Abstract
Plants are constantly exposed to a variety of different environmental stresses, including drought, salinity, and elevated temperatures. These stress cues are assumed to intensify in the future driven by the global climate change scenario which we are currently experiencing. These stressors have largely detrimental effects on plant growth and development and, therefore, put global food security in jeopardy. For this reason, it is necessary to expand our understanding of the underlying mechanisms by which plants respond to abiotic stresses. Especially boosting our insight into the ways by which plants balance their growth and their defense programs appear to be of paramount importance, as this may lead to novel perspectives that can pave the way to increase agricultural productivity in a sustainable manner. In this review, our aim was to present a detailed overview of different facets of the crosstalk between the antagonistic plant hormones abscisic acid (ABA) and auxin, two phytohormones that are the main drivers of plant stress responses, on the one hand, and plant growth, on the other.
1. Introduction
Global warming and environmental pollution are among the major threats that humanity will face in the coming decades. Rising global temperatures and more frequent heat waves accompanied by unforeseeable torrential rains [1,2] are assumed to substantially exacerbate the conditions of abiotic stress in which plants will have to survive and prosper in the future. These unfavorable conditions are expected to have detrimental effects on crop quality and productivity [3]. Abiotic stress cues affect plant morphology, biochemistry, physiology, and anatomy by altering biological processes as diverse as photosynthesis, respiration, growth, and development. Moreover, long-term stress can finally lead to plant death [4]. Plants have developed sophisticated gene regulatory networks and defense mechanisms to face abiotic stresses [5,6]. Drought and increased soil salinity affect plant yields [7,8], and if we add the increasing population that humanity is experiencing, the problem is exacerbated. By 2050, the world population is expected to reach around nine billion [9,10] and, consequently, the availability of arable land will decrease [9]. The global demand for crops will grow 100–110% [11] and one solution we have is to improve plant yields to secure future food supply by developing genetic strategies that allow crops to tolerate abiotic stresses caused by climate change [12].
Plants perceive changes in their environment and, in turn, modulate the content and distribution of a small number of metabolites, referred to as plant hormones, which serve the integration and transmission of internal signals [13,14,15]. ‘Hormone’, from Greek hormān, meaning ‘to stimulate’ or ‘to set in motion’ in English, refers to small bioactive signaling molecules that can exert their action at a very low, submicromolar concentration [16]. Throughout evolution, plants have developed a set of these small molecules that play key roles in the regulation of plant growth and development, as well as in the development of adequate responses to biotic and abiotic stress cues [17,18]. Currently, nine different classes of plant hormones are considered and classified according to their major effect on plant growth. The common classification includes, on the one hand, stress response-related hormones, i.e., abscisic acid (ABA), ethylene, salicylates (SAs), and jasmonates (JAs), and, on the other hand, growth-promoting hormones, including the classes of auxins, gibberellins (GAs), cytokinins (CKs), brassinosteroids (BRs) and strigolactones (SLs) [19,20,21]. For a hormone to be capable of triggering its physiological function, it must first be recognized by its receptor(s); subsequently, its signaling pathway must be activated, which will ultimately result in transcriptional modifications driven by the activation or inactivation of a given number of specific plant hormone-responsive transcription factors [15].
However, many recent studies indicated that the classification of plant hormones, according to their growth-promoting or -repressing effects, is all too simplistic and needs to be readdressed because many interactions between antagonistic plant hormones have been described in the past. Just to name some examples, wound induced formation of JA has been shown to trigger auxin biosynthesis in Arabidopsis thaliana through the induction of YUCCA8 and YUCCA9 gene expression, involving the transcriptional control of target genes by transcription factors from the JA-specific basic helix loop helix family myelocytomatosis oncogene (MYC) [22,23]. Moreover, auxin and JA have been described to collaborate in flower development [24,25], and the signaling pathways of both hormones are known to share some common components, including the Arabidopsis S-PHASE KINASE-ASSOCIATED PROTEIN 1 (ASK1) and TOPLESS (TPL) [17] while auxin and ethylene are interacting in controlling fruit ripening in fruits [26,27]. This review provides updated information on the molecular intricacies by which the main plant growth factor, indole-3-acetic acid (IAA), interacts with the growth repressing phytohormone ABA, thus contributing to orchestrate plant growth under abiotic stress conditions that can arise from the global climate change scenario. The discussed novel insight into the crosstalk between IAA and ABA is assumed to open new possibilities for biotechnological innovations that will lead to sustainable solutions to improve agricultural productivity and help ensure food security in the long run.
2. Auxins
The class of auxins contains the most important phytohormones related to plant growth and development. Auxins are involved in processes ranging from cell expansion growth, control of cell division, vascular tissue differentiation, lateral and adventitious root initiation, tropistic responses (gravitropism and phototropism), maintenance of apical dominance, delay of leaf senescence, and contributions to the control of leaf and fruit abscission, as well as fruit ripening [28,29].
2.1. Auxin Biosynthesis
IAA is considered the most abundant and most important representative of the auxin class in the plant kingdom. Its occurrence in plants was first described nearly a century ago by Frits W. Went and Kenneth V. Thimann [30,31], but despite decades of intense efforts, the biosynthesis of this first identified plant hormone remained enigmatic until a few years ago. For a long time, two different biosynthetic starting points were discussed in the literature. For one thing, auxin biosynthesis pathways dependent on l-tryptophan (l-Trp), including the indole-3-pyruvic acid (IPyA) pathway [32,33,34,35,36], the main auxin biosynthesis pathway in plants and the only biosynthetic route fully disclosed to date leading to auxin in higher plants, were suggested. On the contrary, an l-Trp-independent auxin biosynthesis pathway was introduced more than 30 years ago [37,38], although clear genetic evidence was lacking and mass spectrometric findings were controversially discussed in the literature [39]. However, a more recent study describing the presumably crucial role of the indole synthase complex during embryogenesis revived the discussion of an l-Trp-independent auxin biosynthesis pathway [40].
Regarding the auxin biosynthesis pathways dependent on l-Trp, together with the IPyA pathway for IAA biosynthesis that involves the conversion of l-Trp to IPyA by means of two l-Trp aminotransferases, TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS 1 (TAA1) and TRYPTOPHAN AMINOTRANSFERASE-RELATED 2 (TAR2) [33,41], and the subsequent enzymatic transformation of IPyA into IAA by flavin-containing monooxygenases of the YUCCA family [32,35,42], three additional pathways have been described: the tryptamine (TAM)-, the indole-3-acetamide (IAM)-, and the indole-3-acetaldoxime (IAOx) pathway [43,44,45]. While the TAM pathway does not appear to be of great relevance for auxin de novo biosynthesis, as there is no evidence that the inactivation of the involved aldehyde oxidases in the aba3 mutant translated into altered IAA levels in Arabidopsis [35,36], it must be concluded that the two other pathways may play minor or specialized roles in auxin biosynthesis. The occurrence of IAOx has initially been limited to members of species of the Brassicaceae family, as outside of the Brassicas, no IAOx producing cytochrome P450 monooxygenases (CYP79B2, CYP79B3) have been described [46,47]. Recent publications challenged this notion, reporting either the identification of CYP79 enzymes capable of producing IAOx or the conversion of IAOx into IAA in non-Brassica species, including, for instance, poplar, maize, and Medicago truncatula [48,49,50]. Interestingly, it is suggested that IAOx is the main precursor of IAM in Arabidopsis [51], but the enzyme that converts IAOx to IAM remains elusive. In addition to that, the main role of the IAOx pathway is probably the production of secondary plant metabolites, including indole glucosinolates, camalexin, and indole-3-carboxylic acid, which are crucial for defense reactions in Arabidopsis [52]. The IAM pathway has long been thought to be restricted to bacteria that use this two-step pathway involving a tryptophan 2-monooxygenase (iaaM/tms1/aux1) and an IAM hydrolase (iaaH/tms2/aux2) to convert l-Trp into IAA [53]. Nonetheless, nearly 20 years ago, the first plant IAM hydrolase gene from Arabidopsis, AMIDASE 1 (AMI1), was cloned and several other AMI1-like enzymes have been characterized from a multitude of other plant species since then [54,55,56]. AMI1 belongs to the amidase signature (AS) family of amidohydrolases, of which at least one other member, FATTY ACID AMIDE HYDROLASE (FAAH), is involved in terminating the signaling activity of other signaling molecules, N-acylethanolamines [57,58]. Along with AMI1, a recently described genetic screen suggested the involvement of two additional putative IAM hydrolases, IAMH1 and IAMH2, in the conversion of IAM, but so far no reports on their enzymatic characterization have been provided, and the double mutant does not show significant differences in its IAA content under control conditions but only when IAM is applied exogenously, suggesting a possible unspecific activity of IAMH1 and IAMH2 when IAM levels are artificially increased [59]. The latter observation stands in clear contrast to the recently reported findings for two ami1 alleles that both showed significantly reduced IAA levels and elevated IAM contents under control conditions [60]. Along with the broad distribution of AMI1-like enzymes in the plant kingdom, this implies a more specific role of AMI1-like IAM hydrolases in plants.
2.2. Auxin Transport
For proper plant growth and development, strict control of cellular auxin homeostasis and auxin fluxes along the plant body is imperative. Auxin gradients produced in plant tissues change dynamically due to developmental programs and in response to external stimuli [61,62]. In this context, auxin transport plays a key role. In general, plants possess two different auxin transport systems. On the one hand, plants transport auxin non-directionally in the phloem to recipient organs, together with the photo-assimilates. On the other hand, they possess directional polar auxin transport (PAT), which distributes auxin in a spatially precise manner. The latter transport system is the main contributor to the generation of auxin gradients across plant tissues [63] and fundamental for generating local auxin maxima [64]. The formation of these maxima is of paramount importance for many developmental processes, such as phyllotaxis, as well as flower and lateral root development [65,66,67], or to generate adequate responses to external stimuli, including phototropism [68].
The acidic pH of the apoplast (pH~5.5) [69], in contrast to the neutral pH (pH~7) of the cytoplasm, allows passive diffusion of auxin across cell membranes, although auxin can also enter the cell via specialized transporters of the AUXIN RESISTENT 1/LIKE AUX1 family (AUX1/LAX) [70]. Due to the low pH in the apoplast, IAA occurs in its protonated state (IAAH), while in the cytoplasm IAA dissociates into its ionic form (IAA−), preventing the passive efflux of the molecule from the cell [64]. Auxin efflux depends on auxin transporters of the PIN-formed (PIN) family or P-glycoprotein/ATP-binding cassette B (ABCB) transporters. PIN1-4 and PIN7 are located in the plasma membrane, where they mainly contribute to auxin efflux to the apoplast. In contrast, PIN5 and PIN8 are found in the endoplasmic reticulum together with PIN6, which can be found in both the plasma membrane and the endoplasmic reticulum [71,72]. However, the most remarkable feature of the PINs is their polar distribution in the plasma membrane, which allows for the directional flux of auxin and the formation of auxin maxima.
2.3. Auxin Signaling
Auxin signaling occurs both in the nucleus and in the plasma membrane. The core element that controls the signaling pathway is the E3 ubiquitin ligase complex SCFTIR1/AFB that either contains the F-box protein TRANSPORT INHIBITOR RESPONSE 1 (TIR1) or a TIR1 homolog auxin-related F-box protein (AFB) [73], the co-receptor/repressor proteins auxin/indole-3-acetic acid (Aux/IAA) [74], and the auxin response factor (ARF) transcription factor family [75]. The receptor switches between two different activity states depending on the presence or absence of auxin (Figure 1).
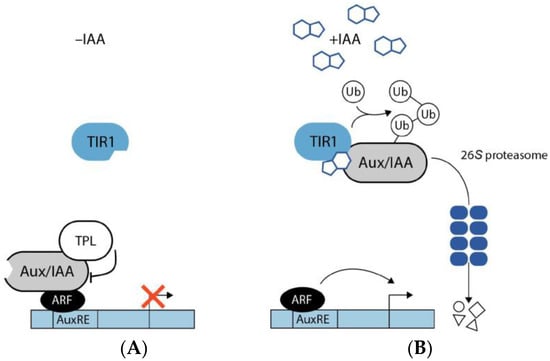
Figure 1.
Auxin signaling pathway. (A) In the absence of IAA, the F-box protein TRANSPORT INHIBITOR RESPONSE 1 (TIR1), which acts as an auxin receptor, comprises no bound IAA molecules. Auxin/indole-3-acetic acid (Aux/IAA) proteins together with the co-repressor TOPLESS (TPL) interact physically with auxin response factor (ARF) transcription factors and, thereby, prevent the transcription of genes that contain auxin responsive elements (AuxRE) in their promoter sequence. (B) In the presence of IAA, the bioactive hormone binds to the binding cavity of the TIR1 receptor. Through the binding of IAA, TIR1 affinity for the Aux/IAA transcriptional repressors increases. Subsequently, Aux/IAA proteins are tagged with ubiquitin (Ub) for their degradation via the 26S proteasome. The ubiquitin-mediated proteolysis of Aux/IAA results in the liberation of the co-repressor TPL and the ARFs, thus, allowing ARFs to promote transcription of AuxRE containing genes.
In the absence of auxin, Aux/IAA repressor proteins heterooligomerize with the ARFs and the co-repressor TOPLESS (TPL), thereby inhibiting the transcriptional activity of the ARF transcription factors. However, in the presence of auxin, which binds at the bottom of an Aux/IAA protein-specific pocket of the F-box protein, the E3 ubiquitin ligase complex SCFTIR1/AFB can recruit the Aux/IAA co-receptor proteins. Following binding, the Aux/IAA proteins are polyubiquitinated and thereby tagged for their subsequent proteolytic degradation by the 26S proteasome. The ARFs, in turn, are liberated from their repression and are free to mediate auxin-responsive transcriptional responses [76,77]. However, some very rapid auxin-triggered effects, including plasma membrane hyperpolarization and protoplast swelling that manifest within minutes or even seconds could not be attributed to the described gene regulatory system. Thus, for almost 50 years, the existence of a second auxin receptor responsible for fast responses has been hypothesized [78], but it was only very recently that the role of the long discussed best candidate, the plasma membrane localized AUXIN BINDING PROTEIN 1 (ABP1), in conferring rapid responses to auxin, was demonstrated beyond doubt [79].
3. Abscisic Acid
ABA is a sesquiterpene isoprenoid (C15H20O4). ABA biosynthesis commences in plastids, while the final steps occur in the cytosol [80]. It is obtained mainly from the cleavage of β-carotene (C40) [80,81,82], a derivative of the 2-C-methyl-d-erytritol (MEP) pathway, through multistep enzymatic catalysis. ABA received its name in 1968 [83], after being previously discovered and named as ‘inhibitor β’ and ‘dormin’, respectively [84], in studies on the control of leaf abscission and bud dormancy. Up to date, the role of ABA as a hormone involved in developmental processes is still being revisited. For example, basal ABA levels seem to play an indispensable role in xylem and leaf development [85], and promote crucial subcellular processes, such as chloroplast biogenesis [86]. For a more thorough review on ABA discovery, refer to [87].
3.1. ABA Biosynthesis
The formation of ABA involves a complex multi-step enzymatic conversion of β-carotene [88,89]. The first committed step in ABA biosynthesis occurs in plastids. Zeaxanthin or antheraxanthin is converted to all-trans-violaxanthin by the enzyme zeaxanthin epoxidase (ZEP), which is known as ABA1 in Arabidopsis, the only ZEP gene in this species. However, it should be noted that aba1 mutants contain some residual ABA, suggesting that there is an alternative minor biosynthesis pathway operative in Arabidopsis [90]. Then, all-trans-violaxanthin is converted into 9-cis-violaxanthin or (via trans-neoxanthin) into 9′-cis-neoxanthin. In Arabidopsis, ABA4 produces cis-neoxanthin and cis-violaxanthin [91,92]. Subsequently, the ABA4 product is converted further to xanthoxin (C15) by 9-cis-epoxy carotenoid dioxygenases (NCEDs) by cleavage of 9-cis-violaxanthin or 9′-cis-neoxanthin. The NCED enzyme family has been found to require iron and oxygen to become catalytically active [93]. In Arabidopsis, NCED3 is thought to play a very prominent role in catalyzing the rate-limiting step in the biosynthesis of ABA [94]. Interestingly, NCED3 gene expression has recently been associated with IAM abundance in Arabidopsis [59]. Xanthoxin is then transported to the cytosol, where the remaining reaction steps of ABA synthesis take place. In a first step, a short-chain alcohol dehydrogenase/reductase, ABA2 in Arabidopsis, converts xanthoxin to ABA-aldehyde [95], before ABA-aldehyde is finally oxidized to ABA by abscisic acid oxidases (AAOs). In Arabidopsis, ABSCISIC ALDEHYDE OXIDASE 3 (AAO3) is described to be the only AAO enzyme involved in ABA biosynthesis [96]. To exert its enzyme activity, AAO3 requires a sulfurated molybdenum cofactor (MoCo-S), which is provided by ABA3 in Arabidopsis [97] and FLACCA in tomato [98], respectively. Mutants in ABA3 or FLACCA present an ABA-deficient phenotype [99,100].
3.2. ABA Transport
Classically, ABA was considered to show a root-to-shoot transport pattern, especially under drought stress. However, more recent studies have put this notion into question, and it is accepted today that ABA biosynthesis also takes place in aerial vascular tissues [101]. Several ABA transporters have recently been discovered, supporting the idea that ABA is transported throughout the plant by its specific transporters [102]. In this regard, proteins belonging to the P-glycoprotein/ATP-binding cassette G (ABCG) family have been described [103], both as exporters, as for instance ABCG25 [104], and importers, such as ABCG40 [105]. Moreover, members of this family participate in shoot-to-root ABA transport [106]. Other families which have been attributed to have an ABA transport activity include the nitrate transporters, e.g., ABA-IMPORTING TRANSPORTER 1/NITRATE TRANSPORTER 1.2(AIT1/NRT1.2) [107] and detoxification efflux carriers (DTX)/multidrug and toxic compound extrusion (MATE), such as DTX50 [108]. The observation that the NRT1/PTR family is involved in the transport of nitrogen-containing compounds, as well as ABA, is of special interest, since this opens new opportunities to study the intricacies of the relationship between shared ABA and nitrogen transport activities [109].
3.3. ABA Signaling
ABA perception and signaling are complex and involve the interaction of several proteins. For this reason, elucidation of ABA perception appeared to be very difficult and took considerable time and effort [110]. As depicted in Figure 2, ABA is sensed by its specific receptors named PYRABACTIN RESISTANCE/PYRABACTIN RESISTANCE-LIKE/REGULATORY COMPONENT OF ABA RECEPTORS (PYR/PYL/RCAR) [111,112]. In the absence of ABA, the receptors are inactive and their target proteins, the Ser/Thr phosphatases of the protein phosphatase 2C (PP2C) family, can block the activity of their downstream component, SUCROSE NONFERMENTING-1 RELATED PROTEIN KINASE 2 (SnRK2), thus preventing activity of the corresponding kinase [113,114], such as OST1 also known as SnRK2.6 [115]. In Arabidopsis, members of the PP2C family include ABI1/2, HAB1/2, HAI1/2/3, and AHG1/3 [116]. However, there is mounting evidence that other regulatory kinases could also be involved, such as the recently described RECEPTOR-LIKE PROTEIN KINASE 1 (RPK1) [117], and it would not be surprising if more pieces of the puzzle could be found in the future.
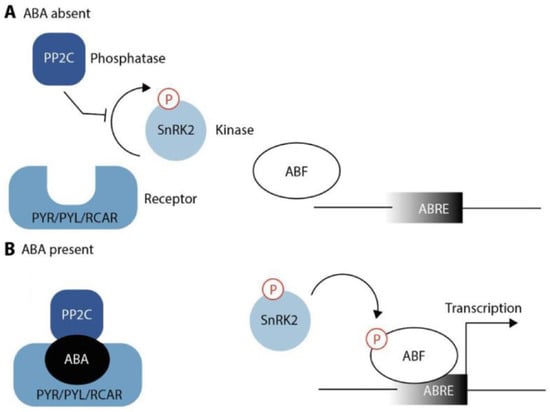
Figure 2.
Schematic model of ABA signaling. (A) In the absence of ABA, the PP2C phosphatase is free to inhibit autophosphorylation of a family of SnRK kinases. (B) ABA presence enables the PYR/PYL/RCAR family of proteins to bind to and sequester PP2C. This releases the SnRK kinases from inhibition, which then become auto-activated and, in turn, obtain the capacity to phosphorylate and activate downstream ABA-responsive transcription factors (ABF) to initiate transcription of genes that contain ABA-responsive promoter elements (ABREs) in their regulatory sequences.
In the presence of ABA, the hormone binds to its receptors. This facilitates the binding of the receptors to PP2Cs, causing their inactivation. Inactivation of PP2C, in turn, leads to the release of SnRK2s from repression. SnRK2s activate themselves through their autophosphorylation loop, which renders them capable of phosphorylating their downstream target proteins, most notably transcription factors from the ABA-responsive element binding protein/ABRE-binding factor (AREB/ABF) families [118]. Furthermore, SnRKs phosphorylate plasma membrane proteins, including SLOW ANION CHANNEL 1 (SLAC1) or POTASSIUM CHANNEL IN ARABIDOPSIS THALIANA 1 (KAT1) [113,114]. Negative effectors modulate the intensity of the response and eventually terminate the signaling cascade to prevent an excessive positive feedback loop [116].
4. Plant Hormone Crosstalk
Plant hormones do not act in entirely isolated manners, but in regulatory networks in which they operate in synergistic or antagonistic ways with each other. Consequently, the final condition of growth and development represents the net effect of the total hormone balance [119]. The term ‘hormone crosstalk’ describes how signal integration from multiple hormone inputs within a response network affects a common biological output [120]. Furthermore, it should be noted that the term ‘crosstalk’ refers to the interaction between two or more hormones or hormone pathways in cell signaling at the molecular level. However, hormone interplay would be an umbrella term that includes crosstalk too, and would encompass any hormonal interaction in the regulation of a physiological function, not necessarily antagonistic or synergistic [15]. Plant hormone crosstalk coordinates a sophisticated regulatory network, in a spatiotemporal fashion, to achieve specific physiological processes, regulate plant growth and development, as well as metabolism and defense [89]. Plant hormone networks are very complex and the change in one hormone, either through genetic- or metabolic changes, can cause alterations in others, whether in anabolism, catabolism, or sensitivity. The connections in these pathways through common elements are referred to as ‘nodes’ in the crosstalk [121]. These nodes are connected amongst each other and with other integrators that help to process internal or external signals, to steer adequate plant responses, and adjust plant growth and development to prevailing conditions. Different plant hormone signaling pathways can display redundancy, functional overlap, and multiple feedback loops combined with direct and indirect regulation between different routes. This complexity makes it extremely difficult to understand the complete outcome of a specific hormone signal [122]. It is well known that several phytohormones collaborate to adjust the developmental growth program to stress conditions, and hormone crosstalk is a novel approach that helps to understand these complex outcomes [123]. Phenotypic analyses of certain mutants suggest that hormones influence each other, both in synthesis and signaling. There have been different approaches to address and systematically understand hormone interactions, or crosstalk, forming complex networks that coordinate general plant growth and development [124]. Although our knowledge has been reviewed and substantially improved in the past decade, the ‘growth-defense trade-off’ phenomenon [125], by which plants allocate their limited resources either to development or defense against abiotic or biotic stresses remains a key concept in explaining how hormone levels are modulated to guarantee the survival of a plant [126]. For this reason, the exploration of the crosstalk between growth promoting and growth repressing plant hormones, such as auxin and ABA, appears to be particularly interesting (Figure 3). A deeper understanding of the intertwined biosynthesis of IAA and ABA will likely further our understanding of how plants prepare to withstand abiotic stresses, which biomolecules are involved in the process, and how they trigger the response to stress on the molecular level. The crosstalk between auxins and ABA might be fundamental in understanding the growth-defense trade-off phenomenon.
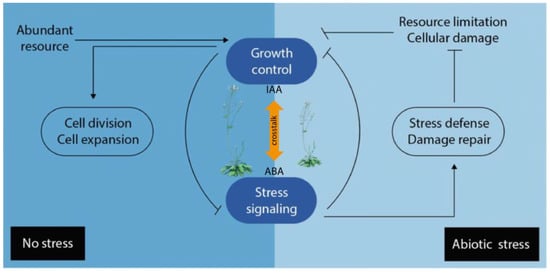
Figure 3.
Simplified overview of the growth-defense trade-off under normal and abiotic stress conditions. When resources are abundant and stress is absent, growth signaling is activated, repressing stress signaling (left panel) in addition to promoting growth. Abiotic stress passively inhibits plant growth (right panel) by causing cellular damage and limiting resources (e.g., carbon dioxide, nutrients, and energy). Arrows indicate positive regulation, and bars indicate negative regulation. ABA: abscisic acid; IAA: indole-3-acetic acid.
4.1. Crosstalk under Abiotic Stress Conditions
The interactions between auxin and ABA regulate a wide variety of developmental processes, such as seed germination, cell expansion, hypocotyl elongation, root elongation, lateral root formation, and cotyledon growth [127]. In addition, there is compelling evidence for the involvement of auxin-ABA crosstalk in the regulation of abiotic stress-related responses. For example, auxin homeostasis influences ABA biosynthesis and associated drought stress responses in rice [128,129]. In white clover, exogenous IAA treatment significantly increases ABA contents, upregulate the expression of drought stress responsive genes, including DEHYDRATION-RESPONSIVE ELEMENT BINDING 2 (DREB2), the transcription factor genes bZIP11, MYB14, MYB48, WRKY2, WRKY56, and RESPONSIVE TO DESICCATION 22 (RD22), along with some auxin responsive genes, such as the Gretchen Hagen 3 genes GH3.1 and GH3.9, as well as the Aux/IAA gene IAA8. At the same time, IAA treatment triggered the downregulation of leaf senescence-associated genes, such as SENESCENCE-ASSOCIATED GENE 101 (SAG101) and SAG102 [130]. Small auxin upregulated RNAs (SAURs) are recognized as auxin-responsive genes involved in the regulation of abiotic stress adaptive growth. In Arabidopsis, SAUR32 was dominantly expressed in roots and was highly induced by ABA and drought [131]. SAUR32 interacts with the clade-A PP2C proteins HAI1 and AIP1 to regulate ABA sensitivity. A barley mutant, nec1, that is defective in an HLM gene that encodes the CYCLIC NUCLEOTIDE-GATED ION CHANNEL 4, has been reported to over accumulate IAA and exhibit changes in stomatal regulation in response to exogenous auxin. The Arabidopsis orthologous mutant dnd2 shows increased salt tolerance, over-accumulation of both IAA and ABA, and displays related phenotypic and physiological changes, such as reduced stomata size, higher stomatal density, and stomatal index [132]. Interestingly, a recent study reported that the increase in IAM content in the ami1 mutant leads to an induction of NCED3 expression and, consequently, to significantly elevated levels of ABA [59]. Furthermore, ami1 mutants were shown to have altered osmotic stress responses. Thus, it must be concluded that IAM is an additional hub that links the biosynthesis pathways of IAA and ABA and, thus, contributes to the crosstalk between these two plant hormones. Furthermore, Ortiz-García et al. [133] provided evidence for the co-regulation of the transcription factor MYB74, which is reported to be involved in osmotic stress responses, by IAM and ABA (Figure 4).
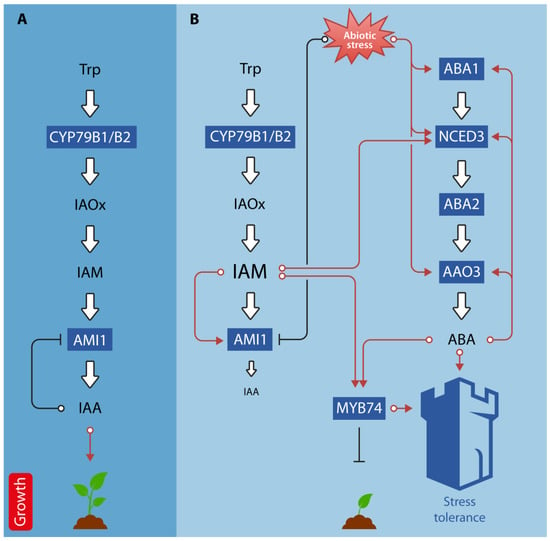
Figure 4.
Schematic representation of auxin-ABA crosstalk under normal and abiotic stress conditions. (A) Under normal conditions, indole-3-acetamide (IAM) is converted to indole-3-acetic acid (IAA) by the virtue of AMI1, which promotes plant growth. (B) Under abiotic stress conditions, AMI1 expression is repressed, which translates into a reduced IAA biosynthesis rate and the elevation of cellular IAM levels. IAM, in turn, is inducing ABA biosynthesis through the transcriptional activation of NCED3. The expression of several ABA biosynthesis-related genes, including NCED3, ABA1, and AAO3, is also directly stimulated by abiotic stress stimuli. Both, IAM and ABA, trigger the expression of the negative growth regulator MYB74. Increased ABA and MYB74 levels promote the abiotic stress tolerance of the plant. Red lines indicate stimulation of gene expression/growth, while black lines refer to the repression of gene expression and plant growth, respectively. The white bold arrows pointing to IAA and ABA, respectively, refer to the biosynthetic pathways of both hormones. Metabolites are given in black letters (letter size of IAM and IAA shows changes in endogenous content). Involved genes are given in dark blue boxes. The figure also shows positive feed-back loops for the regulation of ABA biosynthesis genes and highlights the transcriptional regulation of AMI1 by its substrate, IAM, and reaction product, IAA.
4.2. Further Examples for Auxin-ABA Crosstalk
The extensive interaction of ABA with other hormones in the regulation of stress responses, growth, and development is well documented [134]. Nonetheless, there are still large gaps in our knowledge on the crosstalk of ABA with other phytohormones, and sometimes we lack consensus on a given interaction. Crosstalk between two hormones can exist in a given process but not in another. For example, ABA and ethylene are antagonizing hormones at the germination level, but appear to interact in controlling stomatal closure [135]. In fact, ABA and ethylene have been commonly understood as antagonists, but more recent studies suggest that they could operate in parallel or even interact synergistically [89]. In addition, multiple and seemingly unrelated stimuli can affect the same hormonal pathways, e.g., many sugar germination assays revealed loci allelic to ABA, since they had been already described as ABA-responsive genes, but not all ABA-responsive genes are sugar-responsive [136]. Interactions between ABA and auxins are recognized to control essential plant programs, including seed germination [137] and seedling establishment [138], stomata closure [139], as well as meristematic activity of the main root and lateral root development [140]. Germination is a terminal process, irreversible in nature, and therefore influenced by countless inputs, both exogenous and endogenous [124] and must begin in a favorable environment. Although this field of research is still in its infancy, some nodes have been uncovered. In most of the examples of auxin-ABA crosstalk described so far, ABA seems to act upstream of auxin [127], but there seem to be hints that auxin-ABA crosstalk is more mutual and intertangled, and in some instances the master regulation of ABA has not been unequivocally proven yet. As a classical example, in Arabidopsis, ABA acts upstream of ARF2 which, in turn, directly regulates the expression of the homeodomain gene HOMEOBOX PROTEIN 33 (HB33) in both seed germination and primary root growth [141]. In fact, one of the well-investigated mechanisms in which ABA influences auxin signaling is through the regulation of ARFs, including post-transcriptional modulation [142]. For example, ABA induces ARF6 ubiquitination, leading to protein degradation [143]. ABI3 would constitute an interesting node, because this particular protein is essential for multiple gene networks that control developmental processes [144,145] and is considered the major regulator of seed dormancy in plants. In Arabidopsis, auxin is known to control dormancy through stimulation of ABA signaling by inducing ABI3 expression [146]. Auxin acts upstream of ABI3 by recruiting auxin response factors ARF10 and ARF16. In turn, ARF10 is negatively regulated by miRNA160 [147]. ARF10/16 promote ABI3 transcription, whose transcripts are high in dormant seeds and lowered after germination, regulating the seed dormancy in synergy with ABA [146], but not by direct binding, raising the question of which intermediate transcription factors operate in this pathway. Closing the feedback loop, ABI3 negatively regulates MIR160B, amongst other miRNA genes [148]. Furthermore, auxin-induced ABI3 expression has been reported to be required for the formation of somatic embryogenesis, because it induces the expression of embryo identity genes through, at least, ARF10/16 activation [148]. Regarding its connection with ARFs, ABI3 has also been described to be negatively regulated by the auxin signaling repressor IAA8, whose accumulation promotes seed germination [149]. The authors suggest that the binding of IAA8 to the ABI3 promoter through ARFs suppresses ABI3 transcription during seed germination, but the exact ARF proteins have not yet been identified. Interestingly, IAA8 has been shown to interact with ARF6 and ARF8 in modulating JA levels during flower development [150]. In another example, ABI5 is not only a known hub for ABA-mediated abiotic stress responses and crosstalk with GAs, BRs, and JAs [151], but also an important node connecting ABA and sugar sensing through the TARGET OF RAPAMYCIN (TOR) kinase, which is activated by auxins [152]. In fact, the reciprocal regulation of the TOR kinase and ABA receptors has been proposed to balance plant growth and stress responses [153] and inhibition of TOR alters auxin signaling [154]. For example, at high concentrations of glucose, ABI5 suppresses the accumulation of PIN1 in the root meristem, thus decreasing auxin activity and inhibiting root elongation [155]. ABA also suppresses auxin-mediated primary root elongation under abiotic stress through upregulation of ABI5, which acts by inducing degradation of PIN2 proteins [156]. Complementary, salt stress significantly decreases the expression of the PIN1, PIN3, and PIN7 genes and promotes the stabilization of AUXIN RESISTANT 3 (AXR3)/IAA17 through nitric oxide (NO) [157]. However, although ABI5 is known to be regulated by NO at both the transcriptional and protein levels [158,159] the precise involvement of ABA in this pathway remains unknown. Given its utmost importance, ABI5 expression and the biological half-life of the protein are tightly regulated [151], and other transcription factors fine-tune its transcription, such as the rice APETALA2-type transcription factor SALT AND ABA RESPONSE ERF 1 (OsSAE1), which acts as a positive regulator of seed germination and salt tolerance in rice by suppressing the expression of OsABI5 [160]. In another example, mediator (MED) proteins, such as MED25, positively contribute to auxin signaling, but interact with ABI5 to negatively regulate ABA responses [161,162], while MED16 competes with MED25 in physical interactions with ABI5 and is a positive regulator of ABA responses [163]. ABI4 would serve as another connection point, being stabilized by stress, ABA, and phosphorylation [164], and is also known to receive auxin inputs [165], at least in roots, where its expression is induced by ABA and cytokinin and repressed by auxin [166], but its role in crosstalk needs further exploration [167]. Interestingly, PIN1 levels are reduced under abiotic stress in an ABA-dependent manner, e.g., after mannitol treatment [168], since ABI4 mediates ABA- and the cytokinin-mediated inhibition of lateral root formation by reducing the expression of PIN1, which plays a role in polar auxin transport necessary for root development [166], even over-riding other hormonal signals, such as ethylene [169]. In turn, ABI4 is also regulated, for example, by WRKY46, which integrates ABA-dependent and ABA-independent abiotic stress responses and, thus, controls auxin-related gene expression [170]. ABI4 may also be key to integrating ABA, auxin, and reactive oxygen species (ROS) signaling via ASCORBATE PEROXIDASE 6 (APX6) in seeds [171,172] and controls auxin levels through upstream involvement of YUC4 [173], although the precise mechanisms are not yet clear. In rice, the OsPIN2 mutant wavy root 1 (war1) is defective in auxin transport and auxin distribution at root tips, leading to loss of gravitropic perception, but also shows increased sensitivity to ABA in seed germination, increased ABA levels, and changes in ABA-associated gene expression in roots [174]. A subsequent study later revealed that ABA reduces the amount of PIN2 in the membrane by suppressing the expression of PIN2 rather than accelerating the degradation of PIN2 [140]. In rice as well, OsIAA20 is positively regulated by abiotic stress conditions and exogenous ABA treatment, and contributes positively to abiotic stress tolerance: reduces water loss, improves seed germination, decreases the Na+/K+ ratio, increases the proportion of closed stomata, and also enhances growth in different developmental stages [175]. Remarkably, OsIAA18 also confers salt and drought tolerance, at least by increasing proline biosynthesis and reducing ROS accumulation, through an ABA-dependent pathway, therefore acting upstream of ABA-signaling pathways [176], even the heterologous expression of the rice gene in Arabidopsis [177], and heterologous expression of the grapevine VvIAA18 in tobacco enhances drought tolerance, as measured by upregulation of salt stress-responsive genes, including PYRROLINE-5-CARBOXYLATE SYNTHASE (P5CS), LATE EMBRYOGENESIS ABUNDANT PROTEIN 5 (LEA5), SUPEROXIDE DISMUTASE (SOD) and PEROXIDASE (POD) under drought stress, as well as higher SOD and POD activities [178]. However, whether these Aux/IAAs are directly activated by ABA has not been elucidated yet. The regulatory model in which salt/drought stress regulates Aux/IAAs exists also in Arabidopsis, where under drought conditions, DREB2A and DREB2B directly regulate the expression of the Aux/IAA genes IAA5, IAA6, and IAA19 [179].
5. Open Challenges in Auxin-ABA Crosstalk
Despite recent discoveries that shed light on the intricacies of auxin-ABA crosstalk, there are remaining knowledge gaps that need to be closed. For example, the role of indeterminate domain (IDD) transcription factors in abiotic stress responses have recently been discovered. IDD14 can physically interact with ABF1-4, promoting their transcriptional activities by forming a protein complex that positively regulates drought-stress responses [180]. Numerous rice IDDs can also respond to auxin and ABA [181]. Three Arabidopsis IDD transcription factors, IDD14, IDD15, and IDD16, are known to cooperatively regulate spatial auxin accumulation by directly targeting YUC5, TAA1, and PIN1 to promote auxin biosynthesis and transport [182], but it is not clear how this could influence auxin-ABA crosstalk. Another very recent study demonstrated that the ABA signaling-related transcription factor bZIP46 directly controls the expression levels of YUC8 in rice, and genetic studies provided additional evidence that ABA uses auxin as a downstream signal to modify root elongation and radial expansion [183]. One further work highlighted that the transcription factor WRKY41 is an important regulator of ABI3 expression during seed dormancy, acting not downstream of ABA signaling, but probably in parallel [184]. However, the stimuli that induce WRKY41 are not yet clear, and their elucidation might reveal new interactions. PIN3 expression is also directly inhibited by HAT2, an HD-Zip II transcription factor, whose expression is upregulated by osmotic stress, at least in roots, thus inhibiting meristem growth [185], and its expression is also downregulated in response to NaCl [186], but the exact mechanism remains uncertain. Auxin-ABA interactions remain particularly understudied in relation to cold responses, which are mediated by ABA [187]. Low temperatures inhibit root growth, due to the fact that endosomal trafficking of auxin efflux carriers is inhibited [188], and partially by repressing the expression of PIN1/3/7 and auxin biosynthesis, a possible crosstalk node would be the ARABIDOPSIS RESPONSE REGULATOR 1 (ARR1) transcription factor [189], in which auxin, cytokinin, and ABA signaling seem to converge [190], but the mechanisms remain elusive [191].
Author Contributions
P.O.-G., A.G.O.-V. and F.C.O. performed the literature review. P.O.-G., A.G.O.-V., S.P. and M.M. wrote the manuscript. S.P. provided resources and funding. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by grant PID2020-119441RB-100 to SP funded by MCIN/AEI/10.13039/501100011033 and as appropriate, by “ERDF a way of making Europe” by the “European Union” or by the “European Union Next Generation EU/PRTR”.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data sharing is not applicable. No new data were generated or analyzed in this study.
Acknowledgments
The authors appreciate the thoughtful feedback and highly valuable comments by all members of the CBGP laboratories 132 and 134. We apologize to all colleagues in the field whose work we were not able to cite due to space constrains.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Mazdiyasni, O.; Aghakouchak, A. Substantial Increase in Concurrent Droughts and Heatwaves in the United States. Proc. Natl. Acad. Sci. USA 2015, 112, 11484–11489. [Google Scholar] [CrossRef] [PubMed]
- IPCC. Summary for Policymakers. In Climate Change 2021: The Physical Science Basis. Contribution of Working Group I to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change; Masson-Delmotte, V., Zhai, P., Pirani, A., Connors, S.L., Péan, C., Berger, S., Caud, N., Chen, Y., Goldfarb, L., Gomis, M.I., et al., Eds.; Cambridge University Press: Cambridge, UK, 2021. [Google Scholar]
- Ding, Y.; Shi, Y.; Yang, S. Molecular Regulation of Plant Responses to Environmental Temperatures. Mol. Plant 2020, 13, 544–564. [Google Scholar] [CrossRef] [PubMed]
- Nadarajah, K.K. Ros Homeostasis in Abiotic Stress Tolerance in Plants. Int. J. Mol. Sci. 2020, 21, 5208. [Google Scholar] [CrossRef] [PubMed]
- Hirayama, T.; Shinozaki, K. Research on Plant Abiotic Stress Responses in the Post-Genome Era: Past, Present and Future. Plant J. 2010, 61, 1041–1052. [Google Scholar] [CrossRef]
- Claeys, H.; Inzé, D. The Agony of Choice: How Plants Balance Growth and Survival under Water-Limiting Conditions. Plant Physiol. 2013, 162, 1768–1779. [Google Scholar] [CrossRef]
- Hassani, A.; Azapagic, A.; Shokri, N. Global Predictions of Primary Soil Salinization under Changing Climate in the 21st Century. Nat. Commun. 2021, 12, 6663. [Google Scholar] [CrossRef]
- Mustafa, Y.; Irem, P.; Aslinur, Ç.; Yasin, Ö.; Ramazan, B. Plant Responses to Salt Stress. In Plant Breeding; Abdurakhmonov Ibrokhim, Y., Ed.; IntechOpen: Rijeka, Croatia, 2020; p. 93920. [Google Scholar]
- Godfray, H.C.J.; Beddington, J.R.; Crute, I.R.; Haddad, L.; Lawrence, D.; Muir, J.F.; Pretty, J.; Robinson, S.; Thomas, S.M.; Toulmin, C. Food Security: The Challenge of Feeding 9 Billion People. Science 2010, 327, 812–818. [Google Scholar] [CrossRef]
- United Nations, Department of Economic and Social Affairs. World Population Prospects: The 2017 Revision, Volume I: Comprehensive Tables; (ST/ESA/SER.A/399); United Nations, Department of Economic and Social Affairs: New York, NY, USA, 2017.
- Tilman, D.; Balzer, C.; Hill, J.; Befort, B.L. Global Food Demand and the Sustainable Intensification of Agriculture. Proc. Natl. Acad. Sci. USA 2011, 108, 20260–20264. [Google Scholar] [CrossRef]
- Bailey-Serres, J.; Parker, J.E.; Ainsworth, E.A.; Oldroyd, G.E.D.; Schroeder, J.I. Genetic Strategies for Improving Crop Yields. Nature 2019, 575, 109–118. [Google Scholar] [CrossRef]
- Weiler, E.W. Sensory Principles of Higher Plants. Angew. Chem. Int. Ed. 2003, 42, 392–411. [Google Scholar] [CrossRef]
- Miyakawa, T.; Tanokura, M. Structural Basis for the Regulation of Phytohormone Receptors. Biosci. Biotechnol. Biochem. 2017, 81, 1261–1273. [Google Scholar] [CrossRef] [PubMed]
- Müller, M.; Munné-Bosch, S. Hormonal Impact on Photosynthesis and Photoprotection in Plants. Plant Physiol. 2021, 185, 1500–1522. [Google Scholar] [CrossRef] [PubMed]
- Santner, A.; Calderon-Villalobos, L.I.A.; Estelle, M. Plant Hormones Are Versatile Chemical Regulators of Plant Growth. Nat. Chem. Biol. 2009, 5, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Hentrich, M.; Pollmann, S. Auxin-Oxylipin Crosstalk: Relationship of Antagonists. J. Integr. Plant Biol. 2011, 53, 429–445. [Google Scholar] [CrossRef]
- Pozo, M.J.; López-Ráez, J.A.; Azcón-Aguilar, C.; García-Garrido, J.M. Phytohormones as Integrators of Environmental Signals in the Regulation of Mycorrhizal Symbioses. New Phytol. 2015, 205, 1431–1436. [Google Scholar] [CrossRef]
- Waadt, R.; Seller, C.A.; Hsu, P.-K.; Takahashi, Y.; Munemasa, S.; Schroeder, J.I. Plant Hormone Regulation of Abiotic Stress Responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 680–694. [Google Scholar] [CrossRef]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant Hormone-Mediated Regulation of Stress Responses. BMC Plant Biol. 2016, 16, 86. [Google Scholar] [CrossRef]
- Yu, Z.; Duan, X.; Luo, L.; Dai, S.; Ding, Z.; Xia, G. How Plant Hormones Mediate Salt Stress Responses. Trends Plant Sci. 2020, 25, 1117–1130. [Google Scholar] [CrossRef]
- Hentrich, M.; Böttcher, C.; Düchting, P.; Cheng, Y.; Zhao, Y.; Berkowitz, O.; Masle, J.; Medina, J.; Pollmann, S. The Jasmonic Acid Signaling Pathway Is Linked to Auxin Homeostasis through the Modulation of YUCCA8 and YUCCA9 Gene Expression. Plant J. 2013, 74, 626–637. [Google Scholar] [CrossRef]
- Pérez-Alonso, M.M.; Sánchez-Parra, B.; Ortiz-García, P.; Santamaria, M.E.; Diaz, I.; Pollmann, S. Jasmonic Acid-Dependent Myc Transcription Factors Bind to a Tandem G-Box Motif in the YUCCA8 and YUCCA9 Promoters to Regulate Biotic Stress Responses. Int. J. Mol. Sci. 2021, 22, 9768. [Google Scholar] [CrossRef]
- Szécsi, J.; Joly, C.; Bordji, K.; Varaud, E.; Cock, J.M.; Dumas, C.; Bendahmane, M. Bigpetalp, a Bhlh Transcription Factor Is Involved in the Control of Arabidopsis Petal Size. EMBO J. 2006, 25, 3912–3920. [Google Scholar] [CrossRef] [PubMed]
- Nagpal, P.; Ellis, C.M.; Weber, H.; Ploense, S.E.; Barkawi, L.S.; Guilfoyle, T.J.; Hagen, G.; Alonso, J.M.; Cohen, J.D.; Farmer, E.E.; et al. Auxin Response Factors ARF6 and ARF8 Promote Jasmonic Acid Production and Flower Maturation. Development 2005, 132, 4107–4118. [Google Scholar] [CrossRef] [PubMed]
- Busatto, N.; Tadiello, A.; Moretto, M.; Farneti, B.; Populin, F.; Vrhovsek, U.; Commisso, M.; Sartori, E.; Sonego, P.; Biasioli, F.; et al. Ethylene-Auxin Crosstalk Regulates Postharvest Fruit Ripening Process in Apple. Fruit Res. 2021, 1, 1–13. [Google Scholar] [CrossRef]
- Böttcher, C.; Burbidge, C.A.; Boss, P.K.; Davies, C. Interactions between Ethylene and Auxin Are Crucial to the Control of Grape (Vitis vinifera L.) Berry Ripening. BMC Plant Biol. 2013, 13, 222. [Google Scholar] [CrossRef] [PubMed]
- Perrot-Rechenmann, C.; Napier, R.M. Auxins. Vitam. Horm. 2005, 72, 203–233. [Google Scholar] [PubMed]
- Davies, P.J. Plant Hormones. Biosynthesis, Signal Transduction, Action! 3rd ed.; Springer: Dordrecht, The Netherlands; Boston, MA, USA; London, UK, 2010. [Google Scholar]
- Went, F.W. Auxin, the Plant Growth-Hormone. Bot. Rev. 1935, 1, 162–182. [Google Scholar] [CrossRef]
- Went, F.W.; Thimann, K.V. Phytohormones; Macmillan Company: New York, NY, USA, 1937. [Google Scholar]
- Won, C.; Shen, X.; Mashiguchi, K.; Zheng, Z.; Dai, X.; Cheng, Y.; Kasahara, H.; Kamiya, Y.; Chory, J.; Zhao, Y. Conversion of Tryptophan to Indole-3-Acetic Acid by Tryptophan Aminotransferases of Arabidopsis and YUCCAs in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 18518–18523. [Google Scholar] [CrossRef]
- Stepanova, A.N.; Robertson-Hoyt, J.; Yun, J.; Benavente, L.M.; Xie, D.Y.; Dolezal, K.; Schlereth, A.; Jürgens, G.; Alonso, J.M. TAA1-Mediated Auxin Biosynthesis Is Essential for Hormone Crosstalk and Plant Development. Cell 2008, 133, 177–191. [Google Scholar] [CrossRef]
- Stepanova, A.N.; Yun, J.; Robles, L.M.; Novak, O.; He, W.; Guo, H.; Ljung, K.; Alonso, J.M. The Arabidopsis YUCCA1 Flavin Monooxygenase Functions in the Indole-3-Pyruvic Acid Branch of Auxin Biosynthesis. Plant Cell 2011, 23, 3961–3973. [Google Scholar] [CrossRef]
- Mashiguchi, K.; Tanaka, K.; Sakai, T.; Sugawara, S.; Kawaide, H.; Natsume, M.; Hanada, A.; Yaeno, T.; Shirasu, K.; Yao, H.; et al. The Main Auxin Biosynthesis Pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 18512–18517. [Google Scholar] [CrossRef]
- Tivendale, N.D.; Ross, J.J.; Cohen, J.D. The Shifting Paradigms of Auxin Biosynthesis. Trends Plant Sci. 2014, 19, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Wright, A.D.; Sampson, M.B.; Neuffer, M.G.; Michalczuk, L.; Slovin, J.P.; Cohen, J.D. Indole-3-Acetic Acid Biosynthesis in the Mutant Maize Orange Pericarp, a Tryptophan Auxotroph. Science 1991, 254, 998–1000. [Google Scholar] [CrossRef] [PubMed]
- Normanly, J.; Cohen, J.D.; Fink, G.R. Arabidopsis thaliana Auxotrophs Reveal a Tryptophan-Independent Biosynthetic Pathway for Indole-3-Acetic Acid. Proc. Natl. Acad. Sci. USA 1993, 90, 10355–10359. [Google Scholar] [CrossRef]
- Müller, A.; Weiler, E.W. Indolic Constituents and Indole-3-Acetic Acid Biosynthesis in the Wild-Type and a Tryptophan Auxotroph Mutant of Arabidopsis thaliana. Planta 2000, 211, 855–863. [Google Scholar] [CrossRef]
- Wang, B.; Chu, J.; Yu, T.; Xu, Q.; Sun, X.; Yuan, J.; Xiong, G.; Wang, G.; Wang, Y.; Li, J. Tryptophan-Independent Auxin Biosynthesis Contributes to Early Embryogenesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2015, 112, 4821–4826. [Google Scholar] [CrossRef]
- Tao, Y.; Ferrer, J.L.; Ljung, K.; Pojer, F.; Hong, F.; Long, J.A.; Li, L.; Moreno, J.E.; Bowman, M.E.; Ivans, L.J.; et al. Rapid Synthesis of Auxin Via a New Tryptophan-Dependent Pathway Is Required for Shade Avoidance in Plants. Cell 2008, 133, 164–176. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Christensen, S.K.; Fankhauser, C.; Cashman, J.R.; Cohen, J.D.; Weigel, D.; Chory, J. A Role for Flavin Monooxygenase-Like Enzymes in Auxin Biosynthesis. Science 2001, 291, 306–309. [Google Scholar] [CrossRef] [PubMed]
- Pollmann, S.; Müller, A.; Weiler, E.W. Many Roads Lead to “Auxin”: Of Nitrilases, Synthases, and Amidases. Plant Biol. 2006, 8, 326–333. [Google Scholar] [CrossRef] [PubMed]
- Mano, Y.; Nemoto, K. The Pathway of Auxin Biosynthesis in Plants. J. Exp. Bot. 2012, 63, 2853–2872. [Google Scholar] [CrossRef] [PubMed]
- Kasahara, H. Current Aspects of Auxin Biosynthesis in Plants. Biosci. Biotechnol. Biochem. 2016, 80, 34–42. [Google Scholar] [CrossRef]
- Mikkelsen, M.D.; Hansen, C.H.; Wittstock, U.; Halkier, B.A. Cytochrome P450 Cyp79b2 from Arabidopsis Catalyzes the Conversion of Tryptophan to Indole-3-Acetaldoxime, a Precursor of Indole Glucosinolates and Indole-3-Acetic Acid. J. Biol. Chem. 2000, 275, 33712–33717. [Google Scholar] [CrossRef] [PubMed]
- Hull, A.K.; Vij, R.; Celenza, J.L. Arabidopsis Cytochrome P450s That Catalyze the First Step of Tryptophan-Dependent Indole-3-Acetic Acid Biosynthesis. Proc. Natl. Acad. Sci. USA 2000, 97, 2379–2384. [Google Scholar] [CrossRef] [PubMed]
- Pérez, V.C.; Dai, R.; Bai, B.; Tomiczek, B.; Askey, B.C.; Zhang, Y.; Rubin, G.M.; Ding, Y.; Grenning, A.; Block, A.K.; et al. Aldoximes Are Precursors of Auxins in Arabidopsis and Maize. New Phytol. 2021, 231, 1449–1461. [Google Scholar] [CrossRef] [PubMed]
- Irmisch, S.; Zeltner, P.; Handrick, V.; Gershenzon, J.; Kollner, T.G. The Maize Cytochrome P450 Cyp79a61 Produces Phenylacetaldoxime and Indole-3-Acetaldoxime in Heterologous Systems and Might Contribute to Plant Defense and Auxin Formation. BMC Plant Biol. 2015, 15, 128. [Google Scholar] [CrossRef]
- Buezo, J.; Esteban, R.; Cornejo, A.; López-Gómez, P.; Marino, D.; Chamizo-Ampudia, A.; Gil, M.J.; Martínez-Merino, V.; Moran, J.F. IAOx Induces the Sur Phenotype and Differential Signalling from Iaa under Different Types of Nitrogen Nutrition in Medicago truncatula Roots. Plant Sci. 2019, 287, 110176. [Google Scholar] [CrossRef]
- Sugawara, S.; Hishiyama, S.; Jikumaru, Y.; Hanada, A.; Nishimura, T.; Koshiba, T.; Zhao, Y.; Kamiya, Y.; Kasahara, H. Biochemical Analyses of Indole-3-Acetaldoxime-Dependent Auxin Biosynthesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 5430–5435. [Google Scholar] [CrossRef]
- Müller, T.M.; Böttcher, C.; Glawischnig, E. Dissection of the Network of Indolic Defence Compounds in Arabidopsis thaliana by Multiple Mutant Analysis. Phytochemistry 2019, 161, 11–20. [Google Scholar] [CrossRef]
- Spaepen, S.; Vanderleyden, J. Auxin and Plant-Microbe Interactions. Cold Spring Harb. Perspect. Biol. 2011, 3, a001438. [Google Scholar] [CrossRef]
- Pollmann, S.; Neu, D.; Weiler, E.W. Molecular Cloning and Characterization of an Amidase from Arabidopsis thaliana Capable of Converting Indole-3-Acetamide into the Plant Growth Hormone, Indole-3-Acetic Acid. Phytochemistry 2003, 62, 293–300. [Google Scholar] [CrossRef]
- Sánchez-Parra, B.; Frerigmann, H.; Pérez-Alonso, M.M.; Carrasco-Loba, V.; Jost, R.; Hentrich, M.; Pollmann, S. Characterization of Four Bifunctional Plant Iam/Pam-Amidohydrolases Capable of Contributing to Auxin Biosynthesis. Plants 2014, 3, 324–347. [Google Scholar] [CrossRef]
- Nemoto, K.; Hara, M.; Suzuki, M.; Seki, H.; Muranaka, T.; Mano, Y. The NtAMI1 Gene Functions in Cell Division of Tobacco by-2 Cells in the Presence of Indole-3-Acetamide. FEBS Lett. 2009, 583, 487–492. [Google Scholar] [CrossRef] [PubMed]
- Keereetaweep, J.; Blancaflor, E.B.; Hornung, E.; Feussner, I.; Chapman, K.D. Ethanolamide Oxylipins of Linolenic Acid Can Negatively Regulate Arabidopsis Seedling Development. Plant Cell 2013, 25, 3824–3840. [Google Scholar] [CrossRef] [PubMed]
- Aziz, M.; Chapman, K.D. Fatty Acid Amide Hydrolases: An Expanded Capacity for Chemical Communication? Trends Plant Sci. 2020, 25, 236–249. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Alonso, M.M.; Ortiz-García, P.; Moya-Cuevas, J.; Lehmann, T.; Sánchez-Parra, B.; Björk, R.G.; Karim, S.; Amirjani, M.R.; Aronsson, H.; Wilkinson, M.D.; et al. Endogenous Indole-3-Acetamide Levels Contribute to the Crosstalk between Auxin and Abscisic Acid, and Trigger Plant Stress Responses in Arabidopsis thaliana. J. Exp. Bot. 2021, 72, 459–475. [Google Scholar] [CrossRef]
- Gao, Y.; Dai, X.; Aoi, Y.; Takebayashi, Y.; Yang, L.; Guo, X.; Zeng, Q.; Yu, H.; Kasahara, H.; Zhao, Y. Two Homologous Indole-3-Acetamide (IAM) Hydrolase Genes Are Required for the Auxin Effects of IAM in Arabidopsis. J. Genet. Genom. 2020, 47, 157–165. [Google Scholar] [CrossRef]
- Petrásek, J.; Friml, J.I. Auxin Transport Routes in Plant Development. Development 2009, 136, 2675–2688. [Google Scholar] [CrossRef]
- Woodward, A.W.; Bartel, B. Auxin: Regulation, Action, and Interaction. Ann. Bot. 2005, 95, 707–735. [Google Scholar] [CrossRef]
- Michniewicz, M.; Brewer, P.B.; Friml, J. Polar Auxin Transport and Asymmetric Auxin Distribution. Arab. Book 2007, 5, e0108. [Google Scholar]
- Adamowski, M.; Friml, J. Pin-Dependent Auxin Transport: Action, Regulation, and Evolution. Plant Cell 2015, 27, 20–32. [Google Scholar] [CrossRef]
- Aloni, R.; Aloni, E.; Langhans, M.; Ullrich, C.I. Role of Auxin in Regulating Arabidopsis Flower Development. Planta 2006, 223, 315–328. [Google Scholar] [CrossRef]
- Smith, R.S.; Guyomarc’h, S.; Mandel, T.; Reinhardt, D.; Kuhlemeier, C.; Prusinkiewicz, P. A Plausible Model of Phyllotaxis. Proc. Natl. Acad. Sci. USA 2006, 103, 1301–1306. [Google Scholar] [CrossRef] [PubMed]
- Roychoudhry, S.; Kepinski, S. Auxin in Root Development. Cold Spring Harb. Perspect. Biol. 2022, 14, a039933. [Google Scholar] [CrossRef]
- Goyal, A.; Szarzynska, B.; Fankhauser, C. Phototropism: At the Crossroads of Light-Signaling Pathways. Trends Plant Sci. 2013, 18, 393–401. [Google Scholar] [CrossRef] [PubMed]
- Geilfus, C.-M. The Ph of the Apoplast: Dynamic Factor with Functional Impact under Stress. Mol. Plant 2017, 10, 1371–1386. [Google Scholar] [CrossRef] [PubMed]
- Swarup, R.; Bhosale, R. Developmental Roles of AUX1/LAX Auxin Influx Carriers in Plants. Front. Plant Sci. 2019, 10, 1306. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.-J.; Luo, J. The PIN-Formed Auxin Efflux Carriers in Plants. Int. J. Mol. Sci. 2018, 19, 2759. [Google Scholar] [CrossRef]
- Geisler, M.; Aryal, B.; Di Donato, M.; Hao, P. A Critical View on ABC Transporters and Their Interacting Partners in Auxin Transport. Plant Cell Physiol. 2017, 58, 1601–1614. [Google Scholar] [CrossRef]
- Dharmasiri, N.; Dharmasiri, S.; Estelle, M. The F-Box Protein TIR1 Is an Auxin Receptor. Nature 2005, 435, 441–445. [Google Scholar] [CrossRef]
- Tiwari, S.B.; Wang, X.J.; Hagen, G.; Guilfoyle, T.J. Aux/IAA Proteins Are Active Repressors, and Their Stability and Activity Are Modulated by Auxin. Plant Cell 2001, 13, 2809–2822. [Google Scholar] [CrossRef]
- Chandler, J.W. Auxin Response Factors. Plant Cell Environ. 2016, 39, 1014–1028. [Google Scholar] [CrossRef]
- Quint, M.; Gray, W.M. Auxin Signaling. Curr. Opin. Plant Biol. 2006, 9, 448–453. [Google Scholar] [CrossRef] [PubMed]
- Leyser, O. Auxin Signaling. Plant Physiol. 2017, 176, 465–479. [Google Scholar] [CrossRef] [PubMed]
- Hertel, R.; Thomson, K.S.; Russo, V.E.A. In-Vitro Auxin Binding to Particulate Cell Fractions from Corn Coleoptiles. Planta 1972, 107, 325–340. [Google Scholar] [CrossRef] [PubMed]
- Friml, J.; Gallei, M.; Gelová, Z.; Johnson, A.; Mazur, E.; Monzer, A.; Rodriguez, L.; Roosjen, M.; Verstraeten, I.; Živanović, B.D.; et al. ABP1–TMK Auxin Perception for Global Phosphorylation and Auxin Canalization. Nature 2022, 609, 575–581. [Google Scholar] [CrossRef] [PubMed]
- Xiong, L.; Zhu, J.-K. Regulation of Abscisic Acid Biosynthesis. Plant Physiol. 2003, 133, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Milborrow, B.V. The Pathway of Biosynthesis of Abscisic Acid in Vascular Plants: A Review of the Present State of Knowledge of Aba Biosynthesis. J. Exp. Bot. 2001, 52, 1145–1164. [Google Scholar] [CrossRef]
- Kasahara, H.; Takei, K.; Ueda, N.; Hishiyama, S.; Yamaya, T.; Kamiya, Y.; Yamaguchi, S.; Sakakibara, H. Distinct Isoprenoid Origins of Cis- and Trans-Zeatin Biosyntheses in Arabidopsis. J. Biol. Chem. 2004, 279, 14049–14054. [Google Scholar] [CrossRef]
- Addicott, F.T.; Lyon, J.L.; Ohkuma, K.; Thiessen, W.E.; Carns, H.R.; Smith, O.E.; Cornforth, J.W.; Milborrow, B.V.; Ryback, G.; Wareing, P.F. Abscisic Acid: A New Name for Abscisin II (Dormin). Science 1968, 159, 1493. [Google Scholar] [CrossRef]
- Bennet-Clark, T.A.; Kefford, N.P. Chromatography of the Growth Substances in Plant Extracts. Nature 1953, 171, 645–647. [Google Scholar] [CrossRef]
- Yoshida, T.; Christmann, A.; Yamaguchi-Shinozaki, K.; Grill, E.; Fernie, A.R. Revisiting the Basal Role of ABA—Roles Outside of Stress. Trends Plant Sci. 2019, 24, 625–635. [Google Scholar] [CrossRef]
- Brookbank, B.P.; Patel, J.; Gazzarrini, S.; Nambara, E. Role of Basal ABA in Plant Growth and Development. Genes 2021, 12, 1936. [Google Scholar] [CrossRef] [PubMed]
- Dörffling, K. The Discovery of Abscisic Acid: A Retrospect. J. Plant Growth Regul. 2015, 34, 795–808. [Google Scholar] [CrossRef]
- Nambara, E.; Marion-Poll, A. Abscisic Acid Biosynthesis and Catabolism. Ann. Rev. Plant Biol. 2005, 56, 165–185. [Google Scholar] [CrossRef] [PubMed]
- Müller, M. Foes or Friends: Aba and Ethylene Interaction under Abiotic Stress. Plants 2021, 10, 448. [Google Scholar] [CrossRef]
- Barrero, J.M.; Piqueras, P.; González-Guzmán, M.; Serrano, R.; Rodríguez, P.L.; Ponce, M.R.; Micol, J.L. A Mutational Analysis of the ABA1 Gene of Arabidopsis thaliana Highlights the Involvement of Aba in Vegetative Development. J. Exp. Bot. 2005, 56, 2071–2083. [Google Scholar] [CrossRef]
- Perreau, F.; Frey, A.; Effroy-Cuzzi, D.; Savane, P.; Berger, A.; Gissot, L.; Marion-Poll, A. Abscisic Acid-Deficient4 Has an Essential Function in Both Cis-Violaxanthin and Cis-Neoxanthin Synthesis. Plant Physiol. 2020, 184, 1303–1316. [Google Scholar] [CrossRef]
- North, H.M.; Almeida, A.D.; Boutin, J.-P.; Frey, A.; To, A.; Botran, L.; Sotta, B.; Marion-Poll, A. The Arabidopsis Aba-Deficient Mutant ABA4 Demonstrates That the Major Route for Stress-Induced Aba Accumulation Is Via Neoxanthin Isomers. Plant J. 2007, 50, 810–824. [Google Scholar] [CrossRef]
- Schwartz, S.H.; Tan, B.C.; Gage, D.A.; Zeevaart, J.a.D.; Mccarty, D.R. Specific Oxidative Cleavage of Carotenoids by Vp14 of Maize. Science 1997, 276, 1872–1874. [Google Scholar] [CrossRef]
- Iuchi, S.; Kobayashi, M.; Taji, T.; Naramoto, M.; Seki, M.; Kato, T.; Tabata, S.; Kakubari, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Regulation of Drought Tolerance by Gene Manipulation of 9-Cis-Epoxycarotenoid Dioxygenase, a Key Enzyme in Abscisic Acid Biosynthesis in Arabidopsis. Plant J. 2001, 27, 325–333. [Google Scholar] [CrossRef]
- Cheng, W.-H.; Endo, A.; Zhou, L.; Penney, J.; Chen, H.-C.; Arroyo, A.; Leon, P.; Nambara, E.; Asami, T.; Seo, M.; et al. A Unique Short-Chain Dehydrogenase/Reductase in Arabidopsis Glucose Signaling and Abscisic Acid Biosynthesis and Functions. Plant Cell 2002, 14, 2723–2743. [Google Scholar] [CrossRef]
- González-Guzmán, M.; Abia, D.; Salinas, J.; Serrano, R.; Rodríguez, P.L. Two New Alleles of the Abscisic Aldehyde Oxidase 3 Gene Reveal Its Role in Abscisic Acid Biosynthesis in Seeds. Plant Physiol. 2004, 135, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Bittner, F.; Oreb, M.; Mendel, R.R. ABA3 Is a Molybdenum Cofactor Sulfurase Required for Activation of Aldehyde Oxidase and Xanthine Dehydrogenase in Arabidopsis thaliana. J. Biol. Chem. 2001, 276, 40381–40384. [Google Scholar] [CrossRef] [PubMed]
- Sagi, M.; Scazzocchio, C.; Fluhr, R. The Absence of Molybdenum Cofactor Sulfuration Is the Primary Cause of the Flacca Phenotype in Tomato Plants. Plant J. 2002, 31, 305–317. [Google Scholar] [CrossRef]
- Léon-Kloosterziel, K.M.; Gil, M.A.; Ruijs, G.J.; Jacobsen, S.E.; Olszewski, N.E.; Schwartz, S.H.; Zeevaart, J.a.D.; Koornneef, M. Isolation and Characterization of Abscisic Acid-Deficient Arabidopsis Mutants at Two New Loci. Plant J. 1996, 10, 655–661. [Google Scholar] [CrossRef] [PubMed]
- Sagi, M.; Fluhr, R.; Lips, S.H. Aldehyde Oxidase and Xanthine Dehydrogenase in a Flacca Tomato Mutant with Deficient Abscisic Acid and Wilty Phenotype. Plant Physiol. 1999, 120, 571–578. [Google Scholar] [CrossRef]
- Kuromori, T.; Seo, M.; Shinozaki, K. Aba Transport and Plant Water Stress Responses. Trends Plant Sci. 2018, 23, 513–522. [Google Scholar] [CrossRef]
- Ma, Y.; Cao, J.; He, J.; Chen, Q.; Li, X.; Yang, Y. Molecular Mechanism for the Regulation of ABA Homeostasis During Plant Development and Stress Responses. Int. J. Mol. Sci. 2018, 19, 3643. [Google Scholar] [CrossRef]
- Dahuja, A.; Kumar, R.R.; Sakhare, A.; Watts, A.; Singh, B.; Goswami, S.; Sachdev, A.; Praveen, S. Role of ATP-Binding Cassette Transporters in Maintaining Plant Homeostasis under Abiotic and Biotic Stresses. Physiol. Plant. 2021, 171, 785–801. [Google Scholar] [CrossRef]
- Kuromori, T.; Miyaji, T.; Yabuuchi, H.; Shimizu, H.; Sugimoto, E.; Kamiya, A.; Moriyama, Y.; Shinozaki, K. Abc Transporter AtABCG25 Is Involved in Abscisic Acid Transport and Responses. Proc. Natl. Acad. Sci. USA 2010, 107, 2361–2366. [Google Scholar] [CrossRef]
- Kang, J.; Hwang, J.-U.; Lee, M.; Kim, Y.-Y.; Assmann, S.M.; Martinoia, E.; Lee, Y. Pdr-Type Abc Transporter Mediates Cellular Uptake of the Phytohormone Abscisic Acid. Proc. Natl. Acad. Sci. USA 2010, 107, 2355–2360. [Google Scholar] [CrossRef]
- Zhang, Y.; Kilambi, H.V.; Liu, J.; Bar, H.; Lazary, S.; Egbaria, A.; Ripper, D.; Charrier, L.; Belew, Z.M.; Wulff, N.; et al. ABA Homeostasis and Long-Distance Translocation Are Redundantly Regulated by ACBG ABA Importers. Sci. Adv. 2021, 7, eabf6069. [Google Scholar] [CrossRef] [PubMed]
- Kanno, Y.; Hanada, A.; Chiba, Y.; Ichikawa, T.; Nakazawa, M.; Matsui, M.; Koshiba, T.; Kamiya, Y.; Seo, M. Identification of an Abscisic Acid Transporter by Functional Screening Using the Receptor Complex as a Sensor. Proc. Natl. Acad. Sci. USA 2012, 109, 9653–9658. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhu, H.; Pan, Y.; Yu, Y.; Luan, S.; Li, L. A DTX/Mate-Type Transporter Facilitates Abscisic Acid Efflux and Modulates ABA Sensitivity and Drought Tolerance in Arabidopsis. Mol. Plant 2014, 7, 1522–1532. [Google Scholar] [CrossRef] [PubMed]
- Boursiac, Y.; Léran, S.; Corratgé-Faillie, C.; Gojon, A.; Krouk, G.; Lacombe, B. ABA Transport and Transporters. Trends Plant Sci. 2013, 18, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, P.L.; Lozano-Juste, J.; Albert, A. Pyr/Pyl/Rcar Aba Receptors. In Advances in Botanical Research; Seo, M., Marion-Poll, A., Eds.; Academic Press: Cambridge, MA, USA, 2019; pp. 51–82. [Google Scholar]
- Ma, Y.; Szostkiewicz, I.; Korte, A.; Moes, D.; Yang, Y.; Christmann, A.; Grill, E. Regulators of PP2C Phosphatase Activity Function as Abscisic Acid Sensors. Science 2009, 324, 1064–1068. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-Y.; Fung, P.; Nishimura, N.; Jensen, D.R.; Fujii, H.; Zhao, Y.; Lumba, S.; Santiago, J.; Rodrigues, A.; Chow, T.-F.F.; et al. Abscisic Acid Inhibits Type 2c Protein Phosphatases Via the Pyr/Pyl Family of Start Proteins. Science 2009, 324, 1068–1071. [Google Scholar] [CrossRef]
- Geiger, D.; Scherzer, S.; Mumm, P.; Stange, A.; Marten, I.; Bauer, H.; Ache, P.; Matschi, S.; Liese, A.; Al-Rasheid, K.A.S.; et al. Activity of Guard Cell Anion Channel Slac1 Is Controlled by Drought-Stress Signaling Kinase-Phosphatase Pair. Proc. Natl. Acad. Sci. USA 2009, 106, 21425–21430. [Google Scholar] [CrossRef]
- Brandt, B.; Brodsky, D.E.; Xue, S.; Negi, J.; Iba, K.; Kangasjärvi, J.; Ghassemian, M.; Stephan, A.B.; Hu, H.; Schroeder, J.I. Reconstitution of Abscisic Acid Activation of SLAC1 Anion Channel by CPK6 and OST1 Kinases and Branched ABI1 PP2C Phosphatase Action. Proc. Natl. Acad. Sci. USA 2012, 109, 10593–10598. [Google Scholar] [CrossRef]
- Hsu, P.-K.; Dubeaux, G.; Takahashi, Y.; Schroeder, J.I. Signaling Mechanisms in Abscisic Acid-Mediated Stomatal Closure. Plant J. 2021, 105, 307–321. [Google Scholar] [CrossRef]
- Ali, A.; Pardo, J.M.; Yun, D.-J. Desensitization of Aba-Signaling: The Swing from Activation to Degradation. Front. Plant Sci. 2020, 11, 379. [Google Scholar] [CrossRef]
- Konopka-Postupolska, D.; Dobrowolska, G. Aba Perception Is Modulated by Membrane Receptor-Like Kinases. J. Exp. Bot. 2020, 71, 1210–1214. [Google Scholar] [CrossRef] [PubMed]
- Fujii, H.; Chinnusamy, V.; Rodrigues, A.; Rubio, S.; Antoni, R.; Park, S.-Y.; Cutler, S.R.; Sheen, J.; Rodriguez, P.L.; Zhu, J.-K. In Vitro Reconstitution of an Abscisic Acid Signalling Pathway. Nature 2009, 462, 660–664. [Google Scholar] [CrossRef]
- Leopold, A.C.; Noodén, L.D. Hormonal Regulatory Systems in Plants. In Hormonal Regulation of Development II: The Functions of Hormones from the Level of the Cell to the Whole Plant; Scott, T.K., Ed.; Springer: Berlin/Heidelberg, Germany, 1984; pp. 4–22. [Google Scholar]
- Chandler, J.W. Auxin as Compère in Plant Hormone Crosstalk. Planta 2009, 231, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Parwez, R.; Aftab, T.; Gill, S.S.; Naeem, M. Abscisic Acid Signaling and Crosstalk with Phytohormones in Regulation of Environmental Stress Responses. Environ. Exp. Bot. 2022, 199, 104885. [Google Scholar] [CrossRef]
- Liu, J.; Moore, S.; Chen, C.; Lindsey, K. Crosstalk Complexities between Auxin, Cytokinin, and Ethylene in Arabidopsis Root Development: From Experiments to Systems Modeling, and Back Again. Mol. Plant 2017, 10, 1480–1496. [Google Scholar] [CrossRef]
- Singh, V.P.; Prasad, S.M.; Munné-Bosch, S.; Müller, M. Editorial: Phytohormones and the Regulation of Stress Tolerance in Plants: Current Status and Future Directions. Front. Plant Sci. 2017, 8, 1871. [Google Scholar] [CrossRef]
- Gazzarrini, S.; Mccourt, P. Cross-Talk in Plant Hormone Signalling: What Arabidopsis Mutants Are Telling Us. Ann. Bot. 2003, 91, 605–612. [Google Scholar] [CrossRef]
- Herms, D.A.; Mattson, W.J. The Dilemma of Plants: To Grow or Defend. Quart. Rev. Biol. 1992, 67, 283–335. [Google Scholar] [CrossRef]
- Huot, B.; Yao, J.; Montgomery, B.L.; He, S.Y. Growth-Defense Tradeoffs in Plants: A Balancing Act to Optimize Fitness. Mol. Plant 2014, 7, 1267–1287. [Google Scholar] [CrossRef]
- Emenecker, R.J.; Strader, L.C. Auxin-Abscisic Acid Interactions in Plant Growth and Development. Biomolecules 2020, 10, 281. [Google Scholar] [CrossRef]
- Du, H.; Wu, N.; Fu, J.; Wang, S.; Li, X.; Xiao, J.; Xiong, L. A GH3 Family Member, OsGH3-2, Modulates Auxin and Abscisic Acid Levels and Differentially Affects Drought and Cold Tolerance in Rice. J. Exp. Bot. 2012, 63, 6467–6480. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Liu, H.; Xiong, L. Endogenous Auxin and Jasmonic Acid Levels Are Differentially Modulated by Abiotic Stresses in Rice. Front. Plant Sci. 2013, 4, 397. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, Y.; Hassan, M.J.; Li, Z.; Peng, Y. Indole-3-Acetic Acid Improves Drought Tolerance of White Clover Via Activating Auxin, Abscisic Acid and Jasmonic Acid Related Genes and Inhibiting Senescence Genes. BMC Plant Biol. 2020, 20, 150. [Google Scholar] [CrossRef]
- He, Y.; Liu, Y.; Li, M.; Lamin-Samu, A.T.; Yang, D.; Yu, X.; Izhar, M.; Jan, I.; Ali, M.; Lu, G. The Arabidopsis Small Auxin up RNA32 Protein Regulates ABA-Mediated Responses to Drought Stress. Front. Plant Sci. 2021, 12, 625493. [Google Scholar] [CrossRef]
- Kale, L.; Nakurte, I.; Jalakas, P.; Kunga-Jegere, L.; Brosché, M.; Rostoks, N. Arabidopsis Mutant dnd2 Exhibits Increased Auxin and Abscisic Acid Content and Reduced Stomatal Conductance. Plant Physiol. Biochem. 2019, 140, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-García, P.; Pérez-Alonso, M.M.; González Ortega-Villaizan, A.; Sánchez-Parra, B.; Ludwig-Müller, J.; Wilkinson, M.D.; Pollmann, S. The Indole-3-Acetamide-Induced Arabidopsis Transcription Factor MYB74 Decreases Plant Growth and Contributes to the Control of Osmotic Stress Responses. Front Plant. Sci. 2022, 13, 928386. [Google Scholar] [CrossRef]
- Sirko, A.; Wawrzyńska, A.; Brzywczy, J.; Sieńko, M. Control of Aba Signaling and Crosstalk with Other Hormones by the Selective Degradation of Pathway Components. Int. J. Mol. Sci. 2021, 22, 4638. [Google Scholar] [CrossRef]
- Hugouvieux, V.; Kwak, J.M.; Schroeder, J.I. An mRNA Cap Binding Protein, ABH1, Modulates Early Abscisic Acid Signal Transduction in Arabidopsis. Cell 2001, 106, 477–487. [Google Scholar] [CrossRef]
- Gibson, S.I. Plant Sugar-Response Pathways. Part of a Complex Regulatory Web. Plant Physiol. 2000, 124, 1532–1539. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Shang, L.; Wang, X.; Xing, Y.; Xu, W.; Zhang, Y.; Wang, T.; Li, H.; Zhang, J.; Ye, Z. MAPK11 Regulates Seed Germination and ABA Signaling in Tomato by Phosphorylating SnRKs. J. Exp. Bot. 2020, 72, 1677–1690. [Google Scholar] [CrossRef]
- Ali, F.; Qanmber, G.; Li, F.; Wang, Z. Updated Role of ABA in Seed Maturation, Dormancy, and Germination. J. Adv. Res. 2022, 35, 199–214. [Google Scholar] [CrossRef] [PubMed]
- Bharath, P.; Gahir, S.; Raghavendra, A.S. Abscisic Acid-Induced Stomatal Closure: An Important Component of Plant Defense against Abiotic and Biotic Stress. Front. Plant Sci. 2021, 12, 615114. [Google Scholar] [CrossRef] [PubMed]
- Xie, Q.; Essemine, J.; Pang, X.; Chen, H.; Jin, J.; Cai, W. Abscisic Acid Regulates the Root Growth Trajectory by Reducing Auxin Transporter PIN2 Protein Levels in Arabidopsis thaliana. Front. Plant Sci. 2021, 12, 632676. [Google Scholar] [CrossRef]
- Wang, L.; Hua, D.; He, J.; Duan, Y.; Chen, Z.; Hong, X.; Gong, Z. Auxin Response Factor2 (ARF2) and Its Regulated Homeodomain Gene HB33 Mediate Abscisic Acid Response in Arabidopsis. PLoS Genet. 2011, 7, e1002172. [Google Scholar] [CrossRef] [PubMed]
- Cancé, C.; Martin-Arevalillo, R.; Boubekeur, K.; Dumas, R. Auxin Response Factors Are Keys to the Many Auxin Doors. New Phytol. 2022, 235, 402–419. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Wang, S.; Wu, H.; Wang, H. Protein Levels of Several Arabidopsis Auxin Response Factors Are Regulated by Multiple Factors and ABA Promotes ARF6 Protein Ubiquitination. Int. J. Mol. Sci. 2020, 21, 9437. [Google Scholar] [CrossRef]
- Holdsworth, M.J.; Bentsink, L.; Soppe, W.J.J. Molecular Networks Regulating Arabidopsis Seed Maturation, after-Ripening, Dormancy and Germination. New Phytol. 2008, 179, 33–54. [Google Scholar] [CrossRef]
- Kozaki, A.; Aoyanagi, T. Molecular Aspects of Seed Development Controlled by Gibberellins and Abscisic Acids. Int. J. Mol. Sci. 2022, 23, 1876. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, H.; Zhao, Y.; Feng, Z.; Li, Q.; Yang, H.Q.; Luan, S.; Li, J.; He, Z.H. Auxin Controls Seed Dormancy through Stimulation of Abscisic Acid Signaling by Inducing ARF-Mediated ABI3 Activation in Arabidopsis. Proc. Natl. Acad. Sci. USA 2013, 110, 15485–15490. [Google Scholar] [CrossRef]
- Liu, P.-P.; Montgomery, T.A.; Fahlgren, N.; Kasschau, K.D.; Nonogaki, H.; Carrington, J.C. Repression of Auxin Response Factor10 by microRNA160 Is Critical for Seed Germination and Post-Germination Stages. Plant J. 2007, 52, 133–146. [Google Scholar] [CrossRef]
- Tian, R.; Wang, F.; Zheng, Q.; Niza, V.M.a.G.E.; Downie, A.B.; Perry, S.E. Direct and Indirect Targets of the Arabidopsis Seed Transcription Factor Abscisic Acid Insensitive3. Plant J. 2020, 103, 1679–1694. [Google Scholar] [CrossRef]
- Hussain, S.; Kim, S.H.; Bahk, S.; Ali, A.; Nguyen, X.C.; Yun, D.-J.; Chung, W.S. The Auxin Signaling Repressor IAA8 Promotes Seed Germination through Down-Regulation of ABI3 Transcription in Arabidopsis. Front. Plant Sci. 2020, 11, 111. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yan, D.-W.; Yuan, T.-T.; Gao, X.; Lu, Y.-T. A Gain-of-Function Mutation in IAA8 Alters Arabidopsis Floral Organ Development by Change of Jasmonic Acid Level. Plant Mol. Biol. 2013, 82, 71–83. [Google Scholar] [CrossRef] [PubMed]
- Brocard, I.S.M.; Lynch, T.J.; Finkelstein, R.R. Regulation and Role of the Arabidopsis Abscisic Acid-Insensitive 5 Gene in Abscisic Acid, Sugar, and Stress Response. Plant Physiol. 2002, 129, 1533–1543. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhu, T.; Song, Y.; Feng, L.; Farag, E.a.H.; Ren, M. Abscisic Acid Insensitive5 Interacts with Ribosomal S6 Kinase2 to Mediate Aba Responses During Seedling Growth in Arabidopsis. Front. Plant Sci. 2021, 11, 598654. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Zhao, Y.; Li, Z.; Hsu, C.-C.; Liu, X.; Fu, L.; Hou, Y.-J.; Du, Y.; Xie, S.; Zhang, C.; et al. Reciprocal Regulation of the Tor Kinase and ABA Receptor Balances Plant Growth and Stress Response. Mol. Cell 2018, 69, 100–112.e6. [Google Scholar] [CrossRef] [PubMed]
- Schepetilnikov, M.; Makarian, J.; Srour, O.; Geldreich, A.; Yang, Z.; Chicher, J.; Hammann, P.; Ryabova, L.A. GTPase ROP2 Binds and Promotes Activation of Target of Rapamycin, TOR, in Response to Auxin. EMBO J. 2017, 36, 886–903. [Google Scholar] [CrossRef]
- Yuan, T.-T.; Xu, H.-H.; Zhang, K.-X.; Guo, T.-T.; Lu, Y.-T. Glucose Inhibits Root Meristem Growth Via Aba Insensitive 5, Which Represses PIN1 Accumulation and Auxin Activity in Arabidopsis. Plant Cell Environ. 2014, 37, 1338–1350. [Google Scholar] [CrossRef]
- Wu, Y.; Chang, Y.; Luo, L.; Tian, W.; Gong, Q.; Liu, X. Abscisic Acid Employs NRP-Dependent PIN2 Vacuolar Degradation to Suppress Auxin-Mediated Primary Root Elongation in Arabidopsis. New Phytol. 2022, 233, 297–312. [Google Scholar] [CrossRef]
- Liu, W.; Li, R.-J.; Han, T.-T.; Cai, W.; Fu, Z.-W.; Lu, Y.-T. Salt Stress Reduces Root Meristem Size by Nitric Oxide-Mediated Modulation of Auxin Accumulation and Signaling in Arabidopsis. Plant Physiol. 2015, 168, 343–356. [Google Scholar] [CrossRef]
- Albertos, P.; Romero-Puertas, M.C.; Tatematsu, K.; Mateos, I.; Sánchez-Vicente, I.; Nambara, E.; Lorenzo, O. S-Nitrosylation Triggers ABI5 Degradation to Promote Seed Germination and Seedling Growth. Nat. Commun. 2015, 6, 8669. [Google Scholar] [CrossRef] [PubMed]
- Manrique-Gil, I.; Sánchez-Vicente, I.; Torres-Quezada, I.; Lorenzo, O. Nitric Oxide Function During Oxygen Deprivation in Physiological and Stress Processes. J. Exp. Bot. 2020, 72, 904–916. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhou, J.; Li, Z.; Qiao, J.; Quan, R.; Wang, J.; Huang, R.; Qin, H. Salt and Aba Response ERF1 Improves Seed Germination and Salt Tolerance by Repressing Aba Signaling in Rice. Plant Physiol. 2022, 189, 1110–1127. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Jiang, H.; Li, L.; Zhai, Q.; Qi, L.; Zhou, W.; Liu, X.; Li, H.; Zheng, W.; Sun, J.; et al. The Arabidopsis Mediator Subunit Med25 Differentially Regulates Jasmonate and Abscisic Acid Signaling through Interacting with the MYC2 and ABI5 Transcription Factors. Plant Cell 2012, 24, 2898–2916. [Google Scholar] [CrossRef]
- Ito, J.; Fukaki, H.; Onoda, M.; Li, L.; Li, C.; Tasaka, M.; Furutani, M. Auxin-Dependent Compositional Change in Mediator in ARF7- and ARF19-Mediated Transcription. Proc. Natl. Acad. Sci. USA 2016, 113, 6562–6567. [Google Scholar] [CrossRef]
- Guo, P.; Chong, L.; Wu, F.; Hsu, C.-C.; Li, C.; Zhu, J.-K.; Zhu, Y. Mediator Tail Module Subunits Med16 and Med25 Differentially Regulate Abscisic Acid Signaling in Arabidopsis. J. Integr. Plant Biol. 2021, 63, 802–815. [Google Scholar] [CrossRef]
- Maymon, T.; Eisner, N.; Bar-Zvi, D. The Abcisic Acid Insensitive (ABI) 4 Transcription Factor Is Stabilized by Stress, ABA and Phosphorylation. Plants 2022, 11, 2179. [Google Scholar] [CrossRef]
- Matilla, A.J. Auxin: Hormonal Signal Required for Seed Development and Dormancy. Plants 2020, 9, 705. [Google Scholar] [CrossRef]
- Shkolnik-Inbar, D.; Bar-Zvi, D. ABI4 Mediates Abscisic Acid and Cytokinin Inhibition of Lateral Root Formation by Reducing Polar Auxin Transport in Arabidopsis. Plant Cell 2010, 22, 3560–3573. [Google Scholar] [CrossRef]
- Chandrasekaran, U.; Luo, X.; Zhou, W.; Shu, K. Multifaceted Signaling Networks Mediated By abscisic Acid Insensitive 4. Plant Commun. 2020, 1, 100040. [Google Scholar] [CrossRef]
- Nakayama, N.; Smith, R.S.; Mandel, T.; Robinson, S.; Kimura, S.; Boudaoud, A.; Kuhlemeier, C. Mechanical Regulation of Auxin-Mediated Growth. Curr. Biol. 2012, 22, 1468–1476. [Google Scholar] [CrossRef] [PubMed]
- Rowe, J.H.; Topping, J.F.; Liu, J.; Lindsey, K. Abscisic Acid Regulates Root Growth under Osmotic Stress Conditions Via an Interacting Hormonal Network with Cytokinin, Ethylene and Auxin. New Phytol. 2016, 211, 225–239. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.J.; Yan, J.Y.; Li, C.X.; Li, G.X.; Wu, Y.R.; Zheng, S.J. Transcription Factor WRKY46 Modulates the Development of Arabidopsis Lateral Roots in Osmotic/Salt Stress Conditions Via Regulation of ABA Signaling and Auxin Homeostasis. Plant J. 2015, 84, 56–69. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Letnik, I.; Hacham, Y.; Dobrev, P.; Ben-Daniel, B.-H.; Vanková, R.; Amir, R.; Miller, G. Ascorbate Peroxidase6 Protects Arabidopsis Desiccating and Germinating Seeds from Stress and Mediates Cross Talk between Reactive Oxygen Species, Abscisic Acid, and Auxin. Plant Physiol. 2014, 166, 370–383. [Google Scholar] [CrossRef]
- Chen, C.; Twito, S.; Miller, G. New Cross Talk between ROS, ABA and Auxin Controlling Seed Maturation and Germination Unraveled in APX6 Deficient Arabidopsis Seeds. Plant Signal. Behav. 2014, 9, e976489. [Google Scholar] [CrossRef] [PubMed]
- Munguía-Rodríguez, A.G.; López-Bucio, J.S.; Ruiz-Herrera, L.F.; Ortiz-Castro, R.; Guevara-García, Á.A.; Marsch-Martínez, N.; Carreón-Abud, Y.; López-Bucio, J.; Martínez-Trujillo, M. YUCCA4 Overexpression Modulates Auxin Biosynthesis and Transport and Influences Plant Growth and Development Via Crosstalk with Abscisic Acid in Arabidopsis thaliana. Genet. Mol. Biol. 2020, 43, e20190221. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zhang, M.; Qiao, L.; Chen, Y.; Zhang, D.; Jing, X.; Gan, P.; Huang, Y.; Gao, J.; Liu, W.; et al. Characterization of Wavy Root 1, an Agravitropism Allele, Reveals the Functions of Ospin2 in Fine Regulation of Auxin Transport and Distribution and in Aba Biosynthesis and Response in Rice (Oryza sativa L.). Crop J. 2022, 10, 980–992. [Google Scholar] [CrossRef]
- Zhang, A.; Yang, X.; Lu, J.; Song, F.; Sun, J.; Wang, C.; Lian, J.; Zhao, L.; Zhao, B. OsIAA20, an Aux/IAA Protein, Mediates Abiotic Stress Tolerance in Rice through an Aba Pathway. Plant Sci. 2021, 308, 110903. [Google Scholar] [CrossRef]
- Wang, F.; Niu, H.; Xin, D.; Long, Y.; Wang, G.; Liu, Z.; Li, G.; Zhang, F.; Qi, M.; Ye, Y.; et al. OsIAA18, an Aux/IAA Transcription Factor Gene, Is Involved in Salt and Drought Tolerance in Rice. Front. Plant Sci. 2021, 12, 738660. [Google Scholar] [CrossRef]
- Li, G.; Ye, Y.X.; Ren, X.Q.; Qi, M.Y.; Zhao, H.Y.; Zhou, Q.; Chen, X.H.; Wang, J.; Yuan, C.Y.; Wang, F.B. The Rice Aux/IAA Transcription Factor Gene OsIAA18 Enhances Salt and Osmotic Tolerance in Arabidopsis. Biol. Plant. 2020, 64, 454–464. [Google Scholar] [CrossRef]
- Li, W.; Dang, C.; Ye, Y.; Wang, Z.; Hu, L.; Zhang, F.; Zhang, Y.; Qian, X.; Shi, J.; Guo, Y.; et al. Overexpression of Grapevine VvIAA18 Gene Enhanced Salt Tolerance in Tobacco. Int. J. Mol. Sci. 2020, 21, 1323. [Google Scholar] [CrossRef] [PubMed]
- Shani, E.; Salehin, M.; Zhang, Y.; Sanchez, S.E.; Doherty, C.; Wang, R.; Mangado, C.C.; Song, L.; Tal, I.; Pisanty, O.; et al. Plant Stress Tolerance Requires Auxin-Sensitive Aux/IAA Transcriptional Repressors. Curr. Biol. 2017, 27, 437–444. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Shu, D.; Tan, Z.; Ma, M.; Guo, N.; Gao, S.; Duan, G.; Kuai, B.; Hu, Y.; Li, S.; et al. The Arabidopsis IDD14 Transcription Factor Interacts with bZIP-Type ABFs/AREBs and Cooperatively Regulates ABA-Mediated Drought Tolerance. New Phytol. 2022, 236, 929–942. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Tan, M.; Geng, L.; Li, J.; Xiang, Y.; Zhang, B.; Zhao, Y. New Insight into Comprehensive Analysis of Indeterminate Domain (IDD) Gene Family in Rice. Plant Physiol. Biochem. 2020, 154, 547–556. [Google Scholar] [CrossRef]
- Cui, D.; Zhao, J.; Jing, Y.; Fan, M.; Liu, J.; Wang, Z.; Xin, W.; Hu, Y. The Arabidopsis IDD14, IDD15, and IDD16 Cooperatively Regulate Lateral Organ Morphogenesis and Gravitropism by Promoting Auxin Biosynthesis and Transport. PLoS Genet. 2013, 9, e1003759. [Google Scholar] [CrossRef]
- Qin, H.; Wang, J.; Zhou, J.; Qiao, J.; Li, Y.; Quan, R.; Huang, R. Abscisic Acid Promotes Auxin Biosynthesis to Inhibit Primary Root Elongation in Rice. Plant Physiol. 2022. [Google Scholar] [CrossRef]
- Ding, Z.J.; Yan, J.Y.; Li, G.X.; Wu, Z.C.; Zhang, S.Q.; Zheng, S.J. WRKY41 Controls Arabidopsis Seed Dormancy Via Direct Regulation of ABI3 Transcript Levels Not Downstream of ABA. Plant J. 2014, 79, 810–823. [Google Scholar] [CrossRef]
- Yuan, T.-T.; Xiang, Z.-X.; Li, W.; Gao, X.; Lu, Y.-T. Osmotic Stress Represses Root Growth by Modulating the Transcriptional Regulation of Pin-Formed3. New Phytol. 2021, 232, 1661–1673. [Google Scholar] [CrossRef]
- Cackett, L.; Cannistraci, C.V.; Meier, S.; Ferrandi, P.; Pěnčík, A.; Gehring, C.; Novák, O.; Ingle, R.A.; Donaldson, L. Salt-Specific Gene Expression Reveals Elevated Auxin Levels in Arabidopsis thaliana Plants Grown under Saline Conditions. Front. Plant Sci. 2022, 13, 804716. [Google Scholar] [CrossRef]
- Mega, R.; Meguro-Maoka, A.; Endo, A.; Shimosaka, E.; Murayama, S.; Nambara, E.; Seo, M.; Kanno, Y.; Abrams, S.R.; Sato, Y. Sustained Low Abscisic Acid Levels Increase Seedling Vigor under Cold Stress in Rice (Oryza sativa L.). Sci. Rep. 2015, 5, 13819. [Google Scholar] [CrossRef]
- Ashraf, M.A.; Rahman, A. Cold Stress Response in Arabidopsis thaliana Is Mediated by Gnom ARF-GEF. Plant J. 2019, 97, 500–516. [Google Scholar] [CrossRef] [PubMed]
- Ioio, R.D.; Nakamura, K.; Moubayidin, L.; Perilli, S.; Taniguchi, M.; Morita, M.T.; Aoyama, T.; Costantino, P.; Sabatini, S. A Genetic Framework for the Control of Cell Division and Differentiation in the Root Meristem. Science 2008, 322, 1380–1384. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Zhang, K.-X.; Wang, W.-S.; Gong, W.; Liu, W.-C.; Chen, H.-G.; Xu, H.-H.; Lu, Y.-T. Low Temperature Inhibits Root Growth by Reducing Auxin Accumulation Via ARR1/12. Plant Cell Physiol. 2014, 56, 727–736. [Google Scholar] [CrossRef] [PubMed]
- Vissenberg, K.; Claeijs, N.; Balcerowicz, D.; Schoenaers, S. Hormonal Regulation of Root Hair Growth and Responses to the Environment in Arabidopsis. J. Exp. Bot. 2020, 71, 2412–2427. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).