Abstract
The incidence and mortality of hepatocellular carcinoma (HCC) in Sub-Saharan Africa is projected to increase sharply by 2040 against a backdrop of limited diagnostic and therapeutic options. Two large South African-based case control studies have developed a serum-based miRNome for Hepatitis B-associated hepatocellular carcinoma (HBV-HCC), as well as identifying their gene targets and pathways. Using a combination of RNA sequencing, differential analysis and filters including a unique molecular index count (UMI) ≥ 10 and log fold change (LFC) range > 2: <−0.5 (p < 0.05), 91 dysregulated miRNAs were characterized including 30 that were upregulated and 61 were downregulated. KEGG analysis, a literature review and other bioinformatic tools identified the targeted genes and HBV-HCC pathways of the top 10 most dysregulated miRNAs. The results, which are based on differentiating miRNA expression of cases versus controls, also develop a serum-based miRNA diagnostic panel that indicates 95.9% sensitivity, 91.0% specificity and a Youden Index of 0.869. In conclusion, the results develop a comprehensive African HBV-HCC miRNome that potentially can contribute to RNA-based diagnostic and therapeutic options.
1. Introduction
Hepatocellular carcinoma (HCC) remains an intractable global problem against a background of rising incidence and changing etiology [1]. In 2020, liver cancer mortality was ranked in the top three across 46 countries, with a prediction that 1.3 million people would die from this disease by 2040, indicating a 56.4% increase from 2020. In Sub-Saharan Africa (SSA), 38,600 new cases of liver cancer were estimated, with west Africa as the leading region (17,600/ASR 8.4), followed by east Africa (12,300/ASR 5.0), middle Africa (6 100/ASR 6.1) and southern Africa (2600/ASR 4.6) [2]. Viral infection with hepatitis B remains a primary risk factor in this region [3,4,5] and chronic hepatitis B virus (HBV) infection in SSA has been etiologically implicated in 43% to 80% of total HCC incidence in this region (1,2,3). High HBsAg seroprevalence (>5%) levels also currently persist in multiple SSA countries and South Africa, in particular [6,7].
MicroRNA (miRNA) are often regarded as ancillary epigenetic regulators that act as post-transcriptional gene silencers [8]. These miRNAs collectively repress target mRNA expression in order to ensure homeostasis, and their dynamic fluctuating role is constantly changing as a result of gene transcription and environmental fluctuations [9]. In the HBV-HCC continuum, from asymptomatic HBV infection to the onset of HBV-HCC pathogenesis, an increasing number of miRNAs become dysregulated due to viral infection, epigenetic changes [10], inflammation [11], fibrosis [12], cirrhosis [13] and finally, the onset of HCC. In particular, the HBx protein modulates miRNA expression to influence both its own, as well as its host’s, genome expression in all of the HCC cancer and immune pathways [14].
Despite the early promise of RNA therapeutics and diagnostics as a magic bullet to diagnose early-stage cancer, as well as to modulate aberrant signaling in cancer, this field remains a work-in-progress with regard to their diagnostic and therapeutic potential [15]. Although RNA-based detection of disease is highly predictive, this has not translated into any commercial application, and RNA inhibition-based therapeutics (RNAi) have been confronted with RNA delivery problems and host immunogenic issues [15]. These problems are illustrated by a lack of successful RNAi clinical trials and FDA-approved drugs for the treatment of HCC [16]. Nevertheless, RNAi offers great potential as a therapeutic option for cancer and RNA-based therapeutics are now a reality for the prevention of viral diseases (COVID-19). This paper presents a comprehensive profile of HCC serum miRNA and their gene targets in a South African case study involving high levels of hepatitis B infection and, to the best of our knowledge, is the first comprehensive serum-based mIRNome study on HCC in Africa. Using a sample selected from two case control studies, our specific objectives were to (1) classify significantly dysregulated mappable miRNA, (2) identify their targets, (3) identify the HCC pathways that they modulate and (4) investigate potential miRNA diagnostic panels.
2. Results
The results focus on the top 10 upregulated miRNAs and their validated targets (Table 1) and the top 10 downregulated miRNAs and their validated targets (Table 2) but specifically provide supplementary data with respect to all significantly dysregulated miRNAs above a threshold of a unique molecular index (UMI) ≥ 1, LFC > 2 and log fold change (LFC) < −0.5 (p < 0.05) (Supplementary Table S1) and 91 dysregulated miRNAs and their validated targets above a threshold of unique molecular index (UMI) ≥ 10, LFC > 2 and log fold change (LFC) < −0.5 (p < 0.05) (Supplementary Table S2). The results then separately discuss significantly dysregulated miRNAs in terms of log fold change before illustrating validated KEGG HCC pathways of the top 10 upregulated and downregulated miRNAs in terms of mean UMI (Figure 1 and Figure 2). Finally, the results present selected miRNA panels as diagnostic agents before selecting and presenting the optimum panel based on its sensitivity and specificity, as well as a heatmap of its associations with a range of disease and cellular processes (Table 3, Figure 3 and Figure 4).

Table 1.
Top 10 upregulated miRNA UMI ≥ 10.

Table 2.
Top 10 downregulated miRNAs ≥ 10 UMI.
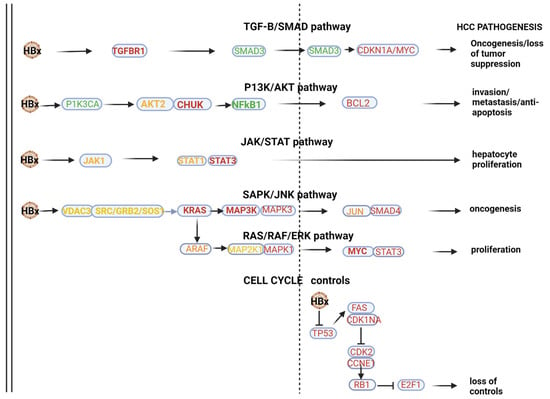
Figure 1.
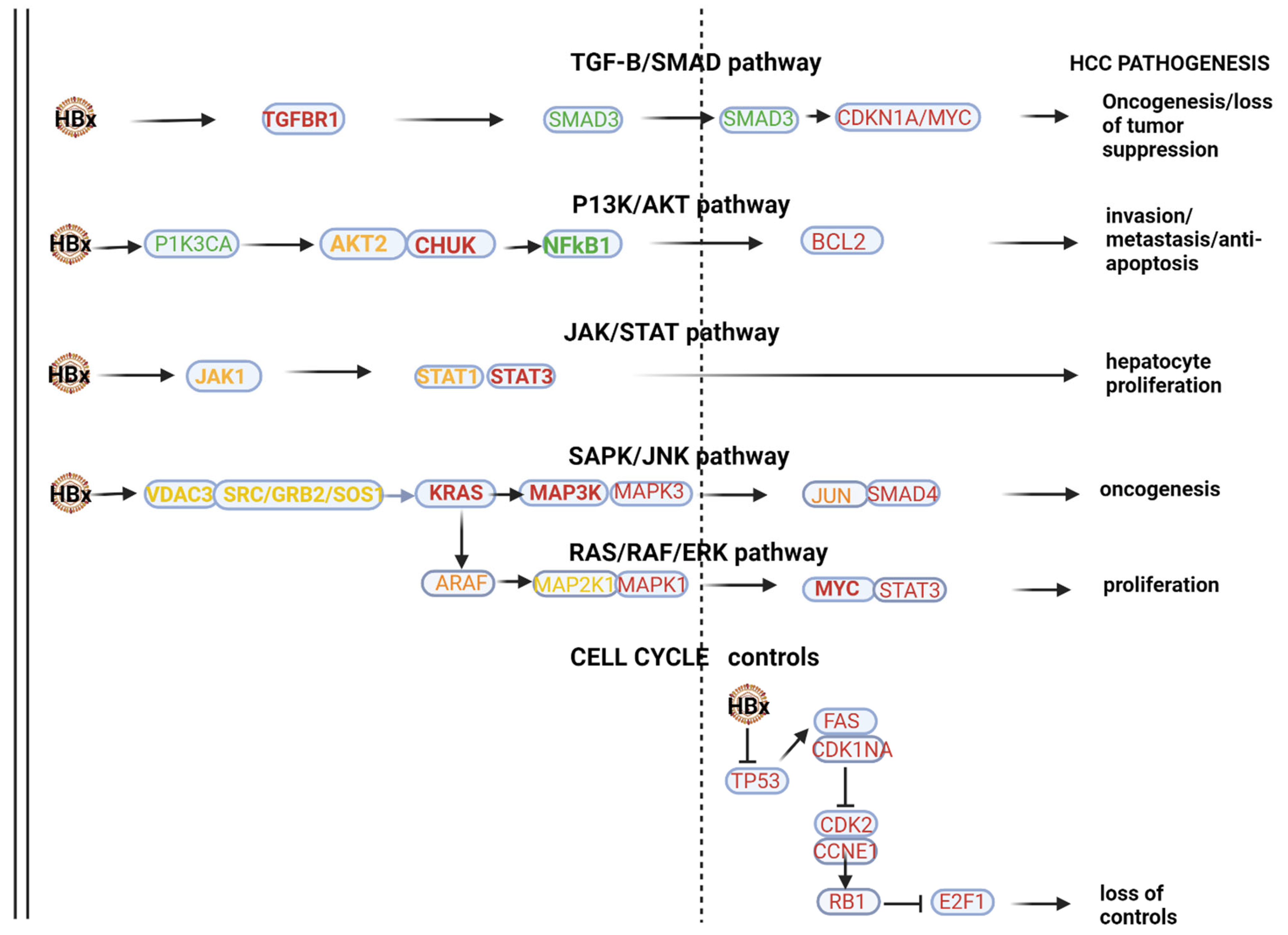
HBV-HCC pathways of top 10 downregulated miRNAs. In the HBx dysregulated HBV-HCC pathways, the top 10 downregulated miRNAs modulate expression in the TGF-B/SMAD pathway by targeting validated gene targets like TGFBR1 and SMAD3/CDKN1A/MYC to influence both oncogenesis and the loss of tumor suppression. In the P13K/AKT pathway, the top 10 downregulated miRNAs target PIK3A and AKT2/CHUK to influence NF-kB1 promotion of BCL2 to influence invasion, metastasis and anti-apoptosis, and in the JAK/STAT pathway, this miRNA panel targets STAT1/3 to influence hepatocyte proliferation. MAPK signaling is targeted by the top 10 downregulated miRNAs by influencing the expression of VDAC3/SRC and GRB2/SOS/KRAS to influence the MAP3K/MAP3/JUN and SMAD4 to influence oncogenesis in the SAPK/JNK. Similarly, MAPK signaling is influenced in the RAS/RAF/ERK pathway by this panel of miRNA targeting ARAF/MAP2KT/MAPK1/MYC and STAT3 to promote cell proliferation. Validated targets in cell cycle controls that are also dysregulated by the HBx protein include TP53/FAS/CDKN1A/CDK2, CCNE1/RBI and E2F1 to influence loss of cell cycle controls. See Table 2 and the Discussion section for further verification of the validated associations in HBV-HCC pathogenesis.
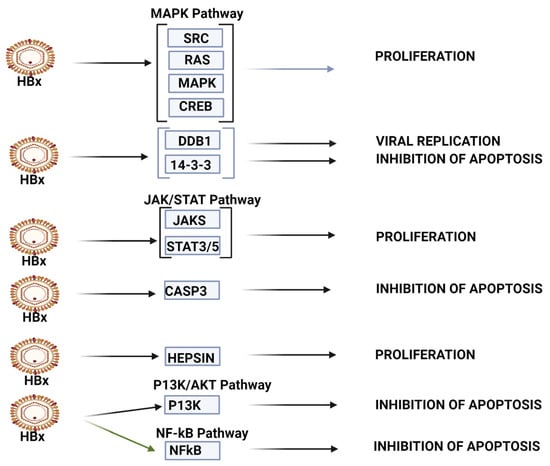
Figure 2.
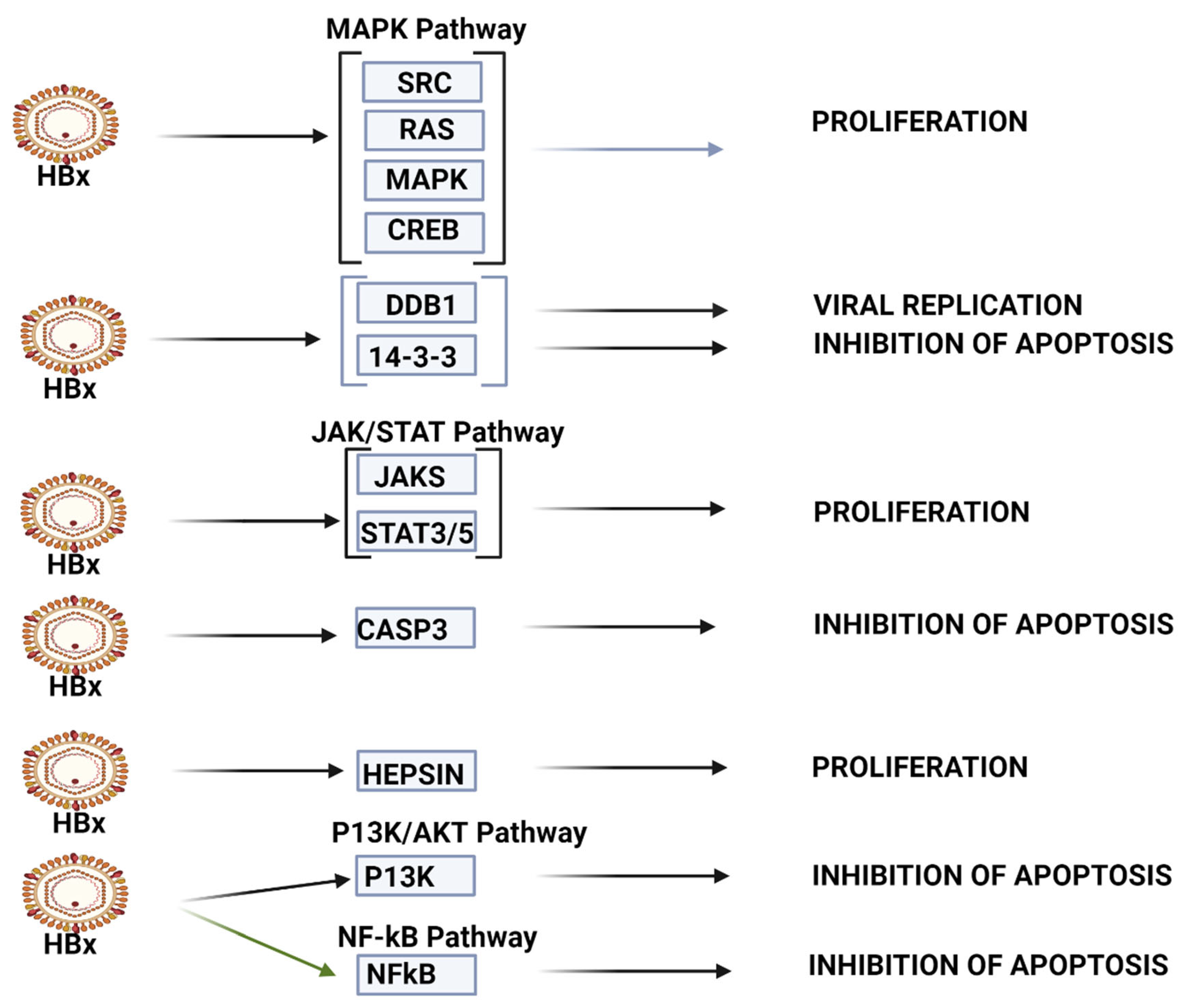
HBV-HCC pathways of top 10 upregulated miRNA. The top 10 upregulated miRNAs influence viral carcinogenesis primarily by modulating cell proliferation and apoptosis. This panel can modulate cell proliferation in MAPK pathways by targeting SRC/RAS/MAPK/CREB, as well as by targeting JAK/STAT3/5 in the JAK/STAT signaling pathway. This panel appears to be able to modulate apoptosis by targeting 14-3-3, CASP3 and p53, as well as P13K in the P13K/AKT pathway and NF-kB in NF-kB signaling (See Table 1 and the Discussion section for further verification of direct HCC role).

Table 3.
Potential diagnostic miRNA panels.
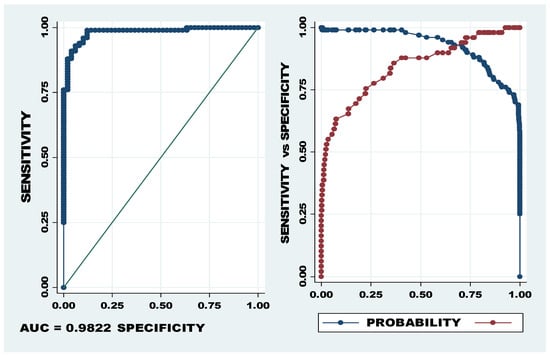
Figure 3.
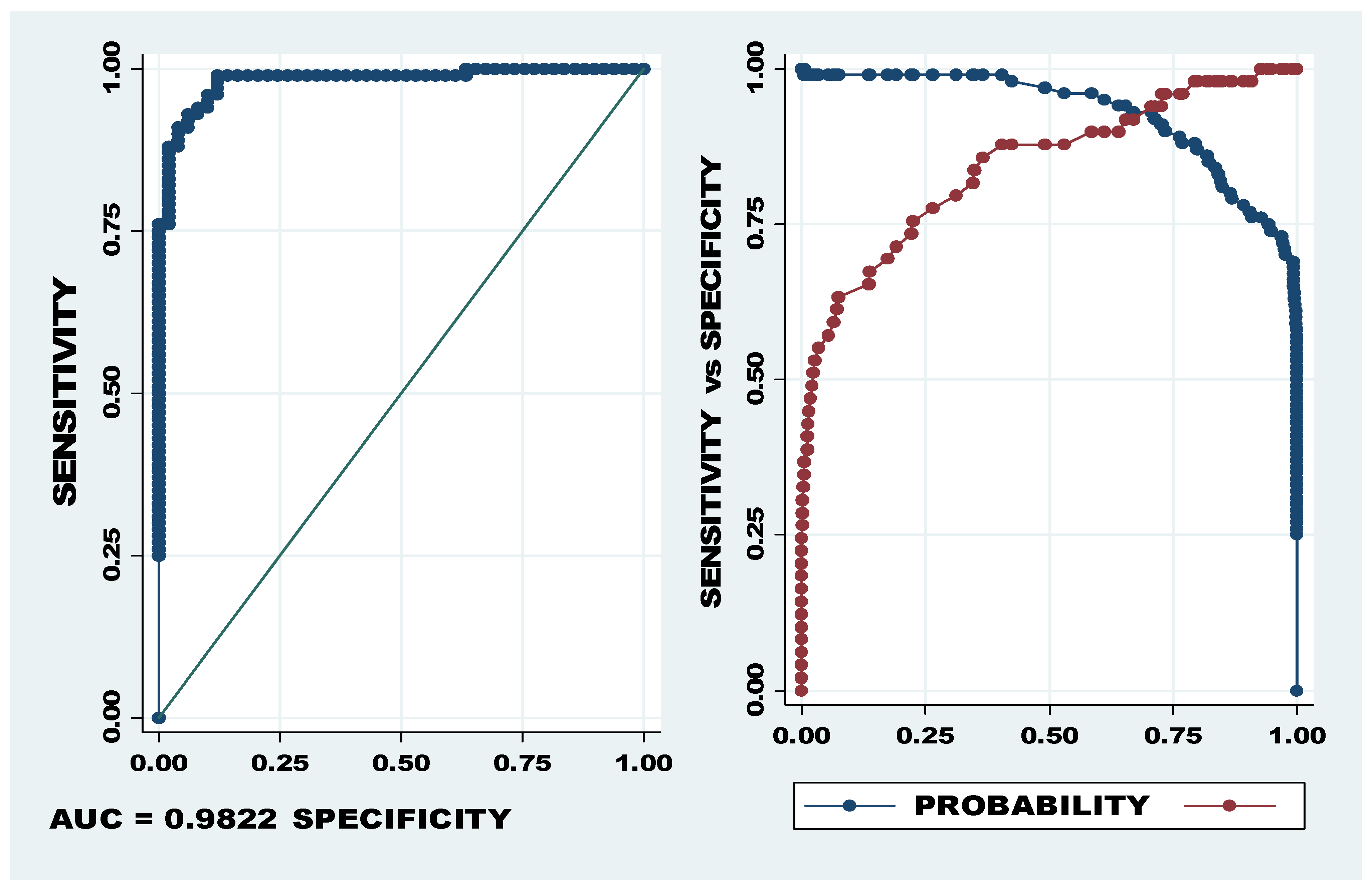
AUC and probability cut-off of let-7a-5p, miR-320b/c. The first graph (LHS) showing sensitivity on the Y axis and specificity on the X axis depicts the AUC of 98.22% and the second graph shows the optimum cut-off point between sensitivity (blue) and specificity (red). The optimum cut-off, indicated as probability cut-off, in the second graph (RHS) indicates a specificity of 95.9% and specificity of 91.0%.
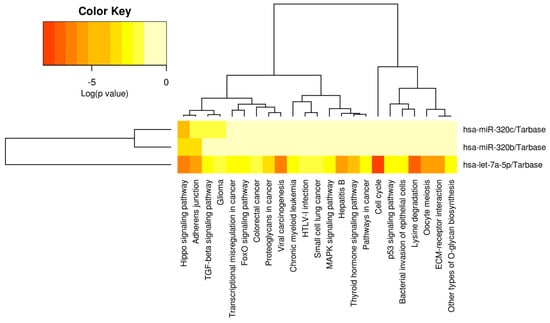
Figure 4.
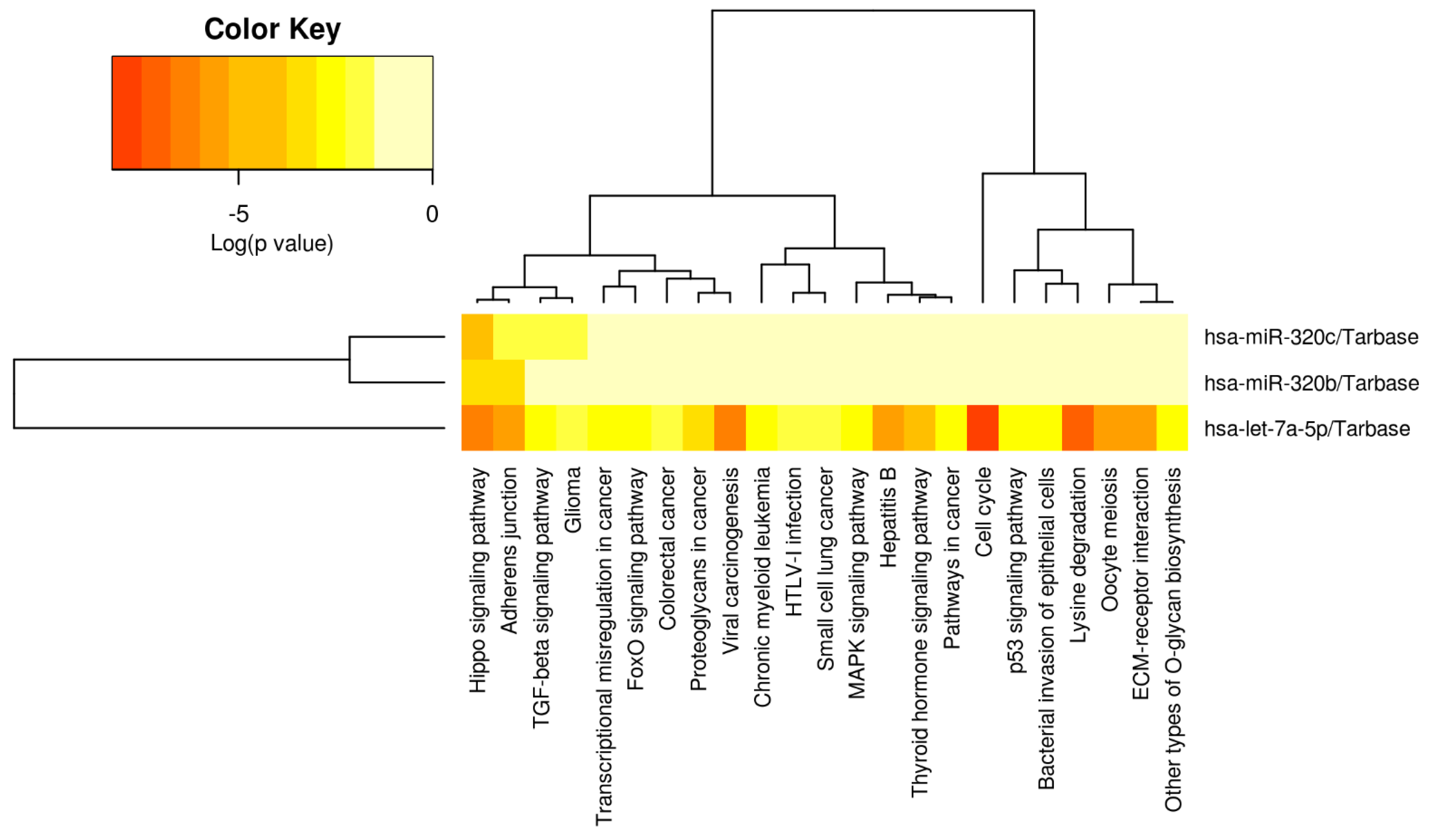
Heatmap of diagnostic miRNA let-7a-5p, miR-320b/c. From left to right, the miRPATHv3 results indicate that let7a-5p is strongly associated with cell cycle, lysine degradation, viral carcinogenesis, the Hippo signaling pathway, Hepatitis B, the Adherens Junction Oocyte meiosis and ECM–receptor interaction (see Table 2/the Discussion section for further validation of direct HCC role). Let-7a-5p has also been associated with the modulation of TGF-B/SMAD signaling, Glioma, transcriptional irregularity in cancer, Fox-O signaling pathway, colorectal cancer, Proteoglycans in cancer, chronic myeloid Leukemia, HTLV-1-infection, small-cell lung cancer, MAPK signaling pathway, p53 signaling, the bacterial invasion of epithelial cells and other types of O-glycan synthesis. The full panel of let-7a-5p, miR-320b/c is only significantly associated with Hippo signaling.
2.1. Top 10 Dysregulated miRNAs Exceeding an UMI ≥ 10
The top 10 upregulated miRNAs with a mean UMI ≥ 10 (see Table 1) ranged between a mean UMI expression of 344 (miR-130b) and 65596 (miR-122-5p), and a mean LFC ranged from 2.018 (miR-320b) to 5.142 (miR-4532). Validated targets have only been included in this section to enhance the reliability of the results but are further developed in the discussion. Validated HCC targets of miR-122-5p, the most widely expressed liver miRNA, include β-CAT/ CCNG1/53 /HNF4α and BCL-W. The second highest upregulated miRNA, miR-483-5p, modulated expression of PPARα/TIMP2 and CDK15 (see Table 1).
The top 10 downregulated miRNAs ranged from a mean UMI of 2691 (miR-191-5p LFC −0.569) to a mean UMI of 57609 (miR-16-5p LFC −0.54). Although the most downregulated miRNAs in terms of absolute UMI expression was miR-16-5p, the let-7-5p family members (a-b-f-i), collectively, were the most downregulated miRNAs in terms of both FC (−0.922 to −1.236) and UMI (9798–17716). Interestingly, other significantly downregulated Let-7-5p members (d/e/g) indicated no direct validated target genes. The most dysregulated miRNAs in terms of both UMI and LFC was miR-483-3p (UMI = 44961; LFC 4.89), suggesting that it could potentially play a major modulating role in HBV-HCC pathogenesis.
In terms of LFC and UMI ≤ 10, significantly upregulated miRNAs not already reported in Table 1 were miR-373-3p (UMI = 6.0; LFC 8.422), miR-1290 (UMI = 9.0; LFC 6.375) and miR-4492 (UMI = 5.0; LFC 5.521). Two of these miRNAs in previous HCC studies demonstrated mIR-373-3p targeting TFAP4/CDH1/CSDC2 and PPP6C [72,73,74,75], and miR-1290 targeting SMEK1/FOXC1/SLU7/GLIPR1. The 10 top downregulated miRNAs in terms of LFC ranged between miR-1306-3p (UMI = 2.0; LFC −2.705) modulating FBXL5 [76] and miR-1275 (UMI = 8.0 LFC −2.007) targeting IGFBP1/2/3/E1F5A2 [77,78,79,80,81] and, in general, were less referenced, except for mIR-491-5p (UMI = 3.0; LFC −2.488) targeting PKM2 and SEC61A1/EGFR [82,83,84] and miR-221-5p (UMI = 2.0; LFC −2.146) targeting ITGB5/GCDH and CD44-TGF-B1 [85,86,87,88] (see Supplementary Table S2).
2.2. Downregulated miRNA Pathways
KEGG pathway analysis (mirPATHv4), which only highlights validated gene targets in specific pathways, indicated that the 10 most downregulated miRNAs (mIR-191-5p, miR-26b-5p, miR-146a-5p, miR-142-3p, miR-126-3p, let-7i-5p, let-7f-5p, let-7a-5p, let-7b-5p and miR-16-5p) indicate viral oncogenesis as the top ranked association. This panel can influence HBx-induced HCC pathogenesis via modulating multiple pathways (see Figure 1). In the HBx dysregulated HBV-HCC pathways, this panel of MIRNAs can influence a number of genes in the TGF-B/SMAD pathway by targeting validated gene targets like TGFBR1/SMAD3/, CDKN1A and MYC to influence both oncogenesis and the loss of tumor suppression. In the P13K/AKT pathway, the top 10 downregulated miRNAs target PIK3A/AKT2/CHUK to influence NF-kB1 promotion of BCL2 to influence invasion, metastasis and anti-apoptosis, and in the JAK/STAT pathway, this miRNA panel targets STAT1/3 to influence hepatocyte proliferation. MAPK signaling is targeted by the top 10 downregulated miRNAs by influencing the expression of VDAC3/SRC/GRB2/SOS and KRAS to influence MAP3K, MAP3/JUN and SMAD4 to influence oncogenesis in the SAPK/JNK. Similarly, MAPK signaling is influenced in the RAS/RAF/ERK pathway by this panel of miRNA targeting ARAF/MAP2KT/MAPK1/MYC and STAT3 to promote cell proliferation. Validated targets in cell cycle controls that are also dysregulated by the HBx protein include TP53/FAS, CDKN1A/CDK2, CCNE1/RBI and E2F1 to influence the loss of cell cycle controls. Our Table 2, which only provides some validated gene targets, does not list many of the validated gene targets highlighted by miRPATHv4 KEGG analysis. It is important to stress that downregulated miRNAs mostly promote oncogenesis by failing to regulate oncoprotein expression (e.g., MYC) but can also moderate oncogenesis by reduced modulation of tumor suppressor expression (e.g., TP53).
The miRPATHv4 results further indicate that the complexity of the interaction of these top 10 downregulated miRNAs is emphasized by their additional role of promoting immune evasion in the innate immune system by targeting the MyD88 dependent and independent pathways. This panel also modulates innate immune evasion by modulating the MYD88 pathway by targeting MYD88/TIRAP/IRAK1/TAB2/MAPK3K7/CHUK/MAPK2K1/MAPK2K3/MAPK2K4/MAPK1/MAPK14/MAPK8/JUN/TLR4/TICAM/MAVS and TRAF3 /TBK1/IRF3 to influence immune evasion.
2.3. Upregulated miRNA Pathways
KEGG bioinformatic analysis (mirPATHv4), which identifies validated targets and pathways in HCC, also indicates that the top 10 upregulated miRNAs (miR-130b-5p, miR-320d, miR-483-3p, miR-1246, miR-320b, miR-192-5p, miR-4532, miR-320c, miR-483-5p and miR-122-5p) also influence expression in multiple HCC pathways (see Figure 2). These miRNAs appear to be less linked with viral carcinogenesis than the top 10 downregulated miRNAs. In the P13K/AKT pathway, this upregulated miRNA panel can target IGF2/IGF1R/PIK3CB/AKT3/RPS6KB to influence survival, while in the p53 signaling pathway, it can target CDK4/CCND1/E2F3 to modulate G1/S progression. In the TGF-B signaling pathway, these miRNAs target SMAD2/4 to influence the loss of inhibitory effect to promote oncogenesis. In the WNT signaling pathway, the miRNA set targets WNT2 and FZD5/LRP6/DVL3/GSK3B /LEF/MYC/CCND1 to influence proliferation and differentiation, and in the MAPK signaling pathway, these miRNAs target PIK3CB/AKT3 and MTOR/RPS6KB to modulate survival. The miRPathv4-identified targets are not necessarily listed in our Table 1; miR-122-5p, for example, has 32 gene targets regarding proteoglycans in cancer and 16 targets regarding TGF-B/SMAD signaling, and our Table 1 only lists 7 targets for this miRNA in HCC studies.
2.4. Diagnostic miRNA
The results, indicating that potential diagnostic miRNA panels differentiate cases from controls, showed that five miRNA panels, namely, let-7a-5p and miR-1246, let-7f-5p and miR-4532, let-7a-5p and miR-320c, miR-122-5p and let-7a-5p, and miR-320c/b and let-7a-5p, all developed an AUC > 90% (see Table 3). All five miRNA panels recorded AUCs > 90%, indicating p-values < 0.01 and an UMI level > 500 (see Table 2). These miRNA panels included miRNAs with the greatest UMI and LFC that ranged between an UMI of 693 and 65596 and an LFC of 2.018 and 5.142 (upregulated) and an LFC of −1.155 and −1.236 (downregulated).
The specificity and sensitivity of the miRNA panel of let-7a-5p, miR-320b/c indicated the most powerful predictive ability, with 95.9% sensitivity and 91.0% specificity at the cut-off of 0.727 (see Figure 3). The Youden Index for this miRNA panel was 0.869184, confirming high levels of sensitivity and specificity.
A heatmap was then developed for the selected miRNA panel (see Figure 4, miRPATHv4), indicating that let-7a-5p has been significantly associated with multiple types of pathogenesis, cell processes and signaling pathways. Only the Hippo signaling pathway is modulated by all three of the miRNA panels, while let-7a-5p is strongly associated with the modulation of the Adherens Junction, viral carcinogenesis, hepatitis B, thyroid hormone signaling, cell cycle, oocyte meiosis, lysine degradation and ECM–receptor interaction.
Further analysis, using miRTargetLink2.0 and miRPATHv4, illustrates 64 strongly validated gene targets (Supplementary Table S3), with hsa-Let-7a-5p targeting 45 genes (<0.001), hsa-miR-320b targeting 10 genes (<0.001) and has-miR-320c targeting 9 genes (<0.001), respectively. Further analysis of the selected diagnostic panel indicates that it strongly targets DICER1, CASP3, MYC, CDK6, NRAS, EZH2, TRIM71, HMGA2, EGFR, STAT3, HRAS and KRAS (https://mpd.bioinf.uni-sb.de/ accessed on 10 October 2023). KEGG pathway analysis indicates that the selected miRNA panel can specifically target SMAD3/CDKN1A/MYC in the TGF-B/SMAD pathway to influence tumor suppression and oncogenesis, as well as target CASP3 in the P13K/AKT pathway to influence apoptosis. This panel also modulates TP53 signaling by targeting CDKN1A, as well as STAT3 in the JAK/STAT pathway to influence proliferation in both of these pathways. In the MAPK pathway, this panel can also influence proliferation by modulating NRAS and MYC/STAT3 expression (https://diana-lab.e-ce.uth.gr/app/miRPathv4 accessed on 10 October 2023) (also see Supplementary Tables S1 and S2 and Table 1 and Table 2).
3. Discussion
The results present a profile of serum miRNA dysregulation in 98 HBV-HCC cases versus 48 controls, of which 30 miRNAs were significantly upregulated and 61 were significantly downregulated. In this section, we briefly compare these results with similar studies in other settings before discussing their modulatory role in the HBV-HCC pathogenesis continuum, specifically examining their role in inflammation, fibrogenesis, carcinogenesis, angiogenesis, apoptosis, HCC pathogenesis, metastasis and migration–invasion, as well as making specific reference to their epigenetic and immune-modulatory functions. Finally, this section discusses the potential role of serum-based miRNAs as diagnostic agents.
Comparative studies: In a comparative serum miRNome study, 214 miRNA families were listed; the let-7/98/4458/4500, miR-320abcd/4429, miR-378/422a/378bcdefhi and miR-17/17-5p/20ab/20b-5p/93/106ab/427/518a-3p/519d families were the most represented families. Similar to our results, this study indicated that miR-122-5p was highly elevated and miR-320a/b/c was among the top ranked families in HCC patients [27]. In a miRNome study focusing on the most abundantly expressed miRNAs in liver tissue, miR-122-5p, let-7a-5p, let-7b-5p and let-7f-5p were listed in the top five [89], thus corresponding with our results which indicated abundant expression in HCC serum. Interestingly, this study also indicates that miR-199a/b-3p, the 11th most downregulated miRNA in our results, is a potential therapeutic agent because it can target tumor-promoting PAK4 to suppress HCC growth by inhibiting the PAK4/Raf/MEK/ERK pathway. Interestingly, in a study involving dysregulated HCC miRNAs in tumor tissue in the absence of viral infection, 86 miRNAs were identified as significantly dysregulated at a cut-off of log2 (FC) > 1 which included 26 upregulated and 60 downregulated miRNAs [90]. In this study, none of the top 10 upregulated or downregulated miRNAs in terms of LFC coincide with our results.
HBV infection and inflammation: The literature demonstrates that two important dysregulated miRNA family members in our results, the let-7 and miR-122 families, play a significant role in modulating both the expression of the HBV virion in host hepatocytes as well inflammation. A number of studies indicate that let-7a/b/c/d/e/f/g/i are frequently downregulated by the HBx protein in early-stage infection in the PME and therefore can fail to modulate inflammatory proteins like Il-6 and IL-10 [91,92]. Since the literature confirms early-stage miRNA dysregulation, our results, which reflect a range of downregulated let-7-5p family members in (late stage) HBV-HCC serum, suggest that these miRNAs might remain consistently dysregulated throughout the HBV-HCC continuum; however, this needs to be validated by additional studies. Similarly, many studies indicate that miR-122 family members can be upregulated in early-stage pathogenesis [93,94], and both miR-122-5p and miR-151a-3p have been identified as biomarkers for chronic hepatitis B infection (CHB) [95]. Our results, reflecting later stage HCC, indicate that miR-122-5p/3p were highly upregulated, posing the question as to whether this miRNA family remains upregulated in both the PME and tumor microenvironment. Similarly, our results suggest that miR-15a [96,97,98], mIR-17 [91] and miR-182 [99] family members can be downregulated in early HBV infection and in late-stage HCC. Our results also indicate that upregulated miRNAs like miR-192-5p could be upregulated in early-stage HBV infection [100,101], as well as in later stage HBV-HCC serum targeting XIAP/TRIM44, SEMA3A and FABP3/YY1/PABPC4 [29,30]. Conversely, miR-151a-3p, which has been cited as a potential biomarker, can be upregulated in early-stage HBV infection [95], whereas our results indicate that this miRNA is significantly downregulated and targets ATM [102]. Interestingly, the same miRNA can select different targets in different stages of pathogenesis, and downregulated miR-15a, for instance, fails to repress TGF-B/SMAD7 signaling in early infection [96,97,98], whereas our results indicate that downregulated miR-15a-5p can also repress GLI2/IGF1/PD1/eIF4E/BDNF/E2F3 in HBV-HCC serum [103,104,105,106,107]. Our results indicate that miRNA dysregulation is dynamic and that it can remain the same or change from early-stage HBV infection and inflammation to the onset of carcinogenesis, with the literature indicating different validated gene targets along the HBV-HCC continuum.
Fibrogenesis: Hepatic fibrosis is characterized by the accumulation of extracellular matrix (ECM) depositions, changes in the hepatic architecture, scarring and increases in fibrous tissue, liver stiffness and alterations in the vasculature in response to a changing microenvironment [108]. A central event initiating liver fibrogenesis is the activation of hepatic stellate cells (HSCs) [109,110]. Multiple miRNAs that modulate fibrogenesis become dysregulated in this pre-malignant environment (PME) and upregulated miR-122-5p, for example, has been correlated with the degree of fibrogenic damage [95,111,112]. Again, our results suggest that this miRNA, which becomes dysregulated in the fibrogenic PME, can remain dysregulated in HBV-HCC serum. Similarly, our results indicate that upregulated miR-140-5p is also upregulated in cirrhosis [113] and downregulated miR-151a-3p is also downregulated in the PME as a response to fibrogenesis [95]. Interestingly, miR-151a-3p is also cited as a biomarker for liver injury caused by CHB [95]. Conversely, upregulated miR-125a-5p targeting F1H1 in fibrogenesis [114,115] is downregulated in our results, targeting PTPN1/MAP3K11/SIRT7/VDR/MACC1/LASP1 and ErbB3 [115,116,117,118,119,120,121], suggesting that miRNA expression can be differentially manipulated in hepatocarcinogenesis. Similarly, miR-142-3p, which has been reported as upregulated in fibrogenesis in the PME [113], is one of our top 10 downregulated miRNAs targeting LDHA/HMGB1/ RAC1, ZEB1/CD133 and SLC3A2 in HBV-HCC [122,123,124,125,126,127]. The results emphasize the dynamic nature and complexity of miRNA-induced regulation in the HBV-HCC continuum, indicating that many of the same miRNAs may be differentially dysregulated by fibrogenesis in the PME vs. the tumor environment, or alternatively, that they remain consistently dysregulated in the same direction but modulate different targets. Further validation of these statements is required.
HBV-HCC pathogenesis: HBV-HCC pathways typically include aberrant miRNA expression in the RB1-TP53 suppressor networks, the WNT pathway, PI3K/MAPK pathways and the JAK/STAT pathway [128,129]. HBV infection dysregulates a wide range of host gene expressions by initiating deletions, amplifications, mutations and epigenetic changes or by targeting microRNA loci or their transcription factors [130]. Typically, dysregulated miRNAs enhance onco-protein expression or repress tumor suppressor activation in the HCC pathways [131] and therefore play a modulatory role in cell proliferation, apoptosis, angiogenesis, invasion, migration and metastasis, as well as modulation of the immune system and the epigenetic machinery. Although multiple miRNAs modulate HBV-HCC pathogenesis, two ubiquitous liver miRNA families in our results, namely, miR-122 and the let-7 family members, illustrate how miRNAs modulate multiple cellular functions and processes in the HBV-HCC continuum. The miR-122 family modulates proliferation, invasion and apoptosis in HCC pathogenesis by targeting PBF, ADAM10/Cyclin G1 and Igf1R/ADAM 17/BCL-W/NDRG3 [39,40,97,132,133], as well by influencing EMT, cell migration, invasion and metastasis [97,134]. Similarly, the tumor suppressor let-7-5p family members play an important modulatory role in multiple human cancers including HCC by repressing oncogenic targets such as EGFR/LIN28B/ HMGA2 and C-MYC that are all important players in oncogenesis [135,136]. Let-7 family members can also modulate angiogenesis/growth/migration and inflammation, as well as modulate TP53 tumor suppressor function by targeting STAT3/RAS/HMGA2/MYC/IL-6/IL-10/TLR-4/COL1A2 and NGF /BCL-XL [64,92,136,137]. Collectively, our results suggest that 91 significantly dysregulated miRNAs with an UMI ≥ 10 probably play a significant modulatory role in HBV-HCC pathogenesis by influencing cell proliferation, angiogenesis, migration and invasion, and metastasis; however, further validation is suggested to test individual miRNA targets and their effect.
Cell proliferation: KEGG pathway analysis indicated that the dysregulated miRNAs in our results can influence cell proliferation in HBV-HCC pathogenesis. The upregulated tumor suppressor miR-9-3p, for instance, can attenuate cell proliferation by repressing oncogenic ERK1/2, AKT and B-CAT expression [138]. Conversely, upregulated miR-452-5p, which is cited as being consistently elevated in HCC tissue [139], can promote cell proliferation and the progression of HCC by targeting COLEC10 [140]. Downregulated miR-142-5p, on the other hand, fails to repress cell proliferation by its reduced modulation of LDHA that is often aberrantly expressed in HCC [122]. Downregulated miR-125a-5p also fails to inhibit cell proliferation by failing to mediate the PI3K/AKT/mTOR pathway [119] and ERBB3 [121]. Conversely, upregulated miR-483-5p inhibits cell proliferation and fibrosis by targeting PPARα and TIMP2 [34]. Other miRNAs in our results that might modulate cell proliferation in HBV-HCC include miR-874-3p [141], miR-451a [142,143,144,145,146,147], miR-182-5p [148,149,150,151,152,153], miR-98-5p [154,155,156,157,158], miR-363-3p [159,160,161,162] and miR-339-3p [163,164,165].
Apoptosis: Normal apoptotic response is frequently dysregulated in HBV-HCC pathogenesis, and miRNA dysregulation can both enhance as well as attenuate apoptosis. Upregulated miR-494-3p, for instance, represses apoptosis by targeting the PTEN tumor suppressor and promoting PI3K/AKT expression [166], while upregulated miR-494-3p can promote apoptosis by targeting TRAF3 to attenuate hepatic stellate cell activation [167]. Downregulated miR-26-b-5p also attenuates apoptosis by failing to modulate KPNA2, thus contributing to the inactivation of p53 signaling [49]. Similarly, upregulated miR-192-5p promotes survival and suppresses apoptosis by repressing PABPC4 [30]. Our results reflect multiple miRNAs that might influence apoptosis in the HBV-HCC continuum including miR-199a-3p [168,169,170,171,172,173,174,175,176,177,178], miR-584-5p [179,180,181,182], miR-98-5p [154,155,156,157,158], miR-454-3p [183,184,185,186], miR-4516 [187,188] and miR-483-3p [20,21], but further investigation should validate this.
Angiogenesis: Multiple miRNAs can regulate angiogenesis in order to nurture or restrict the expansion of the vasculature microenvironment. Upregulated miR-34a-5p, for instance, can attenuate angiogenesis and cell proliferation by regulating VEGFA expression [189]. Conversely, upregulated miR-210-3p promotes angiogenesis by targeting SMAD4 and STAT6 [190] and downregulated miR-26b-5p fails to repress angiogenesis because of its reduced ability to modulate the expression of E-CAD/SNAIL1/MMP2 [48]. In another study ranking the top 10 downregulated miRNAs in HCC with/without vascular invasion, miR-126-3p (log2FC of −3.36) was an important regulator of ADAM9 expression in HBV-HCC [57,191]. Other miRNAs in our results that might influence angiogenesis in the HBV-HCC continuum include miR-494-3p [166,167,192,193,194], miR-126-3p, miR-146a-5p, miR-199a-3p and miR-491 [195].
Migration and invasion: Multiple miRNAs modulate the expansion of the tumor microenvironment that is underpinned by the migration and invasion of cancer cells. Dysregulated miRNAs in our results indicate that downregulated miR-16-5p, for instance, can fail to repress the invasion and migration of HCC cells by its reduced modulation of IGF1R protein expression [68]. Conversely, upregulated miR-34a-5p suppresses the invasion and metastasis of liver cancer by targeting the transcription factor YY1 to mediate MYCT1 upregulation [196]. Interestingly, hypoxia-related miR-210-5p and miR-210-3p can regulate hypoxia-induced migration and epithelial–mesenchymal transition in hepatoma cells [197]. Conversely, downregulated miR-26b-5p fails to repress epithelial–mesenchymal transition (EMT), migration and invasion by its reduced repression of SMAD1 [47]. Other miRNAs in our results that might influence migration and invasion in HBV-HCC pathogenesis include miR-452-3p [139,140,198,199,200], miR-625-3p [201,202] and miR-15a-5p [103,104,105,106,107].
Metastasis: Multiple miRNAs modulate the ability of HBV-HCC-related metastasis. Downregulated miR-142-3p, for instance, fails to inhibit the metastasis of hepatocellular carcinoma cells by its reduced repression of HMGβ1 gene expression [123]. Similarly, downregulated miR-126-3p also fails to suppress tumor metastasis and angiogenesis of hepatocellular carcinoma by its reduced ability to repress LRP6 and PIK3R2 expression [55]. Downregulated miR-451a also fails to repress metastasis and EMT because of its reduced modulation of YWHAZ and ADAM10 [143,144], and upregulated miR-494-3p can enhance HCC metastasis by targeting BMAL1 [192]. Other miRNAs in our results that might potentially influence metastasis in HCC pathogenesis include miR-96-5p [151,203,204,205], miR-1246 [22,23,24,25,26] and miR-210-3p [190,197,206,207].
miRNA as an ancillary epigenetic system: In the HBV-HCC continuum, the HBx protein can influence epigenetic changes like DNA methylation, histone modifications and Polycomb proteins that can dysregulate a specific subset of miRNAs often labeled as epi-miRNAs [8]. In turn, miRNA expression can modulate downstream epigenetic machinery, suggesting epigenetic feedback loops that involve upstream epigenetic manipulation of miRNA expression that can then target downstream epigenetic targets [97,208,209]. In the HBV-HCC continuum, HBx-dysregulated miRNAs, therefore, are ancillary epigenetic regulators that can be subject to upstream epigenetic modulation. In the HBV-HCC continuum, DNA methylation can downregulate miR-1/-122/-124/-132/-/148/-200/-205 [210,211]. Conversely, histone acetylation or HDAC inhibitors can upregulate miR-224/-29/-155/-17-92, and histone methylation can downregulate Let-7c/miR-101/-125b/-139-5p [97]. Our results reflected in Table 1 and Table 2 suggest that a number of these miRNAs interact with the epigenetic machinery, as well as play other roles in HBV-HCC pathogenesis. For example, miR-140-3p can modulate DNA methyltransferase 1 (DNMT1) which results in changes in NF-kB signaling to promote carcinogenesis in HBV-HCC [212]. Interestingly, it has been widely reported that the HBx protein can directly use epigenetic machinery to promote NF-kB signaling and promote hepatocarcinogenesis [213]. Other examples of upstream epigenetic machinery modulating miRNA expression includes the histone H3 lysine 27 (H3K27) tri-methylating enzyme, an enhancer of Zeste homolog 2 (EZH2) that can be recruited by the HBx protein to downregulate miR-139 family members like miR-139-5p [214] (see Supplementary Table S1).
miRNA modulation of immune response: The innate and adaptive immune systems are influenced by an elaborate network of genes whose expression is controlled by extracellular signaling, epigenetic modifiers, transcription and splicing factors, translational protein modifiers and an extensive network of miRNAs [215]. In HBV-HCC pathogenesis, a single miRNA can play multiple roles in both the innate and adaptive immune systems. Two miRNA families in our results that modulate immune response, namely, miR-155 and miR-34a family members, can both be dysregulated by the HBx protein [14]. The miR-155 family is a multifunctional miRNA that plays a crucial role in the modulation of physiological and pathological processes such as hematopoietic lineage differentiation, immunity, inflammation and cancer [216]. This miRNA is expressed in a variety of immune cell types, including B cells, T cells, macrophages, dendritic cells (DCs) and progenitor/stem cell populations. Normally, the miR-155 family is found at low levels in most of these cells types until their activation by immune stimuli, such as antigens, Toll-like receptor (TLR) ligands and inflammatory cytokines, which rapidly increase miR-155 expression [217]. This miRNA, therefore, has an important role in regulating cytokine production and inflammation, as well as in modulating myeloid and lymphoid differentiation [218]. In the immune system, miR-155 is unique in its ability to shape the transcriptome of activated myeloid and lymphoid cells [219]. In virally driven modulation of the immune system, the HBx protein can also repress p53-stimulated miR-34 expression in hepatocytes, leading to the upregulation of a macrophage-derived chemokine (CCL22) that stimulates regulatory T cells (Tregs) which, in turn, represses effector T cell expression, thus allowing HBV expression to increase [97,220]. Upregulated p53-induced miR-34a is also reported to suppress FOXP1, resulting in the inhibition of pro-B cell to pre-B cell transition [221]. Other highly dysregulated miRNAs in our results, like the let-7 family members, play a role in HBV-HCC pathogenesis in the innate immune system [92], and in particular, let-7e and miR-146a can modulate macrophage output, as well as other miRNAs like miR-155 and miR-9 [215]. In addition, miR-146a can modulate Toll-like receptor 4 (TLR4) signaling that has an important role in regulating innate and adaptive immune responses [222]. The most abundantly expressed liver miRNA, miR-122, can also influence immune response by modulating interferon [223], and miR-142-5p can influence T cell response in HCC pathogenesis by modulating PD-L1 expression [224].
miRNAs as diagnostic agents: Our results developed a tentative proposal of alternate miRNA panels that need to be further tested alongside multiple studies that have also profiled diagnostic miRNA panels in HBV-HCC serum. Our results proposed combinations of let-7a-5p and miR-1246, let-7f-5p and miR-4532, miR-320c and let-7a-5p, miR-122-5p and let-7a-5p and the selected panel, namely, miR-320b/c and let-7a-5p, that showed the highest AUC of 0.9822. Of these three miRNAs, only let-7a-5p is supported by multiple studies indicating that let-7a-5p can modulate cell growth, migration and invasion, cell proliferation and apoptosis, and importantly, that it is associated with hepatis B and viral carcinogenesis, as indicated in Figure 4 [60,61,62,63,64]. In a Chinese cohort where 74.8% of patients were HBV positive, three serum-exosome-derived miRNAs (miR-122-5p, let-7d-5p and miR-425-5p) with an AUC of 0.905 were identified as promising biomarkers for identifying HCC patients [225]. In a study in India, let-7a was also listed as a key potential biomarker in a sample where 27% of cases were HBV positive and 24% were HCV positive [226]. Interestingly, let-7d-5p is also listed as a potential biomarker for HCV-HCC and NAFLD [227,228], while miR-425-5p is a well-referenced miRNA in HCC that can promote invasion, metastasis and cell proliferation. This miRNA has also been cited by other studies as a potential diagnostic agent [229,230,231,232]. In our results using miR-122-5p and Let-7a-5p, we developed an ROC of 0.9420, and when we tested for a combination of miR-122-5p, let-7d-5p and miR-425-5p, we developed an ROC of 0.9241, but miR-425-5p was not significant. In other miRNA biomarker studies, miR-122-5p has been listed as a key potential biomarker [19,233]. In a meta study of HBV-HCC miRNA biomarkers combining twenty-three studies from China and two from India, miR-125b emerged as the most promising biomarker for HBV-HCC [234]. Conversely, our results for miR-125b-1-3p, miR-125b-2-3p and miR-125b-5p showed that they are not significant biomarkers, and miR-125b-5p, for instance, indicated an AUC of 0.4340. In the same meta study, miR-126b-3p and miR-142-3p were listed as a potential biomarkers, and this was confirmed by our results that indicated that miR-126b-3p, the 6th most downregulated miRNA, was a significant potential biomarker, with an AUC of 0.8020. This sort of contradiction illustrates the wide number of different outcomes for the same miRNA in different studies attempting to develop definitive serum-based miRNA biomarkers. We also confirmed that miR-142-3p was a potential biomarker, with an ROC of 0.8696, thus illustrating that multiple miRNAs are potential candidates in our results that have been separately tested in other studies [234]. This is echoed in the literature in which highly comparable miRNA studies with the same objectives yield very different results [235] because multiple variables like age, gender, ethnicity, sample preparation, and detection and quantification of miRNAs can differ across identical studies, thus illustrating the dynamic nature of miRNA expression.
4. Materials and Methods
This section outlines the reagents and resources (Table 4), resource availability, the experimental model, the patient details (Table 5), method details and statistical and biostatistical analysis.

Table 4.
Resources and reagents used for methods.

Table 5.
Sample characteristics.
4.1. Resource Availability
4.1.1. Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact.
4.1.2. Material Availability
Serum or RNA generated in this study are available from the Lead Contact with a completed Material Transfer Agreement (some samples are depleted).
4.1.3. Data and Code Availability
The total microRNA-seq datasets illustrating total reads and total UMI per characterized miRNA × 148 sample is currently located in Supplementary Table S2.
4.2. Experimental Model and Patient Details
4.2.1. Patients and Study Sample
The sample of human subjects involved HCC cases (n = 98) that were drawn from two different case control studies, namely, from consenting patients that were diagnosed in the Johannesburg Cancer Case Control Study (JCCCS) [7], a study that recruited self-identified black African (not mixed ancestry) cancer patients in the greater Johannesburg area, Gauteng province, South Africa, and consenting HCC cases that were collected by the oncology departments of Inkosi Albert Luthuli Central Hospital (IALCH) and Greys hospital in Durban and Pietermaritzburg, Kwazulu-Natal, South Africa, respectively. Controls (n-48) included non-HBV infected, otherwise healthy, patients, that were all drawn from the UKZN-based study. Ethics approval for this study was received from the Biomedical Research Ethics Committee (BREC) at the University of KwaZulu-Natal (BREC Reference Number: BE059/15) and the University of the Witwatersrand Human Research Ethics Committee (Medical) (M150239; M140271). All study participants were required to sign an informed consent form before participating in the studies. Patient confidentiality was ensured by recording only case numbers.
4.2.2. Clinical Data Collection
Classification regarding the presence of current HBV infection was not exactly the same in the two case studies. In the JCCCS study, the presence of current infection measured HBV DNA and HBcAb, and its data regarding HBsAg positive and HBeAg positive status was not recorded for many of their HCC cases. Conversely, the UKZN dataset shows HBsAg/HBeAg/HBcAb positive data for every patient but not the presence or absence of HBV-DNA. The two studies thus recorded current HBV infection either by indicating HBsAg or HBV DNA positive status. The evidence of past infection across both studies recorded HBcAb status, as well as well as gender, age and HCV infection. The data indicate that 54% of controls were HBcAb positive and 2% were HBsAg positive (see Table 5). Conversely, the HCC cases indicate that 61% of respondents were either HBsAg or HBV-DNA positive, as well as HBcAb positive, suggesting that a significant majority of HCC patients were either currently infected with hepatitis B or had been exposed to hepatitis B infection that had resolved.
4.3. Method Details
4.3.1. Blood Sample Collection
Blood samples were collected from 146 consenting patients using an 18–20 gauge syringe needle and placed in BD Vacutainer serum collection tubes before being centrifuged at 2500× g for 20 min at room temperature within 60 min. The serum supernatant was extracted and frozen as 500 uL aliquots and stored at −80 °C.
4.3.2. RNA Extraction
We extracted circulating RNA from 200 μL of serum using the miRNeasy Serum/Plasma Advanced Kit (Qiagen, Redwood City, CA, USA) according to the manufacturer’s protocol. For analysis of the complete plasma RNA, buffer RPL was mixed with the plasma sample to ensure efficient lysis. The kit included buffer RPP, which precipitates contaminants such as plasma proteins, and the kit employed a protocol based on spin column technology. The sample was first applied to the spin column to bind total RNA, and any unwanted analytes or contaminants were removed in subsequent washing and centrifugation steps before the samples were eluted in the final centrifugation step. The HCC serum (200 µL) was mixed with buffer RPL to release and stabilize RNA from plasma proteins and extracellular vesicles. The sample was then mixed with buffer RPP and centrifuged to precipitate proteins. Isopropanol was added to the supernatant to provide the appropriate conditions for RNA molecules (>18 nucleotides) to bind to the silica membrane. The sample was then applied to the RNeasy UCP MinElute spin column, where RNA binds to the membrane and other contaminants are washed away in subsequent wash steps. In the final step, total RNA (>18 nucleotides) was eluted using RNase-free water.
4.3.3. miRNA Sequence
The miRNA sequencing libraries were prepared using the QIAseq miRNA Library Kit. Briefly, adapters were ligated sequentially to the 3′ and 5′ ends of miRNAs before universal cDNA synthesis with UMI assignment. This sequencing, using modified oligonucleotides, eliminated the presence of adapter dimers in the sequencing library and effectively removed a major contaminant often observed during sequencing [236]. The miRNA-seq libraries were amplified by 22 cycles of PCR, during which a unique sample index was added to each sample. The amplified DNA fragments were subjected to double size selection using QIAseq magnetic beads to select the molecules with sizes ranging from 150 to 200 bp. The libraries were quantified using Qubit 3.0 HS dsDNA assay, and the library quality was examined using BioAnalyzer. The miRNA-seq libraries were sequenced at 75 bp single-end reads on an Illumina NovaSeq 6000 (Illumina, San Diego, CA, USA). The 146 samples were sequenced on one Novaseq S2 flowcell with a 100 bp single read run, and one Novaseq SP flowcell with a 100 bp single read run and one 50 bp single read run for barcode QC.
4.3.4. miRNA-seq Data Analysis
All sequencing data were demultiplexed and converted into fastq files by Illumina bcl2fastq (v2.20). The reads were mapped and counted using the primary analysis procedure of Qiagen online miRNA-seq analysis software (https://geneglobe.qiagen.com/sg/analyze, accessed on 10 October 2023) (Version 23.1). Briefly, reads were trimmed off the 3′ adaptor and the low-quality bases and those shorter than 16 bp and less than 10 UMI counts were excluded from the analysis. Alignment was performed using Bowtie with a maximum of two mismatches. Aligned reads were annotated using miRBase (v21). After the removal of duplicates, UMI counts with more than 10 reads were used for miRNA differential expression analysis. UMI read counts for each miRNA was used in the analysis to ensure an accurate reflection of the original miRNA template amount before PCR amplification.
4.4. Statistical and Biostatistical Analysis
The mean unique molecular index (UMI) counts and differential miRNA UMI expression for each miRNA for every HCC case (n = 98) and control (n= 48) was initially calculated using Primary QIAseq miRNA Quantification Data Analysis software (version 2) on the basis of classifying and establishing an UMI count for every classified miRNA nucleotide sequence that has been verified and recorded for every known miRNA [237]. The initial results were assembled on an excel spreadsheet indicating UMI counts for 840 classified miRNAs.
The Ideal R/Bioconductor package for interactive differential expression analysis was deployed to ascertain the differential UMI expression between HCC cases and controls of 840 classified serum-based miRNAs (https://bioconductor.org/packages/ideal/ accessed on 10 October 2023) [238]. The initial count matrix produced by the QIASeq mIRNA Library kit software (Version 2), was normalized before using a negative binomial generalized linear model (GLM) in the R/Bioconductor package that filtered the differential expression of UMI to exclude p-value > 0.05. Using a cut-off ≥ 1 for the UMI and an LFC > 2.00 or <−0.50 and with an adjusted p-value < 0.05, we identified 148 dysregulated miRNAs, with 49 (33%) upregulated and 99 (67%) downregulated (see Supplementary Table S1). A further cut-off of ≥ 10 for the UMI reduced this to 30 upregulated miRNAs and 61 downregulated miRNAs (see Table 2).
Further biostatistical analysis of authenticated miRNA targets and HBV-HCC pathways was investigated by deploying miRPATHv4 (https://diana-lab.e-ce.uth.gr/app/miRPathv4, accessed on 10 October 2023) [239,240].
The identification of potential miRNAs as diagnostic agents was developed from the top 10 most upregulated and downregulated miRNAs (UMI and LFC), as well as from testing additional nine miRNAs that were significantly dysregulated in our results based on their being identified in other studies investigating serum-based miRNAs as diagnostic agents for HBV-HCC detection. A total of 29 potential miRNA candidates were selected, including the top 20 dysregulated miRNAs in terms of UMI and LFC, plus 9 other miRNAs identified in HBV-HCC serum studies as potential biomarkers [71,227,228,230]. These miRNAs were all tested individually for their AUC value using a logistic regression model in which UMI count formed the predictor variable. Each candidate was assessed for its z-score, p-value and its relevant AUC score adhering to a p-value ≤ 0.05 and an AUC ≥ 0.80. Panels of miRNAs were then assembled to evaluate the one with the highest AUC, sensitivity, specificity and their Youden Index (p < 0.05) using Stata 12.0 (http://www.stata.com, accessed on 10 October 2023). A heatmap identifying validated associations with specified signaling pathways, disease and cell processes was developed using miRPATHv4 at https://diana-lab.e-ce.uth.gr/app/miRPathv4, accessed on 10 October 2023) [239,240,241].
5. Conclusions and Limitations
This paper provides a comprehensive study of an African miRNome involving dysregulated miRNAs and their targets in HBV-HCC serum drawn from two South African case control studies and, to the best of our knowledge, is the only comprehensive serum-based miRNome study on HCC in Sub-Saharan Africa. In particular, the results focus on the targets and pathways of 30 significantly upregulated miRNAs and 61 downregulated miRNAs (base mean value ≥ 10 UMI, LFC > 2, <−0.5). In addition, the results investigate selected panels of miRNAs as diagnostic biomarkers before selecting the panel with the highest level of specificity and sensitivity. The validated targets of the 91 highly dysregulated miRNAs support the contention that, collectively, miRNAs play a significant role in the modulation of HBV-HCC pathogenesis and that they can be deployed as accurate diagnostic biomarkers.
The primary limitation of this study is that miRNA dysregulation heterogeneity in the face of multiple variables and stages in hepatocarcinogenesis limits the degree to which miRNA results can be generalized. A multitude of diagnostic miRNAs have been developed, for instance, using different miRNAs, even at the same stage of disease. In addition, the usefulness of our diagnostic miRNA panel study is limited because the patient sample involved late-stage HCC rather than early-stage disease. In conclusion, this study acknowledges that despite the undoubted potential of RNA-based therapeutics and diagnostics, this field largely remains a work-in-progress. The problematic nature of miRNA-based therapeutic and diagnostic agents is illustrated by the fact that no results have been published from 34 clinical trials [242]; nevertheless, significant breakthroughs like mRNA-based vaccines illustrate that RNA-based intervention should continue to be pursued. A further limitation is that the patient characteristics across the two case control studies included some different variables like Bilharzia, albumin, ferritin in the UKZN study and not in the JCCCS study, and smoking, drinking, diabetes in the JCCCS study but not in the UKZN study.
Further validation of the gene targets and their effect on gene expression from HCC liver tissue is required for the top dysregulated miRNAs identified in our study, especially miRNAs like let-7a-5p, miR-1246, miR-4532, miR-320b, miR-320c and miR-122-5p.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms25020975/s1.
Author Contributions
K.S. developed the research question, developed the first draft, and coordinated the project and authors. P.A. coordinated all laboratory procedures that characterized miRNAs, as well reviewed and edited every draft of the paper. B.S. was responsible for differential analysis as well as for reviewing and editing each draft. C.W. and A.C. provided technical guidance, as well as reviewed, revised and edited all drafts of the paper. T.-W.S. and Y.Z. were responsible for RNA analysis and coordination of bioinformatic inputs, as well as a review of the final draft. All authors have read and agreed to the published version of the manuscript.
Funding
The funders had no role in the study design, data collection, analysis, decision to publish or preparation of the paper. This project has been funded in part by federal funds from the Frederick National Laboratory for Cancer Research, National Institutes of Health, under contract HHSN261200800001E, and in part by the Intramural Research Program of NIH, National Cancer Institute, Center for Cancer Research. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does the mention of trade names, commercial products or organizations imply endorsement by the U.S. Government.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki. Ethics approval for this study was received from the Biomedical Research Ethics Committee (BREC) at the University of KwaZulu-Natal, Durban, South Africa (BREC Reference Number: BE059/15) and the University of the Witwatersrand Human Research Ethics Committee (Medical), University of the Witwatersrand, Johannesburg, South Africa (M150239; M140271).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study. No patient details are divulged.
Data Availability Statement
All of the data files generated by the differential analysis software including UMI expression of 840 classified miRNAs in both cases and samples are available on request.
Acknowledgments
This is to acknowledge that our African-based study would never have happened without the help of our dear friend and colleague Cherie (Professor Emeritus Cheryl Winkler) from the NCI, USA.
Conflicts of Interest
The funder was not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication, and as a result of this, all of the authors declare no conflicts of interest. In terms of any conflicts of interest regarding author affiliations, all of the authors declare no conflicts of interest.
References
- Akinyemiju, T.; Abera, S.; Ahmed, M.; Alam, N.; Alemayohu, M.A.; Allen, C.; Al-Raddadi, R.; Alvis-Guzman, N.; Amoako, Y.; Artaman, A. The burden of primary liver cancer and underlying etiologies from 1990 to 2015 at the global, regional, and national level: Results from the global burden of disease study 2015. JAMA Oncol. 2017, 3, 1683–1691. [Google Scholar] [PubMed]
- Rumgay, H.; Arnold, M.; Ferlay, J.; Lesi, O.; Cabasag, C.J.; Vignat, J.; Laversanne, M.; McGlynn, K.A.; Soerjomataram, I. Global burden of primary liver cancer in 2020 and predictions to 2040. J. Hepatol. 2022, 77, 1598–1606. [Google Scholar] [CrossRef]
- Kew, M.C. Hepatocellular carcinoma in African Blacks: Recent progress in etiology and pathogenesis. World J. Hepatol. 2010, 2, 65. [Google Scholar] [CrossRef]
- Kew, M.C.; Yu, M.C.; Kedda, M.A.; Coppin, A.; Sarkin, A.; Hodkinson, J. The relative roles of hepatitis B and C viruses in the etiology of hepatocellular carcinoma in southern African blacks. Gastroenterology 1997, 112, 184–187. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.H.; Cheng, Y.; Zhang, S.; Fan, J.; Gao, Q. Changing epidemiology of hepatocellular carcinoma in Asia. Liver Int. 2022, 42, 2029–2041. [Google Scholar] [CrossRef]
- Ginzberg, D.; Wong, R.J.; Gish, R. Global HBV burden: Guesstimates and facts. Hepatol. Int. 2018, 12, 315–329. [Google Scholar] [CrossRef] [PubMed]
- Mak, D.; Babb de Villiers, C.; Chasela, C.; Urban, M.I.; Kramvis, A. Analysis of risk factors associated with hepatocellular carcinoma in black South Africans: 2000–2012. PLoS ONE 2018, 13, e0196057. [Google Scholar] [CrossRef]
- Sartorius, K.; An, P.; Winkler, C.; Chuturgoon, A.; Li, X.; Makarova, J.; Kramvis, A. The epigenetic modulation of cancer and immune pathways in hepatitis B virus-associated hepatocellular carcinoma: The influence of HBx and miRNA dysregulation. Front. Immunol. 2021, 12, 661204. [Google Scholar] [CrossRef]
- Vidigal, J.A.; Ventura, A. The biological functions of miRNAs: Lessons from in vivo studies. Trends Cell Biol. 2015, 25, 137–147. [Google Scholar] [CrossRef]
- Furuta, M.; Kozaki, K.-I.; Tanaka, S.; Arii, S.; Imoto, I.; Inazawa, J. miR-124 and miR-203 are epigenetically silenced tumor-suppressive microRNAs in hepatocellular carcinoma. Carcinogenesis 2009, 31, 766–776. [Google Scholar] [CrossRef]
- Hatziapostolou, M.; Polytarchou, C.; Aggelidou, E.; Drakaki, A.; Poultsides, G.A.; Jaeger, S.A.; Ogata, H.; Karin, M.; Struhl, K.; Hadzopoulou-Cladaras, M. An HNF4α-miRNA inflammatory feedback circuit regulates hepatocellular oncogenesis. Cell 2011, 147, 1233–1247. [Google Scholar] [CrossRef] [PubMed]
- Murakami, Y.; Yasuda, T.; Saigo, K.; Urashima, T.; Toyoda, H.; Okanoue, T.; Shimotohno, K. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene 2006, 25, 2537. [Google Scholar] [CrossRef]
- Jiang, J.; Gusev, Y.; Aderca, I.; Mettler, T.A.; Nagorney, D.M.; Brackett, D.J.; Roberts, L.R.; Schmittgen, T.D. Association of MicroRNA expression in hepatocellular carcinomas with hepatitis infection, cirrhosis, and patient survival. Clin. Cancer Res. 2008, 14, 419–427. [Google Scholar] [CrossRef] [PubMed]
- Sartorius, K.; Swadling, L.; An, P.; Makarova, J.; Winkler, C.; Chuturgoon, A.; Kramvis, A. The multiple roles of hepatitis B virus X protein (HBx) dysregulated microRNA in hepatitis B virus-associated hepatocellular carcinoma (HBV-HCC) and immune pathways. Viruses 2020, 12, 746. [Google Scholar] [CrossRef]
- Sartorius, K.; Antwi, S.O.; Chuturgoon, A.; Roberts, L.R.; Kramvis, A. RNA therapeutic options to manage aberrant signaling pathways in hepatocellular carcinoma: Dream or reality? Front. Oncol. 2022, 12, 891812. [Google Scholar] [CrossRef] [PubMed]
- Kamochi, J.; Tokunaga, T.; Morino, F.; Nagata, J.; Tomii, Y.; Abe, Y.; Hatanaka, H.; Kijima, H.; Yamazaki, H.; Watanabe, N. Ribozyme mediated suppression of vascular endothelial growth factor gene expression enhances matrix metalloproteinase 1 expression in a human hepatocellular carcinoma cell line. Int. J. Oncol. 2002, 21, 81–84. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Hu, Z.; Qiang, Y.; Zhu, X. Circulating miR-130b-and miR-21-based diagnostic markers and therapeutic targets for hepatocellular carcinoma. Mol. Genet. Genom. Med. 2019, 7, e1012. [Google Scholar] [CrossRef]
- Li, W.; Ding, X.; Wang, S.; Xu, L.; Yin, T.; Han, S.; Geng, J.; Sun, W. Downregulation of serum exosomal miR-320d predicts poor prognosis in hepatocellular carcinoma. J. Clin. Lab. Anal. 2020, 34, e23239. [Google Scholar] [CrossRef]
- Jin, Y.; Wong, Y.S.; Goh, B.K.; Chan, C.Y.; Cheow, P.C.; Chow, P.K.; Lim, T.K.; Goh, G.B.; Krishnamoorthy, T.L.; Kumar, R. Circulating microRNAs as potential diagnostic and prognostic biomarkers in hepatocellular carcinoma. Sci. Rep. 2019, 9, 10464. [Google Scholar] [CrossRef]
- Liu, H.; French, B.A.; Li, J.; Tillman, B.; French, S.W. Altered regulation of miR-34a and miR-483-3p in alcoholic hepatitis and DDC fed mice. Exp. Mol. Pathol. 2015, 99, 552–557. [Google Scholar] [CrossRef]
- Shen, J.; Wang, A.; Wang, Q.; Gurvich, I.; Siegel, A.B.; Remotti, H.; Santella, R.M. Exploration of Genome-Wide Circulating MicroRNA in Hepatocellular Carcinoma: MiR-483-5p as a Potential BiomarkerCirculating MiRNAs in Differentiation of Hepatocellular Carcinoma from Control. Cancer Epidemiol. Biomark. Prev. 2013, 22, 2364–2373. [Google Scholar] [CrossRef] [PubMed]
- Moshiri, F.; Salvi, A.; Gramantieri, L.; Sangiovanni, A.; Guerriero, P.; De Petro, G.; Bassi, C.; Lupini, L.; Sattari, A.; Cheung, D. Circulating miR-106b-3p, miR-101-3p and miR-1246 as diagnostic biomarkers of hepatocellular carcinoma. Oncotarget 2018, 9, 15350. [Google Scholar] [CrossRef] [PubMed]
- Chai, S.; Ng, K.Y.; Tong, M.; Lau, E.Y.; Lee, T.K.; Chan, K.W.; Yuan, Y.F.; Cheung, T.T.; Cheung, S.T.; Wang, X.Q. Octamer 4/microRNA-1246 signaling axis drives Wnt/β-catenin activation in liver cancer stem cells. Hepatology 2016, 64, 2062–2076. [Google Scholar] [CrossRef]
- Chuma, M.; Toyoda, H.; Matsuzaki, J.; Saito, Y.; Kumada, T.; Tada, T.; Kaneoka, Y.; Maeda, A.; Yokoo, H.; Ogawa, K. Circulating microRNA-1246 as a possible biomarker for early tumor recurrence of hepatocellular carcinoma. Hepatol. Res. 2019, 49, 810–822. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.-L.; Fu, Y.-P.; Gan, W.; Liu, G.; Zhou, P.-Y.; Zhou, C.; Sun, B.-Y.; Guan, R.-Y.; Zhou, J.; Fan, J. Hepatic stellate cells promote the progression of hepatocellular carcinoma through microRNA-1246-RORα-Wnt/β-Catenin axis. Cancer Lett. 2020, 476, 140–151. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Meng, C.; Wang, S.; Zhou, N.; Guan, M.; Bai, C.; Lu, S.; Han, Q.; Zhao, R.C. MicroRNA-1246 enhances migration and invasion through CADM1 in hepatocellular carcinoma. BMC Cancer 2014, 14, 616. [Google Scholar] [CrossRef] [PubMed]
- Pascut, D.; Krmac, H.; Gilardi, F.; Patti, R.; Calligaris, R.; Crocè, L.S.; Tiribelli, C. A comparative characterization of the circulating miRNome in whole blood and serum of HCC patients. Sci. Rep. 2019, 9, 8265. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Li, P.; Li, J.; Qi, Q.; Sun, Z.; Shi, S.; Xie, Y.; Liu, S.; Wang, Y.; Du, L. Exosomal and intracellular miR-320b promotes lymphatic metastasis in esophageal squamous cell carcinoma. Mol. Ther. Oncol. 2021, 23, 163–180. [Google Scholar] [CrossRef]
- Ren, F.-J.; Yao, Y.; Cai, X.-Y.; Fang, G.-Y. Emerging role of MiR-192-5p in human diseases. Front. Pharmacol. 2021, 12, 614068. [Google Scholar] [CrossRef]
- Gu, Y.; Wei, X.; Sun, Y.; Gao, H.; Zheng, X.; Wong, L.L.; Jin, L.; Liu, N.; Hernandez, B.; Peplowska, K. miR-192-5p silencing by genetic aberrations is a key event in hepatocellular carcinomas with cancer stem cell features. Cancer Res. 2019, 79, 941–953. [Google Scholar] [CrossRef]
- Wang, R.; Hurley, A.M.; Simon, S.M. A novel blood diagnostic method for fibrolamellar hepatocellular carcinoma. Cancer Res. 2023, 83, 1017. [Google Scholar] [CrossRef]
- Yao, J.; Liang, L.-H.; Zhang, Y.; Ding, J.; Tian, Q.; Li, J.-J.; He, X.-H. GNAI1 suppresses tumor cell migration and invasion and is post-transcriptionally regulated by Mir-320a/c/d in hepatocellular carcinoma. Cancer Biol. Med. 2012, 9, 234. [Google Scholar]
- Zhang, Z.; Ge, S.; Wang, X.; Yuan, Q.; Yan, Q.; Ye, H.; Che, Y.; Lin, Y.; Zhang, J.; Liu, P. Serum miR-483-5p as a potential biomarker to detect hepatocellular carcinoma. Hepatol. Int. 2013, 7, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Niture, S.; Gadi, S.; Qi, Q.; Gyamfi, M.A.; Varghese, R.S.; Rios-Colon, L.; Chimeh, U.; Ressom, H.W.; Kumar, D. MicroRNA-483-5p Inhibits Hepatocellular Carcinoma Cell Proliferation, Cell Steatosis, and Fibrosis by Targeting PPARα and TIMP2. Cancers 2023, 15, 1715. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Lin, W.; Bai, Y.; Liao, Y.; Lin, Q.; Chen, L.; Wu, Y. Identification of exosomal hsa-miR-483-5p as a potential biomarker for hepatocellular carcinoma via microRNA expression profiling of tumor-derived exosomes. Exp. Cell Res. 2022, 417, 113232. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.-Y.; Chen, D.; Gu, X.-Y.; Ding, J.; Zhao, Y.-J.; Zhao, Q.; Yao, M.; Chen, Z.; He, X.-H.; Cong, W.-M. Predicting value of ALCAM as a target gene of microRNA-483-5p in patients with early recurrence in hepatocellular carcinoma. Front. Pharmacol. 2018, 8, 973. [Google Scholar] [CrossRef]
- Huang, J.T.; Liu, S.M.; Ma, H.; Yang, Y.; Zhang, X.; Sun, H.; Zhang, X.; Xu, J.; Wang, J. Systematic Review and Meta-Analysis: Circulating miRNAs for Diagnosis of Hepatocellular Carcinoma. J. Cell. Physiol. 2016, 231, 328–335. [Google Scholar] [CrossRef]
- Xu, J.; Wu, C.; Che, X.; Wang, L.; Yu, D.; Zhang, T.; Huang, L.; Li, H.; Tan, W.; Wang, C. Circulating MicroRNAs, miR-21, miR-122, and miR-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol. Carcinog. 2011, 50, 136–142. [Google Scholar] [CrossRef]
- Wang, S.; Qiu, L.; Yan, X.; Jin, W.; Wang, Y.; Chen, L.; Wu, E.; Ye, X.; Gao, G.F.; Wang, F. Loss of microRNA 122 expression in patients with hepatitis B enhances hepatitis B virus replication through cyclin G1-modulated P53 activity. Hepatology 2012, 55, 730–741. [Google Scholar] [CrossRef]
- Li, C.; Wang, Y.; Wang, S.; Wu, B.; Hao, J.; Fan, H.; Ju, Y.; Ding, Y.; Chen, L.; Chu, X. Hepatitis B virus mRNA-mediated miR-122 inhibition upregulates PTTG1-binding protein, which promotes hepatocellular carcinoma tumor growth and cell invasion. J. Virol. 2013, 87, 2193–2205. [Google Scholar] [CrossRef]
- Wu, Q.; Liu, H.-O.; Liu, Y.-D.; Liu, W.-S.; Pan, D.; Zhang, W.-J.; Yang, L.; Fu, Q.; Xu, J.-J.; Gu, J.-X. Decreased expression of hepatocyte nuclear factor 4α (Hnf4α)/microRNA-122 (miR-122) axis in hepatitis B virus-associated hepatocellular carcinoma enhances potential oncogenic GALNT10 protein activity. J. Biol. Chem. 2015, 290, 1170–1185. [Google Scholar] [CrossRef] [PubMed]
- Peng, F.; Xiao, X.; Jiang, Y.; Luo, K.; Tian, Y.; Peng, M.; Zhang, M.; Xu, Y.; Gong, G. HBx down-regulated Gld2 plays a critical role in HBV-related dysregulation of miR-122. PLoS ONE 2014, 9, e92998. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.-G.; Wang, C.-M.; Tian, C.; Wang, Y.; Li, L.; Sun, W.-S.; Li, R.-F.; Liu, Y.-G. miR-122 inhibits viral replication and cell proliferation in hepatitis B virus-related hepatocellular carcinoma and targets NDRG3. Oncol. Rep. 2011, 26, 1281–1286. [Google Scholar]
- Lee, H.M.; Wong, W.K.K.; Fan, B.; Lau, E.S.; Hou, Y.; Luk, A.O.Y.; Chow, E.Y.K.; Ma, R.C.W.; Chan, J.C.N.; Kong, A.P.S. Detection of increased serum miR-122-5p and miR-455-3p levels before the clinical diagnosis of liver cancer in people with type 2 diabetes. Sci. Rep. 2021, 11, 23756. [Google Scholar] [CrossRef]
- Wu, H.-Y.; Li, M.-W.; Li, Q.-Q.; Pang, Y.-Y.; Chen, G.; Lu, H.-P.; Pan, S.-L. Elevation of miR-191-5p level and its potential signaling pathways in hepatocellular carcinoma: A study validated by microarray and in-house qRT-PCR with 1291 clinical samples. Int. J. Clin. Exp. Pathol. 2019, 12, 1439. [Google Scholar]
- Gao, Y.; Luo, T.; Ouyang, X.; Zhu, C.; Zhu, J.; Qin, X. IGF2BP3 and miR191-5p synergistically increase HCC cell invasiveness by altering ZO-1 expression. Oncol. Lett. 2020, 20, 1423–1431. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, B.; Zhao, X.; Zhao, N.; Sun, R.; Zhu, D.; Zhang, Y.; Li, Y.; Gu, Q.; Dong, X. Twist1-related miR-26b-5p suppresses epithelial-mesenchymal transition, migration and invasion by targeting SMAD1 in hepatocellular carcinoma. Oncotarget 2016, 7, 24383–24401. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, B.; Sun, H.; Zhao, X.; Wang, X.; Zhao, N.; Zhang, Y.; Li, Y.; Gu, Q.; Liu, F. Regulation of proliferation, angiogenesis and apoptosis in hepatocellular carcinoma by miR-26b-5p. Tumor Biol. 2016, 37, 10965–10979. [Google Scholar] [CrossRef]
- Tang, G.; Zhao, H.; Xie, Z.; Wei, S.; Chen, G. Long non-coding RNA HAGLROS facilitates tumorigenesis and progression in hepatocellular carcinoma by sponging miR-26b-5p to up-regulate karyopherin α2 (KPNA2) and inactivate p53 signaling. Bioengineered 2022, 13, 7829–7846. [Google Scholar] [CrossRef]
- Han, W.; Li, N.; Liu, J.; Sun, Y.; Yang, X.; Wang, Y. MicroRNA-26b-5p enhances T cell responses by targeting PIM-2 in hepatocellular carcinoma. Cell. Signal. 2019, 59, 182–190. [Google Scholar] [CrossRef]
- Li, J.-F.; Dai, X.-P.; Zhang, W.; Sun, S.-H.; Zeng, Y.; Zhao, G.-Y.; Kou, Z.-H.; Guo, Y.; Yu, H.; Du, L.-Y. Upregulation of microRNA-146a by hepatitis B virus X protein contributes to hepatitis development by downregulating complement factor H. MBio 2015, 6, e02459-14. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Ye, Z.H.; Liang, H.W.; Ren, F.H.; Li, P.; Dang, Y.W.; Chen, G. Down-regulation of miR-146a-5p and its potential targets in hepatocellular carcinoma validated by a TCGA-and GEO-based study. FEBS Open Bio 2017, 7, 504–521. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Sun, Y.; Lin, H.; Chou, F.; Xiao, Y.; Jin, R.A.; Cai, X.; Chang, C. Olaparib and enzalutamide synergistically suppress HCC progression via the AR-mediated miR-146a-5p/BRCA1 signaling. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2020, 34, 5877. [Google Scholar] [CrossRef] [PubMed]
- Zu, Y.; Yang, Y.; Zhu, J.; Bo, X.; Hou, S.; Zhang, B.; Qiu, J.; Zheng, J. MiR-146a suppresses hepatocellular carcinoma by downregulating TRAF6. Am. J. Cancer Res. 2016, 6, 2502. [Google Scholar] [PubMed]
- Du, C.; Lv, Z.; Cao, L.; Ding, C.; Gyabaah, O.-A.K.; Xie, H.; Zhou, L.; Wu, J.; Zheng, S. MiR-126-3p suppresses tumor metastasis and angiogenesis of hepatocellular carcinoma by targeting LRP6 and PIK3R2. J. Transl. Med. 2014, 12, 259. [Google Scholar] [CrossRef]
- Manganelli, M.; Grossi, I.; Ferracin, M.; Guerriero, P.; Negrini, M.; Ghidini, M.; Senti, C.; Ratti, M.; Pizzo, C.; Passalacqua, R. Longitudinal circulating levels of miR-23b-3p, miR-126-3p and lncRNA GAS5 in HCC patients treated with sorafenib. Biomedicines 2021, 9, 813. [Google Scholar] [CrossRef]
- Xiang, L.-Y.; Ou, H.-H.; Liu, X.-C.; Chen, Z.-J.; Li, X.-H.; Huang, Y.; Yang, D.-H. Loss of tumor suppressor miR-126 contributes to the development of hepatitis B virus–related hepatocellular carcinoma metastasis through the upregulation of ADAM9. Tumor Biol. 2017, 39, 1010428317709128. [Google Scholar] [CrossRef]
- Office, E. Erratum to miR-126-3p contributes to sorafenib resistance in hepatocellular carcinoma via downregulating SPRED1. Ann. Transl. Med. 2022, 10, 1076. [Google Scholar] [CrossRef]
- Yang, H.D.; Kim, H.S.; Kim, S.Y.; Na, M.J.; Yang, G.; Eun, J.W.; Wang, H.J.; Cheong, J.Y.; Park, W.S.; Nam, S.W. HDAC6 suppresses Let-7i-5p to elicit TSP1/CD47-mediated anti-tumorigenesis and phagocytosis of hepatocellular carcinoma. Hepatology 2019, 70, 1262–1279. [Google Scholar] [CrossRef]
- Yang, L.; Si, H.; Ma, M.; Fang, Y.; Jiang, Y.; Wang, J.; Zhang, C.; Xiao, H. LINC00221 silencing prevents the progression of hepatocellular carcinoma through let-7a-5p-targeted inhibition of MMP11. Cancer Cell Int. 2021, 21, 202. [Google Scholar] [CrossRef]
- Liu, L.; Zhao, J.; Peng, Y.; Yang, M.; Zhang, L.; Jin, X. miR-let-7a-5p Inhibits Invasion and migration of hepatoma cells by regulating BZW2 expression. OncoTargets Ther. 2020, 13, 12269–12279. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.-Y.; Tao, S.-Q.; Wang, X.-N.; Lobie, P.E.; Wu, Z.-S. XIAP 3′-untranslated region serves as a competitor for HMGA2 by arresting endogenous let-7a-5p in human hepatocellular carcinoma. Tumor Biol. 2017, 39, 1010428317719578. [Google Scholar] [CrossRef] [PubMed]
- Waly, A.A.; El-Ekiaby, N.; Assal, R.A.; Abdelrahman, M.M.; Hosny, K.A.; El Tayebi, H.M.; Esmat, G.; Breuhahn, K.; Abdelaziz, A.I. Methylation in MIRLET7A3 gene induces the expression of IGF-II and its mRNA binding proteins IGF2BP-2 and 3 in hepatocellular carcinoma. Front. Physiol. 2019, 9, 1918. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lu, Y.; Toh, S.T.; Sung, W.-K.; Tan, P.; Chow, P.; Chung, A.Y.; Jooi, L.L.; Lee, C.G. Lethal-7 is down-regulated by the hepatitis B virus x protein and targets signal transducer and activator of transcription 3. J. Hepatol. 2010, 53, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Peng, F.; Ning, Y.; Jiang, P.; Peng, J.; Ding, X.; Zhang, J.; Jiang, T.; Xiang, S. SNHG16 as the miRNA let-7b-5p sponge facilitates the G2/M and epithelial-mesenchymal transition by regulating CDC25B and HMGA2 expression in hepatocellular carcinoma. J. Cell. Biochem. 2020, 121, 2543–2558. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Ma, Y.; Liu, X.; Yu, P.; Huang, N.; Song, L.; Xu, R.; Huo, Z.; Zhu, T.; Tang, X. Targeted delivery of glypican 3 (GPC3) antibody-modified microRNA (miR let-7b-5p) polymer nanoparticles to sorafenib-resistant hepatsocellular carcinoma cells. J. Biomed. Nanotechnol. 2021, 17, 677–690. [Google Scholar] [CrossRef] [PubMed]
- Ge, W.; Yu, D.-C.; Li, Q.-G.; Chen, X.; Zhang, C.-Y.; Ding, Y.-T. Expression of serum miR-16, let-7f, and miR-21 in patients with hepatocellular carcinoma and their clinical significances. Clin. Lab. 2014, 60, 427–434. [Google Scholar] [CrossRef] [PubMed]
- Cheng, B.; Ding, F.; Huang, C.; Xiao, H.; Fei, F.; Li, J. Role of miR-16-5p in the proliferation and metastasis of hepatocellular carcinoma. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 137–145. [Google Scholar]
- Liu, Z.; Wang, Y.; Wang, L.; Yao, B.; Sun, L.; Liu, R.; Chen, T.; Niu, Y.; Tu, K.; Liu, Q. Long non-coding RNA AGAP2-AS1, functioning as a competitive endogenous RNA, upregulates ANXA11 expression by sponging miR-16-5p and promotes proliferation and metastasis in hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2019, 38, 194. [Google Scholar] [CrossRef]
- Shen, S.; Lin, Y.; Yuan, X.; Shen, L.; Chen, J.; Chen, L.; Qin, L.; Shen, B. Biomarker MicroRNAs for diagnosis, prognosis and treatment of hepatocellular carcinoma: A functional survey and comparison. Sci. Rep. 2016, 6, 38311. [Google Scholar] [CrossRef]
- Fang, Y.; Yan, D.; Wang, L.; Zhang, J.; He, Q. Circulating microRNAs (miR-16, miR-22, miR-122) expression and early diagnosis of hepatocellular carcinoma. J. Clin. Lab. Anal. 2022, 36, e24541. [Google Scholar] [CrossRef]
- Li, H.; Wang, N.; Xu, Y.; Chang, X.; Ke, J.; Yin, J. Upregulating microRNA-373-3p promotes apoptosis and inhibits metastasis of hepatocellular carcinoma cells. Bioengineered 2022, 13, 1304–1319. [Google Scholar] [CrossRef]
- Arzumanyan, A.; Friedman, T.; Kotei, E.; Ng, I.O.; Lian, Z.; Feitelson, M.A. Epigenetic repression of E-cadherin expression by hepatitis B virus x antigen in liver cancer. Oncogene 2012, 31, 563. [Google Scholar] [CrossRef] [PubMed]
- Wu, N.; Liu, X.; Xu, X.; Fan, X.; Liu, M.; Li, X.; Zhong, Q.; Tang, H. MicroRNA-373, a new regulator of protein phosphatase 6, functions as an oncogene in hepatocellular carcinoma. FEBS J. 2011, 278, 2044–2054. [Google Scholar] [CrossRef] [PubMed]
- Cairo, S.; Wang, Y.; de Reyniès, A.; Duroure, K.; Dahan, J.; Redon, M.-J.; Fabre, M.; McClelland, M.; Wang, X.W.; Croce, C.M. Stem cell-like micro-RNA signature driven by Myc in aggressive liver cancer. Proc. Natl. Acad. Sci. USA 2010, 107, 20471–20476. [Google Scholar] [CrossRef]
- He, Z.-J.; Li, W.; Chen, H.; Wen, J.; Gao, Y.-F.; Liu, Y.-J. miR-1306–3p targets FBXL5 to promote metastasis of hepatocellular carcinoma through suppressing snail degradation. Biochem. Biophys. Res. Commun. 2018, 504, 820–826. [Google Scholar] [CrossRef] [PubMed]
- Fawzy, I.O.; Hamza, M.T.; Hosny, K.A.; Esmat, G.; El Tayebi, H.M.; Abdelaziz, A.I. miR-1275: A single microRNA that targets the three IGF2-mRNA-binding proteins hindering tumor growth in hepatocellular carcinoma. FEBS Lett. 2015, 589, 2257–2265. [Google Scholar] [CrossRef]
- Shaalan, Y.M.; Handoussa, H.; Youness, R.A.; Assal, R.A.; El-Khatib, A.H.; Linscheid, M.W.; El Tayebi, H.M.; Abdelaziz, A.I. Destabilizing the interplay between miR-1275 and IGF2BPs by Tamarix articulata and quercetin in hepatocellular carcinoma. Nat. Prod. Res. 2018, 32, 2217–2220. [Google Scholar] [CrossRef]
- Yang, J.; Qin, T.; Liu, S.; Tang, H.; Liu, M.; Wang, Q. Interaction analysis of miR-1275/IGF2BP1/IGF2BP3 with the susceptibility to hepatocellular carcinoma. Biomark. Med. 2020, 14, 283–292. [Google Scholar] [CrossRef]
- Yang, X.; Jiang, W.; Kong, X.; Zhou, X.; Zhu, D.; Kong, L. Genistein Restricts the Epithelial Mesenchymal Transformation (EMT) and Stemness of Hepatocellular Carcinoma via Upregulating miR-1275 to Inhibit the EIF5A2/PI3K/Akt Pathway. Biology 2022, 11, 1383. [Google Scholar] [CrossRef]
- Pascut, D.; Cavalletto, L.; Pratama, M.Y.; Bresolin, S.; Trentin, L.; Basso, G.; Bedogni, G.; Tiribelli, C.; Chemello, L. Serum miRNA are promising biomarkers for the detection of early hepatocellular carcinoma after treatment with direct-acting antivirals. Cancers 2019, 11, 1773. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Dou, C.; Liu, X.; Yang, L.; Ni, C.; Wang, J.; Guo, Y.; Yang, W.; Tong, X.; Huang, D. Oviductus ranae protein hydrolysate (ORPH) inhibits the growth, metastasis and glycolysis of HCC by targeting miR-491-5p/PKM2 axis. Biomed. Pharmacother. 2018, 107, 1692–1704. [Google Scholar] [CrossRef] [PubMed]
- Fa, X.; Song, P.; Fu, Y.; Deng, Y.; Liu, K. Long non-coding RNA VPS9D1-AS1 facilitates cell proliferation, migration and stemness in hepatocellular carcinoma. Cancer Cell Int. 2021, 21, 131. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Zhu, Q.; Shen, J. miRNA-491-5p Inhibited Cell Proliferation in Human Hepatocellular Carcinoma Through Phosphatidylinositol 3-Kinase/Protein Kinase B Signaling Pathway. J. Biomater. Tissue Eng. 2021, 11, 567–572. [Google Scholar] [CrossRef]
- Huang, C.; Li, K.; Huang, R.; Zhu, J.; Yang, J. RNF185-AS1 promotes hepatocellular carcinoma progression through targeting miR-221-5p/integrin β5 axis. Life Sci. 2021, 267, 118928. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Xu, W.; Chen, T. Identification of onco-miRNAs in hepatocellular carcinoma and analysis of their regulatory network. Nan Fang Yi Ke Da Xue Xue Bao = J. South. Med. Univ. 2022, 42, 45–54. [Google Scholar]
- Park, N.R.; Cha, J.H.; Cho, S.W.; Jang, J.W.; Choi, J.Y.; Yoon, S.K.; Bae, S.H. miRNA-221-5p promotes epithelial-mesenchymal transition in hepatocellular carcinoma through regulation of CD44/TGF-beta1. J. Gastroenterol. Hepatol. 2018, 33, 72. [Google Scholar]
- Markovic, J.; Sharma, A.D.; Balakrishnan, A. MicroRNA-221: A fine tuner and potential biomarker of chronic liver injury. Cells 2020, 9, 1767. [Google Scholar] [CrossRef]
- Hou, J.; Lin, L.; Zhou, W.; Wang, Z.; Ding, G.; Dong, Q.; Qin, L.; Wu, X.; Zheng, Y.; Yang, Y. Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell 2011, 19, 232–243. [Google Scholar] [CrossRef]
- Zheng, J.; Sadot, E.; Vigidal, J.A.; Klimstra, D.S.; Balachandran, V.P.; Kingham, T.P.; Allen, P.J.; D’Angelica, M.I.; DeMatteo, R.P.; Jarnagin, W.R. Characterization of hepatocellular adenoma and carcinoma using microRNA profiling and targeted gene sequencing. PLoS ONE 2018, 13, e0200776. [Google Scholar] [CrossRef]
- Li, G.; Cai, G.; Li, D.; Yin, W. MicroRNAs and liver disease: Viral hepatitis, liver fibrosis and hepatocellular carcinoma. Postgrad. Med. J. 2014, 90, 106–112. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Kanda, T.; Wu, S.; Nakamura, M.; Miyamura, T.; Nakamoto, S.; Banerjee, A.; Yokosuka, O. Regulation of microRNA by hepatitis B virus infection and their possible association with control of innate immunity. World J. Gastroenterol. WJG 2014, 20, 7197. [Google Scholar] [CrossRef] [PubMed]
- Akamatsu, S.; Hayes, C.N.; Tsuge, M.; Miki, D.; Akiyama, R.; Abe, H.; Ochi, H.; Hiraga, N.; Imamura, M.; Takahashi, S. Differences in serum microRNA profiles in hepatitis B and C virus infection. J. Infect. 2015, 70, 273–287. [Google Scholar] [CrossRef] [PubMed]
- Waidmann, O.; Bihrer, V.; Pleli, T.; Farnik, H.; Berger, A.; Zeuzem, S.; Kronenberger, B.; Piiper, A. Serum microRNA-122 levels in different groups of patients with chronic hepatitis B virus infection. J. Viral Hepat. 2012, 19, e58–e65. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.-L.; Zhao, H.; Yang, S.-G.; Chen, E.-M.; Chen, W.-Q.; Li, L.-J. Plasma miRNA-122-5p and miRNA-151a-3p identified as potential biomarkers for liver injury among CHB patients with PNALT. Hepatol. Int. 2018, 12, 277–287. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Jiang, L.; Ji, X.; Yang, B.; Zhang, Y.; Fu, X.-D. Hepatitis B viral RNA directly mediates down-regulation of the tumor suppressor microRNA miR-15a/miR-16-1 in hepatocytes. J. Biol. Chem. 2013, 288, 18484–18493. [Google Scholar] [CrossRef]
- Xie, K.-L.; Zhang, Y.-G.; Liu, J.; Zeng, Y.; Wu, H. MicroRNAs associated with HBV infection and HBV-related HCC. Theranostics 2014, 4, 1176. [Google Scholar] [CrossRef]
- Liu, N.; Jiao, T.; Huang, Y.; Liu, W.; Li, Z.; Ye, X. Hepatitis B virus regulates apoptosis and tumorigenesis through the microRNA-15a-Smad7-transforming growth factor beta pathway. J. Virol. 2015, 89, 2739–2749. [Google Scholar] [CrossRef]
- Riazalhosseini, B.; Mohamed, R.; Apalasamy, Y.D.; Langmia, I.M.; Mohamed, Z. Circulating microRNA as a marker for predicting liver disease progression in patients with chronic hepatitis B. Rev. Soc. Bras. Med. Trop. 2017, 50, 161–166. [Google Scholar] [CrossRef]
- Nielsen, K.O.; Jacobsen, K.S.; Mirza, A.H.; Winther, T.N.; Størling, J.; Glebe, D.; Pociot, F.; Hogh, B. Hepatitis B virus upregulates host microRNAs that target apoptosis-regulatory genes in an in vitro cell model. Exp. Cell Res. 2018, 371, 92–103. [Google Scholar] [CrossRef]
- Brunetto, M.R.; Cavallone, D.; Oliveri, F.; Moriconi, F.; Colombatto, P.; Coco, B.; Ciccorossi, P.; Rastelli, C.; Romagnoli, V.; Cherubini, B. A serum microRNA signature is associated with the immune control of chronic hepatitis B virus infection. PLoS ONE 2014, 9, e110782. [Google Scholar] [CrossRef]
- Jiao, W.-P.; Jiao, W.-J.; Guo, X.-J.; Feng, J.-H.; Zhang, J.-Y. Study on the mechanism of action of MiR-151a-3p in alcohol-related liver cancer. J. Hebei Med. Univ. 2021, 42, 1272. [Google Scholar]
- He, Y.; Huang, H.; Jin, L.; Zhang, F.; Zeng, M.; Wei, L.; Tang, S.; Chen, D.; Wang, W. CircZNF609 enhances hepatocellular carcinoma cell proliferation, metastasis, and stemness by activating the Hedgehog pathway through the regulation of miR-15a-5p/15b-5p and GLI2 expressions. Cell Death Dis. 2020, 11, 358. [Google Scholar] [CrossRef] [PubMed]
- Qiu, R.; Zeng, Z. Hsa_circ_0006988 promotes sorafenib resistance of hepatocellular carcinoma by modulating IGF1 using miR-15a-5p. Can. J. Gastroenterol. Hepatol. 2022, 2022, 1206134. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.-Y.; Liang, H.-X.; Wu, S.-H.; Jiang, H.-Q.; Wang, Q.; Yu, Z.-J. Overexpressed tumor suppressor exosomal miR-15a-5p in cancer cells inhibits PD1 expression in CD8+ T cells and suppresses the hepatocellular carcinoma progression. Front. Oncol. 2021, 11, 622263. [Google Scholar] [CrossRef] [PubMed]
- Long, J.; Jiang, C.; Liu, B.; Fang, S.; Kuang, M. MicroRNA-15a-5p suppresses cancer proliferation and division in human hepatocellular carcinoma by targeting BDNF. Tumor Biol. 2016, 37, 5821–5828. [Google Scholar] [CrossRef]
- Li, Y.; Li, D.; Yang, Y.; Wang, J. miR-15a-5p regulates liver cancer cell migration, apoptosis and cell cycle progression by targeting transcription factor E2F3. Crit. Rev. Eukaryot. Gene Expr. 2022, 32, 1–10. [Google Scholar] [CrossRef]
- Dhar, D.; Baglieri, J.; Kisseleva, T.; Brenner, D.A. Mechanisms of liver fibrosis and its role in liver cancer. Exp. Biol. Med. 2020, 245, 96–108. [Google Scholar] [CrossRef]
- Hernandez-Gea, V.; Friedman, S.L. Pathogenesis of liver fibrosis. Annu. Rev. Pathol. Mech. Dis. 2011, 6, 425–456. [Google Scholar] [CrossRef]
- Seki, E.; Schwabe, R.F. Hepatic inflammation and fibrosis: Functional links and key pathways. Hepatology 2015, 61, 1066–1079. [Google Scholar] [CrossRef]
- Wang, W.J.; Lai, R.T.; Lu, J.; Xiang, X.G.; Zhao, G.D.; Tang, W.L.; Cai, W.; Wang, H.; Zhou, H.J.; Xie, Q. Correlation between circulating miR-122 and prognosis of chronic HBV-related liver failure. J. Dig. Dis. 2016, 17, 334–339. [Google Scholar] [CrossRef]
- Tan, Y.; Lin, B.; Ye, Y.; Wen, D.; Chen, L.; Zhou, X. Differential expression of serum microRNAs in cirrhosis that evolve into hepatocellular carcinoma related to hepatitis B virus. Oncol. Rep. 2015, 33, 2863–2870. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yan, X.-L.; Guo, X.-X.; Shi, M.-J.; Lu, Y.-Y.; Zhou, Q.-M.; Chen, Q.-L.; Hu, Y.-Y.; Xu, L.-M.; Huang, S. MiR-27a as a predictor for the activation of hepatic stellate cells and hepatitis B virus-induced liver cirrhosis. Oncotarget 2018, 9, 1075. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Li, J.; Li, C.; Qi, H.; Dong, P.; Zheng, J.; Yu, F. MicroRNA-125a-5p contributes to hepatic stellate cell activation through targeting FIH1. Cell. Physiol. Biochem. 2016, 38, 1544–1552. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Zhou, Z.; Xu, Z.; Li, G.; Dong, P.; Chen, Z.; Lin, D.; Chen, B.; Yu, F. Serum microRNA-125a-5p, a useful biomarker in liver diseases, correlates with disease progression. Mol. Med. Rep. 2015, 12, 1584–1590. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Tao, Y.; Niu, Y.; Wang, Z.; Zhang, C.; Yu, Y.; Ma, L. miR-125a-5p inhibits tumorigenesis in hepatocellular carcinoma. Aging 2019, 11, 7639. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.K.; Noh, J.H.; Jung, K.H.; Eun, J.W.; Bae, H.J.; Kim, M.G.; Chang, Y.G.; Shen, Q.; Park, W.S.; Lee, J.Y. Sirtuin7 oncogenic potential in human hepatocellular carcinoma and its regulation by the tumor suppressors MiR-125a-5p and MiR-125b. Hepatology 2013, 57, 1055–1067. [Google Scholar] [CrossRef]
- Xu, J.; Wang, Y.; Zhang, Y.; Dang, S.; He, S. Astemizole promotes the anti-tumor effect of vitamin D through inhibiting miR-125a-5p-meidated regulation of VDR in HCC. Biomed. Pharmacother. 2018, 107, 1682–1691. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, Q.; Chang, L.; Liu, J. miR-34a and miR-125a-5p inhibit proliferation and metastasis but induce apoptosis in hepatocellular carcinoma cells via repressing the MACC1-mediated PI3K/AKT/mTOR pathway. Neoplasma 2020, 67, 1042–1053. [Google Scholar] [CrossRef]
- Liu, Y.; Geng, X. Long non-coding RNA (lncRNA) CYTOR promotes hepatocellular carcinoma proliferation by targeting the microRNA-125a-5p/LASP1 axis. Bioengineered 2022, 13, 3666–3679. [Google Scholar] [CrossRef]
- Li, G.; Zhang, W.; Gong, L.; Huang, X. MicroRNA 125a-5p inhibits cell proliferation and induces apoptosis in hepatitis B virus-related hepatocellular carcinoma by downregulation of ErbB3. Oncol. Res. 2019, 27, 449. [Google Scholar] [CrossRef] [PubMed]
- Hua, S.; Liu, C.; Liu, L.; Wu, D. miR-142-3p inhibits aerobic glycolysis and cell proliferation in hepatocellular carcinoma via targeting LDHA. Biochem. Biophys. Res. Commun. 2018, 496, 947–954. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Sun, L.-Q.; Huang, Y.; Quan, J.; Hu, X.; Tang, D.; Kang, R.; Li, N.; Fan, X.-G. miR-142-3p inhibits the metastasis of hepatocellular carcinoma cells by regulating HMGB1 gene expression. Curr. Mol. Med. 2018, 18, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Shan, W.-F.; Jin, T.-T.; Wu, G.-Q.; Xiong, X.-X.; Jin, H.-Y.; Zhu, S.-M. Propofol exerts anti-hepatocellular carcinoma by microvesicle-mediated transfer of miR-142-3p from macrophage to cancer cells. J. Transl. Med. 2014, 12, 279. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Liu, Z.; Jin, L.; Zhang, F.; Peng, X.; Xiao, Y.; Wang, X.; Lyu, Q.; Cai, X. lncRNA TUG1-mediated Mir-142-3p downregulation contributes to metastasis and the epithelial-to-mesenchymal transition of hepatocellular carcinoma by targeting ZEB1. Cell. Physiol. Biochem. 2018, 48, 1928–1941. [Google Scholar] [CrossRef] [PubMed]
- Chai, S.; Tong, M.; Ng, K.Y.; Kwan, P.S.; Chan, Y.P.; Fung, T.M.; Lee, T.K.; Wong, N.; Xie, D.; Yuan, Y.-F. Regulatory role of miR-142-3p on the functional hepatic cancer stem cell marker CD133. Oncotarget 2014, 5, 5725. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Yin, Y.; Jiang, J.; Yan, C.; Wang, Y.; Wang, D.; Li, L. Exosomal miR-142-3p secreted by hepatitis B virus (HBV)-hepatocellular carcinoma (HCC) cells promotes ferroptosis of M1-type macrophages through SLC3A2 and the mechanism of HCC progression. J. Gastrointest. Oncol. 2022, 13, 754. [Google Scholar] [CrossRef]
- Schulz, W.A. Molecular Biology of Human Cancers: An Advanced Student’s Textbook; Springer Science & Business Media: Dordrecht, The Netherlands, 2005. [Google Scholar]
- Garzon, R.; Marcucci, G.; Croce, C.M. Targeting microRNAs in cancer: Rationale, strategies and challenges. Nat. Rev. Drug Discov. 2010, 9, 775–789. [Google Scholar] [CrossRef]
- Croce, C.M. Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 2009, 10, 704. [Google Scholar] [CrossRef]
- Sartorius, K.; Makarova, J.; Sartorius, B.; An, P.; Winkler, C.; Chuturgoon, A.; Kramvis, A. The regulatory role of microRNA in hepatitis-B virus-associated hepatocellular carcinoma (HBV-HCC) pathogenesis. Cells 2019, 8, 1504. [Google Scholar] [CrossRef]
- Gramantieri, L.; Ferracin, M.; Fornari, F.; Veronese, A.; Sabbioni, S.; Liu, C.-G.; Calin, G.A.; Giovannini, C.; Ferrazzi, E.; Grazi, G.L. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res. 2007, 67, 6092–6099. [Google Scholar] [CrossRef] [PubMed]
- Coulouarn, C.; Factor, V.M.; Andersen, J.B.; Durkin, M.E.; Thorgeirsson, S.S. Loss of miR-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. Oncogene 2009, 28, 3526–3536. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.-W.; Wang, N.; Wang, Y.; Wang, F.; Fu, Z.; Yan, X.; Zhu, H.; Diao, W.; Ding, Y.; Chen, X. Hepatitis B virus-human chimeric transcript HBx-LINE1 promotes hepatic injury via sequestering cellular microRNA-122. J. Hepatol. 2016, 64, 278–291. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Dutta, A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007, 21, 1025–1030. [Google Scholar] [CrossRef] [PubMed]
- Takata, A.; Otsuka, M.; Ohno, M.; Kishikawa, T.; Yoshikawa, T.; Koike, K. Mutual antagonism between hepatitis B viral mRNA and host microRNA let-7. Sci. Rep. 2016, 6, 23237. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Huang, P.; Ju, X.; Li, Z.; Wang, Y. Lin28B over-expression mediates the repression of let-7 by hepatitis B virus X protein in hepatoma cells. Int. J. Clin. Exp. Med. 2015, 8, 15108. [Google Scholar] [PubMed]
- Higashi, T.; Hayashi, H.; Ishimoto, T.; Takeyama, H.; Kaida, T.; Arima, K.; Taki, K.; Sakamoto, K.; Kuroki, H.; Okabe, H. miR-9-3p plays a tumour-suppressor role by targeting TAZ (WWTR1) in hepatocellular carcinoma cells. Br. J. Cancer 2015, 113, 252–258. [Google Scholar] [CrossRef]
- Rong, M.-H.; Cai, K.-T.; Lu, H.-P.; Guo, Y.-N.; Huang, X.-Y.; Zhu, Z.-H.; Tang, W.; Huang, S.-N. Overexpression of MiR-452-5p in hepatocellular carcinoma tissues and its prospective signaling pathways. Int. J. Clin. Exp. Pathol. 2019, 12, 4041. [Google Scholar]
- Zheng, J.; Cheng, D.; Wu, D.; Wang, L.; Qu, F.; Wu, X.; Cheng, L.; Wei, Y.; Liu, X. MiR-452-5p mediates the proliferation, migration and invasion of hepatocellular carcinoma cells via targeting COLEC10. Pers. Med. 2021, 18, 97–106. [Google Scholar] [CrossRef]
- Leong, K.-W.; Cheng, C.-W.; Wong, C.-M.; Oi-Lin, I.N.; Kwong, Y.-L.; Tse, E. miR-874-3p is down-regulated in hepatocellular carcinoma and negatively regulates PIN1 expression. Oncotarget 2017, 8, 11343. [Google Scholar] [CrossRef]
- Zhao, S.; Li, J.; Zhang, G.; Wang, Q.; Wu, C.; Zhang, Q.; Wang, H.; Sun, P.; Xiang, R.; Yang, S. Exosomal miR-451a functions as a tumor suppressor in hepatocellular carcinoma by targeting LPIN1. Cell. Physiol. Biochem. 2019, 53, 19–35. [Google Scholar] [PubMed]
- Wei, G.-Y.; Hu, M.; Zhao, L.; Guo, W.-S. MiR-451a suppresses cell proliferation, metastasis and EMT via targeting YWHAZ in hepatocellular carcinoma. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 5158–5167. [Google Scholar] [PubMed]
- Xu, Y.; Lai, Y.; Cao, L.; Li, Y.; Chen, G.; Chen, L.; Weng, H.; Chen, T.; Wang, L.; Ye, Y. Human umbilical cord mesenchymal stem cells-derived exosomal microRNA-451a represses epithelial–mesenchymal transition of hepatocellular carcinoma cells by inhibiting ADAM10. RNA Biol. 2021, 18, 1408–1423. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Li, H.; Zhao, S.; Wang, E.; Zhu, J.; Feng, D.; Zhu, Y.; Dou, W.; Fan, Q.; Hu, J. Epigenetic silencing of miR-144/451a cluster contributes to HCC progression via paracrine HGF/MIF-mediated TAM remodeling. Mol. Cancer 2021, 20, 46. [Google Scholar] [CrossRef]
- Mu, W.; Gu, P.; Song, W.; Zhu, T.; Wang, W.; Zhou, Y. Comprehensive analysis and identification of the circ_0084615/miR-451a/MEF2D axis in benzo (a) pyrene exposed tumor cells in hepato-carcinogenesis. Food Chem. Toxicol. 2023, 176, 113810. [Google Scholar] [CrossRef] [PubMed]
- Lv, C.; Wan, Q.; Shen, C.; Wu, H.; Zhou, B.; Wang, W. Long non-coding RNA ZSCAN16-AS1 promotes the malignant properties of hepatocellular carcinoma by decoying microRNA-451a and consequently increasing ATF2 expression. Mol. Med. Rep. 2021, 24, 780. [Google Scholar] [CrossRef]
- Cao, M.-Q.; You, A.-B.; Zhu, X.-D.; Zhang, W.; Zhang, Y.-Y.; Zhang, S.-Z.; Zhang, K.-W.; Cai, H.; Shi, W.-K.; Li, X.-L. miR-182-5p promotes hepatocellular carcinoma progression by repressing FOXO3a. J. Hematol. Oncol. 2018, 11, 12. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Xu, M.; Sun, Z.; Yu, X.; Deng, Y.; Chang, H. LINC01018 confers a novel tumor suppressor role in hepatocellular carcinoma through sponging microRNA-182-5p. Am. J. Physiol.-Gastrointest. Liver Physiol. 2019, 317, G116–G126. [Google Scholar] [CrossRef]
- Compagnoni, C.; Capelli, R.; Zelli, V.; Corrente, A.; Vecchiotti, D.; Flati, I.; Di Vito Nolfi, M.; Angelucci, A.; Alesse, E.; Zazzeroni, F. MiR-182-5p Is Upregulated in Hepatic Tissues from a Diet-Induced NAFLD/NASH/HCC C57BL/6J Mouse Model and Modulates Cyld and Foxo1 Expression. Int. J. Mol. Sci. 2023, 24, 9239. [Google Scholar] [CrossRef]
- Assal, R.A.; El Tayebi, H.M.; Hosny, K.A.; Esmat, G.; Abdelaziz, A.I. A pleiotropic effect of the single clustered hepatic metastamiRs miR-96-5p and miR-182-5p on insulin-like growth factor II, insulin-like growth factor-1 receptor and insulin-like growth factor-binding protein-3 in hepatocellular carcinoma. Mol. Med. Rep. 2015, 12, 645–650. [Google Scholar] [CrossRef]
- Zuo, Z.; Li, Y.; Zeng, C.; Xi, Y.; Tao, H.; Guo, Y. Integrated analyses identify key molecules and reveal the potential mechanism of miR-182-5p/FOXO1 axis in alcoholic liver disease. Front. Med. 2021, 8, 767584. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Su, J.; He, H.; Zhan, Y.; Liu, H. Hsa_circ_0070269 inhibits hepatocellular carcinoma progression through modulating miR-182/NPTX1 axis. Biomed. Pharmacother. 2019, 120, 109497. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Li, M.; Li, Q.; Guo, Z.; Sun, X.; Zhang, X.; Liu, Y.; Yao, W.; Xiao, P. MicroRNA-98-5p inhibits cell proliferation and induces cell apoptosis in hepatocellular carcinoma via targeting IGF2BP1. Oncol. Res. 2017, 25, 1117. [Google Scholar] [CrossRef] [PubMed]
- Shen, Q.; Jiang, S.; Wu, M.; Zhang, L.; Su, X.; Zhao, D. LncRNA HEIH confers cell sorafenib resistance in hepatocellular carcinoma by regulating miR-98-5p/PI3K/AKT pathway. Cancer Manag. Res. 2020, 12, 6585–6595. [Google Scholar] [CrossRef] [PubMed]
- Ji, P.-t.; Wang, X.-y. Clinical application study on miR-98–5p as a prognostic biomarker in hepatocellular carcinoma. Clin. Res. Hepatol. Gastroenterol. 2023, 47, 102077. [Google Scholar] [CrossRef] [PubMed]
- Fei, X.; Zhang, P.; Pan, Y.; Liu, Y. MicroRNA-98-5p inhibits tumorigenesis of hepatitis B virus-related hepatocellular carcinoma by targeting NF-κB-inducing kinase. Yonsei Med. J. 2020, 61, 460. [Google Scholar] [CrossRef]
- Shi, J.; Ci, Y.; Zheng, Y.; Chen, W.; Chen, X. Submicron silica particles have cytotoxicities on hepatocellular carcinoma, non-small cell lung cancer and breast cancer by unified regulating the XLOC_001659/miR-98-5p/MAP3K2-mediated pathway. Toxicol. Res. 2021, 10, 824–834. [Google Scholar] [CrossRef]
- Wang, J.; Tang, Q.; Lu, L.; Luo, Z.; Li, W.; Lu, Y.; Pu, J. LncRNA OIP5-AS1 interacts with miR-363-3p to contribute to hepatocellular carcinoma progression through up-regulation of SOX4. Gene Ther. 2020, 27, 495–504. [Google Scholar] [CrossRef]
- Hu, P.-A.; Miao, Y.-Y.; Yu, S.; Guo, N. Long non-coding RNA SNHG5 promotes human hepatocellular carcinoma progression by regulating miR-363-3p/RNF38 axis. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 3592–3604. [Google Scholar]
- Ying, J.; Yu, X.; Ma, C.; Zhang, Y.; Dong, J. MicroRNA-363-3p is downregulated in hepatocellular carcinoma and inhibits tumorigenesis by directly targeting specificity protein 1. Mol. Med. Rep. 2017, 16, 1603–1611. [Google Scholar] [CrossRef]
- Lu, Y.-B.; Jiang, Q.; Yang, M.-Y.; Zhou, J.-X.; Zhang, Q. Long noncoding RNA NNT-AS1 promotes hepatocellular carcinoma progression and metastasis through miR-363/CDK6 axis. Oncotarget 2017, 8, 88804. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-L.; Chen, C.-M.; Wang, X.-M.; Wang, L. Effects of miR-339-5p on invasion and prognosis of hepatocellular carcinoma. Clin. Res. Hepatol. Gastroenterol. 2016, 40, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; Zheng, J.; Wen, S.; Luo, J.; Shao, G.; Zhang, Y. MicroRNA-339 inhibits human hepatocellular carcinoma proliferation and invasion via targeting ZNF689. Drug Des. Dev. Ther. 2019, 13, 435–445. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Guo, J.; Ye, A.; Zhou, J.; Zheng, L.; Cai, J. Knockdown of Long Non-Coding RNA (LncRNA) MAFG Divergent Transcript Inhibits the Tumorigenesis of Hepatocellular Carcinoma Through miR-339-5p/CDC25A Axis. J. Biomed. Nanotechnol. 2023, 19, 195–205. [Google Scholar] [CrossRef]
- Lin, H.; Huang, Z.-P.; Liu, J.; Qiu, Y.; Tao, Y.-P.; Wang, M.-C.; Yao, H.; Hou, K.-Z.; Gu, F.-M.; Xu, X.-F. MiR-494-3p promotes PI3K/AKT pathway hyperactivation and human hepatocellular carcinoma progression by targeting PTEN. Sci. Rep. 2018, 8, 10461. [Google Scholar] [CrossRef]
- Li, H.; Zhang, L.; Cai, N.; Zhang, B.; Sun, S. MicroRNA-494-3p prevents liver fibrosis and attenuates hepatic stellate cell activation by inhibiting proliferation and inducing apoptosis through targeting TRAF3. Ann. Hepatol. 2021, 23, 100305. [Google Scholar] [CrossRef]
- Henry, J.C.; Park, J.-K.; Jiang, J.; Kim, J.H.; Nagorney, D.M.; Roberts, L.R.; Banerjee, S.; Schmittgen, T.D. miR-199a-3p targets CD44 and reduces proliferation of CD44 positive hepatocellular carcinoma cell lines. Biochem. Biophys. Res. Commun. 2010, 403, 120–125. [Google Scholar] [CrossRef]
- Fornari, F.; Milazzo, M.; Chieco, P.; Negrini, M.; Calin, G.A.; Grazi, G.L.; Pollutri, D.; Croce, C.M.; Bolondi, L.; Gramantieri, L. MiR-199a-3p regulates mTOR and c-Met to influence the doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res. 2010, 70, 5184–5193. [Google Scholar] [CrossRef]
- Yin, J.; Hou, P.; Wu, Z.; Wang, T.; Nie, Y. Circulating miR-375 and miR-199a-3p as potential biomarkers for the diagnosis of hepatocellular carcinoma. Tumor Biol. 2015, 36, 4501–4507. [Google Scholar] [CrossRef]
- Ren, K.; Li, T.; Zhang, W.; Ren, J.; Li, Z.; Wu, G. miR-199a-3p inhibits cell proliferation and induces apoptosis by targeting YAP1, suppressing Jagged1-Notch signaling in human hepatocellular carcinoma. J. Biomed. Sci. 2016, 23, 79. [Google Scholar] [CrossRef]
- Callegari, E.; D’Abundo, L.; Guerriero, P.; Simioni, C.; Elamin, B.K.; Russo, M.; Cani, A.; Bassi, C.; Zagatti, B.; Giacomelli, L. miR-199a-3p modulates MTOR and PAK4 pathways and inhibits tumor growth in a hepatocellular carcinoma transgenic mouse model. Mol. Ther.-Nucleic Acids 2018, 11, 485–493. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Zhang, R.; Hou, P.; Wang, J.; Wu, L.; Li, J. Circ-ZEB1 promotes PIK3CA expression by silencing miR-199a-3p and affects the proliferation and apoptosis of hepatocellular carcinoma. Mol. Cancer 2022, 21, 72. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Cheng, Q.; Xia, M.; Huang, X.; He, X.; Liao, J. Hypoxia-induced lncRNA-NEAT1 sustains the growth of hepatocellular carcinoma via regulation of miR-199a-3p/UCK2. Front. Oncol. 2020, 10, 998. [Google Scholar] [CrossRef]
- Kim, J.H.; Badawi, M.; Park, J.-K.; Jiang, J.; Mo, X.; Roberts, L.R.; Schmittgen, T.D. Anti-invasion and anti-migration effects of miR-199a-3p in hepatocellular carcinoma are due in part to targeting CD151. Int. J. Oncol. 2016, 49, 2037–2045. [Google Scholar] [CrossRef] [PubMed]
- Guan, J.; Liu, Z.; Xiao, M.; Hao, F.; Wang, C.; Chen, Y.; Lu, Y.; Liang, J. MicroRNA-199a-3p inhibits tumorigenesis of hepatocellular carcinoma cells by targeting ZHX1/PUMA signal. Am. J. Transl. Res. 2017, 9, 2457. [Google Scholar] [PubMed]
- Li, Z.; Zhou, Y.; Zhang, L.; Jia, K.; Wang, S.; Wang, M.; Li, N.; Yu, Y.; Cao, X.; Hou, J. microRNA-199a-3p inhibits hepatic apoptosis and hepatocarcinogenesis by targeting PDCD4. Oncogenesis 2020, 9, 95. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Dasgupta, D.; Ghosh, A.; Roychoudhury, S.; Kumar, D.; Gorain, M.; Butti, R.; Datta, S.; Agarwal, S.; Gupta, S. MiRNA199a-3p suppresses tumor growth, migration, invasion and angiogenesis in hepatocellular carcinoma by targeting VEGFA, VEGFR1, VEGFR2, HGF and MMP2. Cell Death Dis. 2017, 8, e2706. [Google Scholar] [CrossRef]
- Wei, H.; Wang, J.; Xu, Z.; Lu, Y.; Wu, X.; Zhuo, C.; Tan, C.; Tang, Q.; Pu, J. miR-584-5p regulates hepatocellular carcinoma cell migration and invasion through targeting KCNE2. Mol. Genet. Genom. Med. 2019, 7, e702. [Google Scholar] [CrossRef]
- Liu, Q.; Wang, C.; Jiang, Z.; Li, S.; Li, F.; Tan, H.B.; Yue, S.Y. circRNA 001306 enhances hepatocellular carcinoma growth by up-regulating CDK16 expression via sponging miR-584-5p. J. Cell. Mol. Med. 2020, 24, 14306–14315. [Google Scholar] [CrossRef]
- Liu, Z.; Lu, J.; Fang, H.; Sheng, J.; Cui, M.; Yang, Y.; Tang, B.; Zhang, X. m6A modification-mediated DUXAP8 regulation of malignant phenotype and chemotherapy resistance of hepatocellular carcinoma through miR-584-5p/MAPK1/ERK pathway axis. Front. Cell Dev. Biol. 2021, 9, 783385. [Google Scholar] [CrossRef]
- Shao, Z.; Pan, Q.; Zhang, Y. Hepatocellular carcinoma cell-derived extracellular vesicles encapsulated microRNA-584-5p facilitates angiogenesis through PCK1-mediated nuclear factor E2-related factor 2 signaling pathway. Int. J. Biochem. Cell Biol. 2020, 125, 105789. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Jiao, Y.; Fu, Z.; Luo, Z.; Su, J.; Li, Y. High miR-454-3p expression predicts poor prognosis in hepatocellular carcinoma. Cancer Manag. Res. 2019, 11, 2795–2802. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Wang, Q.; Sun, J.; Liu, H.; Wang, H. Long non-coding RNA TPTEP1 exerts inhibitory effects on hepatocellular carcinoma by impairing microRNA-454-3p-mediated DLG5 downregulation. Dig. Liver Dis. 2022, 54, 268–279. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zheng, J.; Yang, S.; Zong, Q.; Wang, Z.; Liao, X. Mir-454-3p induced WTX deficiency promotes hepatocellular carcinoma progressions through regulating TGF-β signaling pathway. J. Cancer 2022, 13, 1820. [Google Scholar] [CrossRef] [PubMed]
- Shi, D.; Li, H.; Zhang, J.; Li, Y. CircGDI2 regulates the proliferation, migration, invasion and apoptosis of OSCC via miR-454-3p/FOXF2 axis. Cancer Manag. Res. 2021, 13, 1371–1382. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wang, W.; Yang, T.; Li, D.; Huang, Y.; Bai, G.; Li, Q. LINC00520 up-regulates SOX5 to promote cell proliferation and invasion by miR-4516 in human hepatocellular carcinoma. Biol. Chem. 2022, 403, 665–678. [Google Scholar] [CrossRef] [PubMed]
- Li, O.; Li, Z.; Tang, Q.; Li, Y.; Yuan, S.; Shen, Y.; Zhang, Z.; Li, N.; Chu, K.; Lei, G. Long stress induced non-coding transcripts 5 (LSINCT5) promotes hepatocellular carcinoma progression through interaction with high-mobility group AT-hook 2 and MiR-4516. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2018, 24, 8510. [Google Scholar] [CrossRef]
- Niu, X.; Wei, N.; Peng, L.; Li, X.; Zhang, X.; Wang, C. miR-34a-5p plays an inhibitory role in hepatocellular carcinoma by regulating target gene VEGFA. Malays. J. Pathol. 2022, 44, 39–52. [Google Scholar]
- Lin, X.-J.; Fang, J.-H.; Yang, X.-J.; Zhang, C.; Yuan, Y.; Zheng, L.; Zhuang, S.-M. Hepatocellular carcinoma cell-secreted exosomal microRNA-210 promotes angiogenesis in vitro and in vivo. Mol. Ther.-Nucleic Acids 2018, 11, 243–252. [Google Scholar] [CrossRef]
- Moirangthem, A.; Gondaliya, P.; Yan, I.K.; Sayyed, A.A.; Driscoll, J.; Patel, T. Extracellular vesicle-mediated miR-126-3p transfer contributes to inter-cellular communication in the liver tumor microenvironment. Int. J. Oncol. 2023, 62, 31. [Google Scholar] [CrossRef]
- Yang, Y.; Yang, T.; Zhao, Z.; Zhang, H.; Yuan, P.; Wang, G.; Zhao, Z.; An, J.; Lyu, Z.; Xing, J. Down-regulation of BMAL1 by MiR-494-3p Promotes Hepatocellular Carcinoma Growth and Metastasis by Increasing GPAM-mediated Lipid Biosynthesis. Int. J. Biol. Sci. 2022, 18, 6129. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Ma, X.; Mi, Z.; He, Z.; Rong, P. Extracellular polysaccharide from Rhizopus nigricans inhibits hepatocellular carcinoma via miR-494-3p/TRIM36 axis and cyclin E ubiquitination. J. Clin. Transl. Hepatol. 2022, 10, 608. [Google Scholar] [CrossRef] [PubMed]
- Pollutri, D.; Patrizi, C.; Marinelli, S.; Giovannini, C.; Trombetta, E.; Giannone, F.A.; Baldassarre, M.; Quarta, S.; Vandewynckel, Y.-P.; Vandierendonck, A. The epigenetically regulated miR-494 associates with stem-cell phenotype and induces sorafenib resistance in hepatocellular carcinoma. Cell Death Dis. 2018, 9, 4. [Google Scholar] [CrossRef] [PubMed]
- Giuffrè, M.; Patti, R.; Pascut, D.; Sukowati, C.H.C.; Masutti, F.; Cristiana, A.; Fabio, T.; Macor, D.; Buonocore, M.R.; Colombo, A. MicroRNAs as regulators of neo-angiogenesis in hepatocellular carcinoma. Ann. Gastroenterol. Dig. Disord. 2018, 1, 9–16. [Google Scholar]
- Xu, X.-P.; Peng, X.-Q.; Yin, X.-M.; Liu, Y.; Shi, Z.-Y. miR-34a-5p suppresses the invasion and metastasis of liver cancer by targeting the transcription factor YY1 to mediate MYCT1 upregulation. Acta Histochem. 2020, 122, 151576. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wu, H.; Wu, M.; Feng, Y.; Wu, S.; Shen, X.; He, J.; Luo, X. Hypoxia-related miR-210-5p and miR-210-3p regulate hypoxia-induced migration and epithelial-mesenchymal transition in hepatoma cells. Int. J. Clin. Exp. Med 2019, 12, 5096–5104. [Google Scholar]
- Zhu, L.; Yang, N.; Chen, J.; Zeng, T.; Yan, S.; Liu, Y.; Yu, G.; Chen, Q.; Du, G.; Pan, W. LINC00052 upregulates EPB41L3 to inhibit migration and invasion of hepatocellular carcinoma by binding miR-452-5p. Oncotarget 2017, 8, 63724. [Google Scholar] [CrossRef]
- Hu, Z.; Chen, J.; Zhao, Y.; Yan, C.; Wang, Y.; Zhu, J.; Li, L. Exosomal miR-452-5p induce M2 macrophage polarization to accelerate hepatocellular carcinoma progression by targeting TIMP3. J. Immunol. Res. 2022, 2022, 1032106. [Google Scholar]
- Yang, W.; Ju, H.-Y.; Tian, X.-F. Circular RNA-ABCB10 suppresses hepatocellular carcinoma progression through upregulating NRP1/ABL2 via sponging miR-340-5p/miR-452-5p. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 2347–2357. [Google Scholar]
- Wang, J.; Zhu, Y.; Ai, X.; Wan, H.; Jia, W.; Chu, J.; Xu, B.; Kong, X.; Kong, L. Long noncoding RNA 02027 inhibits proliferation, migration and invasion of hepatocellular carcinoma via miR-625-3p/PDLIM5 pathway. J. Gene Med. 2023, 25, e3485. [Google Scholar] [CrossRef]
- Chen, S. LINC00852 regulates cell proliferation, invasion, migration and apoptosis in hepatocellular carcinoma Via the miR-625/E2F1 Axis. Cell. Mol. Bioeng. 2022, 15, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Iwai, N.; Yasui, K.; Tomie, A.; Gen, Y.; Terasaki, K.; Kitaichi, T.; Soda, T.; Yamada, N.; Dohi, O.; Seko, Y. Oncogenic miR-96-5p inhibits apoptosis by targeting the caspase-9 gene in hepatocellular carcinoma. Int. J. Oncol. 2018, 53, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Yuan, F.; Tang, Y.; Cao, M.; Ren, Y.; Li, Y.; Yang, G.; Ou, Q.; Tustumi, F.; Sandri, G.B.L.; Raissi, D. Identification of the hsa_circ_0039466/miR-96-5p/FOXO1 regulatory network in hepatocellular carcinoma by whole-transcriptome analysis. Ann. Transl. Med. 2022, 10, 769. [Google Scholar] [CrossRef] [PubMed]
- Matsui, T.; Hamada-Tsutsumi, S.; Naito, Y.; Nojima, M.; Iio, E.; Tamori, A.; Kubo, S.; Ide, T.; Kondo, Y.; Eguchi, Y. Identification of microRNA-96-5p as a postoperative, prognostic microRNA predictor in nonviral hepatocellular carcinoma. Hepatol. Res. 2022, 52, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Morishita, A.; Fujita, K.; Iwama, H.; Chiyo, T.; Fujihara, S.; Oura, K.; Tadokoro, T.; Mimura, S.; Nomura, T.; Tani, J. Role of microRNA-210-3p in hepatitis B virus-related hepatocellular carcinoma. Am. J. Physiol.-Gastrointest. Liver Physiol. 2020, 318, G401–G409. [Google Scholar] [CrossRef]
- You, R.; Yang, Y.; Yin, G.; Jiang, H.; Lu, Y.; Gui, L.; Bao, J.; Xu, Q.; Feng, L. CPEB2 Suppresses Hepatocellular Carcinoma Epithelial–Mesenchymal Transition and Metastasis through Regulating the HIF-1α/miR-210-3p/CPEB2 Axis. Pharmaceutics 2023, 15, 1887. [Google Scholar] [CrossRef]
- Dandri, M. Epigenetic modulation in chronic hepatitis B virus infection. Semin. Immunopathol. 2020, 42, 173–185. [Google Scholar] [CrossRef]
- Wahid, B.; Ali, A.; Rafique, S.; Idrees, M. New insights into the epigenetics of hepatocellular carcinoma. BioMed Res. Int. 2017, 2017, 1609575. [Google Scholar] [CrossRef]
- Wei, X.; Xiang, T.; Ren, G.; Tan, C.; Liu, R.; Xu, X.; Wu, Z. miR-101 is down-regulated by the hepatitis B virus x protein and induces aberrant DNA methylation by targeting DNA methyltransferase 3A. Cell. Signal. 2013, 25, 439–446. [Google Scholar] [CrossRef]
- Wei, X.; Tan, C.; Tang, C.; Ren, G.; Xiang, T.; Qiu, Z.; Liu, R.; Wu, Z. Epigenetic repression of miR-132 expression by the hepatitis B virus x protein in hepatitis B virus-related hepatocellular carcinoma. Cell. Signal. 2013, 25, 1037–1043. [Google Scholar] [CrossRef]
- Takata, A.; Otsuka, M.; Yoshikawa, T.; Kishikawa, T.; Hikiba, Y.; Obi, S.; Goto, T.; Kang, Y.J.; Maeda, S.; Yoshida, H. MicroRNA-140 acts as a liver tumor suppressor by controlling NF-κB activity by directly targeting DNA methyltransferase 1 (Dnmt1) expression. Hepatology 2013, 57, 162–170. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.-H.; Choi, H.S.; Park, Y.K.; Park, E.-S.; Shin, G.C.; Kim, D.H.; Ahn, S.H.; Kim, K.-H. HBx-induced NF-κB signaling in liver cells is potentially mediated by the ternary complex of HBx with p22-FLIP and NEMO. PLoS ONE 2013, 8, e57331. [Google Scholar] [CrossRef] [PubMed]
- Au, S.L.K.; Wong, C.C.L.; Lee, J.M.F.; Fan, D.N.Y.; Tsang, F.H.; Ng, I.O.L.; Wong, C.M. Enhancer of zeste homolog 2 epigenetically silences multiple tumor suppressor microRNAs to promote liver cancer metastasis. Hepatology 2012, 56, 622–631. [Google Scholar] [CrossRef] [PubMed]
- Mehta, A.; Baltimore, D. MicroRNAs as regulatory elements in immune system logic. Nat. Rev. Immunol. 2016, 16, 279. [Google Scholar] [CrossRef] [PubMed]
- Faraoni, I.; Antonetti, F.R.; Cardone, J.; Bonmassar, E. miR-155 gene: A typical multifunctional microRNA. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2009, 1792, 497–505. [Google Scholar] [CrossRef] [PubMed]
- O’connell, R.M.; Rao, D.S.; Chaudhuri, A.A.; Baltimore, D. Physiological and pathological roles for microRNAs in the immune system. Nat. Rev. Immunol. 2010, 10, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, A.; Vigorito, E.; Clare, S.; Warren, M.V.; Couttet, P.; Soond, D.R.; Van Dongen, S.; Grocock, R.J.; Das, P.P.; Miska, E.A. Requirement of bic/microRNA-155 for normal immune function. Science 2007, 316, 608–611. [Google Scholar] [CrossRef]
- Vigorito, E.; Kohlhaas, S.; Lu, D.; Leyland, R. miR-155: An ancient regulator of the immune system. Immunol. Rev. 2013, 253, 146–157. [Google Scholar] [CrossRef]
- Yang, P.; Li, Q.-J.; Feng, Y.; Zhang, Y.; Markowitz, G.J.; Ning, S.; Deng, Y.; Zhao, J.; Jiang, S.; Yuan, Y. TGF-β-miR-34a-CCL22 signaling-induced Treg cell recruitment promotes venous metastases of HBV-positive hepatocellular carcinoma. Cancer Cell 2012, 22, 291–303. [Google Scholar] [CrossRef]
- Rao, D.S.; O’Connell, R.M.; Chaudhuri, A.A.; Garcia-Flores, Y.; Geiger, T.L.; Baltimore, D. MicroRNA-34a perturbs B lymphocyte development by repressing the forkhead box transcription factor Foxp1. Immunity 2010, 33, 48–59. [Google Scholar] [CrossRef]
- Wang, H.; Li, X.; Li, T.; Wang, L.; Wu, X.; Liu, J.; Xu, Y.; Wei, W. Multiple roles of microRNA-146a in immune responses and hepatocellular carcinoma. Oncol. Lett. 2019, 18, 5033–5042. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Qian, J.; He, J.; Zhang, Q.; Zhai, A.; Song, W.; Li, Y.; Ling, H.; Zhong, Z.; Zhang, F. Modulation of miR-122 expression affects the interferon response in human hepatoma cells. Mol. Med. Rep. 2013, 7, 585–590. [Google Scholar] [CrossRef] [PubMed]
- Gramantieri, L.; Giovannini, C.; Piscaglia, F.; Fornari, F. MicroRNAs as modulators of tumor metabolism, microenvironment, and immune response in hepatocellular carcinoma. J. Hepatocell. Carcinoma 2021, 8, 369–385. [Google Scholar] [CrossRef] [PubMed]
- Rui, T.; Zhang, X.; Guo, J.; Xiang, A.; Tang, N.; Liu, J.; Mao, Z. Serum-Exosome-Derived miRNAs Serve as Promising Biomarkers for HCC Diagnosis. Cancers 2022, 15, 205. [Google Scholar] [CrossRef] [PubMed]
- Yousuf, T.; Dar, S.B.; Bangri, S.A.; Choh, N.A.; Rasool, Z.; Shah, A.; Rather, R.A.; Rah, B.; Bhat, G.R.; Ali, S. Diagnostic implication of a circulating serum-based three-microRNA signature in hepatocellular carcinoma. Front. Genet. 2022, 13, 929787. [Google Scholar] [CrossRef] [PubMed]
- Aly, D.M.; Gohar, N.A.-H.; Abd El-Hady, A.A.; Khairy, M.; Abdullatif, M.M. Serum microRNA let-7a-1/let-7d/let-7f and miRNA 143/145 gene expression profiles as potential biomarkers in HCV induced hepatocellular carcinoma. Asian Pac. J. Cancer Prev. 2020, 21, 555. [Google Scholar] [CrossRef]
- Infante-Menéndez, J.; López-Pastor, A.R.; González-Illanes, T.; González-López, P.; Huertas-Lárez, R.; Rey, E.; González-Rodríguez, Á.; García-Monzón, C.; Patil, N.P.; Vega de Céniga, M. Increased let-7d-5p in non-alcoholic fatty liver promotes insulin resistance and is a potential blood biomarker for diagnosis. Liver Int. 2023, 43, 1714–1728. [Google Scholar] [CrossRef]
- Fang, F.; Song, T.; Zhang, T.; Cui, Y.; Zhang, G.; Xiong, Q. MiR-425-5p promotes invasion and metastasis of hepatocellular carcinoma cells through SCAI-mediated dysregulation of multiple signaling pathways. Oncotarget 2017, 8, 31745. [Google Scholar] [CrossRef]
- Rao, D.; Guan, S.; Huang, J.; Chang, Q.; Duan, S. miR-425-5p acts as a molecular marker and promoted proliferation, migration by targeting RNF11 in hepatocellular carcinoma. BioMed Res. Int. 2020, 2020, 6530973. [Google Scholar] [CrossRef]
- Wu, S.; Liu, S.; Cao, Y.; Chao, G.; Wang, P.; Pan, H. Downregulation of ZC3H13 by miR-362-3p/miR-425-5p is associated with a poor prognosis and adverse outcomes in hepatocellular carcinoma. Aging 2022, 14, 2304. [Google Scholar] [CrossRef]
- Wu, H.; Shang, J.; Zhan, W.; Liu, J.; Ning, H.; Chen, N. miR-425-5p promotes cell proliferation, migration and invasion by directly targeting FOXD3 in hepatocellular carcinoma cells. Mol. Med. Rep. 2019, 20, 1883–1892. [Google Scholar] [CrossRef] [PubMed]
- Qi, P.; Cheng, S.-Q.; Wang, H.; Li, N.; Chen, Y.-F.; Gao, C.-F. Serum microRNAs as biomarkers for hepatocellular carcinoma in Chinese patients with chronic hepatitis B virus infection. PLoS ONE 2011, 6, e28486. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Cai, C.; Qiu, Y. Diagnostic value of circulating microRNAs in hepatitis B virus-related hepatocellular carcinoma: A systematic review and meta-analysis. J. Cancer 2019, 10, 4754. [Google Scholar] [CrossRef] [PubMed]
- Witwer, K.W. Circulating microRNA biomarker studies: Pitfalls and potential solutions. Clin. Chem. 2015, 61, 56–63. [Google Scholar] [CrossRef] [PubMed]
- El-Daly, S.M.; Gouhar, S.A.; Abd Elmageed, Z.Y. Circulating microRNAs as reliable tumor biomarkers: Opportunities and Challenges facing Clinical Application. J. Pharmacol. Exp. Ther. 2023, 384, 35–51. [Google Scholar] [CrossRef] [PubMed]
- Heinicke, F.; Zhong, X.; Zucknick, M.; Breidenbach, J.; Sundaram, A.Y.; Flåm, S.T.; Leithaug, M.; Dalland, M.; Farmer, A.; Henderson, J.M. Systematic assessment of commercially available low-input miRNA library preparation kits. RNA Biol. 2020, 17, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Khamina, K.; Diendorfer, A.B.; Skalicky, S.; Weigl, M.; Pultar, M.; Krammer, T.L.; Fournier, C.A.; Schofield, A.L.; Otto, C.; Smith, A.T. A MicroRNA next-generation-sequencing discovery assay (miND) for genome-scale analysis and absolute quantitation of circulating microRNA biomarkers. Int. J. Mol. Sci. 2022, 23, 1226. [Google Scholar] [CrossRef]
- Marini, F.; Linke, J.; Binder, H. ideal: An R/Bioconductor package for interactive differential expression analysis. BMC Bioinform. 2020, 21, 565. [Google Scholar] [CrossRef]
- Tastsoglou, S.; Skoufos, G.; Miliotis, M.; Karagkouni, D.; Koutsoukos, I.; Karavangeli, A.; Kardaras, F.S.; Hatzigeorgiou, A.G. DIANA-miRPath v4. 0: Expanding target-based miRNA functional analysis in cell-type and tissue contexts. Nucleic Acids Res. 2023, 51, W154–W159. [Google Scholar] [CrossRef]
- Kern, F.; Aparicio-Puerta, E.; Li, Y.; Fehlmann, T.; Kehl, T.; Wagner, V.; Ray, K.; Ludwig, N.; Lenhof, H.-P.; Meese, E. miRTargetLink 2.0—Interactive miRNA target gene and target pathway networks. Nucleic Acids Res. 2021, 49, W409–W416. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).