and 30 housekeeping genes. Analyzed genes are reported in Supplementary Table S1.
4.4. Statistical Analysis
Demographic and clinical data were tabulated using descriptive statistics. PFS was calculated from the start of treatment with anti-PD-1 to the evidence of progressive disease or death, whichever occurred first; OS was calculated from the start of treatment with anti-PD-1 to death or censoring at the last follow-up. Survival times for groups were analyzed by univariate analysis and expressed using the Kaplan–Meier method; differences among curves were assessed by the log-rank test. Continuous interactions were analyzed by multivariable analysis using a Cox regression model, and hazard ratios (HRs) and their 95% CIs were estimated. Spearman’s Rho analysis was used to evaluate the associations between variables [32].
To identify the mRNA gene signatures associated with the patient’s response groups, a discriminant analysis for sparse data was performed via the partial least squares procedure. The sparse variant of the partial least squares discriminant analysis (PLS-DA) allows for the selection of the most predictive or discriminative features in the data to classify the samples [15,33].
The identified gene expression signature scores were used as synthetic risk indices; to define the subgroups of low and high risk, an optimal cut-point was selected by maximizing the area under the curve of the receiver operating characteristic performed by a cross-validation approach.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms252212439/s1.
Author Contributions
Study conception and design: D.M. and P.A.A.; collection and interpretation of data: D.M., M.O., M.G.V., M.M. and M.P.; statistical analysis: D.M. and M.F.; manuscript drafting: D.M., M.F., M.O., G.M., M.G.V., M.M., T.D.C., T.M., M.P., P.S., R.D.F., A.C., E.C., C.C., S.W., A.B., E.S. and P.A.A.; manuscript editing: D.M., M.F., M.O., G.M., M.G.V., M.M., M.C. (Mariagrazia Capasso), M.C. (Mariaelena Capone), T.D.C., T.M., M.P., P.S., R.D.F., A.C., E.C., C.C., S.W., A.B., E.S. and P.A.A.; approval to submit: D.M., M.F., M.O., G.M., M.G.V., M.M., M.C. (Mariagrazia Capasso), M.C. (Mariaelena Capone), T.D.C., T.M., M.P., P.S., R.D.F., A.C., E.C., C.C., S.W., A.B., E.S. and P.A.A. All authors have read and agreed to the published version of the manuscript.
Funding
This study received a grant from the Italian Ministry of Health (IT-MOH).
Institutional Review Board Statement
All procedures performed were in accordance with the 1964 Helsinki Declaration and its later amendments. The present study was approved by the Ethics Committee of Istituto Nazionale Tumori IRCCS “Fondazione G. Pascale”, Naples, Italy, [protocol no. 32/22 oss]. All subjects provided informed written consent prior to enrollment in the study.
Informed Consent Statement
Participants provided consent to the publication of anonymous data.
Data Availability Statement
Availability of data and material: https://doi.org/10.5281/zenodo.13838853.
Acknowledgments
All subjects provided informed written consent prior to enrollment in the study. All procedures performed were in accordance with the 1964 Helsinki Declaration and its later amendments. This study was approved by the Ethics Committee of Istituto Nazionale Tumori—IRCCS—Fondazione “G. Pascale”, Naples, Italy, protocol number 32/22 oss. All patients provided informed consent to participate. Editorial assistance was provided by Valentina Attanasio, Laura Brogelli, Massimiliano Pianta, and Aashni Shah (Polistudium SRL, Milan, Italy).
Conflicts of Interest
Alessandra Cesano and Sarah Warren were employed at ESSA Pharma. The authors declare that this research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Topalian, S.L.; Hodi, F.S.; Brahmer, J.R.; Gettinger, S.N.; Smith, D.C.; McDermott, D.F.; Powderly, J.D.; Sosman, J.A.; Atkins, M.B.; Leming, P.D.; et al. Five-Year Survival and Correlates Among Patients with Advanced Melanoma, Renal Cell Carcinoma, or Non-Small Cell Lung Cancer Treated with Nivolumab. JAMA Oncol. 2019, 5, 1411–1420. [Google Scholar] [CrossRef]
- Robert, C.; Hwu, W.J.; Hamid, O.; Ribas, A.; Weber, J.S.; Daud, A.I.; Hodi, F.S.; Wolchok, J.D.; Mitchell, T.C.; Hersey, P.; et al. Long-term safety of pembrolizumab monotherapy and relationship with clinical outcome: A landmark analysis in patients with advanced melanoma. Eur. J. Cancer 2021, 144, 182–191. [Google Scholar] [CrossRef]
- Schachter, J.; Ribas, A.; Long, G.V.; Arance, A.; Grob, J.J.; Mortier, L.; Daud, A.; Carlino, M.S.; McNeil, C.; Lotem, M.; et al. Pembrolizumab versus ipilimumab for advanced melanoma: Final overall survival results of a multicentre, randomised, open-label phase 3 study (KEYNOTE-006). Lancet 2017, 390, 1853–1862. [Google Scholar] [CrossRef]
- Tang, Q.; Zhao, S.; Zhou, N.; He, J.; Zu, L.; Liu, T.; Song, Z.; Chen, J.; Peng, L.; Xu, S. PD-1/PD-L1 immune checkpoint inhibitors in neoadjuvant therapy for solid tumors (Review). Int. J. Oncol. 2023, 62, 49. [Google Scholar] [CrossRef]
- Versluis, J.M.; Menzies, A.M.; Sikorska, K.; Rozeman, E.A.; Saw, R.P.M.; van Houdt, W.J.; Eriksson, H.; Klop, W.M.C.; Ch’ng, S.; van Thienen, J.V.; et al. Survival update of neoadjuvant ipilimumab plus nivolumab in macroscopic stage III melanoma in the OpACIN and OpACIN-neo trials. Ann. Oncol. 2023, 34, 420–430. [Google Scholar] [CrossRef]
- Wolchok, J.D.; Chiarion-Sileni, V.; Gonzalez, R.; Grob, J.J.; Rutkowski, P.; Lao, C.D.; Cowey, C.L.; Schadendorf, D.; Wagstaff, J.; Dummer, R.; et al. Long-Term Outcomes with Nivolumab Plus Ipilimumab or Nivolumab Alone Versus Ipilimumab in Patients with Advanced Melanoma. J. Clin. Oncol. 2022, 40, 127–137. [Google Scholar] [CrossRef]
- Eljilany, I.; Castellano, E.; Tarhini, A.A. Adjuvant Therapy for High-Risk Melanoma: An In-Depth Examination of the State of the Field. Cancers 2023, 15, 4125. [Google Scholar] [CrossRef]
- Kinane, D.F.; Gabert, J.; Xynopoulos, G.; Guzeldemir-Akcakanat, E. Strategic approaches in oral squamous cell carcinoma diagnostics using liquid biopsy. Periodontol. 2000 2024, Online ahead of print. [Google Scholar] [CrossRef]
- Jamshidi, A.; Liu, M.C.; Klein, E.A.; Venn, O.; Hubbell, E.; Beausang, J.F.; Gross, S.; Melton, C.; Fields, A.P.; Liu, Q.; et al. Evaluation of cell-free DNA approaches for multi-cancer early detection. Cancer Cell. 2022, 40, 1537–1549.e12. [Google Scholar] [CrossRef]
- Oliveira, K.C.S.; Ramos, I.B.; Silva, J.M.C.; Barra, W.F.; Riggins, G.J.; Palande, V.; Pinho, C.T.; Frenkel-Morgenstern, M.; Santos, S.E.B.; Assumpcao, P.P.; et al. Current perspectives on circulating tumor DNA, precision medicine, and personalized clinical management of cancer. Mol. Cancer Res. 2020, 18, 517–528. [Google Scholar] [CrossRef]
- Lukacova, E.; Hanzlikova, Z.; Podlesnyi, P.; Sedlackova, T.; Szemes, T.; Grendar, M.; Samec, M.; Hurtova, T.; Malicherova, B.; Leskova, K.; et al. Novel liquid biopsy CNV biomarkers in malignant melanoma. Sci. Rep. 2024, 14, 15786. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mallardo, D.; Simeone, E.; Vanella, V.; Vitale, M.G.; Palla, M.; Scarpato, L.; Paone, M.; De Cristofaro, T.; Borzillo, V.; Cortellini, A.; et al. Concomitant medication of cetirizine in advanced melanoma could enhance anti-PD-1 efficacy by promoting M1 macrophages polarization. J. Transl. Med. 2022, 20, 436. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yunna, C.; Mengru, H.; Lei, W.; Weidong, C. Macrophage M1/M2 polarization. Eur. J. Pharmacol. 2020, 877, 173090. [Google Scholar] [CrossRef] [PubMed]
- Mallardo, D.; Giannarelli, D.; Vitale, M.G.; Galati, D.; Trillò, G.; Esposito, A.; Isgrò, M.A.; D’Angelo, G.; Festino, L.; Vanella, V.; et al. Nivolumab serum concentration in metastatic melanoma patients could be related to outcome and enhanced immune activity: A gene profiling retrospective analysis. J. Immunother. Cancer 2022, 10, e005132. [Google Scholar] [CrossRef]
- Mallardo, D.; Cortellini, A.; Capone, M.; Madonna, G.; Pinato, D.; Warren, S.; Simeone, E.; Ascierto, P. Concomitant type 2 diabetes mellitus (T2DM) in metastatic melanoma patients could be related to lower level of LAG-3: A transcriptomic analysis of a retrospective cohort. Ann. Oncol. 2022, 33, 445–447. [Google Scholar] [CrossRef]
- Lee, C.J.; An, H.J.; Cho, E.S.; Kang, H.C.; Lee, J.Y.; Lee, H.S.; Cho, Y.Y. Stat2 stability regulation, an intersection between immunity and carcinogenesis. Exp. Mol. Med. 2020, 52, 1526–1536. [Google Scholar] [CrossRef]
- Addeo, A.; Friedlaender, A.; Banna, G.L.; Weiss, G.J. TMB or not TMB as a biomarker: That is the question. Crit. Rev. Oncol. Hematol. 2021, 163, 103374. [Google Scholar] [CrossRef]
- Mo, S.F.; Cai, Z.Z.; Kuai, W.H.; Li, X.; Chen, Y.T. Universal cutoff for tumor mutational burden in predicting the efficacy of anti-PD-(L)1 therapy for advanced cancers. Front. Cell Dev. Biol. 2023, 11, 1209243. [Google Scholar] [CrossRef]
- Maio, M.; Ascierto, P.A.; Manzyuk, L.; Motola-Kuba, D.; Penel, N.; Cassier, P.A.; Bariani, G.M.; De Jesus Acosta, A.; Doi, T.; Longo, F.; et al. Pembrolizumab in microsatellite instability high or mismatch repair deficient cancers: Updated analysis from the phase II KEYNOTE-158 study. Ann. Oncol. 2022, 33, 929–938. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, A.; Li, D.; Yuan, Q.; Zhu, A.; Deng, J.; Wang, Y.; Liu, J.; Liang, C.; Li, W.; et al. Development of T follicular helper cell-independent nanoparticle vaccines for SARS-CoV-2 or HIV-1 by targeting ICOSL. NPJ Vaccines 2024, 9, 176. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Khayyamian, S.; Hutloff, A.; Büchner, K.; Gräfe, M.; Henn, V.; Kroczek, R.A.; Mages, H.W. ICOS-ligand, expressed on human endothelial cells, costimulates Th1 and Th2 cytokine secretion by memory CD4+ T cells. Proc. Natl. Acad. Sci. USA 2002, 99, 6198–6203. [Google Scholar] [CrossRef]
- Martin-Orozco, N.; Li, Y.; Wang, Y.; Liu, S.; Hwu, P.; Liu, Y.J.; Dong, C.; Radvanyi, L. Melanoma cells express ICOS ligand to promote the activation and expansion of T-regulatory cells. Cancer Res. 2010, 70, 9581–9590. [Google Scholar] [CrossRef]
- Strauss, L.; Bergmann, C.; Szczepanski, M.J.; Lang, S.; Kirkwood, J.M.; Whiteside, T.L. Expression of ICOS on human melanoma-infiltrating CD4+CD25highFoxp3+ T regulatory cells: Implications and impact on tumor-mediated immune suppression. J. Immunol. 2008, 180, 2967–2980. [Google Scholar] [CrossRef]
- Gigliotti, C.L.; Dianzani, C.; Stoppa, I.; Monge, C.; Sutti, S.; Sblattero, D.; Puricelli, C.; Rolla, R.; Dianzani, U.; Boggio, E. Differential Modulation of Human M1 and M2 Macrophage Activity by ICOS-Mediated ICOSL Triggering. Int. J. Mol. Sci. 2023, 24, 2953. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kanungo, R.; Hippargi, S.B. CD64 Expression on Neutrophils (nCD64) as a Biomarker in Adult Patients with Sepsis: A Cross-Sectional Study. Cureus 2024, 16, e71912. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Takai, T. A novel recognition system for MHC class I molecules constituted by PIR. Adv. Immunol. 2005, 88, 161–192. [Google Scholar] [CrossRef] [PubMed]
- Hawthorne, B.C.; Engel, S.; McCarthy, M.B.R.; Cote, M.C.; Mazzocca, A.D.; Coyner, K.J. Biologic Adjuvants to Rotator Cuff Repairs Induce Anti-inflammatory Macrophage 2 Polarization and Reduce Inflammatory Macrophage 1 Polarization In Vitro. Arthroscopy 2024. Epub ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.B.; Sullivan, R.J.; Ott, P.A.; Carlino, M.S.; Khushalani, N.I.; Ye, F.; Guminski, A.; Puzanov, I.; Lawrence, D.P.; Buchbinder, E.I.; et al. Ipilimumab Therapy in Patients with Advanced Melanoma and Preexisting Autoimmune Disorders. JAMA Oncol. 2016, 2, 234–240. [Google Scholar] [CrossRef] [PubMed]
- Maurer, D.M.; Adamik, J.; Santos, P.M.; Shi, J.; Shurin, M.R.; Kirkwood, J.M.; Storkus, W.J.; Butterfield, L.H. Dysregulated NF-κB-Dependent ICOSL Expression in Human Dendritic Cell Vaccines Impairs T-cell Responses in Patients with Melanoma. Cancer Immunol. Res. 2020, 8, 1554–1567. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Müller, E.; Christopoulos, P.F.; Halder, S.; Lunde, A.; Beraki, K.; Speth, M.; Øynebråten, I.; Corthay, A. Toll-Like receptor ligands and interferon-γ synergize for induction of antitumor M1 macrophages. Front. Immunol. 2017, 8, 1383. [Google Scholar] [CrossRef]
- Khunger, A.; Piazza, E.; Warren, S.; Smith, T.H.; Ren, X.; White, A.; Elliott, N.; Cesano, A.; Beechem, J.M.; Kirkwood, J.M.; et al. CTLA-4 blockade and interferon-α induce proinflammatory transcriptional changes in the tumor immune landscape that correlate with pathologic response in melanoma. PLoS ONE 2021, 16, e0245287. [Google Scholar] [CrossRef] [PubMed]
- Hazra, A.; Gogtay, N. Biostatistics Series Module 6: Correlation and Linear Regression. Indian J. Dermatol. 2016, 61, 593–601. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lê Cao, K.A.; Boitard, S.; Besse, P. Sparse PLS discriminant analysis: Biologically relevant feature selection and graphical displays for multiclass problems. BMC Bioinform. 2011, 12, 253. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).