Multiplex Immunofluorescence: A Powerful Tool in Cancer Immunotherapy
Abstract
1. Introduction
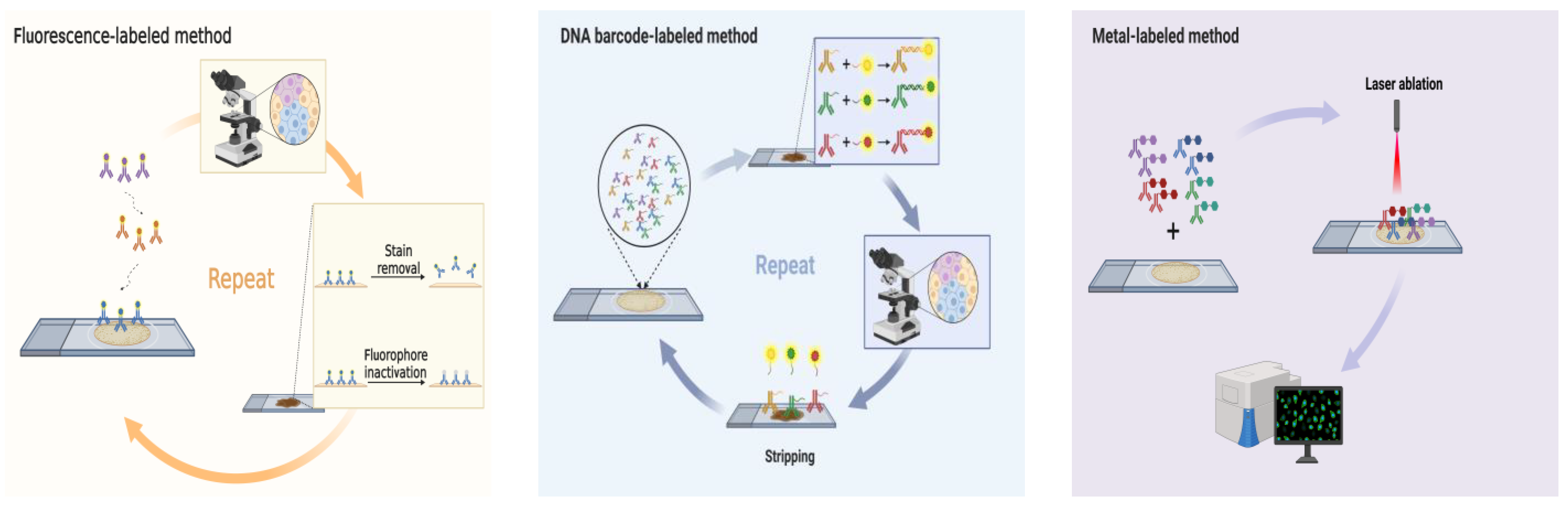
2. Fluorescent Immunohistochemistry
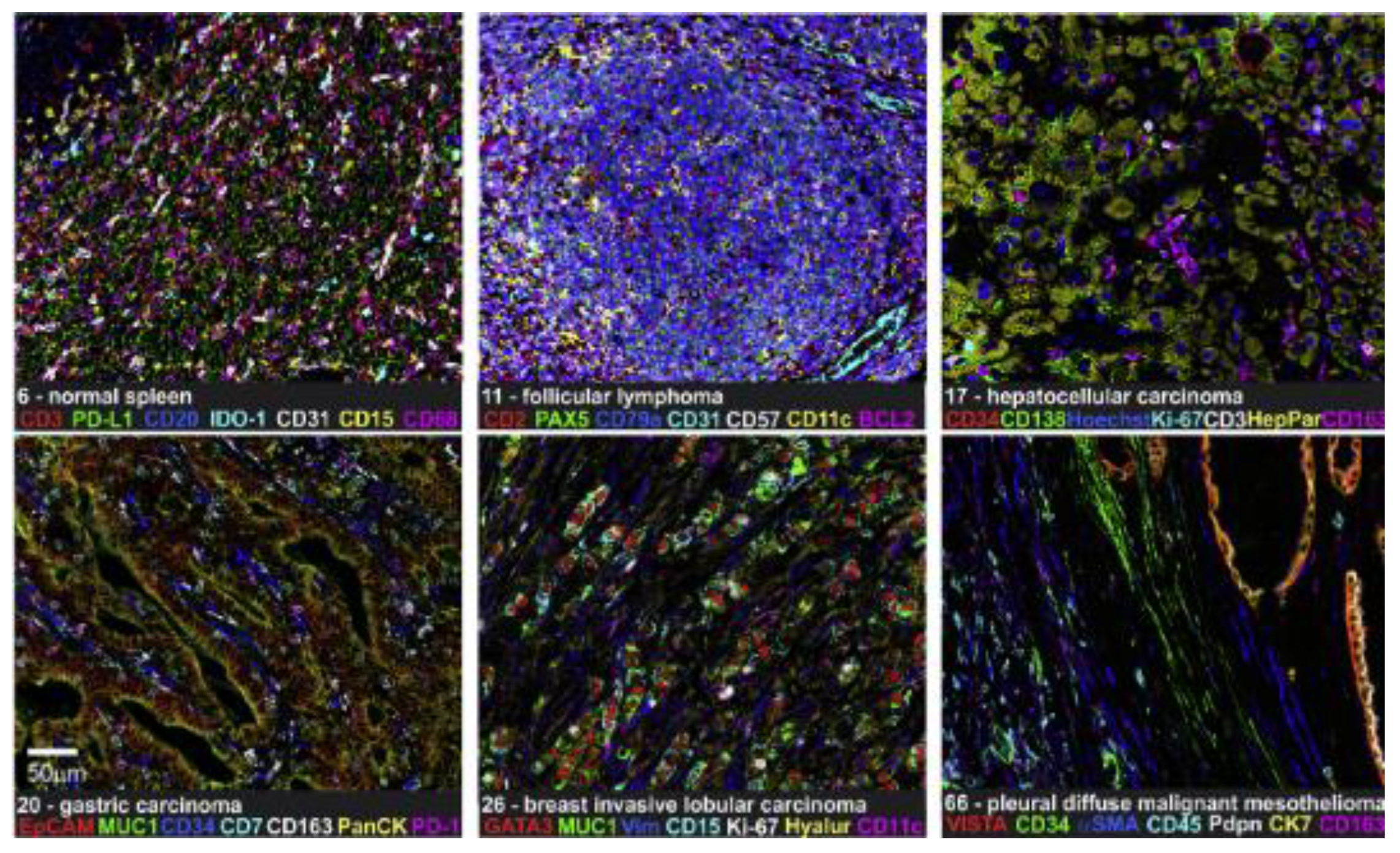
3. Multiplex Fluorescent Immunohistochemistry
3.1. Stain Removal Technologies
3.1.1. Multiepitope-Ligand Cartography
3.1.2. Sequential Immuno-Peroxidase Labelling and Erasing
3.1.3. Iterative Bleaching Extends Multiplexity
3.2. Fluorophore Inactivation Technologies
3.2.1. Multiplexed Fluorescence Microscopy
3.2.2. Cyclic Immunofluorescence
3.2.3. ChipCytometry
3.3. Multiplexed Signal Amplification
3.3.1. Multiplex Modified Hapten-Based
3.3.2. Tyramide Signal Amplification (TSA)
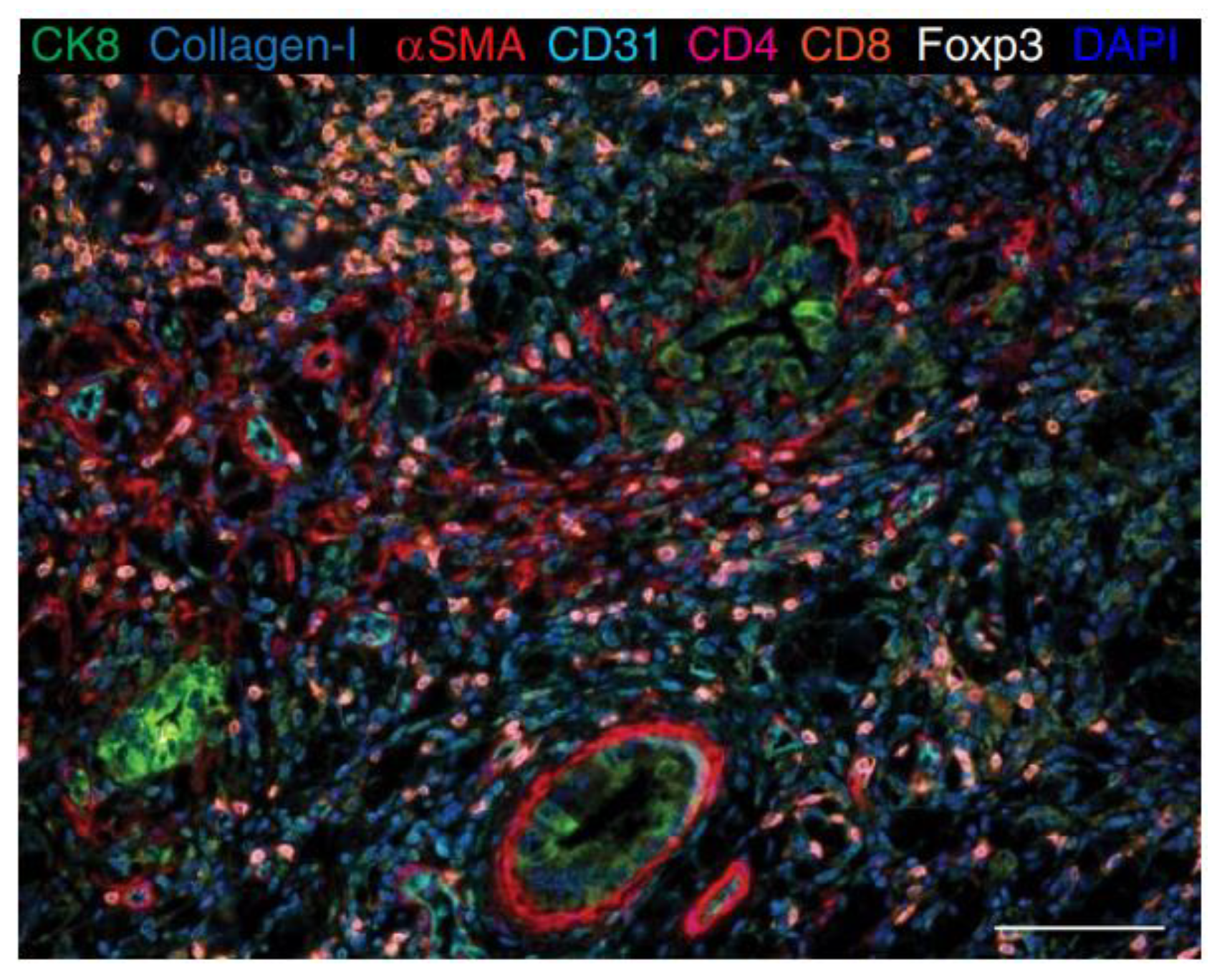
3.3.3. Nanocrystal Quantum Dots
3.4. DNA Barcoding Technologies
3.4.1. DNA Exchange Imaging
3.4.2. Codetection by Indexing
3.4.3. Signal Amplification by Exchange Reaction
3.4.4. Digital Spatial Profiling (DSP)
3.4.5. InSituPlex®
3.5. Mass Cytometry
3.5.1. Imaging Mass Cytometry
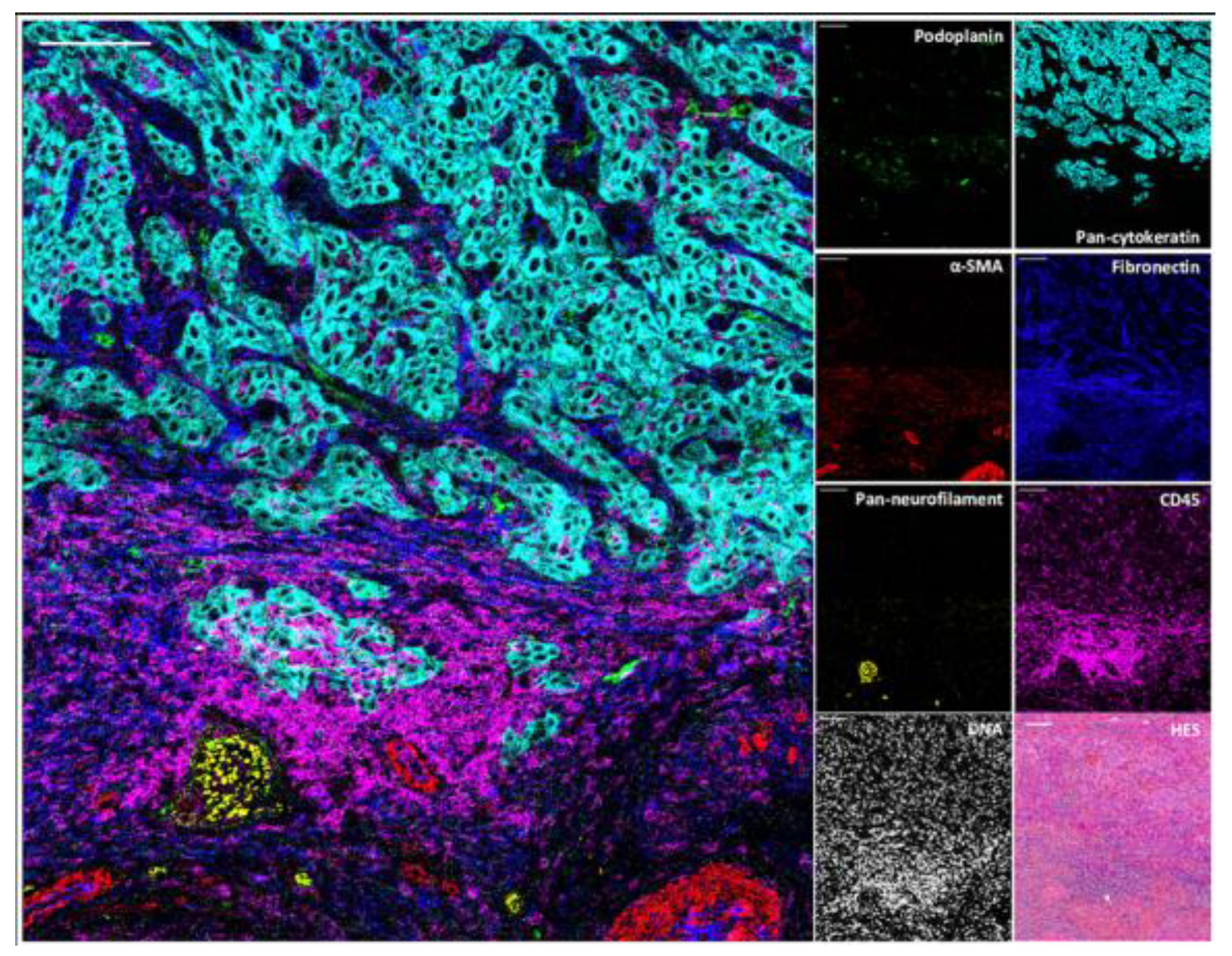
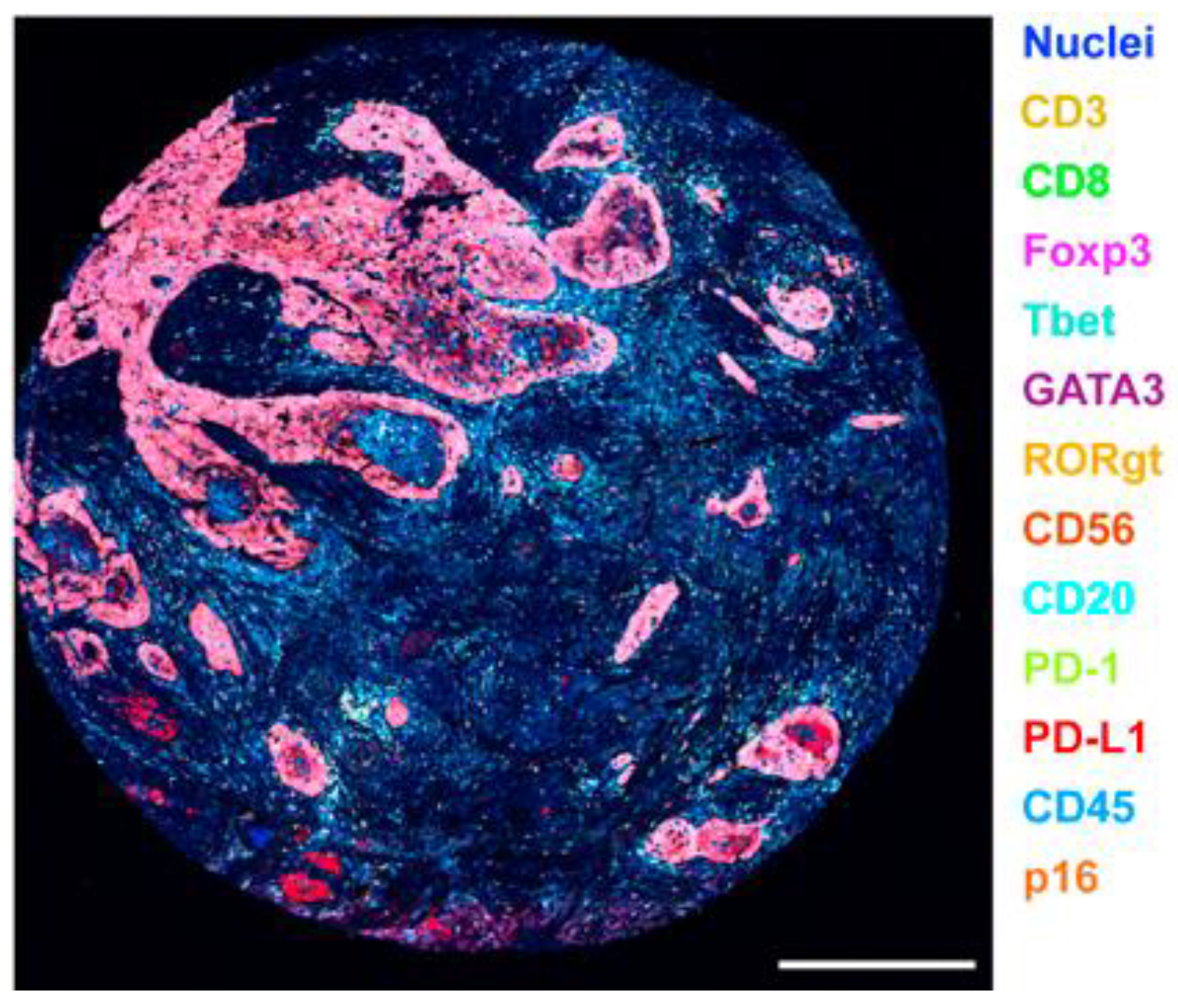
3.5.2. Multiplexed Ion Beam Imaging
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Coons, A.H.; Creech, H.J.; Jones, R.N. Immunological properties of an antibody containing a fluorescent group. Proc. Soc. Exp. Biol. Med. 1941, 47, 200–202. [Google Scholar] [CrossRef]
- Roach, C.; Zhang, N.; Corigliano, E.; Jansson, M.; Toland, G.; Ponto, G.; Dolled-Filhart, M.; Emancipator, K.; Stanforth, D.; Kulangara, K. Development of a Companion Diagnostic PD-L1 Immunohistochemistry Assay for Pembrolizumab Therapy in Non-Small-cell Lung Cancer. Appl. Immunohistochem. Mol. Morphol. 2016, 24, 392–397. [Google Scholar] [CrossRef] [PubMed]
- Lechner, M.G.; Karimi, S.S.; Barry-Holson, K.; Angell, T.E.; Murphy, K.A.; Church, C.H.; Ohlfest, J.R.; Hu, P.; Epstein, A.L. Immunogenicity of murine solid tumor models as a defining feature of in vivo behavior and response to immunotherapy. J. Immunother. 2013, 36, 477–489. [Google Scholar] [CrossRef] [PubMed]
- Paret, C.; Simon, P.; Vormbrock, K.; Bender, C.; Kolsch, A.; Breitkreuz, A.; Yildiz, O.; Omokoko, T.; Hubich-Rau, S.; Hartmann, C.; et al. CXorf61 is a target for T cell based immunotherapy of triple-negative breast cancer. Oncotarget 2015, 6, 25356–25367. [Google Scholar] [CrossRef] [PubMed]
- Harel, M.; Ortenberg, R.; Varanasi, S.K.; Mangalhara, K.C.; Mardamshina, M.; Markovits, E.; Baruch, E.N.; Tripple, V.; Arama-Chayoth, M.; Greenberg, E.; et al. Proteomics of Melanoma Response to Immunotherapy Reveals Mitochondrial Dependence. Cell 2019, 179, 236–250.e218. [Google Scholar] [CrossRef]
- Parra, E.R.; Uraoka, N.; Jiang, M.; Cook, P.; Gibbons, D.; Forget, M.A.; Bernatchez, C.; Haymaker, C.; Wistuba, I.I.; Rodriguez-Canales, J. Validation of multiplex immunofluorescence panels using multispectral microscopy for immune-profiling of formalin-fixed and paraffin-embedded human tumor tissues. Sci. Rep. 2017, 7, 13380. [Google Scholar] [CrossRef]
- Tan, W.C.C.; Nerurkar, S.N.; Cai, H.Y.; Ng, H.H.M.; Wu, D.; Wee, Y.T.F.; Lim, J.C.T.; Yeong, J.; Lim, T.K.H. Overview of multiplex immunohistochemistry/immunofluorescence techniques in the era of cancer immunotherapy. Cancer Commun. 2020, 40, 135–153. [Google Scholar] [CrossRef]
- Lu, S.; Stein, J.E.; Rimm, D.L.; Wang, D.W.; Bell, J.M.; Johnson, D.B.; Sosman, J.A.; Schalper, K.A.; Anders, R.A.; Wang, H.; et al. Comparison of Biomarker Modalities for Predicting Response to PD-1/PD-L1 Checkpoint Blockade: A Systematic Review and Meta-analysis. JAMA Oncol. 2019, 5, 1195–1204. [Google Scholar] [CrossRef]
- Yeong, J.; Tan, T.; Chow, Z.L.; Cheng, Q.; Lee, B.; Seet, A.; Lim, J.X.; Lim, J.C.T.; Ong, C.C.H.; Thike, A.A.; et al. Multiplex immunohistochemistry/immunofluorescence (mIHC/IF) for PD-L1 testing in triple-negative breast cancer: A translational assay compared with conventional IHC. J. Clin. Pathol. 2020, 73, 557–562. [Google Scholar] [CrossRef]
- Allam, M.; Cai, S.; Coskun, A.F. Multiplex bioimaging of single-cell spatial profiles for precision cancer diagnostics and therapeutics. NPJ Precis. Oncol. 2020, 4, 11. [Google Scholar] [CrossRef]
- De Smet, F.; Martinez, A.A.; Bosisio, F.M. Next-generation pathology by multiplexed immunohistochemistry. Trends Biochem. Sci. 2021, 46, 80–82. [Google Scholar] [CrossRef] [PubMed]
- Robertson, D.; Savage, K.; Reis-Filho, J.S.; Isacke, C.M. Multiple immunofluorescence labelling of formalin-fixed paraffin-embedded (FFPE) tissue. BMC Cell Biol. 2008, 9, 13. [Google Scholar] [CrossRef] [PubMed]
- Stack, E.C.; Wang, C.; Roman, K.A.; Hoyt, C.C. Multiplexed immunohistochemistry, imaging, and quantitation: A review, with an assessment of Tyramide signal amplification, multispectral imaging and multiplex analysis. Methods 2014, 70, 46–58. [Google Scholar] [CrossRef] [PubMed]
- Angelo, M.; Bendall, S.C.; Finck, R.; Hale, M.B.; Hitzman, C.; Borowsky, A.D.; Levenson, R.M.; Lowe, J.B.; Liu, S.D.; Zhao, S.; et al. Multiplexed ion beam imaging of human breast tumors. Nat. Med. 2014, 20, 436–442. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.C.T.; Yeong, J.P.S.; Lim, C.J.; Ong, C.C.H.; Wong, S.C.; Chew, V.S.P.; Ahmed, S.S.; Tan, P.H.; Iqbal, J. An automated staining protocol for seven-colour immunofluorescence of human tissue sections for diagnostic and prognostic use. Pathology 2018, 50, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Parra, E.R. Novel platforms of multiplexed immunofluorescence for study of paraffin tumor tissues. J. Cancer Treat. Diagn. 2018, 2, 43–53. [Google Scholar] [CrossRef]
- Schubert, W. Automated determining and measuring device and method. U.S. Patent No. 6,150,173, 21 November 2000. [Google Scholar]
- Schubert, W. Topological proteomics, toponomics, MELK-technology. Adv. Biochem. Eng. Biotechnol. 2003, 83, 189–209. [Google Scholar] [CrossRef] [PubMed]
- Schubert, W.; Bonnekoh, B.; Pommer, A.J.; Philipsen, L.; Bockelmann, R.; Malykh, Y.; Gollnick, H.; Friedenberger, M.; Bode, M.; Dress, A.W. Analyzing proteome topology and function by automated multidimensional fluorescence microscopy. Nat. Biotechnol. 2006, 24, 1270–1278. [Google Scholar] [CrossRef]
- Schubert, W. A three-symbol code for organized proteomes based on cyclical imaging of protein locations. Cytom. A 2007, 71, 352–360. [Google Scholar] [CrossRef]
- Schubert, W.; Gieseler, A.; Krusche, A.; Serocka, P.; Hillert, R. Next-generation biomarkers based on 100-parameter functional super-resolution microscopy TIS. N. Biotechnol. 2012, 29, 599–610. [Google Scholar] [CrossRef]
- Berndt, U.; Philipsen, L.; Bartsch, S.; Wiedenmann, B.; Baumgart, D.C.; Hammerle, M.; Sturm, A. Systematic high-content proteomic analysis reveals substantial immunologic changes in colorectal cancer. Cancer Res. 2008, 68, 880–888. [Google Scholar] [CrossRef]
- Glass, G.; Papin, J.A.; Mandell, J.W. SIMPLE: A sequential immunoperoxidase labeling and erasing method. J. Histochem. Cytochem. 2009, 57, 899–905. [Google Scholar] [CrossRef]
- Tsujikawa, T.; Kumar, S.; Borkar, R.N.; Azimi, V.; Thibault, G.; Chang, Y.H.; Balter, A.; Kawashima, R.; Choe, G.; Sauer, D.; et al. Quantitative Multiplex Immunohistochemistry Reveals Myeloid-Inflamed Tumor-Immune Complexity Associated with Poor Prognosis. Cell Rep. 2017, 19, 203–217. [Google Scholar] [CrossRef] [PubMed]
- Radtke, A.J.; Chu, C.J.; Yaniv, Z.; Yao, L.; Marr, J.; Beuschel, R.T.; Ichise, H.; Gola, A.; Kabat, J.; Lowekamp, B.; et al. IBEX: An iterative immunolabeling and chemical bleaching method for high-content imaging of diverse tissues. Nat. Protoc. 2022, 17, 378–401. [Google Scholar] [CrossRef] [PubMed]
- Vaughan, J.C.; Jia, S.; Zhuang, X. Ultrabright photoactivatable fluorophores created by reductive caging. Nat. Methods 2012, 9, 1181–1184. [Google Scholar] [CrossRef] [PubMed]
- Yaniv, Z.; Lowekamp, B.C.; Johnson, H.J.; Beare, R. SimpleITK Image-Analysis Notebooks: A Collaborative Environment for Education and Reproducible Research. J. Digit. Imaging 2018, 31, 290–303. [Google Scholar] [CrossRef]
- Radtke, A.J.; Kandov, E.; Lowekamp, B.; Speranza, E.; Chu, C.J.; Gola, A.; Thakur, N.; Shih, R.; Yao, L.; Yaniv, Z.R.; et al. IBEX: A versatile multiplex optical imaging approach for deep phenotyping and spatial analysis of cells in complex tissues. Proc. Natl. Acad. Sci. USA 2020, 117, 33455–33465. [Google Scholar] [CrossRef] [PubMed]
- Arroyo-Mejías, A.J.; Ichise, H.; Chu, C.; Hor, J.L.; Yaniv, Z.; Kabat, J.; Croteau, J.; Lowekamp, B.; Radtke, A.J.; Germain, R.N. 3D-IBEX: Achieving multiplex 3-dimensional imaging for deep phenotyping of cells in tissues. J. Immunol. 2022, 208, 116123. [Google Scholar] [CrossRef]
- Gerdes, M.J.; Sevinsky, C.J.; Sood, A.; Adak, S.; Bello, M.O.; Bordwell, A.; Can, A.; Corwin, A.; Dinn, S.; Filkins, R.J.; et al. Highly multiplexed single-cell analysis of formalin-fixed, paraffin-embedded cancer tissue. Proc. Natl. Acad. Sci. USA 2013, 110, 11982–11987. [Google Scholar] [CrossRef]
- Li, C.; Ma, H.; Wang, Y.; Cao, Z.; Graves-Deal, R.; Powell, A.E.; Starchenko, A.; Ayers, G.D.; Washington, M.K.; Kamath, V.; et al. Excess PLAC8 promotes an unconventional ERK2-dependent EMT in colon cancer. J. Clin. Invest. 2014, 124, 2172–2187. [Google Scholar] [CrossRef]
- Lin, J.R.; Fallahi-Sichani, M.; Sorger, P.K. Highly multiplexed imaging of single cells using a high-throughput cyclic immunofluorescence method. Nat. Commun. 2015, 6, 8390. [Google Scholar] [CrossRef]
- Lin, J.R.; Fallahi-Sichani, M.; Chen, J.Y.; Sorger, P.K. Cyclic Immunofluorescence (CycIF), A Highly Multiplexed Method for Single-cell Imaging. Curr. Protoc. Chem. Biol. 2016, 8, 251–264. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.R.; Izar, B.; Wang, S.; Yapp, C.; Mei, S.; Shah, P.M.; Santagata, S.; Sorger, P.K. Highly multiplexed immunofluorescence imaging of human tissues and tumors using t-CyCIF and conventional optical microscopes. Elife 2018, 7, 1–46. [Google Scholar] [CrossRef] [PubMed]
- Guerriero, J.L.; Lin, J.-R.; Pastorello, R.G.; Du, Z.; Mei, S.; Taneja, K.; Schnitt, S.J.; Dillon, D.A.; Sorger, P.K.; Santagata, S. Abstract PS18-02: Highly multiplexed tissue-based cyclic immunofluorescence (t-CyCIF) for precision oncology identifies novel patterns of HER2 heterogeneity in breast cancer. Cancer Res. 2021, 81, PS18-02-PS18-02. [Google Scholar] [CrossRef]
- McMahon, N.P.; Jones, J.A.; Kwon, S.; Chin, K.; Nederlof, M.A.; Gray, J.W.; Gibbs, S.L. Oligonucleotide conjugated antibodies permit highly multiplexed immunofluorescence for future use in clinical histopathology. J. Biomed. Opt. 2020, 25, 056004. [Google Scholar] [CrossRef]
- Jarosch, S.; Kohlen, J.; Sarker, R.S.J.; Steiger, K.; Janssen, K.P.; Christians, A.; Hennig, C.; Holler, E.; D’Ippolito, E.; Busch, D.H. Multiplexed imaging and automated signal quantification in formalin-fixed paraffin-embedded tissues by ChipCytometry. Cell Rep. Methods 2021, 1, 100104. [Google Scholar] [CrossRef] [PubMed]
- Teo, J.; Mirenska, A.; Tan, M.; Lee, Y.; Oh, J.; Hong, L.Z.; Wnek, R.; Yap, Y.S.; Shih, S.J.; AA, S.B.; et al. A preliminary study for the assessment of PD-L1 and PD-L2 on circulating tumor cells by microfluidic-based chipcytometry. Future Sci. OA 2017, 3, FSO244. [Google Scholar] [CrossRef]
- Levin, M.; Flor, A.C.; Snyder, H.; Kron, S.J.; Schwartz, D. UltraPlex Hapten-Based Multiplexed Fluorescent Immunohistochemistry. Methods Mol. Biol. 2021, 2350, 267–287. [Google Scholar] [CrossRef]
- Tighe-Snyder, H.; Levin, M.; Akhananaghdam, Z.; Jiang, Y.; Schwartz, D. Abstract LB238: Multiplex immunohistochemistry profiling with UltraPlex IHC on FFPE lung cancer provides a fast and robust staining platform compatible with clinical laboratory workflows. Cancer Res. 2021, 81, LB238. [Google Scholar] [CrossRef]
- Bobrow, M.N.; Shaughnessy, K.J.; Litt, G.J. Catalyzed reporter deposition, a novel method of signal amplification. II. Application to membrane immunoassays. J. Immunol. Methods 1991, 137, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Parra, E.R.; Jiang, M.; Solis, L.; Mino, B.; Laberiano, C.; Hernandez, S.; Gite, S.; Verma, A.; Tetzlaff, M.; Haymaker, C.; et al. Procedural Requirements and Recommendations for Multiplex Immunofluorescence Tyramide Signal Amplification Assays to Support Translational Oncology Studies. Cancers 2020, 12, 255. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, S.; Rojas, F.; Laberiano, C.; Lazcano, R.; Wistuba, I.; Parra, E.R. Multiplex Immunofluorescence Tyramide Signal Amplification for Immune Cell Profiling of Paraffin-Embedded Tumor Tissues. Front. Mol. Biosci. 2021, 8, 667067. [Google Scholar] [CrossRef] [PubMed]
- Yeong, J.; Lum, H.Y.J.; Teo, C.B.; Tan, B.K.J.; Chan, Y.H.; Tay, R.Y.K.; Choo, J.R.; Jeyasekharan, A.D.; Miow, Q.H.; Loo, L.H.; et al. Choice of PD-L1 immunohistochemistry assay influences clinical eligibility for gastric cancer immunotherapy. Gastric Cancer 2022, 25, 741–750. [Google Scholar] [CrossRef] [PubMed]
- Willemsen, M.; Krebbers, G.; Bekkenk, M.W.; Teunissen, M.B.M.; Luiten, R.M. Improvement of Opal Multiplex Immunofluorescence Workflow for Human Tissue Sections. J. Histochem. Cytochem. 2021, 69, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Carstens, J.L.; Correa de Sampaio, P.; Yang, D.; Barua, S.; Wang, H.; Rao, A.; Allison, J.P.; LeBleu, V.S.; Kalluri, R. Spatial computation of intratumoral T cells correlates with survival of patients with pancreatic cancer. Nat. Commun. 2017, 8, 15095. [Google Scholar] [CrossRef] [PubMed]
- Mahvi, D.A.; Liu, R.; Grinstaff, M.W.; Colson, Y.L.; Raut, C.P. Local Cancer Recurrence: The Realities, Challenges, and Opportunities for New Therapies. CA Cancer J. Clin. 2018, 68, 488–505. [Google Scholar] [CrossRef]
- Bruchez, M., Jr.; Moronne, M.; Gin, P.; Weiss, S.; Alivisatos, A.P. Semiconductor nanocrystals as fluorescent biological labels. Science 1998, 281, 2013–2016. [Google Scholar] [CrossRef]
- Yukawa, H.; Watanabe, M.; Kaji, N.; Baba, Y. Influence of Autofluorescence Derived From Living Body on In Vivo Fluorescence Imaging Using Quantum Dots. Cell Med. 2015, 7, 75–82. [Google Scholar] [CrossRef]
- Peng, C.W.; Liu, X.L.; Chen, C.; Liu, X.; Yang, X.Q.; Pang, D.W.; Zhu, X.B.; Li, Y. Patterns of cancer invasion revealed by QDs-based quantitative multiplexed imaging of tumor microenvironment. Biomaterials 2011, 32, 2907–2917. [Google Scholar] [CrossRef]
- Liu, X.L.; Peng, C.W.; Chen, C.; Yang, X.Q.; Hu, M.B.; Xia, H.S.; Liu, S.P.; Pang, D.W.; Li, Y. Quantum dots-based double-color imaging of HER2 positive breast cancer invasion. Biochem. Biophys. Res. Commun. 2011, 409, 577–582. [Google Scholar] [CrossRef]
- Jungmann, R.; Avendano, M.S.; Woehrstein, J.B.; Dai, M.; Shih, W.M.; Yin, P. Multiplexed 3D cellular super-resolution imaging with DNA-PAINT and Exchange-PAINT. Nat. Methods 2014, 11, 313–318. [Google Scholar] [CrossRef]
- Wang, Y.; Woehrstein, J.B.; Donoghue, N.; Dai, M.; Avendano, M.S.; Schackmann, R.C.J.; Zoeller, J.J.; Wang, S.S.H.; Tillberg, P.W.; Park, D.; et al. Rapid Sequential in Situ Multiplexing with DNA Exchange Imaging in Neuronal Cells and Tissues. Nano Lett. 2017, 17, 6131–6139. [Google Scholar] [CrossRef] [PubMed]
- Goltsev, Y.; Samusik, N.; Kennedy-Darling, J.; Bhate, S.; Hale, M.; Vazquez, G.; Black, S.; Nolan, G.P. Deep Profiling of Mouse Splenic Architecture with CODEX Multiplexed Imaging. Cell 2018, 174, 968–981 e915. [Google Scholar] [CrossRef] [PubMed]
- Schurch, C.M.; Bhate, S.S.; Barlow, G.L.; Phillips, D.J.; Noti, L.; Zlobec, I.; Chu, P.; Black, S.; Demeter, J.; McIlwain, D.R.; et al. Coordinated Cellular Neighborhoods Orchestrate Antitumoral Immunity at the Colorectal Cancer Invasive Front. Cell 2020, 183, 838. [Google Scholar] [CrossRef] [PubMed]
- Black, S.; Phillips, D.; Hickey, J.W.; Kennedy-Darling, J.; Venkataraaman, V.G.; Samusik, N.; Goltsev, Y.; Schurch, C.M.; Nolan, G.P. CODEX multiplexed tissue imaging with DNA-conjugated antibodies. Nat. Protoc. 2021, 16, 3802–3835. [Google Scholar] [CrossRef]
- Phillips, D.; Schurch, C.M.; Khodadoust, M.S.; Kim, Y.H.; Nolan, G.P.; Jiang, S. Highly Multiplexed Phenotyping of Immunoregulatory Proteins in the Tumor Microenvironment by CODEX Tissue Imaging. Front. Immunol. 2021, 12, 687673. [Google Scholar] [CrossRef]
- Saka, S.K.; Wang, Y.; Kishi, J.Y.; Zhu, A.; Zeng, Y.; Xie, W.; Kirli, K.; Yapp, C.; Cicconet, M.; Beliveau, B.J.; et al. Immuno-SABER enables highly multiplexed and amplified protein imaging in tissues. Nat. Biotechnol. 2019, 37, 1080–1090. [Google Scholar] [CrossRef]
- Kishi, J.Y.; Lapan, S.W.; Beliveau, B.J.; West, E.R.; Zhu, A.; Sasaki, H.M.; Saka, S.K.; Wang, Y.; Cepko, C.L.; Yin, P. SABER amplifies FISH: Enhanced multiplexed imaging of RNA and DNA in cells and tissues. Nat. Methods 2019, 16, 533–544. [Google Scholar] [CrossRef]
- Zhou, W.; Han, Y.; Beliveau, B.J.; Gao, X. Combining Qdot Nanotechnology and DNA Nanotechnology for Sensitive Single-Cell Imaging. Adv. Mater. 2020, 32, e1908410. [Google Scholar] [CrossRef]
- Snyder, M.P.; Lin, S.; Posgai, A.; Atkinson, M.; Regev, A.; Rood, J.; Rozenblatt-Rosen, O.; Gaffney, L.; Hupalowska, A.; Satija, R.; et al. The human body at cellular resolution: The NIH Human Biomolecular Atlas Program. Nature 2019, 574, 187–192. [Google Scholar]
- Merritt, C.R.; Ong, G.T.; Church, S.E.; Barker, K.; Danaher, P.; Geiss, G.; Hoang, M.; Jung, J.; Liang, Y.; McKay-Fleisch, J.; et al. Multiplex digital spatial profiling of proteins and RNA in fixed tissue. Nat. Biotechnol. 2020, 38, 586–599. [Google Scholar] [CrossRef] [PubMed]
- Blank, C.U.; Rozeman, E.A.; Fanchi, L.F.; Sikorska, K.; van de Wiel, B.; Kvistborg, P.; Krijgsman, O.; van den Braber, M.; Philips, D.; Broeks, A.; et al. Neoadjuvant versus adjuvant ipilimumab plus nivolumab in macroscopic stage III melanoma. Nat. Med. 2018, 24, 1655–1661. [Google Scholar] [CrossRef] [PubMed]
- Toki, M.I.; Merritt, C.R.; Wong, P.F.; Smithy, J.W.; Kluger, H.M.; Syrigos, K.N.; Ong, G.T.; Warren, S.E.; Beechem, J.M.; Rimm, D.L. High-Plex Predictive Marker Discovery for Melanoma Immunotherapy-Treated Patients Using Digital Spatial Profiling. Clin. Cancer Res. 2019, 25, 5503–5512. [Google Scholar] [CrossRef] [PubMed]
- Manesse, M.; Patel, K.K.; Bobrow, M.; Downing, S.R. The InSituPlex® staining method for multiplexed immunofluorescence cell phenotyping and spatial profiling of tumor FFPE samples. In Biomarkers for Immunotherapy of Cancer; Springer: New York, NY, USA, 2020; pp. 585–592. [Google Scholar]
- Singhal, S.K.; Byun, J.S.; Park, S.; Yan, T.; Yancey, R.; Caban, A.; Hernandez, S.G.; Hewitt, S.M.; Boisvert, H.; Hennek, S.; et al. Kaiso (ZBTB33) subcellular partitioning functionally links LC3A/B, the tumor microenvironment, and breast cancer survival. Commun. Biol. 2021, 4, 150. [Google Scholar] [CrossRef] [PubMed]
- Bandura, D.R.; Baranov, V.I.; Ornatsky, O.I.; Antonov, A.; Kinach, R.; Lou, X.; Pavlov, S.; Vorobiev, S.; Dick, J.E.; Tanner, S.D. Mass cytometry: Technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Anal. Chem. 2009, 81, 6813–6822. [Google Scholar] [CrossRef] [PubMed]
- Spitzer, M.H.; Nolan, G.P. Mass Cytometry: Single Cells, Many Features. Cell 2016, 165, 780–791. [Google Scholar] [CrossRef]
- Giesen, C.; Wang, H.A.; Schapiro, D.; Zivanovic, N.; Jacobs, A.; Hattendorf, B.; Schuffler, P.J.; Grolimund, D.; Buhmann, J.M.; Brandt, S.; et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat. Methods 2014, 11, 417–422. [Google Scholar] [CrossRef]
- Gerdtsson, E.; Pore, M.; Thiele, J.A.; Gerdtsson, A.S.; Malihi, P.D.; Nevarez, R.; Kolatkar, A.; Velasco, C.R.; Wix, S.; Singh, M.; et al. Multiplex protein detection on circulating tumor cells from liquid biopsies using imaging mass cytometry. Converg. Sci. Phys. Oncol. 2018, 4, 015002. [Google Scholar] [CrossRef]
- Jackson, H.W.; Fischer, J.R.; Zanotelli, V.R.T.; Ali, H.R.; Mechera, R.; Soysal, S.D.; Moch, H.; Muenst, S.; Varga, Z.; Weber, W.P.; et al. The single-cell pathology landscape of breast cancer. Nature 2020, 578, 615–620. [Google Scholar] [CrossRef]
- Martinez-Morilla, S.; Villarroel-Espindola, F.; Wong, P.F.; Toki, M.I.; Aung, T.N.; Pelekanou, V.; Bourke-Martin, B.; Schalper, K.A.; Kluger, H.M.; Rimm, D.L. Biomarker Discovery in Patients with Immunotherapy-Treated Melanoma with Imaging Mass Cytometry. Clin. Cancer Res. 2021, 27, 1987–1996. [Google Scholar] [CrossRef]
- Hoch, T.; Schulz, D.; Eling, N.; Gomez, J.M.; Levesque, M.P.; Bodenmiller, B. Multiplexed imaging mass cytometry of the chemokine milieus in melanoma characterizes features of the response to immunotherapy. Sci. Immunol. 2022, 7, eabk1692. [Google Scholar] [CrossRef] [PubMed]
- Alnajar, H.; Ravichandran, H.; Figueiredo Rendeiro, A.; Ohara, K.; Al Zoughbi, W.; Manohar, J.; Greco, N.; Sigouros, M.; Fox, J.; Muth, E.; et al. Tumor-immune microenvironment revealed by Imaging Mass Cytometry in a metastatic sarcomatoid urothelial carcinoma with a prolonged response to pembrolizumab. Cold Spring Harb. Mol. Case Stud. 2022, 8, a006151. [Google Scholar] [CrossRef]
- Elaldi, R.; Hemon, P.; Petti, L.; Cosson, E.; Desrues, B.; Sudaka, A.; Poissonnet, G.; Van Obberghen-Schilling, E.; Pers, J.O.; Braud, V.M.; et al. High Dimensional Imaging Mass Cytometry Panel to Visualize the Tumor Immune Microenvironment Contexture. Front. Immunol. 2021, 12, 666233. [Google Scholar] [CrossRef]
- Keren, L.; Bosse, M.; Thompson, S.; Risom, T.; Vijayaragavan, K.; McCaffrey, E.; Marquez, D.; Angoshtari, R.; Greenwald, N.F.; Fienberg, H.; et al. MIBI-TOF: A multiplexed imaging platform relates cellular phenotypes and tissue structure. Sci. Adv. 2019, 5, eaax5851. [Google Scholar] [CrossRef]
- Keren, L.; Bosse, M.; Marquez, D.; Angoshtari, R.; Jain, S.; Varma, S.; Yang, S.R.; Kurian, A.; Van Valen, D.; West, R.; et al. A Structured Tumor-Immune Microenvironment in Triple Negative Breast Cancer Revealed by Multiplexed Ion Beam Imaging. Cell 2018, 174, 1373–1387.e1319. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, F.J.; Mrdjen, D.; McCaffrey, E.; Glass, D.R.; Greenwald, N.F.; Bharadwaj, A.; Khair, Z.; Verberk, S.G.S.; Baranski, A.; Baskar, R.; et al. Single-cell metabolic profiling of human cytotoxic T cells. Nat. Biotechnol. 2021, 39, 186–197. [Google Scholar] [CrossRef]
- Rovira-Clave, X.; Jiang, S.; Bai, Y.; Zhu, B.; Barlow, G.; Bhate, S.; Coskun, A.F.; Han, G.; Ho, C.K.; Hitzman, C.; et al. Subcellular localization of biomolecules and drug distribution by high-definition ion beam imaging. Nat. Commun. 2021, 12, 4628. [Google Scholar] [CrossRef] [PubMed]


| Method Name | Vendor Name | Sample | Maximal Labeling | Direct/ Indirect Detection | Antibody Conjugation | Stain Removal Method | Time Consuming (Per Cycle) * | Resolution |
|---|---|---|---|---|---|---|---|---|
| Stain removal technologies | ||||||||
| MELC (Toponome imaging systems) | ToposNomos GmbH | FFPE | 100 | Direct | Fluorescent based | Bleaching | two proteins per hour (one tag/one dye per cycle) | <40 nm |
| SIMPLE | NA | FFPE | 12 | Indirect | Fluorescent based | Alcohol-soluble red peroxidase substrate AEC method | 3 h | 15–20 μm |
| IBEX | NA | Frozen/FFPE | >65 | Direct/indirect | Fluorescent based | LiBH4-based bleaching | Manual ~3.5 h/automated ~1.5 h | 160 nm |
| Fluorophore inactivation technologies | ||||||||
| MxIF | Cell IDx | FFPE | 60 | Indirect | Fluorescent based | Alkaline oxidation chemistry inactivation | 1 h 15 min | 1 μm |
| CycIF | NA | FFPE | 60 | Direct | Fluorescent based | Hydrogen peroxide and light inactivation | ~24 h | 5 μm |
| Chip Cytometry | Zellsafe™ | Cell suspensions /frozen/FFPE | 60 | Direct | Fluorescent based | Chemical bleaching or light photobleaching | ~1 h | 5 μm |
| Multiplexed signal amplification | ||||||||
| Multiplex modified hapten-based | UltraPlex™ | FFPE | 4 | Indirect | Fluorescent based | Antibody stripping | 2 h | NA |
| TSA | Roche and Akoya Biosciences | Cell suspensions /FFPE | 9 | Indirect | Fluorescent based | Antibody stripping | 1 h | 0.25–0.9 μm |
| QDs | NA | FFPE | 5 | Direct/Indirect | Fluorescent based | Chemical bleaching | 6 h | Super resolution |
| DNA barcoding technologies | ||||||||
| DEI | NA | FFPE | 8 | Indirect | DNA-barcoding based | NA | 2–3 h | 20 nm |
| CODEX | Akoya Biosciences | Cell suspensions/frozen/FFPE | 60 | Indirect | DNA-barcoding based | NA | <1 day (Whole slide imaging) | 260 nm |
| Immuno-SABER | NA | Cell suspensions/frozen/FFPE | 10 | Indirect | DNA-barcoding based | NA | 1 h | Super resolution |
| DSP | NanoString | Frozen/FFPE | 96 | Indirect | DNA-barcoding based | NA | 1–2 h | 10 μm |
| InSituPlex® | Ultivue | FFPE | 15 | Indirect | DNA-barcoding based | NA | 5.5 h | NA |
| Mass cytometry | ||||||||
| IMC | Hyperion | Cell suspensions /frozen/FFPE | >40 | Direct | Metal-based | NA | 2 weeks (0.5 mm × 0.5 mm ROI takes ~3.5 h with a slide scanner) | 1 μm |
| MIBI | Ionpath | Cell suspensions /FFPE | 40–100 | Direct | Metal-based | NA | 2 weeks (Whole slide imaging) | 260 nm |
| Method Name | Cancer Type | Biomarkers Studies | Refs. |
|---|---|---|---|
| Stain removal technologies | |||
| MELC (Toponome imaging systems) | Colorectal cancer | CD3, CD4, CD25, CD29, CD44, human lymphocyte antigen (HLA)-DR | [22] |
| SIMPLE | HNSCC PDAC | CD3, CD4, CD8, CD46, CD68, PD-1, Ki67, Eomes-odermin, GrzB, IDO, Tbet | [24] |
| IBEX | NA | NA | NA |
| Fluorophore inactivation technologies | |||
| MxIF | Colon cancer | ER, androgen receptor (AR), p53, Her2, PLAC8 | [30,31] |
| CycIF | Breast cancer | Her2, ER, PR | [35] |
| ChipCytometry | Breast cancer | PD-L1, PD-L2 | [38] |
| Multiplexed signal amplification | |||
| Multiplex modified hapten-based | NSCLC | CD8, PD-L1, and panCK | [40] |
| TSA | Metastatic gastric cancer (GC) | PD-L1 | [44] |
| QDs | Gastric cancer/ breast cancer | type IV collagen, macrophages, matrix metalloproteinase 9 (MMP9), CD105 | [50] |
| Breast cancer | type IV collagen, Her2 | [51] | |
| DNA barcoding technologies | |||
| DEI | N | NA | NA |
| CODEX | Cutaneous T cell lymphoma (CTCL) | ICOS, IDO-1, LAG-3, PD-1, PD-L1, OX40, Tim-3, VISTA | [57] |
| Immuno-SABER | NA | NA | NA |
| DSP | NSCLC | CD3, CD4, CD8, CD20, PD-L1 | [64] |
| InSituPlex® | Breast cancer | Kaiso | [66] |
| Mass cytometry | |||
| IMC | Melanoma | MHC-I, HMB45, S100, IFNGR1, IRF1, CD45RO, PD-L1, CD163, B7-H3, LAG3, TIM3, FOXP3, CD4, B7-H4, CD68, PD-1, CD20, CD8, PD-1H, Ki67, B2M, CD3a, CSF1R, PD-L2, Granzyme B, MHC-II, CXCL9, CXCL10, CXCL13 | [72,73] |
| SUC | PD-1, PD-L1 | [74] | |
| MIBI | Breast cancer | double-stranded DNA (dsDNA), ERα, PR, E-cadherin, Ki-67, vimentin, actin, keratin, HER2, PD-1, PD-L1 | [14,77] |
| Method Name | Advantage | Disadvantage |
|---|---|---|
| Stain removal technologies | ||
| MELC (Toponome imaging systems) | Detects hundreds of proteins and high resolution | The multiprobe image is limited to a single microscopic medium-to-high power field and high cost |
| SIMPLE | Easy to perform by whole-slide scanner and can be labeled primary antibodies from same species | Up to 12 biomarkers |
| IBEX | Allows over 65 biomarkers to detect and compatible with over 250 commercial antibodies | Not commercialized and few studies |
| Fluorophore inactivation technologies | ||
| MxIF | Up to 60 biomarkers | Time-consuming and relatively expensive |
| CycIF | Use commonly reagents and instruments | Before the next staining, coverslip should be removed and time-consuming |
| ChipCytometry | Detects unlimited number of biomarkers, long-storage samples, removes autofluorescence and instrument automaticity | Damage the tissue adherence and photobleachable dyes may generate weak signals during imaging processing |
| Multiplexed signal amplification | ||
| Multiplex modified hapten-based | Two-hour fast staining and cocktail antibodies are used in a single slide | Maximal four biomarkers can be labeled per slide and not applied widely |
| TSA | Avoids antibody cross-reactivity and may realize an automated protocol | Nine biomarkers can be labeled per slide |
| QDs | Removes autofluorescence and has much stronger signals | Big size relatively, has toxicity and limited nanocrystals |
| DNA barcoding technologies | ||
| DEI | Short-time staining and applies for most microscopy platforms | Lack of signal amplification, few studies |
| CODEX | Allows 60 biomarkers labeled and can be imaged by conventional fluorescence microscopy, also keeps the morphology of normal and diseased tissues | Longer scanning and lack of signal amplification |
| Immuno-SABER | High multiplexing, sensitivity and 5–180-fold signal amplification | Up to 10-plex and few publications |
| DSP | No-damage staining protocol and performs high multiplexing image on FFPE samples | Chooses ROI manually and is not able to reconstruct images |
| InSituPlex® | Good signal in low-expression antigen, 5.5 h workflow and relatively cheap | Few studies |
| Mass cytometry | ||
| IMC | Removes autofluorescence, reveals the quantity of proteins in subcellular level | Lack of signal amplification, the rate of image acquisition is slow and relatively low resolution in subcellular level |
| MIBI | A large number of metal-antibodies can be labeled spectral overlap and high resolution | Time-consuming, instrument and metal-antibodies are expensive |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sheng, W.; Zhang, C.; Mohiuddin, T.M.; Al-Rawe, M.; Zeppernick, F.; Falcone, F.H.; Meinhold-Heerlein, I.; Hussain, A.F. Multiplex Immunofluorescence: A Powerful Tool in Cancer Immunotherapy. Int. J. Mol. Sci. 2023, 24, 3086. https://doi.org/10.3390/ijms24043086
Sheng W, Zhang C, Mohiuddin TM, Al-Rawe M, Zeppernick F, Falcone FH, Meinhold-Heerlein I, Hussain AF. Multiplex Immunofluorescence: A Powerful Tool in Cancer Immunotherapy. International Journal of Molecular Sciences. 2023; 24(4):3086. https://doi.org/10.3390/ijms24043086
Chicago/Turabian StyleSheng, Wenjie, Chaoyu Zhang, T. M. Mohiuddin, Marwah Al-Rawe, Felix Zeppernick, Franco H. Falcone, Ivo Meinhold-Heerlein, and Ahmad Fawzi Hussain. 2023. "Multiplex Immunofluorescence: A Powerful Tool in Cancer Immunotherapy" International Journal of Molecular Sciences 24, no. 4: 3086. https://doi.org/10.3390/ijms24043086
APA StyleSheng, W., Zhang, C., Mohiuddin, T. M., Al-Rawe, M., Zeppernick, F., Falcone, F. H., Meinhold-Heerlein, I., & Hussain, A. F. (2023). Multiplex Immunofluorescence: A Powerful Tool in Cancer Immunotherapy. International Journal of Molecular Sciences, 24(4), 3086. https://doi.org/10.3390/ijms24043086









