Abstract
This review paper provides an extensive analysis of the rapidly evolving convergence of deep learning and long non-coding RNAs (lncRNAs). Considering the recent advancements in deep learning and the increasing recognition of lncRNAs as crucial components in various biological processes, this review aims to offer a comprehensive examination of these intertwined research areas. The remarkable progress in deep learning necessitates thoroughly exploring its latest applications in the study of lncRNAs. Therefore, this review provides insights into the growing significance of incorporating deep learning methodologies to unravel the intricate roles of lncRNAs. By scrutinizing the most recent research spanning from 2021 to 2023, this paper provides a comprehensive understanding of how deep learning techniques are employed in investigating lncRNAs, thereby contributing valuable insights to this rapidly evolving field. The review is aimed at researchers and practitioners looking to integrate deep learning advancements into their lncRNA studies.
1. Introduction
In the evolving landscape of machine learning, deep learning has revolutionized our understanding and application of technology, paving the way for breakthroughs and novel explorations in a plethora of fields [1,2,3,4]. It has spurred the advent of innovative methods, such as image generation [5,6,7] and natural language processing techniques [8,9,10,11], each contributing to expanding the horizons of deep learning applications. This emerging area of study has also found its relevance in molecular biology, particularly in understanding long non-coding RNAs (lncRNAs) [12,13,14].
Our focus in this review is on lncRNAs, which are a category of RNAs that lack protein-coding potential and consist of nucleotide sequences longer than 200 nucleotides, which are transcribed and processed mostly from intergenic regions, introns with or without some exons, or enhancer regions of the genome [12], yet have emerged as significant contributors to numerous biological processes. lncRNAs, with their intricate and diverse functions, have gained increasing attention from the scientific community as their aberrations have been implicated in various diseases. The fast-paced research dedicated to lncRNAs, in parallel with the rapid advancements in deep learning, necessitates a comprehensive overview of the intersection of these two vital areas of study.
As signal molecules, lncRNAs play a crucial role in the transcription of downstream genes, often exhibiting a high degree of context specificity [15,16,17]. Recent studies have shown lncRNAs to be highly organized in their transcription processes, responding and adapting to different environmental stimuli to influence particular signaling pathways [18,19,20]. This detailed orchestration of transcription by lncRNAs, often in association with specific proteins, such as transcription factors, underscores the integral role they play in transcriptional regulation.
In their capacity as decoy molecules, lncRNAs perform a variety of functions, including interference in various molecular pathways. These RNAs interact directly with specific protein molecules after their transcription, leading to a disruption in the normal functioning of these proteins [21,22]. This interaction with transcription regulators inhibits the transcription factors’ functionality, thus suppressing downstream gene transcription. Furthermore, lncRNAs can impede protein functionality, affecting their ability to regulate mRNA expression. Furthermore, lncRNAs have been found to play vital roles in tumor progression, participating in the regulation of gene expression at the epigenetic, transcriptional, and post-transcriptional levels [23,24]. Some lncRNAs influence gene expression by altering the chromatin structure, histone modification status, and DNA methylation status [25,26].
In the current technological landscape, deep learning has rapidly evolved to influence a range of scientific fields significantly. This remarkable evolution, combined with the emerging importance of lncRNAs, underscores the necessity to explore the confluence of these two spheres critically. Recognizing this need, we aim to deliver an exhaustive analysis of how deep learning is employed in the study of lncRNAs, thereby providing fresh insights into this fast-growing intersection.
This review is a testament to our commitment to keeping abreast with the most recent advancements in the field, specifically those from 2021 to 2023. As deep learning continues to evolve at a rapid pace, it is imperative to stay up-to-date with the latest research developments. The significance of such a review cannot be overstated, as it will be invaluable to researchers and practitioners who strive to integrate the advancements of deep learning into their lncRNA studies.
2. Literature Analysis
2.1. Paper Selection Process
The primary aim of the paper selection process was to ensure the inclusion of high-quality, relevant research within the domain. To achieve this, we adopted an algorithmic approach centered around the academic search engine, Web of Science (WOS). Carefully selected search keywords were used, focusing on central themes, such as deep learning, lncRNA, and neural networks, to identify pertinent articles for review.
While recognizing the existence of preprints and conference papers, we chose to concentrate solely on peer-reviewed journal articles. This selection criterion enhances the reliability and validity of the review by ensuring the inclusion of studies that have undergone a stringent review process. This decision was driven by two principal factors. First, the peer-review process serves as a crucial mechanism for maintaining the quality and reliability of scientific literature by subjecting research to thorough scrutiny by domain experts. Second, peer-reviewed journals are traditionally esteemed as trustworthy and credible sources for publishing scientifically sound and influential research.
To maintain the novelty and uniqueness of the review, certain categories of articles, such as review articles and perspective pieces, were intentionally excluded. This approach aimed to emphasize the incorporation of primary research-focused studies, in alignment with the purpose of this review.
The review’s temporal scope was restricted to articles published within the last three years, from 2021 to 2023. This period was selected to assure the relevance and contemporaneity of the review, and to provide a comprehensive understanding of the latest developments and trends in deep learning for lncRNA research. It is worth noting that data collection for 2023 was carried out until May. This was done to ensure that the review’s currency aligns with the most recent advancements in the field. The selected studies are summarized in Table 1.

Table 1.
Summary of investigations employing deep learning approaches in lncRNA studies between 2021 and 2023.
An exploration of previous deep learning methodologies utilized in the study of lncRNA prior to 2021, as compared to the more recent developments from 2021 to 2023 will certainly provide a comprehensive understanding. A wealth of knowledge on the subject can be found in several representative articles. For example, Baek et al. [102] developed a deep learning-based approach, lncRNAnet, to identify lncRNAs, incorporating recurrent neural networks for RNA sequence modeling and convolutional neural networks for detecting stop codons to obtain an open reading frame indicator. Another study by Fan et al. [103] constructed a powerful predictor, lncRNA–MFDL, to identify lncRNAs by fusing multiple features of the open reading frame, k-mer, the secondary structure, and the most likely coding domain sequence, using deep learning classification algorithms. In 2019, Liu et al. [104] developed a deep learning model, which included a bidirectional long short-term memory model layer and a convolutional layer with three additional hidden layers for distinguishing lncRNAs from mRNAs. These methodologies provided significant advancements in the field. Moreover, the detailed review by Shaath et al. [105] further discusses the interplay between lncRNAs and RBPs and their involvement in epigenetic regulation via histone modifications, highlighting the potential for RNA-based therapeutics in cancer treatment. It is anticipated that the recent surge in research during 2021–2023 has built upon these foundational methodologies and will continue to push the frontier in our understanding and utilization of lncRNA in deep learning.
During the data collection process, we compiled information on the citation count and publication log for each selected article. These details played a significant role in evaluating the scope, impact, and acceptance of the research within the scientific community.
To provide a structured overview of the deep learning methodologies utilized in lncRNA research, the selected papers were categorized, based on the specific objectives of the studies. This categorization contributes to a comprehensive understanding of the deep learning landscape for lncRNA research, by enhancing the understanding of the diverse methodologies employed in the field. The objectives of the studies are summarized in Table 1.
2.2. Brief Analysis of Deep Learning Approaches in lncRNA Research
The application of deep learning methodologies to the study of lncRNAs has surged in recent years, as is reflected by the varied research areas illustrated in Table 1. This analysis provides a comprehensive view of the considerable strides made in this emerging field, emphasizing the necessity and relevance of this review.
From the assortment of categories that have emerged, one of the most extensively researched topics is the lncRNA–disease association. Multiple studies have endeavored to harness the predictive capabilities of various deep learning architectures to ascertain the relationship between lncRNAs and diseases [29,34,48]. The prodigious quantity of investigations in this domain highlights the compelling implications these associations could potentially have on clinical diagnostics and therapeutics.
Another critical domain in lncRNA research pertains to lncRNA–protein interactions. In an attempt to uncover the myriad roles that lncRNAs play in molecular biology, numerous researchers have applied deep learning models to predict potential interactions between lncRNAs and proteins [62,70]. By illuminating these interactions, we can elucidate the complex regulatory networks that underpin biological systems.
Similarly, the prediction of lncRNA–miRNA interactions has also gained substantial interest. These investigations capitalize on the ability of deep learning models to discern patterns and predict interactions [72,75], providing vital insights into the modulation of gene expression by lncRNAs.
A further significant facet of lncRNA research that has reaped the benefits of deep learning involves the identification and prediction of characteristics inherent to lncRNAs [88,100]. By leveraging the ability of deep learning models to learn complex representations from data, researchers have made inroads in understanding the fundamental properties that define lncRNAs.
An interesting development has been the application of deep learning in predicting lncRNA subcellular localization. This growing area of study is vital in comprehending the functional mechanisms of lncRNAs since the location of a lncRNA within a cell often indicates its potential role and function.
The investigation of the role lncRNAs play in immune responses using deep learning methods is less explored, yet equally important. While such studies are currently limited in number, they reflect an area that is ripe for further exploration, given the significant implications for immunology and disease pathology. Moreover, identifying lncRNA–protein-coding gene (PCG) associations is another emerging area in lncRNA research where deep learning has been applied.
The diverse application of deep learning in lncRNA research signals an exciting convergence of computational and biological sciences. The rapid development and wide-reaching applicability of deep learning models have significantly enriched our understanding of lncRNAs, highlighting the need to assess and review the state-of-the-art in this field continually. By providing a comprehensive overview of recent studies and emerging trends, this review aspires to be an invaluable resource for researchers exploring the promising intersection of deep learning and lncRNA research.
2.3. Distributive Analysis of Publications across Various Journals
In the quest to examine the widespread diffusion of selected articles in a multitude of academic journals, a conspicuous pattern was brought to the fore. The compendium of selected literature exhibits an extensive array of publication outlets, which emphasizes the interdisciplinarity of the topic at hand, i.e., deep learning for lncRNA.
The journal wielding the most considerable influence, as measured by the sheer number of published papers, appears to be Briefings in Bioinformatics. This esteemed periodical has been the host to no fewer than 21 of the 75 papers evaluated, thus constituting an impressive 28% of the corpus.
BMC Bioinformatics follows on the leaderboard, which accounts for approximately 17% of the selected papers, with a tally of 13. Furthermore, the IEEE-ACM Transactions on Computational Biology and Bioinformatics journal has been instrumental in disseminating seven papers within our studied selection.
This distribution implicitly suggests that specific journals maintain a predilection for publishing research in the domain of deep learning applied to lncRNA. Figure 1 provides a comprehensive analysis, illustrating the allocation of papers among diverse journals.
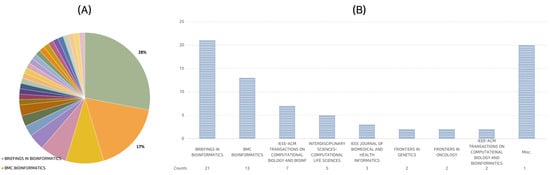
Figure 1.
Journal distribution of 75 research papers. (A) Proportional representation of papers in various journals, where the distinct journals represented by different colors; (B) journals with multiple publications. The Misc. category represents 20 journals with single publications.
3. Deep Learning Approaches in the Prediction of lncRNA–Disease Associations
LncRNAs have been increasingly recognized as critical components in numerous biological functions and disease processes [106]. The application of deep learning to elucidate lncRNA–disease associations provides an avenue for understanding the complex interactions of lncRNAs with other biological components, including genes and proteins [13]. The richness and complexity of these interactions necessitate computational models that can handle high-dimensional, context-dependent data, a challenge ideally suited to deep learning methodologies.
Notably, trends in lncRNA–disease association research are veering toward the integration of diverse biological data into predictive models. The incorporation of lncRNA sequence data, gene expression data, protein interaction data, and disease phenotype data into deep learning models is enriching the understanding of lncRNA–disease associations [107].
Additionally, the introduction of attention mechanisms has enhanced these models’ ability to focus on the most informative parts of input data. This trend is particularly valuable given the context-specific nature of certain lncRNAs or disease phenotypes. Furthermore, the field is seeing an increased adoption of unsupervised and semi-supervised learning techniques to leverage the abundant amount of unlabeled data in the learning process. The summary of recent studies is provided in Table 2.

Table 2.
Summary of recent studies regarding the prediction of lncRNA–disease association. It is important to acknowledge that each study utilized diverse datasets, cross-validation methods, and simulation settings to assess accuracy, thus rendering direct comparisons potentially inconclusive. The best accuracy was selected if the model was assessed with various datasets.
3.1. Recent Advances from 2021 to 2023
The field of lncRNA–disease associations has seen an influx of diverse prediction models, with the recent trend leaning heavily toward deep learning approaches. The deep learning paradigm, through its effective non-linear data processing capabilities, enables the extraction of complex features and dependencies within the lncRNA–disease association data. The typical models employed in this field incorporate varied architectures, such as deep belief networks (DBNs), convolutional neural networks (CNNs), and attention mechanisms. These methodologies provide improved performance metrics, including the area under the receiver operating characteristic curve (AUC), the area under the precision–recall curve (AUPR), and accuracy values. Notably, graph-based methodologies and multiomics data incorporation are often employed due to their ability to capture complex lncRNA–disease associations and provide comprehensive insights into the role of lncRNA in diseases. However, the choice of the prediction model is largely contingent on the specific requirements of each research, thereby carrying distinct merits and challenges.
In several studies, autoencoders and CNNs were used to improve prediction performance and explore potential disease associations [27,28,38,42,43,44,45,53]. Despite achieving high accuracy rates, a shared disadvantage among these studies is the complexity introduced by the use of autoencoders and CNN. These models require careful parameter tuning and have difficulties integrating information from diverse data types or topologies. Some studies also noted issues with noise in the data, such as [42], which could not effectively remove noisy and irrelevant information.
The second group of studies implemented GNNs to predict lncRNA–disease associations [35,36,38,39,40,41,46,47,50]. These studies demonstrated superior performance in making predictions, and were able to handle multi-view data and efficiently fuse node features, topological structures, and semantic information. However, they have a sensitivity to different datasets and can be complex due to the implementation of multiple attention mechanisms and the integration of multi-layer graph convolutional networks.
The final group of studies utilized matrix factorization or other machine learning techniques, such as support vector machines (SVM) and extreme learning machines (ELM) [33,34,37,49,51,52,54]. They were able to handle complex relationships and diverse features and achieved high accuracy rates. However, they were not without challenges. Some methods required optimal parameter selection, and there were concerns about the quality of the negative samples or graph-based information used in these studies.
In recent studies, while all of these studies have shown promising results in predicting lncRNA–disease associations, each approach has its advantages and potential areas for improvement. Future studies should consider these points when developing prediction models for lncRNA–disease associations.
3.2. Emerging Research Trends in Recent Studies
A major shift in recent studies revolves around the application of diverse network architectures, such as DBN, CNN, and attention mechanisms. The primary objective of these trends is to exploit the non-linear data processing potential of deep learning algorithms for complex feature extraction and elucidating the intricate dependencies in lncRNA–disease association data. From a quantitative perspective, models implementing these techniques have persistently demonstrated superior performance metrics.
Another trend gaining momentum is the utilization of graph-based methodologies, with notable contributions by Guo et al. [43], Zeng et al. [33], and Zhao et al. [39]. The inherent strength of these methods lies in their capability to effectively leverage the graph structure of lncRNA–disease associations, thereby capturing the complex interrelationships between lncRNAs and diseases with a higher degree of accuracy. This improved representation of data facilitates more accurate and robust prediction models.
Lastly, the incorporation of multiomics data in lncRNA–disease association prediction, as exemplified by Yuan et al. [45], stands out as a promising trajectory. Quantitatively, this comprehensive approach enables an increased understanding of lncRNA’s role in diseases, substantially enhancing the quality of prediction. While these trends offer numerous advantages, it is important to consider the possible trade-offs. The complexity of these methods can lead to increased computational costs and complexities, and the need for large, high-quality datasets. The benefits and limitations of these trends need to be carefully balanced to maximize their potential in real-world applications.
4. Deep Learning Approaches in the Prediction of lncRNA–Protein Interactions
The prediction of lncRNA–protein interactions, a cornerstone in understanding lncRNA functionality, has seen a surge in the use of deep learning models [108]. Predominantly, these models integrate biological features to discern complex patterns. Ensemble learning and hybrid frameworks are often the preferred choices due to their robustness and resilience to overfitting. Moreover, novel methods for quantifying lncRNA gene essentiality are gaining prominence, allowing researchers to further investigate the implications of these interactions in gene functionality. Lastly, the incorporation of cutting-edge techniques, such as serial fusion, capsule networks, and graph autoencoders, signifies the continued evolution of prediction models in lncRNA–protein interaction prediction.
LncRNA–protein interaction (LPI) predictions often start with sequence-based prediction models, which transform the primary sequences of lncRNAs and proteins into feature vectors. Deep learning has shown great potential in predicting lncRNA–protein interactions, with models capable of automatically extracting meaningful features from raw sequence data, including the amino acids of proteins. To manage proteins of different lengths in these models, a process known as zero-padding is utilized, where zeros are added to each sequence up to a common length [109]. This technique is crucial when using raw amino acid sequences as input, as these models require input with the same shape. Furthermore, to tackle the complexity of the three-dimensional structures of proteins, various methods have been proposed, including those that integrate sequence and structure features of the lncRNA and protein, as well as those that apply machine learning algorithms to extract features from sequences [110]. Despite the complexity of plant genome structures, these techniques could be instrumental in predicting lncRNA–protein interactions in plants. A summary of recent studies can be found in Table 3.

Table 3.
Summary of recent studies regarding the prediction of lncRNA–protein interaction. It is important to acknowledge that each study utilized diverse datasets, cross-validation methods, and simulation settings to assess accuracy, thus rendering direct comparisons potentially inconclusive. The best accuracy was selected if the model was assessed with various datasets.
4.1. Recent Advances from 2021 to 2023
Recent studies can approximately be categorized into three groups: The first cluster comprises methods that apply GNNs, demonstrated in [58] and [63]. They harnessed the power of GNN to predict lncRNA–protein interactions, achieving high AUC and AUPR scores. For instance, BiHo–GNN, a bipartite graph-embedding method based on GNN, reported an impressive AUC of 0.950 and AUPR of 0.899 [58]. iEssLnc, another graph neural network, leveraged meta-path-guided random walks on the lncRNA–protein interaction network to attain an AUC of 0.912 and AUPR of 0.921 [63]. Despite the clear advantages of these methods in terms of accuracy and recall, certain disadvantages exist. For instance, iEssLnc is specialized for essential lncRNA genes and is not generalized for all lncRNA–protein interactions.
In the second group, we see models that have integrated multiple features of lncRNAs and proteins to predict interactions, such as capsule–LPI [59], EnANNDeep [62], LGFC–CNN [64], and LPI–CSFFR [65]. These models demonstrated superior performance by integrating multimodal features or combining raw sequence composition, hand-designed, and structure features. For instance, the capsule–LPI’s multimodal features and multichannel capsule network framework achieved an AUC of 0.951 and AUPR of 0.932, while LGFC–CNN delivered an exceptional AUC of 0.976 and AUPR of 0.970 [59,64]. However, the main limitation of these methods lies in their increased complexity, especially when fusing diverse features. Additionally, some methods, such as capsule–LPI, lack detailed evaluations for each feature [59].
The third group is characterized by models that use deep learning frameworks with different architectures or hybrid approaches. These include DeepLPI [60], DFRPI [61], LPI–deepGBDT [66], LPI–DLDN [67], LPI–HyADBS [68], and RLF–LPI [71]. These models demonstrated high performance due to their unique approaches. For instance, DeepLPI incorporates interactions between lncRNAs and protein isoforms with a hybrid framework of deep neural networks, while LPI–DLDN utilizes a deep learning framework with a dual-net neural architecture. However, these advanced models often require complex integration or feature dimension reduction.
4.2. Emerging Research Trends in Recent Studies
The analysis of the recent studies on lncRNA–protein interactions, as summarized in Table 3, reveals several notable trends. Firstly, the integration of deep learning models and various biological features stands out as a common and effective strategy in the field. The studies of Zhou et al. [70], Peng et al. [62,67], and Huang et al. [64] are characteristic of this approach. Their contributions attest to the powerful role deep learning algorithms play in discerning complex patterns, especially when combined with biological features that enhance the models’ understanding of lncRNA–protein interactions.
Secondly, the recent trend of employing ensemble learning and hybrid frameworks is noticeable in studies, such as in the works by Zhou et al. [68] and Song et al. [71]. These studies capitalize on the strength of diverse learning models, making the prediction of lncRNA–protein interactions more robust and less prone to overfitting.
Moreover, the exploration of novel methods for quantifying lncRNA gene essentiality, such as in the study by Zhang et al. [63], further expands the scope of lncRNA research. This signifies a shift toward more comprehensive studies that not only predict interactions but also investigate the implications of these interactions in gene functionality.
Lastly, the efforts of Huang et al. [65], Li et al. [59], and Zhao et al. [69] epitomize the usage of novel techniques, such as serial fusion, capsule networks, and graph autoencoders. These cutting-edge methods contribute to the evolution of prediction models, further advancing the state-of-the-art in lncRNA–protein interaction prediction.
5. Deep Learning Approaches in the Prediction of lncRNA–miRNA Interactions
Recent studies on lncRNA–miRNA interactions are primarily centered around the development of deep learning frameworks due to their superior accuracy and cross-species applicability. These interactions, central to post-transcriptional gene regulation, are effectively predicted using sophisticated models such as BoT–Net [72]. The integration of attention mechanisms and neural networks into prediction models is becoming increasingly common, contributing to their improved performance. Hybrid feature mining networks are gaining traction due to their ability to extract useful feature information. Moreover, certain studies are exploring the potential of lncRNA–miRNA interactions in predicting miRNA–disease associations, thereby extending the scope of lncRNA–miRNA research into biomedical applications. However, the model choice largely depends on the specificity and sensitivity requirements of the study, each presenting unique advantages and challenges. A summary of recent studies can be found in Table 4.

Table 4.
Summary of recent studies regarding the prediction of the lncRNA–miRNA interaction. It is important to acknowledge that each study utilized diverse datasets, cross-validation methods, and simulation settings to assess accuracy, thus rendering direct comparisons potentially inconclusive. The best accuracy was selected if the model was assessed with various datasets.
5.1. Recent Advances from 2021 to 2023
In the domain of predicting lncRNA–miRNA interaction, numerous methodologies have been utilized, each demonstrating varied performance metrics and innovative methodological elements.
Firstly, deep learning models exploiting recurrent neural network (RNN) structures coupled with other strategies have shown substantial performance in predicting lncRNA–miRNA interactions. The authors of [72] utilized a hybrid model consisting of a bidirectional transformer (BoT–Net) and LSTM with DropConnect, in addition to feature pooling. Despite not specifically indicating any disadvantages, the study successfully optimized the lncRNA sequence length, thus improving specificity. Moreover, an optimized ensemble deep learning model, which employs independent RNNs (IndRNNs) and CNNs, was developed by [77]. While the authors have not pointed out any disadvantages, their model’s advantages lie in the improved accuracy achieved through optimal hyperparameter tuning, suitable for large-scale data.
In the second group of studies, graph-based models were significantly utilized, exploiting the potential of deep learning and attention mechanisms. A representative study [73] implemented DWLMI, using the DeepWalk algorithm on an lncRNA–miRNA–disease–protein–drug graph, achieving high accuracy; however, it did not discuss the evaluation of each feature’s influence. Similarly, GCNCRF [74] applied a graph convolutional network (GCN) coupled with a conditional random field (CRF) and an attention mechanism. Despite not mentioning specific disadvantages, the method managed to integrate an lncRNA-miRNA similarity network to achieve high AUC scores. The study by [76] applied a graph neural network-based RNA representation technique, termed ncRNAInter, and demonstrated robust performance and broad applicability across different species.
The third group of studies incorporated hybrid feature mining networks and multi-level information enhancement models to forecast lncRNA–miRNA interaction. PmliHFM [78] uses a hybrid feature mining network tailored for predicting plant miRNA–lncRNA interactions. Despite not specifying any disadvantages, this model uniquely integrated different encodings for miRNA and lncRNA, along with ensemble modules. PmliPEMG [79] employs an ensemble deep learning model, leveraging multi-level information enhancement and a greedy fuzzy decision, thereby incorporating complex fusion features and multi-scale convolutional LSTM networks.
Finally, some studies have developed models that utilize word vector representation and deep feature mining mechanisms. Notably, preMLI [80] is a deep learning model based on rna2vec pre-training and a deep feature mining mechanism. The approach uses rna2vec for RNA word vector representation and displays exceptional cross-species prediction capabilities.
5.2. Emerging Research Trends in Recent Studies
As we delve into the intricacies of lncRNA–miRNA associations, one can observe a clear trend toward leveraging advanced machine learning models for predicting these interactions, as evidenced by the studies summarized in Table 4. A noteworthy focus has been on developing deep learning frameworks that offer both improved accuracy and applicability across various species [75,76,77,80].
BoT–Net, a network approach that utilizes long short-term memory networks, provides a good illustration of the potential of these methods [72]. Furthermore, a substantial number of recent studies have shown a strong preference for incorporating attention mechanisms and neural networks into their models, leading to higher performance metrics [74,75].
Moreover, there is an emerging trend of integrating hybrid feature mining networks, which are known to effectively extract useful feature information, thus improving predictive accuracy for lncRNA–miRNA interactions [78]. The commitment to prediction accuracy is further underpinned by the DeepWalk-based method proposed by Yang et al., which offers a high average prediction accuracy [73]. Beyond the application of machine learning in prediction models, some studies have explored the promise of using these interactions to forecast potential miRNA–disease associations, hence pushing the boundaries of lncRNA–miRNA research into biomedical applications.
6. Deep Learning Approaches in the Classification and Prediction of lncRNA Characteristics
In the domain of lncRNA characteristics, deep learning has emerged as an effective tool for predicting lncRNA functions, identifying novel lncRNAs, and discovering lncRNA–disease associations. The recent trend is largely geared toward the utilization of machine learning and deep learning models, such as CNN and LSTM, to analyze lncRNA expression profiles, predict lncRNA interactions, and classify different RNA types. Concurrently, the design and application of ensemble methods are growing popular due to their superior predictive performance. Investigations into lncRNA stability and the factors influencing it also provide a more comprehensive understanding of lncRNA biology. However, each approach carries its own strengths and weaknesses, and the choice often depends on the specific objectives of the study.
lncRNAs, in contrast to their protein-coding counterparts, exhibit less conservation, thereby posing considerable challenges for computational models. However, certain unique properties of lncRNAs provide advantageous elements for deep learning. Large lncRNA datasets offer a wealth of data suitable for the training of deep learning algorithms. Additionally, the multi-level regulatory roles of lncRNAs provide multi-modal data (sequence, structure, interactions, expression levels), enabling a comprehensive analysis through multi-modal learning approaches. Importantly, lncRNAs can originate from various genomic regions, including intergenic regions, intronic regions, and antisense transcripts, with each category potentially bearing distinct functional implications. In terms of the prediction of lncRNA characteristics and their source genomic regions, several recent methods have emerged. For instance, multiple studies proposed CNN structures to handle these challenges [82,83,89]. Additionally, LSTM structures were commonly employed for the prediction of lncRNA characteristics [88].
6.1. Recent Advances from 2021 to 2023
Deep learning has paved the way for significant advancements in understanding lncRNA characteristics, primarily focusing on the prediction of lncRNA functions and lncRNA identification. Numerous studies have developed models to decipher the hidden complexities of lncRNA biology.
In the domain of predicting lncRNA functions, Zhang et al. [87] utilized an ensemble deep learning model, lncIBTP, for predicting interactions between lncRNAs and different types of biomolecules. The model demonstrated an impressive degree of effectiveness, presenting the potential of deep learning to provide insights into lncRNA functionality. The identification of lncRNAs is another crucial aspect in which deep learning approaches have been employed. Lin and Wichadakul [89] proposed Xlnc1DCNN, a one-dimensional convolutional neural network-based tool. It distinguishes lncRNAs from protein-coding transcripts and provides a rationalization for its predictions. This tool outperformed several others in accuracy and F1-score, demonstrating the effective application of convolutional neural networks in lncRNA identification. Another notable contribution came from Wang et al. [85], who developed LncDLSM, a deep learning-based framework capable of differentiating lncRNAs from protein-coding transcripts without requiring prior biological knowledge. This model excelled in lncRNA identification and exhibited a promising potential for transfer learning.
A unique approach was taken by Zhang et al. [81], who designed a class similarity network for classifying coding and long non-coding RNA. The network explores relationships between input samples and those from the same or different classes, thus obtaining high-level features. The method consistently achieved high accuracy, precision, and F1 scores, signifying its proficiency in lncRNA classification. Ritu et al. [83] also proposed a novel bimodal CNN-based deep learning system, DeepPlnc, which integrated both sequence and structural properties for the identification of plant lncRNAs. DeepPlnc outperformed other tools even when dealing with ambiguous boundaries and incomplete sequences, solidifying its superior applicability in genome and transcriptome annotation tasks.
Promising developments were also made in understanding the stability of lncRNAs. Shi et al. [84] performed a genome-wide RNA-seq study on human lung adenocarcinoma cells and used deep learning-based regression to identify a non-linear relationship between the half-lives of lncRNAs and associated factors. This research illuminated a comprehensive understanding of lncRNA stability, showing the powerful potential of deep learning in elucidating ncRNA characteristics.
Deep learning’s contributions extend to the identification of dual-functional lncRNAs as well; Liu et al. [86] developed LncReader, a deep learning model with a multi-head self-attention mechanism. LncReader outperformed various classical machine learning methods, reiterating the superior performance of deep learning in lncRNA research.
6.2. Emerging Research Trends in Recent Studies
In recent research trends, as encapsulated in Table 5, several innovative approaches and methodologies have emerged in the study of lncRNA–disease associations. Various machine learning and deep learning models are increasingly being employed to analyze lncRNA expression profiles, predict lncRNA interactions, and classify different RNA types. These computational tools are crucial in enabling more accurate, efficient, and scalable analyses of lncRNA data.

Table 5.
Summary of recent studies regarding the classification and prediction of lncRNA characteristics. It is important to acknowledge that each study utilized diverse datasets, cross-validation methods, and simulation settings to assess accuracy, thus rendering direct comparisons potentially inconclusive.
One remarkable trend observed is the utilization of CNN and LSTM in deep learning models for the prediction and classification of lncRNA [82,83,88,89]. These sophisticated tools offer high accuracy rates and demonstrate robust performance across various datasets. Moreover, they provide an advantage over traditional bioinformatics approaches, which may rely heavily on prior biological knowledge.
Another noteworthy development is the focus on the design and application of ensemble methods, which integrate multiple learning algorithms to obtain better predictive performance [87]. These models, such as WGAN–psoNN and lncIBTP, incorporate advanced concepts, such as NAS for optimal parameter tuning, thus alleviating data imbalance issues.
Furthermore, studies that examine the stability of lncRNAs, and the factors influencing their stability, are gaining traction [84]. These investigations provide a more comprehensive understanding of lncRNA biology and may inform future therapeutic strategies for diseases associated with lncRNA dysfunction.
7. Other Deep Learning Research Domains and Utilization of lncRNA-Related Data as Deep Learning Inputs
The exploration of lncRNAs has seen a significant shift toward the use of advanced deep learning models to predict lncRNA subcellular localization and distinguish different lncRNA types. Moreover, studies are increasingly focusing on the role of lncRNAs in immune responses and disease processes, particularly cancer. Novel frameworks, such as deep learning and graph neural networks, are being employed to handle the complex nature of lncRNA sequences and structures. Moreover, deep learning has also found utility in studies investigating the role of lncRNAs in immune responses and disease processes. Researchers are employing sophisticated machine learning techniques to predict lncRNA behavior and correlate it with disease states, offering insights into the complexities of immune responses and disease pathogenesis. A summary of recent studies can be found in Table 6.

Table 6.
Summary of other recent studies and utilizations of lncRNA-related data as deep learning inputs.
7.1. Recent Advances from 2021 to 2023
The exploration and prediction of lncRNAs have been widely adopted through advanced computational methods. Among them, DeepLncLoc is a noteworthy tool that uses a subsequence embedding method to retain the order information of lncRNA sequences and incorporates a text convolutional neural network for feature extraction and prediction, proving its effectiveness for lncRNA subcellular localization prediction [90]. Along similar lines, the GM–lncLoc model combines sequence information with graph structure information, leveraging a graph neural network with meta-learning for prediction [92]. This approach demonstrates high accuracy and a promising solution for handling limited sample sizes often seen in lncRNA subcellular localization studies.
Further, GraphLncLoc and PlncRNA–HDeep have shown promising results in lncRNA prediction. The former employs graph convolutional networks and transforms lncRNA sequences into de Bruijn graphs, unveiling sequence patterns and motifs [93]. PlncRNA–HDeep implements a hybrid deep learning model using LSTM and CNN trained on RNA sequences encoded by p-nucleotide and one-hot encodings, achieving high accuracy on the Zea mays dataset and outperforming traditional machine learning methods [94].
Turning to the functional roles of lncRNAs in immune response pathways, the study by [95] utilizes lncRNAs to develop a prognostic model, predicting the survival rates of breast cancer (BC) patients. The model constructs ceRNA networks correlated with the infiltration of CD8 T cells, providing valuable insights into the role of lncRNA in BC. Similarly, the authors of [96] employed a combination of logistic regression and multilayered neural networks to identify lncRNAs related to Bovine Johne’s Disease, revealing potential lncRNA targets in host immunity against Mycobacterium avium infection.
Deep learning applications utilizing lncRNA input data have significantly contributed to the biomedical field. MVMTMDA, a multi-view multitask learning method, predicts miRNA–disease associations from lncRNA–miRNA interactions with an average AUC of 0.8410 ± 0.018 [97]. Another study employed a multimodal deep learning model by integrating histopathological and molecular data to evaluate microsatellite instability in colorectal cancer [98]. This model achieved a high AUC of 0.952 when combining H&E image with DNA methylation data. Moreover, a model built on graph convolution networks with a multichannel attention mechanism predicted miRNA–disease associations based on lncRNA–miRNA interactions, achieving high average AUC values in different cross-validation setups [99].
One particularly innovative approach, WGAN–psoNN, combines the WGAN and particle swarm optimization neural network for predicting lymph node metastasis events using lncRNA expression profiles [100]. Finally, for the identification of lncRNA–PCG associations, the GAE–LGA model utilizes graph autoencoders to integrate multiomics data. This method shows robust capacity in capturing lncRNA–PCG associations, outperforming other machine learning-based approaches [101].
7.2. Emerging Research Trends in Recent Studies
The prevailing trend in recent studies, as encapsulated in Table 6, distinctly manifests the increasing significance of computational and deep learning models in the domain of lncRNAs research. A distinct transformation in methodological approaches and predictive models can be observed, mirroring the swift expansion of artificial intelligence and deep learning into biological and medical research fields.
Predominantly, it is seen that many recent studies are oriented toward exploring the subcellular localization of lncRNAs, reflecting the importance of this aspect in understanding the functional roles of lncRNAs. In this quest, researchers are investing significant efforts in constructing robust and sophisticated models that can predict lncRNA localization with high accuracy.
Moreover, the utilization of novel frameworks, such as deep learning and graph neural networks, underscores a consistent theme in these investigations. Such models capitalize on the intricate and complex nature of lncRNA sequences and structures, offering an exciting promise of outperforming traditional machine learning techniques.
Additionally, the studies reflect an increasing interest in the role of lncRNAs in relation to diseases. This manifests in developing models for predicting lncRNA associations with various cell types and disease conditions, in particular, cancer. Such investigations are integral for deepening our understanding of the role of lncRNAs in disease pathogenesis and potential therapeutic interventions.
8. Challenges and Future Prospects
LncRNAs have shown substantial promise as novel biomarkers and therapeutic targets in numerous diseases. Notwithstanding, there are still several significant challenges to address and opportunities to seize in the application of deep learning in lncRNA research.
8.1. Challenges
Firstly, the scarcity of large, well-annotated datasets is a significant obstacle [111]. Many existing lncRNA-related databases are either relatively small, lack comprehensive annotation, or focus on a particular category of lncRNAs, which restricts the variety and volume of data available for model training and validation. Given the importance of large datasets in deep learning, this poses a substantial challenge to the development of highly accurate and robust models. Moreover, these databases often contain biases, which can inadvertently be learned by the model, leading to biased predictions [112].
Secondly, the heterogeneous nature of lncRNAs themselves presents a significant challenge. LncRNAs are known for their diversity in terms of their biogenesis, structure, function, and localization, which can complicate the feature extraction process in model development [113]. Current methods may not fully capture the complexity and diversity of lncRNAs, leading to the potential loss of valuable predictive information.
Finally, interpretability is a persistent problem in deep learning. Many deep learning models, particularly those with many hidden layers, are often criticized as “black boxes” due to their complex internal computations. This lack of transparency is particularly problematic in biological and medical applications, where understanding the rationale behind predictions is critical [114]. In lncRNA research, this could mean difficulty in identifying key lncRNA features that drive the model’s predictions, impeding the biological understanding of lncRNA functions.
8.2. Future Prospects and Directions
Despite these challenges, the application of deep learning in lncRNA research holds significant potential for future advancements. Novel strategies for data augmentation, such as transfer learning, can help to mitigate the issue of data scarcity and improve model performance [115]. Furthermore, the development of more advanced feature extraction techniques that can better capture the complex characteristics of lncRNAs will likely enhance model accuracy and robustness.
Regarding interpretability, recent advancements in model interpretability, such as the development of attention mechanisms and interpretation algorithms, provide promising directions for improving the transparency of deep learning models [116]. In lncRNA research, these tools can aid in identifying important lncRNA features and elucidating their biological significance.
Finally, with the rapid advancement of sequencing technologies and the consequent explosion of genomic data, there is enormous potential for the application of deep learning in lncRNA research. The integration of multiomics data, including transcriptomics, proteomics, and metabolomics, can provide a comprehensive understanding of the complex roles of lncRNAs in biological systems. Developing a multiomics-based deep learning model, especially an advanced multi-attention type encompassing lncRNA, miRNA, mRNA, protein, and their pair matrices, for classification and prediction purposes, would ideally provide a comprehensive understanding of the biological systems. However, there exist substantial challenges in developing such a model, including the requirement for extensive computational resources, the need for expertise in bioinformatics to gather and integrate multiomics data, the difficulty in developing an efficient and effective feature selection strategy due to the high-dimensionality and complexity of the data, and the need to accommodate for the inherent high noise and heterogeneity present in biological data. Despite these challenges, such a direction holds immense promise for future research. For instance, in a recent study [117], mRNA and miRNA data were integrated and used in the analysis. Although lncRNA was not involved in this particular instance, it is anticipated that deep learning methods with multiomics data sources, such as the approach employed in this study, will become a prevalent direction in the future.
9. Conclusions
The journey through the landscape of deep learning applications in lncRNA research, as charted in this review, reaches its culmination in this concluding section, where we attempt to knit together the strands of discussions that have threaded their way through the preceding discourse.
An unmistakable trend, revealed through a diligent combination of the literature, is the considerable quantity of investigations that have concentrated on the association between lncRNAs and diseases. This serves as a testament to the recognition within the research community of the profound influence that lncRNAs wield on our physiological and pathological states. These studies have exploited the power of deep learning architectures to decipher the intricate interconnections between lncRNAs and various maladies.
From this multitude of lncRNA–disease association studies, it is clear that the tools and techniques of deep learning are being increasingly harnessed to delve deeper into the complexities of lncRNA functions and their roles in health and disease. This has undoubtedly added significant depth and breadth to our understanding of lncRNA dynamics.
However, it is important to emphasize that this represents only a fraction of the immense potential that deep learning holds for the further exploration of lncRNA. With the accelerating advancements in deep learning methodologies and increasing availability of high-throughput lncRNA data, there is much to look forward to in the realm of lncRNA research.
While the progress achieved thus far in lncRNA research via deep learning is certainly commendable, the journey has just begun. The horizon is replete with possibilities waiting to be uncovered, and it is our hope that this review has inspired further intellectual curiosity and will act as a catalyst for novel studies in this rapidly evolving field.
Author Contributions
Conceptualization, M.L.; validation, Y.K. and M.L.; formal analysis, Y.K. and M.L.; investigation, Y.K. and M.L.; writing—original draft preparation, Y.K. and M.L.; writing—review and editing, Y.K. and M.L.; visualization, M.L.; supervision, M.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by a grant from the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2021R1I1A1A01044255).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study.
Acknowledgments
In the preparation of this paper, the authors utilized Grammarly, an AI-assisted language editing tool, to enhance the grammatical correctness and fluency of our sentences.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Greener, J.G.; Kandathil, S.M.; Moffat, L.; Jones, D.T. A guide to machine learning for biologists. Nat. Rev. Mol. Cell Biol. 2022, 23, 40–55. [Google Scholar] [CrossRef] [PubMed]
- Carleo, G.; Cirac, I.; Cranmer, K.; Daudet, L.; Schuld, M.; Tishby, N.; Vogt-Maranto, L.; Zdeborová, L. Machine learning and the physical sciences. Rev. Mod. Phys. 2019, 91, 045002. [Google Scholar] [CrossRef]
- Liu, B.; Ding, M.; Shaham, S.; Rahayu, W.; Farokhi, F.; Lin, Z. When machine learning meets privacy: A survey and outlook. ACM Comput. Surv. (CSUR) 2021, 54, 1–36. [Google Scholar] [CrossRef]
- Lee, M. The Geometry of Feature Space in Deep Learning Models: A Holistic Perspective and Comprehensive Review. Mathematics 2023, 11, 2375. [Google Scholar] [CrossRef]
- Ko, K.; Yeom, T.; Lee, M. Superstargan: Generative adversarial networks for image-to-image translation in large-scale domains. Neural Netw. 2023, 162, 330–339. [Google Scholar] [CrossRef]
- Ku, H.; Lee, M. TextControlGAN: Text-to-Image Synthesis with Controllable Generative Adversarial Networks. Appl. Sci. 2023, 13, 5098. [Google Scholar] [CrossRef]
- Kim, J.; Lee, M. Class-Continuous Conditional Generative Neural Radiance Field. arXiv 2023, arXiv:2301.00950. [Google Scholar]
- Lee, M. A Mathematical Investigation of Hallucination and Creativity in GPT Models. Mathematics 2023, 11, 2320. [Google Scholar] [CrossRef]
- Brown, T.; Mann, B.; Ryder, N.; Subbiah, M.; Kaplan, J.D.; Dhariwal, P.; Neelakantan, A.; Shyam, P.; Sastry, G.; Askell, A.; et al. Language models are few-shot learners. Adv. Neural Inf. Process. Syst. 2020, 33, 1877–1901. [Google Scholar]
- Radford, A.; Narasimhan, K.; Salimans, T.; Sutskever, I. Improving Language Understanding by Generative Pre-Training; OpenAI Technical Report; OpenAI Inc.: San Francisco, CA, USA, 2018. [Google Scholar]
- Radford, A.; Wu, J.; Child, R.; Luan, D.; Amodei, D.; Sutskever, I. Language Models Are Unsupervised Multitask Learners; OpenAI Technical Report; OpenAI Inc.: San Francisco, CA, USA, 2019. [Google Scholar]
- Ma, L.; Bajic, V.B.; Zhang, Z. On the classification of long non-coding RNAs. RNA Biol. 2013, 10, 924–933. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef] [PubMed]
- Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Fatemi, R.P.; Velmeshev, D.; Faghihi, M.A. De-repressing LncRNA-targeted genes to upregulate gene expression: Focus on small molecule therapeutics. Mol. Ther.-Nucleic Acids 2014, 3, e196. [Google Scholar] [CrossRef]
- Peng, W.X.; Koirala, P.; Mo, Y.Y. LncRNA-mediated regulation of cell signaling in cancer. Oncogene 2017, 36, 5661–5667. [Google Scholar] [CrossRef]
- Liu, X.; Xu, M.; Li, P.; Zhang, W.; Zeng, L.h.; Yang, Y.; Yang, G. Roles of lncRNAs in the transcription regulation of HIV-1. Biomed. J. 2022, 45, 580–593. [Google Scholar] [CrossRef]
- Zhou, H.; Simion, V.; Pierce, J.B.; Haemmig, S.; Chen, A.F.; Feinberg, M.W. LncRNA-MAP3K4 regulates vascular inflammation through the p38 MAPK signaling pathway and cis-modulation of MAP3K4. FASEB J. 2021, 35, e21133. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Chen, Y.; Nie, X. Regulatory mechanisms of lncRNAs and their target gene signaling pathways in laryngeal squamous cell carcinoma. Front. Pharmacol. 2020, 11, 1140. [Google Scholar] [CrossRef]
- Farzaneh, M.; Najafi, S.; Anbiyaee, O.; Azizidoost, S.; Khoshnam, S.E. LncRNA MALAT1-related signaling pathways in osteosarcoma. Clin. Transl. Oncol. 2023, 25, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Dykes, I.M.; Emanueli, C. Transcriptional and post-transcriptional gene regulation by long non-coding RNA. Genom. Proteom. Bioinform. 2017, 15, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.K.; Xi, Y.; McCarthy, R.; Allton, K.; Akdemir, K.C.; Patel, L.R.; Aronow, B.; Lin, C.; Li, W.; Yang, L.; et al. LncPRESS1 is a p53-regulated LncRNA that safeguards pluripotency by disrupting SIRT6-mediated de-acetylation of histone H3K56. Mol. Cell 2016, 64, 967–981. [Google Scholar] [CrossRef]
- Guo, R.; Zou, B.; Liang, Y.; Bian, J.; Xu, J.; Zhou, Q.; Zhang, C.; Chen, T.; Yang, M.; Wang, H.; et al. LncRNA RCAT1 promotes tumor progression and metastasis via miR-214-5p/E2F2 axis in renal cell carcinoma. Cell Death Dis. 2021, 12, 689. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, M.; Liang, L.; Li, J.; Chen, Y.X. Over-expression of lncRNA DANCR is associated with advanced tumor progression and poor prognosis in patients with colorectal cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 11480. [Google Scholar] [PubMed]
- Merry, C.R.; Forrest, M.E.; Sabers, J.N.; Beard, L.; Gao, X.H.; Hatzoglou, M.; Jackson, M.W.; Wang, Z.; Markowitz, S.D.; Khalil, A.M. DNMT1-associated long non-coding RNAs regulate global gene expression and DNA methylation in colon cancer. Hum. Mol. Genet. 2015, 24, 6240–6253. [Google Scholar] [CrossRef] [PubMed]
- Böhmdorfer, G.; Wierzbicki, A.T. Control of chromatin structure by long noncoding RNA. Trends Cell Biol. 2015, 25, 623–632. [Google Scholar] [CrossRef]
- Xuan, P.; Gong, Z.; Cui, H.; Li, B.; Zhang, T. Fully connected autoencoder and convolutional neural network with attention-based method for inferring disease-related lncRNAs. Brief. Bioinform. 2022, 23, bbac089. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Xu, J.; Wu, Y.; Liu, N.; Wang, Y.; Liang, Y. CapsNet-LDA: Predicting lncRNA-disease associations using attention mechanism and capsule network based on multi-view data. Brief. Bioinform. 2023, 24, bbac531. [Google Scholar] [CrossRef]
- Madhavan, M.; Gopakumar, G. DBNLDA: Deep Belief Network based representation learning for lncRNA-disease association prediction. Appl. Intell. 2022, 52, 5342–5352. [Google Scholar] [CrossRef]
- Madhavan, M.; Gopalakrishnan, G. Long Non-coding RNAs in Heart Failure: A Deep Belief Network based Cluster Analysis. Curr. Bioinform. 2021, 16, 983–991. [Google Scholar] [CrossRef]
- Ma, Y. DeepMNE: Deep Multi-Network Embedding for lncRNA-Disease Association Prediction. IEEE J. Biomed. Health Inform. 2022, 26, 3539–3549. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Yang, Z.; Song, J.; Dai, Q.; Duan, X. DHNLDA: A Novel Deep Hierarchical Network Based Method for Predicting lncRNA-Disease Associations. IEEE-ACM Trans. Comput. Biol. Bioinform. 2022, 19, 3395–3403. [Google Scholar] [CrossRef]
- Zeng, M.; Lu, C.; Fei, Z.; Wu, F.X.; Li, Y.; Wang, J.; Li, M. DMFLDA: A Deep Learning Framework for Predicting lncRNA-Disease Associations. IEEE-ACM Trans. Comput. Biol. Bioinform. 2021, 18, 2353–2363. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yu, Y.; Zhao, S. Dual Attention Mechanisms and Feature Fusion Networks Based Method for Predicting LncRNA-Disease Associations. Interdiscip. Sci.-Comput. Life Sci. 2022, 14, 358–371. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Chen, M.; Pan, X. GCRFLDA: Scoring lncRNA-disease associations using graph convolution matrix completion with conditional random field. Brief. Bioinform. 2022, 23, bbab361. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhong, C. gGATLDA: LncRNA-disease association prediction based on graph-level graph attention network. BMC Bioinform. 2022, 23, 11. [Google Scholar] [CrossRef]
- Xuan, P.; Wang, S.; Cui, H.; Zhao, Y.; Zhang, T.; Wu, P. Learning global dependencies and multi-semantics within heterogeneous graph for predicting disease-related lncRNAs. Brief. Bioinform. 2022, 23, bbac361. [Google Scholar] [CrossRef]
- Xuan, P.; Zhan, L.; Cui, H.; Zhang, T.; Nakaguchi, T.; Zhang, W. Graph Triple-Attention Network for Disease-Related LncRNA Prediction. IEEE J. Biomed. Health Inform. 2022, 26, 2839–2849. [Google Scholar] [CrossRef]
- Zhao, X.; Zhao, X.; Yin, M. Heterogeneous graph attention network based on meta-paths for lncRNA-disease association prediction. Brief. Bioinform. 2022, 23, bbab407. [Google Scholar] [CrossRef]
- Shi, H.; Zhang, X.; Tang, L.; Liu, L. Heterogeneous graph neural network for lncRNA-disease association prediction. Sci. Rep. 2022, 12, 17519. [Google Scholar] [CrossRef]
- Jha, A.; Quesnel-Vallieres, M.; Wang, D.; Thomas-Tikhonenko, A.; Lynch, K.W.; Barash, Y. Identifying common transcriptome signatures of cancer by interpreting deep learning models. Genome Biol. 2022, 23, 117. [Google Scholar] [CrossRef]
- Wei, H.; Liao, Q.; Liu, B. iLncRNAdis-FB: Identify lncRNA-Disease Associations by Fusing Biological Feature Blocks Through Deep Neural Network. IEEE-ACM Trans. Comput. Biol. Bioinform. 2021, 18, 1946–1957. [Google Scholar] [CrossRef]
- Guo, Z.H.; Chen, Z.H.; You, Z.H.; Wang, Y.B.; Yi, H.C.; Wang, M.N. A learning-based method to predict LncRNA-disease associations by combining CNN and ELM. BMC Bioinform. 2022, 22, 622. [Google Scholar] [CrossRef] [PubMed]
- Lan, W.; Lai, D.; Chen, Q.; Wu, X.; Chen, B.; Liu, J.; Wang, J.; Chen, Y.P.P. LDICDL: LncRNA-Disease Association Identification Based on Collaborative Deep Learning. IEEE-ACM Trans. Comput. Biol. Bioinform. 2022, 19, 1715–1723. [Google Scholar] [CrossRef]
- Yuan, L.; Zhao, J.; Sun, T.; Shen, Z. A machine learning framework that integrates multi-omics data predicts cancer-related LncRNAs. BMC Bioinform. 2021, 22, 332. [Google Scholar] [CrossRef] [PubMed]
- Kang, C.; Zhang, H.; Liu, Z.; Huang, S.; Yin, Y. LR-GNN: A graph neural network based on link representation for predicting molecular associations. Brief. Bioinform. 2022, 23, bbab513. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Zhang, Z.Q.; Liu, N.N.; Wu, Y.N.; Gu, C.L.; Wang, Y.L. MAGCNSE: Predicting lncRNA-disease associations using multi-view attention graph convolutional network and stacking ensemble model. BMC Bioinform. 2022, 23, 189. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ye, F.; Gao, X. MCA-Net: Multi-Feature Coding and Attention Convolutional Neural Network for Predicting lncRNA-Disease Association. IEEE-ACM Trans. Comput. Biol. Bioinform. 2022, 19, 2907–2919. [Google Scholar] [CrossRef]
- Ai, C.; Yang, H.; Ding, Y.; Tang, J.; Guo, F. A multi-layer multi-kernel neural network for determining associations between non-coding RNAs and diseases. Neurocomputing 2022, 493, 91–105. [Google Scholar] [CrossRef]
- Wu, Q.W.; Cao, R.F.; Xia, J.F.; Ni, J.C.; Zheng, C.H.; Su, Y.S. Extra Trees Method for Predicting LncRNA-Disease Association Based On Multi-Layer Graph Embedding Aggregation. IEEE-ACM Trans. Comput. Biol. Bioinform. 2022, 19, 3171–3178. [Google Scholar] [CrossRef]
- Al Mamun, A.; Tanvir, R.B.; Sobhan, M.; Mathee, K.; Narasimhan, G.; Holt, G.E.; Mondal, A.M. Multi-Run Concrete Autoencoder to Identify Prognostic lncRNAs for 12 Cancers. Int. J. Mol. Sci. 2021, 22, 11919. [Google Scholar] [CrossRef] [PubMed]
- Wei-Na, L.; Xiao-Nan, F.; Shao-Wu, Z. NELDA: Prediction of LncRNA-disease Associations With Network Embedding. Prog. Biochem. Biophys. 2022, 49, 1369–1380. [Google Scholar]
- Yang, W.; Zheng, X.; Huang, Q.; Liu, Y.; Chen, Y.; Song, Z. Combining BPSO and ELM Models for Inferring Novel lncRNA- Disease Associations. Int. J. Data Warehous. Min. 2023, 19, 1–18. [Google Scholar] [CrossRef]
- Silva, V.A.B.O.; Spinosa, E.J. Graph Convolutional Auto-Encoders for Predicting Novel lncRNA-Disease Associations. IEEE-ACM Trans. Comput. Biol. Bioinform. 2022, 19, 2264–2271. [Google Scholar] [CrossRef]
- Wang, J.; Wu, S.; Zhang, J.; Chen, J. Potential Prognosis and Diagnostic Value of AKT3, LSM12, MEF2C, and RAB30 in Exosomes in Colorectal Cancer on Spark Framework. J. Healthc. Eng. 2021, 2021, 8218043. [Google Scholar] [CrossRef]
- Sheng, N.; Cui, H.; Zhang, T.; Xuan, P. Attentional multi-level representation encoding based on convolutional and variance autoencoders for lncRNA-disease association prediction. Brief. Bioinform. 2021, 22, bbaa067. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Zhang, H.; Jin, C.; Quan, X.; Yin, Y. A representation learning model based on variational inference and graph autoencoder for predicting lncRNA-disease associations. BMC Bioinform. 2021, 22, 136. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Zhang, H.; Jin, C.; Kang, C. Predicting lncRNA-protein interactions with bipartite graph embedding and deep graph neural networks. Front. Genet. 2023, 14, 1136672. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Sun, H.; Feng, S.; Zhang, Q.; Han, S.; Du, W. Capsule-LPI: A LncRNA-protein interaction predicting tool based on a capsule network. BMC Bioinform. 2021, 22, 246. [Google Scholar] [CrossRef]
- Shaw, D.; Chen, H.; Xie, M.; Jiang, T. DeepLPI: A multimodal deep learning method for predicting the interactions between lncRNAs and protein isoforms. BMC Bioinform. 2021, 22, 24. [Google Scholar] [CrossRef]
- Teng, Z.; Zhang, Y.; Dai, Q.; Wu, C.; Li, D. Constructing discriminative feature space for LncRNA-protein interaction based on deep autoencoder and marginal fisher analysis. Comput. Biol. Med. 2023, 157, 106711. [Google Scholar] [CrossRef]
- Peng, L.; Tan, J.; Tian, X.; Zhou, L. EnANNDeep: An Ensemble-based lncRNA-protein Interaction Prediction Framework with Adaptive k-Nearest Neighbor Classifier and Deep Models. Interdiscip. Sci.-Comput. Life Sci. 2022, 14, 209–232. [Google Scholar] [CrossRef]
- Zhang, Y.Y.; Liang, D.M.; Du, P.F. iEssLnc: Quantitativeestimation of lncRNA gene essentialities with meta- path-guided random walks on the lncRNA-protein interaction network. Brief. Bioinform. 2023, 24, bbad097. [Google Scholar] [CrossRef]
- Huang, L.; Jiao, S.; Yang, S.; Zhang, S.; Zhu, X.; Guo, R.; Wang, Y. LGFC-CNN: Prediction of lncRNA-Protein Interactions by Using Multiple Types of Features through Deep Learning. Genes 2021, 12, 1689. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Shi, Y.; Yan, J.; Qu, W.; Li, X.; Tan, J. LPI-CSFFR: Combining serial fusion with feature reuse for predicting LncRNA-protein interactions. Comput. Biol. Chem. 2022, 99, 107718. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Wang, Z.; Tian, X.; Peng, L. LPI-deepGBDT: A multiple-layer deep framework based on gradient boosting decision trees for lncRNA-protein interaction identification. BMC Bioinform. 2021, 22, 479. [Google Scholar] [CrossRef]
- Peng, L.; Wang, C.; Tian, X.; Zhou, L.; Li, K. Finding lncRNA-Protein Interactions Based on Deep Learning With Dual-Net Neural Architecture. IEEE-ACM Trans. Comput. Biol. Bioinform. 2022, 19, 3456–3468. [Google Scholar]
- Zhou, L.; Duan, Q.; Tian, X.; Xu, H.; Tang, J.; Peng, L. LPI-HyADBS: A hybrid framework for lncRNA-protein interaction prediction integrating feature selection and classification. BMC Bioinform. 2021, 22, 568. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Sun, J.; Shuai, S.C.; Zhao, Q.; Shuai, J. Predicting potential interactions between lncRNAs and proteins via combined graph auto-encoder methods. Brief. Bioinform. 2023, 24, bbac527. [Google Scholar] [CrossRef]
- Zhou, H.; Wekesa, J.S.; Luan, Y.; Meng, J. PRPI-SC: An ensemble deep learning model for predicting plant lncRNA-protein interactions. BMC Bioinform. 2021, 22, 415. [Google Scholar] [CrossRef]
- Song, J.; Tian, S.; Yu, L.; Yang, Q.; Dai, Q.; Wang, Y.; Wu, W.; Duan, X. RLF-LPI: An ensemble learning framework using sequence information for predicting lncRNA-protein interaction based on AE-ResLSTM and fuzzy decision. Math. Biosci. Eng. 2022, 19, 4749–4764. [Google Scholar] [CrossRef]
- Asim, M.N.; Ibrahim, M.A.; Zehe, C.; Trygg, J.; Dengel, A.; Ahmed, S. BoT-Net: A lightweight bag of tricks-based neural network for efficient LncRNA-miRNA interaction prediction. Interdiscip. Sci.-Comput. Life Sci. 2022, 14, 841–862. [Google Scholar] [CrossRef]
- Yang, L.; Li, L.P.; Yi, H.C. DeepWalk based method to predict lncRNA-miRNA associations via lncRNA-miRNA-disease-protein-drug graph. BMC Bioinform. 2022, 22, 621. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Zhang, L.; Sun, J.; Zhao, Q.; Shuai, J. Predicting the potential human lncRNA-miRNA interactions based on graph convolution network with conditional random field. Brief. Bioinform. 2022, 23, bbac463. [Google Scholar] [CrossRef]
- Song, J.; Tian, S.; Yu, L.; Yang, Q.; Xing, Y.; Zhang, C.; Dai, Q.; Duan, X. MD-MLI: Prediction of miRNA-lncRNA Interaction by Using Multiple Features and Hierarchical Deep Learning. IEEE-ACM Trans. Comput. Biol. Bioinform. 2022, 19, 1724–1733. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, Y.; Pan, Z.; Sun, X.; Mou, M.; Zhang, B.; Li, Z.; Li, H.; Zhu, F. ncRNAInter: A novel strategy based on graph neural network to discover interactions between lncRNA and miRNA. Brief. Bioinform. 2022, 23, bbac411. [Google Scholar] [CrossRef]
- Hamdy, W.; Ismail, A.; Awad, W.A.; Ibrahim, A.H.; Hassanien, A.E. An Optimized Ensemble Deep Learning Model for Predicting Plant miRNA-IncRNA Based on Artificial Gorilla Troops Algorithm. Sensors 2023, 23, 2219. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Sun, Z.L. PmliHFM: Predicting Plant miRNA-lncRNA Interactions with Hybrid Feature Mining Network. Interdiscip. Sci.-Comput. Life Sci. 2023, 15, 44–54. [Google Scholar] [CrossRef]
- Kang, Q.; Meng, J.; Shi, W.; Luan, Y. Ensemble Deep Learning Based on Multi-level Information Enhancement and Greedy Fuzzy Decision for Plant miRNA-lncRNA Interaction Prediction. Interdiscip. Sci.-Comput. Life Sci. 2021, 13, 603–614. [Google Scholar] [CrossRef]
- Yu, X.; Jiang, L.; Jin, S.; Zeng, X.; Liu, X. preMLI: A pre-trained method to uncover microRNA-lncRNA potential interactions. Brief. Bioinform. 2022, 23, bbab470. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Long, Y.; Kwoh, C.K. Class similarity network for coding and long non-coding RNA classification. BMC Bioinform. 2021, 22, 609. [Google Scholar] [CrossRef]
- Zhang, T.; Tang, Q.; Nie, F.; Zhao, Q.; Chen, W. DeepLncPro: An interpretable convolutional neural network model for identifying long non-coding RNA promoters. Brief. Bioinform. 2022, 23, bbac447. [Google Scholar] [CrossRef]
- Ritu; Gupta, S.; Sharma, N.K.; Shankar, R. DeepPlnc: Bi-modal deep learning for highly accurate plant lncRNA discovery. Genomics 2022, 114, 110443. [Google Scholar]
- Shi, K.; Liu, T.; Fu, H.; Li, W.; Zheng, X. Genome-wide analysis of lncRNA stability in human. PLoS Comput. Biol. 2021, 17, e1008918. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, P.; Du, H.; Cao, Y.; Peng, Q.; Fu, L. LncDLSM: Identification of Long Non-Coding RNAs With Deep Learning-Based Sequence Model. IEEE J. Biomed. Health Inform. 2023, 27, 2117–2127. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Zou, B.; He, M.; Hu, Y.; Dou, Y.; Cui, T.; Tan, P.; Li, S.; Rao, S.; Huang, Y.; et al. LncReader: Identification of dual functional long noncoding RNAs using a multi-head self-attention mechanism. Brief. Bioinform. 2023, 24, bbac579. [Google Scholar] [CrossRef]
- Zhang, Y.; Jia, C.; Kwoh, C.K. Predicting the interaction biomolecule types for lncRNA: An ensemble deep learning approach. Brief. Bioinform. 2021, 22, bbaa228. [Google Scholar] [CrossRef]
- AlZubi, A.A.; Alanazi, J.M. An Optimized Technique for RNA Prediction Based on Neural Network. Intell. Autom. Soft Comput. 2023, 35, 3599–3611. [Google Scholar] [CrossRef]
- Lin, R.; Wichadakul, D. Interpretable Deep Learning Model Reveals Subsequences of Various Functions for Long Non-Coding RNA Identification. Front. Genet. 2022, 13, 876721. [Google Scholar] [CrossRef] [PubMed]
- Zeng, M.; Wu, Y.; Lu, C.; Zhang, F.; Wu, F.X.; Li, M. DeepLncLoc: A deep learning framework for long non-coding RNA subcellular localization prediction based on subsequence embedding. Brief. Bioinform. 2022, 23, bbab360. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Ding, M.; Feng, J.; Ji, B.; Huang, P.; Zhang, J.; Yu, X.; Cao, Z.; Yang, Y.; Zhou, Y.; et al. EVlncRNA-Dpred: Improved prediction of experimentally validated lncRNAs by deep learning. Brief. Bioinform. 2023, 24, bbac583. [Google Scholar] [CrossRef]
- Cai, J.; Wang, T.; Deng, X.; Tang, L.; Liu, L. GM-lncLoc: LncRNAs subcellular localization prediction based on graph neural network with meta-learning. BMC Genom. 2023, 24, 52. [Google Scholar] [CrossRef]
- Li, M.; Zhao, B.; Yin, R.; Lu, C.; Guo, F.; Zeng, M. GraphLncLoc: Long non-coding RNA subcellular localization prediction using graph convolutional networks based on sequence to graph transformation. Brief. Bioinform. 2023, 24, bbac565. [Google Scholar] [CrossRef] [PubMed]
- Meng, J.; Kang, Q.; Chang, Z.; Luan, Y. PlncRNA-HDeep: Plant long noncoding RNA prediction using hybrid deep learning based on two encoding styles. BMC Bioinform. 2021, 22, 242. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Feng, R.; Kahlert, U.D.; Chen, Z.; Torres-dela Roche, L.A.; Soliman, A.; Miao, C.; De Wilde, R.L.; Shi, W. Construction of ceRNA Networks Associated With CD8 T Cells in Breast Cancer. Front. Oncol. 2022, 12, 883197. [Google Scholar] [CrossRef]
- Marete, A.; Ariel, O.; Ibeagha-Awemu, E.; Bissonnette, N. Identification of Long Non-coding RNA Isolated From Naturally Infected Macrophages and Associated With Bovine Johne’s Disease in Canadian Holstein Using a Combination of Neural Networks and Logistic Regression. Front. Vet. Sci. 2021, 8, 639053. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.A.; Chan, K.C.C.; You, Z.H.; Hu, P.; Wang, L.; Huang, Z.A. Predicting microRNA-disease associations from lncRNA-microRNA interactions via Multiview Multitask Learning. Brief. Bioinform. 2021, 22, bbaa133. [Google Scholar] [CrossRef]
- Qiu, W.; Yang, J.; Wang, B.; Yang, M.; Tian, G.; Wang, P.; Yang, J. Evaluating the Microsatellite Instability of Colorectal Cancer Based on Multimodal Deep Learning Integrating Histopathological and Molecular Data. Front. Oncol. 2022, 12, 3011. [Google Scholar] [CrossRef]
- Wang, W.; Chen, H. Predicting miRNA-disease associations based on lncRNA-miRNA interactions and graph convolution networks. Brief. Bioinform. 2023, 24, bbac495. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, S. Prediction of Tumor Lymph Node Metastasis Using Wasserstein Distance-Based Generative Adversarial Networks Combing with Neural Architecture Search for Predicting. Mathematics 2023, 11, 729. [Google Scholar] [CrossRef]
- Gao, M.; Liu, S.; Qi, Y.; Guo, X.; Shang, X. GAE-LGA: Integration of multi-omics data with graph autoencoders to identify lncRNA-PCG associations. Brief. Bioinform. 2022, 23, bbac452. [Google Scholar] [CrossRef]
- Baek, J.; Lee, B.; Kwon, S.; Yoon, S. LncRNAnet: Long non-coding RNA identification using deep learning. Bioinformatics 2018, 34, 3889–3897. [Google Scholar] [CrossRef]
- Fan, X.N.; Zhang, S.W. lncRNA-MFDL: Identification of human long non-coding RNAs by fusing multiple features and using deep learning. Mol. Biosyst. 2015, 11, 892–897. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.Q.; Li, B.X.; Zeng, G.R.; Liu, Q.Y.; Ai, D.M. Prediction of long non-coding RNAs based on deep learning. Genes 2019, 10, 273. [Google Scholar] [CrossRef] [PubMed]
- Shaath, H.; Vishnubalaji, R.; Elango, R.; Kardousha, A.; Islam, Z.; Qureshi, R.; Alam, T.; Kolatkar, P.R.; Alajez, N.M. Long non-coding RNA and RNA-binding protein interactions in cancer: Experimental and machine learning approaches. In Seminars in Cancer Biology; Elsevier: Amsterdam, The Netherlands, 2022. [Google Scholar]
- Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef]
- Yuan, L.; Xu, Z.Y.; Ruan, S.M.; Mo, S.; Qin, J.J.; Cheng, X.D. Long non-coding RNAs towards precision medicine in gastric cancer: Early diagnosis, treatment, and drug resistance. Mol. Cancer 2020, 19, 1–22. [Google Scholar] [CrossRef]
- Alipanahi, B.; Delong, A.; Weirauch, M.T.; Frey, B.J. Predicting the sequence specificities of DNA-and RNA-binding proteins by deep learning. Nat. Biotechnol. 2015, 33, 831–838. [Google Scholar] [CrossRef] [PubMed]
- Lopez-del Rio, A.; Martin, M.; Perera-Lluna, A.; Saidi, R. Effect of sequence padding on the performance of deep learning models in archaeal protein functional prediction. Sci. Rep. 2020, 10, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Heward, J.A.; Lindsay, M.A. Long non-coding RNAs in the regulation of the immune response. Trends Immunol. 2014, 35, 408–419. [Google Scholar] [CrossRef] [PubMed]
- Percha, B.; Altman, R.B. Learning the structure of biomedical relationships from unstructured text. PLoS Comput. Biol. 2015, 11, e1004216. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Zhang, Y.; Williams, J.; Antoniou, E.; McCombie, W.R.; Wu, S.; Zhu, W.; Davidson, N.O.; Denoya, P.; Li, E. Parallel comparison of Illumina RNA-Seq and Affymetrix microarray platforms on transcriptomic profiles generated from 5-aza-deoxy-cytidine treated HT-29 colon cancer cells and simulated datasets. BMC Bioinform. 2013, 14, S1. [Google Scholar] [CrossRef]
- Rudin, C. Stop explaining black box machine learning models for high stakes decisions and use interpretable models instead. Nat. Mach. Intell. 2019, 1, 206–215. [Google Scholar] [CrossRef] [PubMed]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Zeiler, M.D.; Fergus, R. Visualizing and understanding convolutional networks. In Proceedings of the Computer Vision–ECCV 2014: 13th European Conference, Zurich, Switzerland, 6–12 September 2014; Proceedings, Part I 13. Springer: Cham, Switzerland, 2014; pp. 818–833. [Google Scholar]
- Choi, S.R.; Lee, M. Estimating the Prognosis of Low-Grade Glioma with Gene Attention Using Multi-Omics and Multi-Modal Schemes. Biology 2022, 11, 1462. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).