Abstract
The Ylistrum japonicum is a commercially valuable scallop known for its long-distance swimming abilities. Despite its economic importance, genetic and genomic research on this species is limited. This study presents the first complete mitochondrial genome of Y. japonicum. The mitochondrial genome is 19,475 bp long and encompasses 13 protein-coding genes, three ribosomal RNA genes, and 23 transfer RNA genes. Two distinct phylogenetic analyses were used to explore the phylogenetic position of the Y. japonicum within the family Pectinidae. Based on one mitochondrial phylogenetic analysis by selecting 15 Pectinidae species and additional outgroup taxa and one single gene phylogenetic analysis by 16S rRNA, two phylogenetic trees were constructed to provide clearer insights into the evolutionary placement of Y. japonicum within the family Pectinidae. Our analysis reveals that Ylistrum is a basal lineage to the Pectininae clade, distinct from its previously assigned tribe, Amusiini. This study offers critical insights into the genetic makeup and evolutionary history of Y. japonicum, enhancing our knowledge of this economically vital species.
1. Introduction
The genus Ylistrum, a member of the phylum Mollusca, class Bivalvia, order Pectinoida, family Pectinidae, was established by Mynhardt et al. [1] in 2014 and is derived from the Greek word “ylistro”, meaning “glide”, reflecting the gliding habits of this group, and dividing the original genus Amusium into two genera, Ylistrum and Amusium. Both Ylistrum and Amusium are primarily found in the Indian–Pacific region and exhibit similar lifestyles and morphological characteristics [1,2,3]. Despite these similarities, the evolutionary relationships and placement of Ylistrum within the family Pectinidae are not conclusively determined. Molecular phylogenetic studies have indicated that the Ylistrum species is genetically distinct from its close relatives in the genus Amusium studied by Alejandrino et al. [4], Mynhardt et al. [1], and Sherratt et al. [5] who support this genetic separation. However, the precise phylogenetic placement of Ylistrum within the family remains a topic of debate. Serb [6] proposed that the Ylistrum might belong to the tribe Decatopectinini, but this suggestion has not been strongly supported by available evidence due to insufficient molecular data [1].
Ylistrum species are known for their long-distance swimming or gliding abilities, and their distinctive features include colorful valves and radiating interior ribs on both valves [1,3,7]. Two extant species within the Ylistrum genus are found across the globe: Y. japonicum (Gmelin, 1971) was originally discovered in Japan [8], and Ylistrum balloti (Bernardi, 1861), which is predominantly encountered in Australia’s western, eastern, and southern regions [3,9], and New Caledonia [10,11]. There is also a known fossil species from Morgan Limestone [12], Ylistrum morganense (Beu and Darragh, 2001). Species Y. balloti is pivotal to the commercial trawl fisheries in Australia, warranting extensive research. In contrast, studies on Y. japonicum are scarcer, with most research originating from Japan and South Korea. Okada [13] has delved into the ecology and morphology aspects of the species, and Kanmizutaru and Anraku [14] have investigated the effects of MgCl2 injection into the adductor muscle for shell opening. Kanmizutaru et al. [15] have evaluated the light perception capabilities of the pallial eyes through electroretinogram tests. In South Korea, studies have illuminated its reproductive cycle [16], the development of its gonads, the age at first sexual maturity, and the sex ratio [17], as well as the correlation between age and growth [18]. In China, there is limited research available, focusing primarily on ecology and the possibilities of artificial breeding [19,20].
The molecular dataset of Y. japonicum remains insufficient [1]. Before this study, there were nine nucleotides’ data about Y. japonicum in the NCBI GenBank database, including ribosomal RNA (12S rRNA, 16S rRNA, 18S rRNA, and 28S rRNA), partial sequences of protein-coding genes (cox1, nad1), and histone H3 gene. The taxonomic placement of Y. japonicum remains unsolved due to a lack of molecular data. A recent taxonomic investigation by Dijkstra and Beu [3] has provisionally maintained Ylistrum within the Amusiini tribe, awaiting a conclusive molecular phylogenetic analysis of the Pectinidae family.
In this study, the complete mitochondrial genome of Y. japonicum was analyzed, supplemented by the molecular data, and based on this result, phylogenetic studies were conducted to clarify the phylogenetic status of Y. japonicum within the family Pectinidae.
2. Results
2.1. Mitochondrial Genome Composition
The mitochondrial genome structure of Y. japonicum is depicted in Figure 1 and further described in Table 1. The complete mitochondrial genome sequence has been submitted to the NCBI GenBank database (Accession number: PP571649). The genome is characterized by a typical circular, closed, double-stranded structure with a total length of 19,475 bp. It encodes 39 genes, which include 13 protein-coding genes (PCGs), three ribosomal RNAs, and 23 transfer RNAs. The nucleotide composition is as follows: (A) constitutes 21.9%, thymine (T) 36%, guanine (G) 29%, and cytosine (C) 13.1%. The mitochondrial genome exhibits an A + T content of 57.9% and a G + C content of 42.1%.
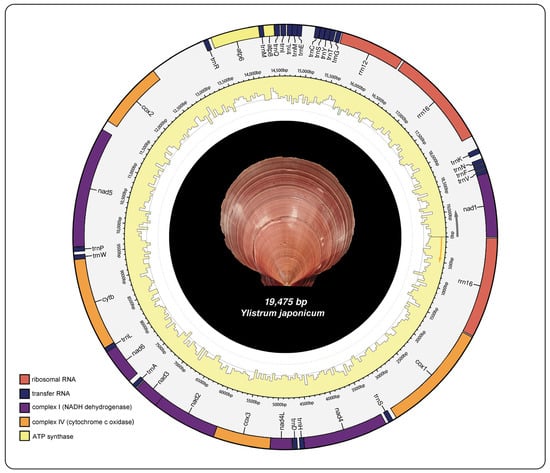
Figure 1.
Mitochondrial genome map of Ylistrum japonicum.

Table 1.
Structural features of the mitochondrial genome of Ylistrum japonicum.
2.2. Protein-Coding Genes
There are a total of 13 protein-coding genes; 12 out of the 13 protein-coding genes (PCGs) of Y. japonicum are commonly found in the majority of most pectinid species [21,22]. All genes are transcribed from the same strand and code in the same direction. Notably, gene atp8, which is typically absent in most mitogenomes of most bivalves [23,24], is present in Y. japonicaum. The total length of the PCGs is 11884 bp, comprising approximately 61% of the complete genome. Among the genes, only four (atp6, cox1, cox2, nad4L) utilize the standard start codon ATG. The remaining nine have alternative start codons, six genes (atp8, cox3, nad1, nad2, nad4, nad6) have GTG, two TTG (cytb, nad5), and nad3 has ATC. Seven genes have the TAG stop codon, five have TAA, and cytb is terminated by T.
2.3. rRNA and tRNA Genes
The rrnS (rrn12) gene spans 957 bp (from position 15459 to 16415), while the rrnL (rrn16) gene has two copies with lengths of 1492 bp and 1486 bp (18-1509, 16453-17939, respectively). The mitochondrial genome of Y. japonicum contains 23 tRNA genes ranging in length from 65 to 72 nucleotides. Three tRNA genes are present in two copies; all three tRNA genes are found with distinct anticodons. Two trnS (rRNA-Ser) have UCU or UGA, two trnL (rRNA-Leu) have UAA or UAG, and two trnM (rRNA-Met) have UAU or CAU. The occurrence of multiple trnM genes in the mitochondrial genomes of bivalves is common [21], and in the majority of mitochondrial genomes of animals, the occurrence of two copies of trnS is frequently noted, as reported by Malkócs et al. [25].
2.4. Control Region
The mitogenome of Y. japonicum, akin to the majority of bivalve species, contains a substantial complement of unassigned nucleotides. In contrast to other scallop species, Y. japonicum exhibits a great number of control regions despite having a shorter major sequence length. Specifically, the mitochondrial genome of Y. japonicum contains a total of 38 control regions, with 5764 bp of intergenic control region nucleotides in which the longest continuous sequence (855bp) is between the cox2 and trnR genes, accounting for approximately 14.83% of all unassigned nucleotides.
2.5. Gene Order
The occurrence of mitogenome rearrangements is prevalent among mollusks [26]; the arrangement observed in the mitogenome of Y. japonicum is also a novel configuration for the family Pectinidae, with no matching gene junctions found in other Pectinidae species (Figure 2). Species with higher gene order similarity were selected for comparison and newly annotated atp8 genes, according to Malkócs et al. [25]. Due to the lack of annotation of rRNA sequence, Mizuhopecten yessoensis (FJ595959) is excluded from the gene order analysis, and based on the high similarity of gene order between three Argopecten species [27], only one species was selected as a representative.
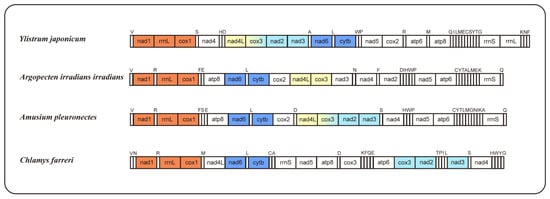
Figure 2.
Gene orders of Ylistrum japonicum, Argopecten irradians irradians, Amusium pleuronectes, and Chlamys farreri, with newly annotated atp8 genes. The same color indicates identical gene junctions (excluding the tRNA genes).
Comparing gene arrangements of four selected species, a conserved gene cluster, “nad6-trnL-cytb,” was identified as common to all. When excluding the tRNA genes, an additional shared gene cluster, “nad1-rrnL-cox1,” was observed among the four species. Gene cluster “nad4L-cox3” is present in Y. japonicum, Argopecten irradians irradians, and Amusium pleuronectes, while gene cluster “cox3-nad2-nad3” is shared by Y. japonicum, A. pleuronectes, and Chlamys farreri. The “nad5-atp6” cluster in Y. japonicum is split by the insertion of cox2, distinguishing it from the other two Pectininae species. The “nad5-atp6-rrnS” cluster, excluding variable tRNA genes, is also shared between A. irradians irradians and A. pleuronectes, indicating a close evolutionary relationship between the two species.
2.6. Gene Collinearity
Gene collinearity analysis using the progressiveMauve algorithm in Mauve has identified seven locally collinear blocks (LCBs) across the complete mitochondrial genome of five Pectininae species (Figure 3). These LCBs are conserved across all mitogenomes analyzed, although variations in the sequence order are evident among the different species. The arrangement of LCBs demonstrated a high degree of similarity among the three Argopecten species, indicating their close evolutionary relationship. In contrast, Y. japonicum exhibited a significantly dissimilar LBCs arrangement compared to three Argopecten species and A. pleuronectes.
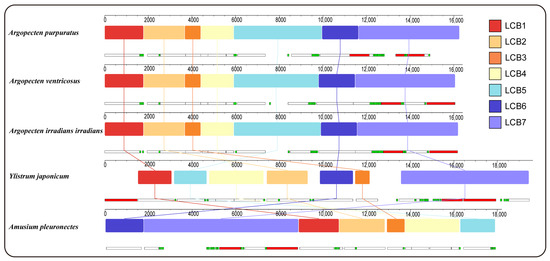
Figure 3.
Gene collinearity analysis of 5 Pectininae species. The level of similarity at each position is shown in the blocks. The white, red, and green boxes represent protein-coding, rRNA, and tRNA genes.
2.7. Phylogenetic Analysis
To delve deeper into the phylogenetic position of Y. japonicum and the taxonomic status within the family Pectinidae, a phylogenetic tree (Figure 4) was constructed based on complete or nearly complete mitochondrial genome data of various Pectinidae species and outgroup taxa. The results of the phylogenetic analysis were found to be comparable with previous studies by Smedley et al. [28], Yao et al. [29], and Malkócs et al. [25] and largely adhered to the taxonomic framework established by Waller [30,31]. Phylogenies based on two methods (Maximum Likelihood and Bayesian inference) of the concatenated protein sequences show almost complete agreement, with high bootstrap values or posterior probabilities supporting all nodes. The systematic arrangement, as proposed by Waller [31], subdivides the family Pectindae into four subfamilies: Pectininae, Chlamydinae, Pallioline, and Camptonectinae. The outgroup Mytilinae and Crassotreinae are found to be consistent with the phylogenetic position proposed by Xu et al. [21], where the clade Mytilinae forms a sister group with the clade Osteridae + Pectinidae.
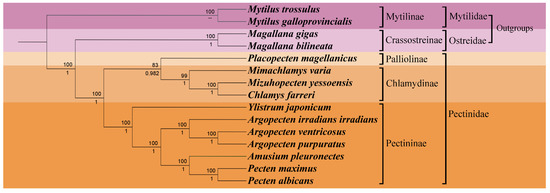
Figure 4.
Phylogenetic tree derived from Maximum likelihood (ML) and Bayesian inference (BI) based on the sequences of mitochondrial protein-coding genes (PCGs). Numbers above the branches indicate bootstrap support; numbers below branches are Bayesian posterior probability. A dash indicates no support for that node.
Our study has validated the earlier proposed hypothesis regarding the monophyly of Pectinidae, as initially concluded by Waller [32]. Nevertheless, the phylogenetic analysis in our study was unable to include any representatives from the Camptonectinae subfamily due to the lack of complete mitochondrial genome sequences available for this group. The Pectinidae species were effectively categorized into three subfamilies: Palliolinae, Chlamydinae, and Pectininae. Placopecten magellanicus, serving as the representative of the Palliolinae, was positioned at the basal position of the branch Palliolinae + Chlamydinae. The clade Palliolinae + Chlamydinae was well supported as the sister group to the Pectininae clade [33,34]. Within the subfamily Chlamydinae, M. yessonesis and C. farreri were found to be the most related, forming a sister taxon relationship with Mimachlamys, which aligns with the findings of Xu et al. [21]. Ylistrum was identified as a basal lineage to the clade Pectininae, separated from its previously assigned tribe Amusiini, a conclusion that is in agreement with the work of Alejandrino et al. [4], Sherratt et al. [5], and Serb [6]. Argopecten species were clustered on the same branch, forming a sister group with the clade composed of Amusium + Pecten. The close relationship between A. pleuronectes and the clade of Pecten maximus + Pecten albicans was also consistent with the research conducted by Barucca et al. [35], Alejandrino et al. [4], and Feng et al. [34].
Another ML tree (Figure 5) was constructed based on 16S rRNA sequences, incorporating three specimens of Y. japonicum from China (PP571649) and Japan (HM622707, KF982785) [4,36]. The genus Amusium and Pecten form a sister group again. Antillipecten antillarum is positioned as a basal lineage, forming a sister group with the clade consisting of Anguipecten + Ylistrum. The two Ylistrum species form a sister clade and are well separated from Amusium. This analysis aligns with the findings of Mynhardt et al. [1]. The Japanese specimens HM622707 and KF982785 are grouped on the same branch, indicating a shared ancestry with the Chinese individuals. All Y. japonicum finally converged into the same branch, showing the close genetic distance between its individuals.
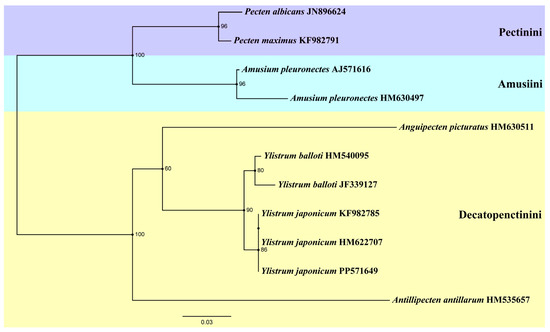
Figure 5.
Phylogenetic tree of genus Ylistrum and some species from three tribes of Pectininae inferred by Maximum likelihood (ML) of 16S rRNA sequences. Numbers indicate bootstrap support.
2.8. Systematic Descriptions
Order Pectinida Gray, 1854
Superfamily Pectinoidea Rafnesque, 1815
Family Pectinidae Rafnesque, 1815
Subfamily Pectininae Rafnesque, 1815
Tribe Decatopectinini Waller, 1986
Genus Ylistrum Mynhardt and Alejandrino, 2014
Ylistrum japonicum (Gmelin, 1791)
Type locality: Japan.
Distribution: Japan (South of central Honshu Island), Korea (Jeju Island), China (Taiwan Province, Guangdong Province, Guangxi Province, and Hainan Province).
Morphological description: Shell large, round, smooth, and glossy. The left valve is dark red to reddish-brown, covered with concentrically arranged dark brown fine lines and spots. The color is slightly lighter at the umbo, with small light-colored spots. The right valve is slightly flat, pale yellow to light tan, white near the umbo, with concentrically arranged brown spots on the surface. Two small auricles are slightly different in size; the color of the auricles on the left valve is darker. The inner surface of the shell is white, with the left valve having a yellow to light brown edge; sometimes, the inner edge of the left valve is pale brown. Interior radial ribbing is on both valves; the specimen was collected from Hailing Island with 33–43 ribs on the left valve and 42–49 ribs on the right valve.
Remarks: In the original description, the species group from China was recorded as a subspecies Amusium japonicum taiwanicum Habe, 1992 [37], and now it has a synonymized name of Y. japonicum. Unlike individuals from Japan, the color of specimens from China are not bright, and the concentrically arranged brown spots are present on the right valve; these are the morphological differences between the individuals from the above two producing areas. Despite their morphological differences, their molecular biological evidence indicates that they are the same species. The counting of the internal ribs by different authors is not always the same (e.g., Zhang et al. [38], Wang [7], Mynhardt et al. [1]). In addition, counts had completely overlapping ranges, and they could not be used to differentiate between the two Ylistrum species [1]. Overall, the most significant difference between Y. japonicum and Y. balloti is the color of their auricles on the right valve and the spots in a concentric pattern on both two valves. To the former species, the auricles on the right valve are generally darker, and spots always appear along with their valve repair marks.
When mixing all the scallops obtained from trawl nets, 5 out of 52 scallops are Y. japonicum in one trawl, with sandy bottom sediment, and others are A. pleuronectes. The average shell length of Y. japonicum in the population is 84.27 ± 10.63 mm (n = 15), the average shell height is 82.97 ± 9.48 mm, and the average shell width is 16.70 ± 2.23 mm.
3. Discussion
The family Pectinidae, as a clade within Bivalvia, exhibits a significant range of morphological and behavioral variations, rendering it of great importance in ecology, evolution, and commercial activities. Nevertheless, the taxonomy of Pectinidae has long been a source of debate within the scientific community.
Waller’s [30] hypothesis for the classification and evolution of Pectinidae is based on morphology, particularly focusing on pre-radial stage shell microsculpture and incorporating fossil data and geological evidence. In 2006, Waller updated his Pectinidae phylogenetic hypothesis in conjunction with previous molecular genetic studies about the phylogenies analysis of the Pectinidae, and a stable classification method was successfully established. Much of the contemporary taxonomic research on Pectinidae is grounded in the taxonomy framework developed by Waller, as evidenced by the work of Serb [6] and Smedley et al. [28]. With the continuous development of molecular technology and the expansion of fossil evidence, phylogenetic studies of Pectinidae have been further improved; however, controversies over the correct classification of this family remain.
In molecular phylogenetic studies, discrepancies in research can arise from numerous sources, including the selection of a single genetic sequence [39] or the combination of multiple sequences [40,41]. Moreover, the precision and constraints of methods used to construct phylogenetic trees [6,42,43], the quantity and diversity of species examined, the choice of outgroups, and the handling of sequences all play a role in shaping the final results. A prime example of such inconsistencies is seen in the phylogenetic positioning of the Palliolinae, a monophyletic subgroup within the family Pectinidae. Various studies have placed the Palliolinae in different clades, either with the Pectininae or the Chlamyinae, illustrating the persistent challenges in achieving phylogenetic resolution within this family (e.g., Alejandrino et al. [4], Sherratt et al. [5], Xu et al. [21], Lin et al. [22], Malkócs et al. [25], Li et al. [27], Smedley et al. [28], Saavedra and Peña [33], Feng et al. [34], and Malkowsky and Klussmann-Kolb [40]). In contrast, Waller’s hypothesis proposed that the Palliolinae are sisters to the Pectininae [31]. On the mito-phylogenomics level, differences in sequence selection and methodologies can lead to varying results, as evidenced by Lin et al. [22], Malkócs et al. [25], and Li et al. [27]. The divergence time estimation analysis conducted by Lin et al. [22] based on concatenated mitochondrial protein-coding gene sequences produced a phylogenetic structure akin to our own findings, hinting that variations in outcomes might be linked to the selection of samples. In conclusion, the discrepancies observed in phylogenetic studies can be ascribed to the diversity in sequence choices, analytical techniques, and the species included in the sampling.
Although the robustness of a phylogenetic tree can be affected by a variety of factors, it is noteworthy that Ylistrum and Amusium have consistently been distinguished in previous molecular phylogenetic studies. The placement of Ylistrum within the subfamily Pectininae is well-supported, as evidenced by the study of Matsumoto and Hayami [39] and subsequent research [4,28]. Nevertheless, due to the significant morphological and distributional similarities between Y. japonicum and A. pleuronectes, Ylistrum has historically been grouped with the Amusiini, even though molecular genetic studies have consistently pointed to its distinctiveness from Amusium. Our phylogenetic study based on the complete mitochondrial genome indicates that Ylistrum has an ancient origin, but its precise placement within the subfamily Pectininae remains ambiguous due to insufficient sample data. Alejandrino et al. [4] analyzed the phylogeny of 81 extant taxa from the Pectinidae based on the nuclear Histone H3, 12S rRNA, 16S rRNA data, and 28S rRNA data, and the result shows that Ylistrum was placed among the species of the tribe Decatopectinini, and Aumsium were nested in a different clade (Pectinini). Subsequently, Smedley et al. [28] expanded the dataset to 62 Pectinidae species on the basis of Alejandrino et al. [4] into a new phylogenetic analysis, and clade Ylistrum was once again placed in the Decatopectinini. Interestingly, it forms a sister clade with two Annachlamys species belonging to the tribe Pectinini, but the phylogenetic location of Annachlamys is still debated [3]. Mynhardt et al. [1] focused on the phylogenetic analysis of Aumsium and Ylistrum, restored their respective monophyletic clades, and described Ylistrum as a new genus. In this study, Ylistrum also forms a sister group with a Decatopectinini species (A. antillarum). Our phylogenetic analysis based on 16S rRNA indicates the same result as that of former studies: two Ylistrum taxa are still nested in the tribe Decatopectinini. In light of these findings and previous studies, we adhere to Serb’s classification [6], positioning Ylistrum within the Decatopectinini tribe.
The complete mitochondrial genome sequence data for Ylistrum remain inadequate. The available data for Y. balloti (accession number ON041136) may be based on an erroneous identification. NCBI-BLAST analysis of this sequence, with an alignment length exceeding 1200 nucleotides, reveals a similarity greater than 98.99% with Y. japonicum but less than 94.10% with Y. balloti. This indicates that ON041136 is likely a misidentified Y. japonicum rather than Y. balloti. The researcher who submitted this sample has not published their findings, and morphological verification is not possible. Additionally, the collection location of ON041136 is Beihai, Guangxi, China, which is problematic since there are no documented distributions of Y. balloti within China. Generally speaking, the right valve of Y. balloti is white or pale brown, with concentric, irregularly sized violet–brown spots [1], rather than a white, unspotted right valve, so the mention of Y. balloti in the Chinese Zoology book [7] is considered a misidentification, highlighting the need for a critical revision of this information.
4. Materials and Methods
4.1. Sample Collection
A total of 50 specimens were collected from 2022 to 2023 from Hailing Island, Yangjiang City, Guangdong Province, China (21.61 N, 111.93 E) (Figure 6). Morphometric measurements were performed using an electronic vernier caliper (0.1 mm), and body measurement traits (shell length, shell height, shell width, shell weight, etc.) were recorded for further investigation. All collected samples were intended for commercial purposes, and there were no concerns regarding animal ethics. The morphological characteristics of these specimens were categorized and compared in accordance with Zhang et al. [38] and Zhang [44].
Figure 6.
Sampling location of Ylistrum japonicum (modified from d-maps: https://d-maps.com, accessed on 10 May 2024).
4.2. DNA Extraction, Library Preparation, and Next Generation Sequence
One specimen had its adductor muscles extracted (5 g) for DNA extraction; gDNA was extracted by the MagPure Bacterial DNA Kit (Magen, Guangzhou, China) following pre-grinding in liquid nitrogen. The Qubit dsDNA HS assay kit (Sangon, Shanghai, China) was used to test the concentration and 1% agarose gel electrophoresis to confirm integrity. The library preparation and next-generation sequence were finished by Sangon Biotech (Shanghai) Co., Ltd. First, 500 ng quantified DNA was randomly fragmented by Covaris (Woburn, MA, USA). Next, Hieff NGS®®MaxUp II DNA Library Prep Kit for Illumina®® (YEASEN, Shanghai, China) was used for the next steps. Briefly, Endprep enzyme was added to repair the end and 3’ end A tail ligation. Then, the adaptor was ligated by an enhancer and Fast T4 DNA ligase. Index primer was added by PCR, and the amplified product, about 400 bp, was selected using DNA selection beads. The library concentration and size were confirmed by Qubit 4.0 (Thermo, Waltham, MA, USA) and 2% agarose gel electrophoresis, respectively.
Then, the libraries were pooled and loaded on the Novaseq 6000 (Illumina. San Diego, CA, USA)/DNBseq-T7 (BGI, Shenzhen, China) sequencer using the 2×150 bp paired-end sequence kit according to the manufacturer´s instructions.
4.3. Sequence Assembly and Annotation
Raw bases yielded at least 6 GB and were used for downstream analysis. First, all of the raw reads were trimmed by Fastqc v0.11.2 [45]. The software SPAdes v3.15 [46] was used to assemble the raw sequence reads into contigs. tBLASTn v2.6.0 and GeneWise were used to obtain the CDS gene boundary by reverse alignment with the near-source reference database; the tRNA sequence annotation was obtained by MiTFi, Rfam used cmsearch alignment to identify non-coding rRNA, and the final summary was put into complete annotation results. The circular gene maps of the species Y. japonicum were drawn by Circos v0.69.
4.4. Gene Collinearity
Gene collinearity among complete mitochondrial sequences of five Pectininae species was explored to assess their phylogenetic relationship with Y. japonicum, using the progressiveMauve algorithm and default parameters (including default seed weight, determine locally collinear blocks and full alignment) in the Mauve v2.4.0 [47].
4.5. Phylogenetic Analysis with Mitochondrial Genome
Two phylogenetic analyses were conducted based on the complete mitochondrial sequences of Y. japonicum in this study. Following the methodologies established in previous studies by Malkowsky and Klussmann-Kolb [39], Xu et al. [21], and Malkócs et al. [25], a total of 15 mitochondrial sequences of Pectinidae species and outgroup taxa were selected to construct phylogenetic trees. This selection included 11 Pectinidae species across three subfamilies: Pectininae, Palliolinae, and Chlamydinae. The available mitochondrial genome sequences were obtained from GeneBank, incorporating 13 complete mitochondrial genomes and 2 incomplete sequences (KP900974, KP900975), each over 16,000 base pairs in length. Two Ostreidae species, Magallana bilineata and Magallana gigas, and two Mytilidae species, Mytilus galloprovincialis and Mytilus trossulus, were used as outgroups.
PhyloSuite v1.2.3 [48] was utilized to extract the protein-coding genes (PCGs) from each sequence. All sequences were aligned in batches with MAFFT v7.505 [49]. The alignments were refined using the codon-aware program MACSE v2.06 [50], which preserves the reading frame and allows the incorporation of sequencing errors or sequences with frameshifts. Ambiguously aligned fragments of the alignments were removed in batches using Gblocks 0.91b [51]. ModelFinder v2.2.0 [52] was used to select the best-fit partition model. The phylogenetic tree was subsequently constructed using both the Maximum likelihood (ML) method in IQ-TREE v2.2.0 [53,54] and Bayesian inference (BI) in MrBayes v3.2.7a [55]. Branch support was determined with 5000 bootstrap iterations for the best-scoring ML tree. Markov Chain Monte Carlo (MCMC) analyses were run for 1,000,000 generations (sampling every 1000 generations), in which an initial 50% of sampled data were discarded as burn-in. The result was beautified with FigTree v1.4.4.
To explore in more detail the monophyletic development of Y. japonicum and its taxonomic position in Pectininae, another ML tree was constructed using 16S rRNA by PhyloSuite v1.2.3 [48]; three Y. japonicum specimens from different regions (China and Japan) and the other 8 specimens of subfamily Pectininae were selected. All Sequences were aligned in batches with MAFFT v7.505 [49] and pruned by Gblocks 0.91b [51]. Branch support was determined with 5000 bootstrap iterations for the best-scoring ML tree. A list of specimens included in molecular studies is shown in Table 2.

Table 2.
List of specimens included in the molecular studies.
Author Contributions
Conceptualization, Y.T.; Methodology, Y.T.; Investigation, Y.H., Y.X. and Y.T.; Resources, Y.T.; Funding acquisition, Y.T. and Z.H.; Writing—original draft, Y.H. and Y.X.; Writing—review and editing, Y.T., J.M. and X.W.; Supervision, Y.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the National Key Research and Development Program of China (2022YFD2400302), the Central Government Subsidy Project for Liaoning Fisheries (2023), and the Science and Technology Foundation of Dalian (2021JB11SN035).
Institutional Review Board Statement
The experimental protocol was designed in accordance with the recommendations of the Regulations of the Laboratory Animal Guideline for Ethical Review of Animal Welfare (National Standards of China, GB/T 35823—2018) and reviewed and approved by the animal care and use committee of Dalian Ocean University (DLOU-2023009).
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Mynhardt, G.; Alejandrino, A.; Puslednik, L.; Corrales, J.; Serb, J.M. Shell shape convergence masks biological diversity in gliding scallops: Description of Ylistrum n. gen. (Pectinidae) from the Indo-Pacifc Ocean. J. Mollus. Stud. 2014, 80, 400–411. [Google Scholar] [CrossRef]
- Dijkstra, H.H. Pectinoidea (Bivalvia: Propeamussiidae and Pectinidae) from the Panglao region, Philippine Islands. Vita Malacologica. 2013, 10, 1–108. [Google Scholar]
- Dijkstra, H.H.; Beu, A.G. Living scallops of Australia and adjacent waters (Mollusca: Bivalvia: Pectinoidea: Propeamussiidae, Cyclochlamydidae and Pectinidae). Rec. Aust. Mus. 2018, 70, 113–330. [Google Scholar] [CrossRef]
- Alejandrino, A.; Puslednik, L.; Serb, J.M. Convergent and parallel evolution in life habit of the scallops (Bivalvia: Pectinidae). BMC Evol. Biol. 2011, 11, 164. [Google Scholar] [CrossRef]
- Sherratt, E.; Alejandrino, A.; Kraemer, A.C.; Serb, J.M.; Adams, D.C. Trends in the sand: Directional evolution in the shell shape of recessing scallops (Bivalvia: Pectinidae). Evolution 2016, 70, 2061–2073. [Google Scholar] [CrossRef]
- Serb, J.M. Reconciling morphological and molecular approaches in developing a phylogeny for the Pectinidae (Mollusca: Bivalvia). In Scallops Biology, Ecology, Aquaculture, and Fisheries, 3rd ed.; Shumway, S.E., Parsons, G.J., Eds.; Elsevier: Amsterdam, The Netherlands, 2016; pp. 1–29. [Google Scholar]
- Wang, Z.R. Fauna Sinica Invertebrate Vol. 31 Mollusca Bivalvia; Science Press: Beijing, China, 2002; pp. 1–374. (In Chinese) [Google Scholar]
- Gmelin, J.F. (Ed.) Caroli a Linnaei Systema Naturae per Regna Tria Naturae, 13th ed.; Tom. 1, Pars 6; Georg. Emanuel Deer: Lipsiae, Germany, 1791; pp. 3021–3910. [Google Scholar]
- Bernardi, M. Description d’espéces nouvellas. J. Conchyl. 1861, 9, 46–49. [Google Scholar]
- Abbott, R.T.; Dance, S.P. Compendium of Seashells: A Colour Guide to More Than 4200 of the World’s Marine Shells; EP Dutton Inc.: New York, NY, USA, 1982; pp. 1–411. [Google Scholar]
- Carpenter, K.E.; Niem, V.H. The living marine resources of the Western Central Pacifc. In Volume 2: Cephalopods, Crustaceans, Holothurians and Sharks; Food and Agriculture Organization of the United Nations: Rome, Italy, 1998; pp. 1–686. [Google Scholar]
- Beu, A.G.; Darragh, T.A. Revision of southern Australian Cenozoic fossil Pectinidae (Mollusca: Bivalvia). Proc. R. Soc. Vic. 2001, 113, 1–205. [Google Scholar]
- Okada, Y.; Uchida, S.; Uchida, T. New Illustrated Encyclopediaof the Fauna of Japan; II. Hokuryukan: Tokyo, Japan, 1998; pp. 1–803. (In Japanese) [Google Scholar]
- Kanmizutaru, T.; Anraku, K. Inducement of Shell Opening of Amusium japonicum by MgCl2 Injection into the Adductor Muscle. Nippon Suisan Gakk. 1999, 65, 856–859. [Google Scholar] [CrossRef]
- Kanmizutaru, T.; Anraku, K.; Toyoda, S. Light perception capability of pallial eyes in Japanese moon scallop Amusium japonicum as determined by electroretinogram. Nippon Suisan Gakk. 2005, 71, 928–934. [Google Scholar] [CrossRef][Green Version]
- Son, P.W.; Chung, E.Y. Annual Reproductive Cycle and Size at First Sexual Maturity of the Sun and Moon Scallop Amusium Japonicum Japonicum (Gmelin, 1791) (Bivalvia: Pectinidae) in the Coastal Waters of Jejudo, Korea. Malacologia 1994, 51, 119–129. [Google Scholar] [CrossRef]
- Son, P.W.; Chung, E.Y. Gonadal Development, First Sexual Maturity and Sex Ratio of the Sun and Moon Scallop Amusium japonicum japonicum on the Coastal Waters of Jejudo, Korea. Dev. Reprod. 2005, 9, 95–103. [Google Scholar]
- Son, P.W.; Ha, D.S.; Rho, S.; Chang, D.S. Studies on the Age and Growth fo Sun and Moon Scallop, Amusium japonicum japonicum (GMELIN). J. Aquac. 1996, 9, 409–417. (In Korean) [Google Scholar]
- Ye, W.J.; Liang, G.Y. A proliminary study on artificial breeding of Amussium japonica (Gmelin). JOL 1989, 4, 86–93. (In Chinese) [Google Scholar]
- Ye, W.J.; Liang, G.Y. A preliminary observation on the ecology of Amussium japonica (Gmelin). Chin. J. Zool. 1990, 25, 5–7. (In Chinese) [Google Scholar]
- Xu, K.; Kanno, M.; Yu, H.; Li, Q.; Kijima, A. Complete mitochondrial DNA sequence and phylogenetic analysis of Zhikong scallop Chlamys farreri (Bivalvia: Pectinidae). Mol. Biol. Rep. 2011, 38, 3067–3074. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.T.; Li, Y.X.; Sun, Y.; Tao, J.; Qiu, J.W. A new species of the genus Catillopecten (Bivalvia: Pectinoidea: Propeamussiidae): Morphology, mitochondrial genome, and phylogenetic relationship. Front. Mar. Sci. 2023, 10, 1168991. [Google Scholar] [CrossRef]
- Boore, J.L.; Medina, M.; Rosenberg, L.A. Complete sequences of the highly rearranged molluscan mitochondrial genomes of the scaphopod Graptacme eborea and the bivalve Mytilus edulis. Mol. Biol. Evol. 2004, 21, 1492–1503. [Google Scholar] [CrossRef]
- Smith, D.R.; Snyder, M. Complete mitochondrial DNA sequence of the scallop Placopecten magellanicus: Evidence of transposition leading to an uncharacteristically large mitochondrial genome. J. Mol. Evol. 2007, 65, 380–391. [Google Scholar] [CrossRef]
- Malkócs, T.; Viricel, A.; Becquet, V.; Evin, L.; Dubillot, E.; Pante, E. Complex mitogenomic rearrangements within the Pectinidae (Mollusca: Bivalvia). BMC Ecol. Evol. 2022, 22, 29. [Google Scholar] [CrossRef]
- Boore, J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef]
- Li, F.; Zhang, Y.; Zhong, T.; Heng, X.; Ao, T.; Gu, Z.; Wang, A.; Liu, C.; Yang, Y. The Complete Mitochondrial Genomes of Two Rock Scallops (Bivalvia: Spondylidae) Indicate Extensive Gene Rearrangements and Adaptive Evolution Compared with Pectinidae. Int. J. Mol. Sci. 2023, 24, 13844. [Google Scholar] [CrossRef] [PubMed]
- Smedley, G.D.; Audino, J.A.; Grula, C.; Porath-Krause, A.; Pairett, A.N.; Alejandrino, A.; Serb, J.M. Molecular phylogeny of the Pectinoidea (Bivalvia) indicates Propeamussiidae to be a non-monophyletic family with one clade sister to the scallops (Pectinidae). Mol. Phylogenet. Evol. 2019, 137, 293–299. [Google Scholar] [CrossRef] [PubMed]
- Yao, L.J.; Yu, H.W.; Liu, Y.R.; Zhang, Y.H.; Li, Y.L. The complete mitochondrial genome and phylogenetic analysis of Amusium pleuronectes. Mitochondrial DNA B 2020, 5, 2318–2319. [Google Scholar] [CrossRef] [PubMed]
- Waller, T.R. Evolutionary relationships among commercial scallops (Mollusca: Bivalvia: Pectinidae). In Scallops: Biology, Ecology and Aquaculture; Shumway, S.E., Ed.; Elsevier: New York, NY, USA, 1991; pp. 1–73. [Google Scholar]
- Waller, T.R. New phylogenies of the Pectinidae (Mollusca: Bivalvia): Reconciling morphological and molecular approaches. In Scallops: Biology, Ecology and Aquaculture; Shumway, S.E., Parsons, G.J., Eds.; Elsevier B.V.: Amsterdam, The Netherlands, 2006; pp. 1–44. [Google Scholar]
- Waller, T.R. The ctenolium of scallop shells: Functional morphology and evolution of a key family-level character in the Pectiniacea (Mollusca: Bivalvia). Malacologia 1984, 25, 20. [Google Scholar]
- Saavedra, C.; Peña, J.B. Phylogenetics of American scallops (Bivalvia: Pectinidae) based on partial 16S and 12S ribosomal RNA gene sequences. Mar. Biol. 2006, 150, 111–119. [Google Scholar] [CrossRef]
- Feng, Y.W.; Li, Q.; Kong, L.F.; Zheng, X.D. DNA barcoding and phylogenetic analysis of Pectinidae (Mollusca: Bivalvia) based on mitochondrial COI and 16S rRNA genes. Mol. Biol. Rep. 2011, 38, 291–299. [Google Scholar] [CrossRef]
- Barucca, M.; Olmo, E.; Schiaparelli, S.; Canapa, A. Molecular phylogeny of the family Pectinidae (Mollusca: Bivalvia) based on mitochondrial 16S and 12S rRNA genes. Mol. Phylogenet. Evol. 2004, 31, 89–95. [Google Scholar] [CrossRef]
- Marin, A.; Fujimoto, T.; Arai, K. The variable 5′end of the 16S rRNA gene as a novel barcoding tool for scallops (Bivalvia, Pectinidae). Fish. Sci. 2015, 81, 73–81. [Google Scholar] [CrossRef]
- Habe, T. New name for Amusium japonicum formosum. Venus 1992, 50, 235. [Google Scholar]
- Zhang, X.; Qi, Z.Y.; Li, J.M.; Ma, X.T.; Wang, Z.R.; Huang, X.M.; Zhuang, Q.Q. Bivalve mollusca of the South China Sea; Science Press: Beijing, China, 1960; pp. 1–274. (In Chinese) [Google Scholar]
- Matsumoto, M.; Hayami, I. Phylogenetic analysis of the family Pectinidae (Bivalvia) based on mitochondrial cytochrome C oxidase subunit I. J. Mollus. Stud. 2000, 66, 477–488. [Google Scholar] [CrossRef]
- Malkowsky, Y.; Klussmann-Kolb, A. Phylogeny and spatio-temporal distribution of European Pectinidae (Mollusca: Bivalvia). Syst. Biodivers. 2012, 10, 233–242. [Google Scholar] [CrossRef]
- Puslednik, L.; Serb, J.M. Molecular phylogenetics of the Pectinidae (Mollusca: Bivalvia) and the effect of outgroup selection and increased taxon sampling on tree topology. Mol. Phylogenet. Evol. 2008, 48, 1178–1188. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.-X.; Li, Y.; Hittinger, C.T.; Chen, X.; Rokas, A. An investigation of irreproducibility in maximum likelihood phylogenetic inference. Nat. Commun. 2020, 11, 6096. [Google Scholar] [CrossRef] [PubMed]
- Haag, J.; Höhler, D.; Bettisworth, B.; Stamatakis, A. From Easy to Hopeless-Predicting the Difficulty of Phylogenetic Analyses. Mol. Biol. Evol. 2022, 39, msac254. [Google Scholar] [CrossRef]
- Zhang, S.P. Atlas of Marine Mollusks of China; China Ocean Press: Beijing, China, 2008; pp. 1–383. (In Chinese) [Google Scholar]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Darling, A.C.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004, 14, 1394–1403. Available online: http://www.genome.org/cgi/doi/10.1101/gr.2289704 (accessed on 16 April 2004). [CrossRef] [PubMed]
- Xiang, C.; Gao, F.; Jakovlić, I.; Lei, H.; Hu, Y.; Zhang, H.; Zou, H.; Wang, G.; Zhang, D. Using PhyloSuite for Molecular Phylogeny and Tree-based Analyses. iMeta 2023, 2, e87. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Ranwez, V.; Douzery, E.J.P.; Cambon, C.; Chantret, N.; Delsuc, F. MACSE v2: Toolkit for the alignment of coding sequences accounting for frameshifts and stop codons. Mol. Biol. Evol. 2018, 35, 2582–2584. [Google Scholar] [CrossRef] [PubMed]
- Talavera, G.; Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Nguyen, M.A.; von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Hohna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).