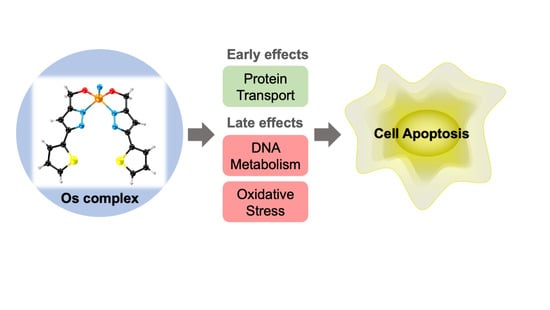
A Pyrazolate Osmium(VI) Nitride Exhibits Anticancer Activity through Modulating Protein Homeostasis in HepG2 Cells
Abstract
1. Introduction
2. Results and Discussion
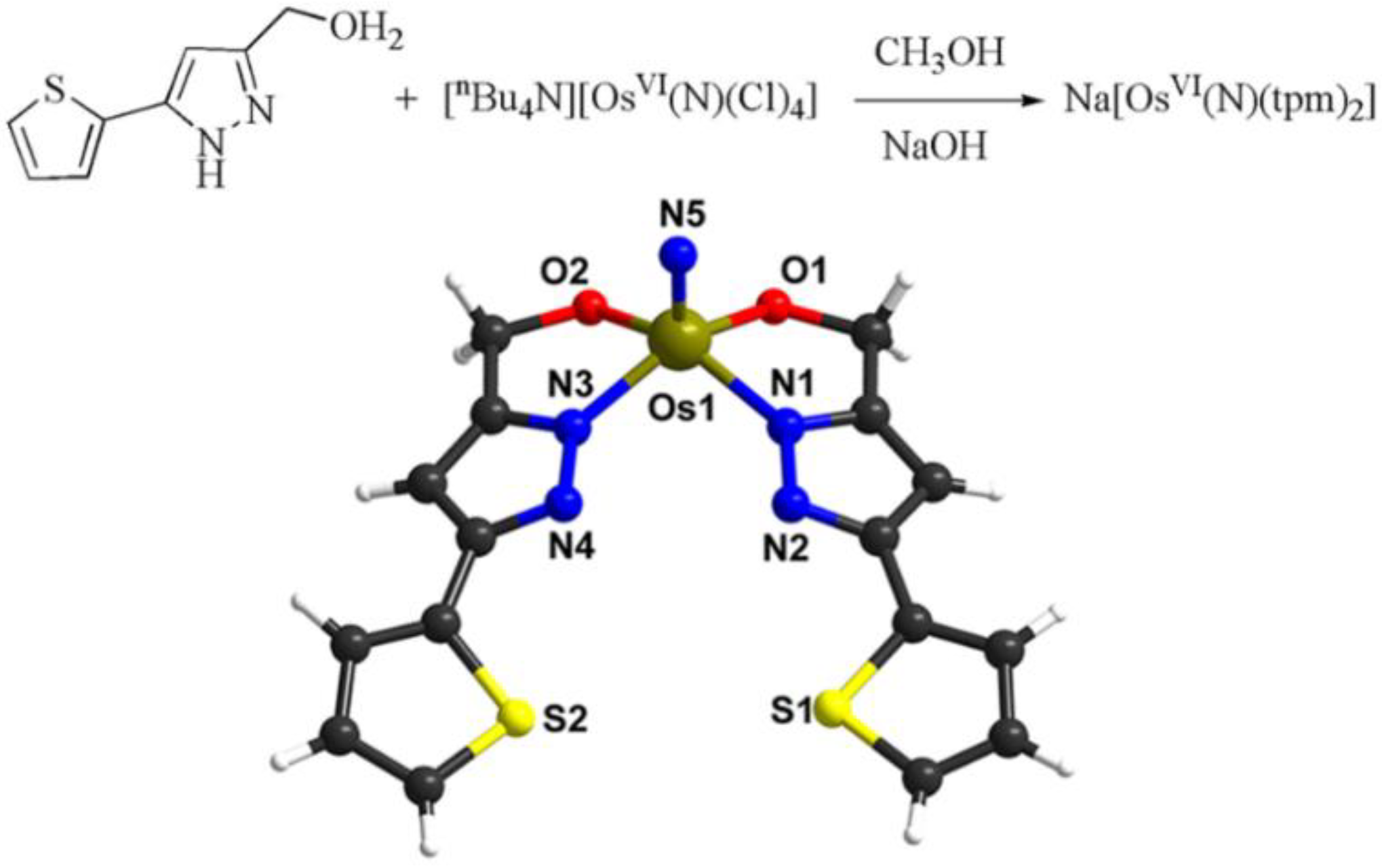
2.1. Synthesis and Characterization of the Na[OsVI(N)(tpm)2] Complex
2.2. Anticancer Ability of the Na[OsVI(N)(tpm)2]
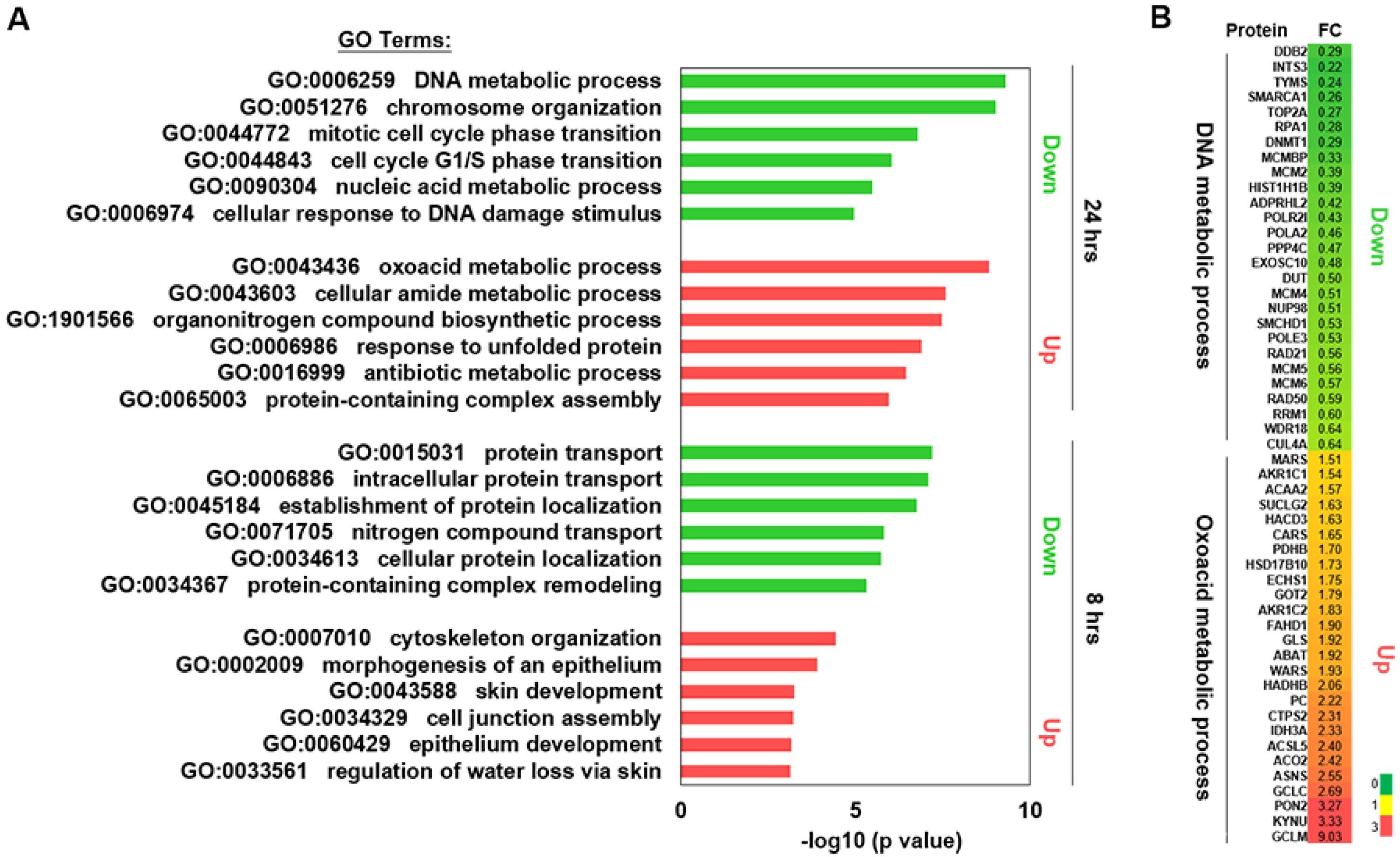
2.3. Proteomics Analysis
2.4. Effects of Na[OsVI(N)(tpm)2] on Cell Cycle and Apoptosis
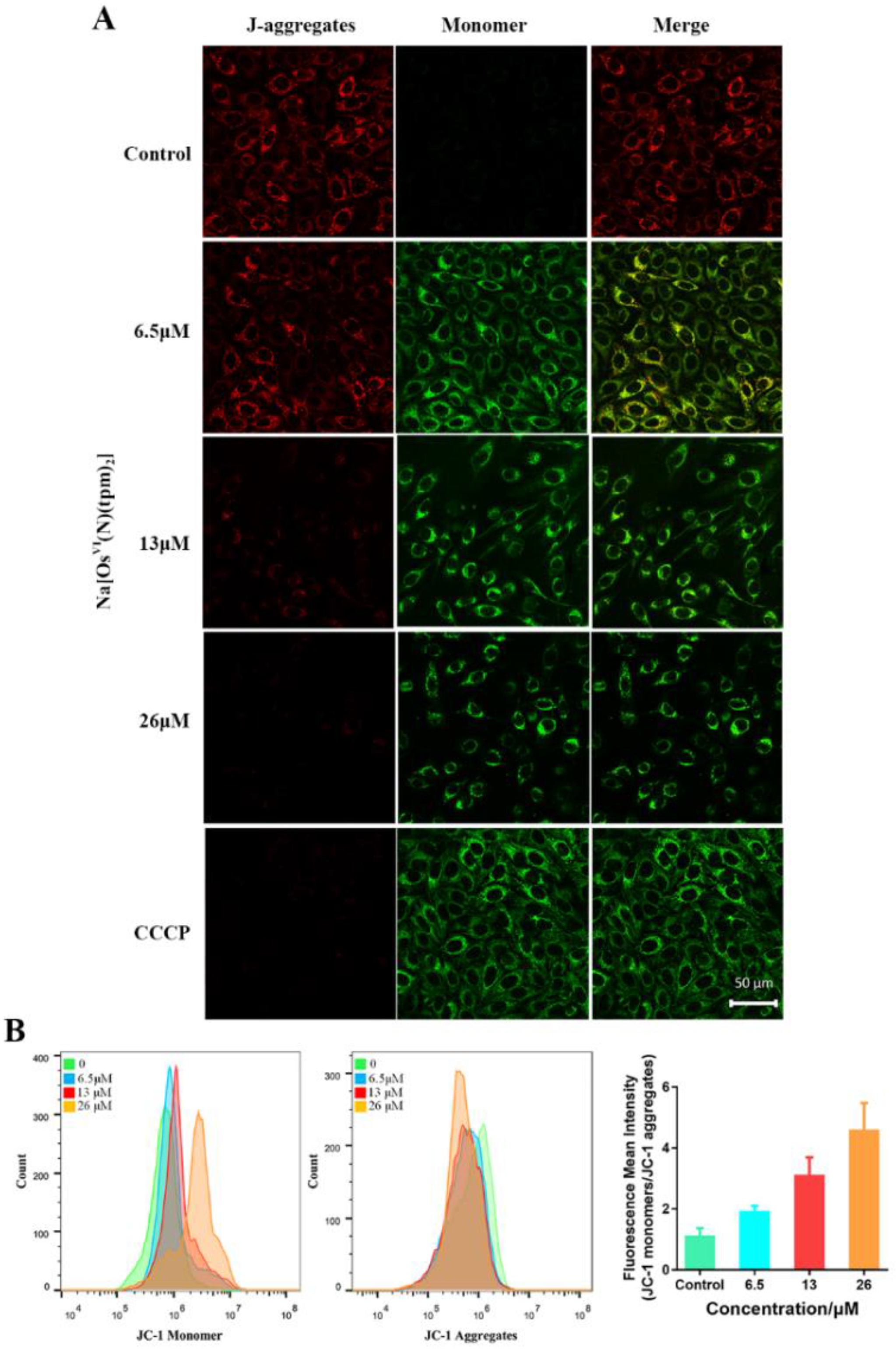
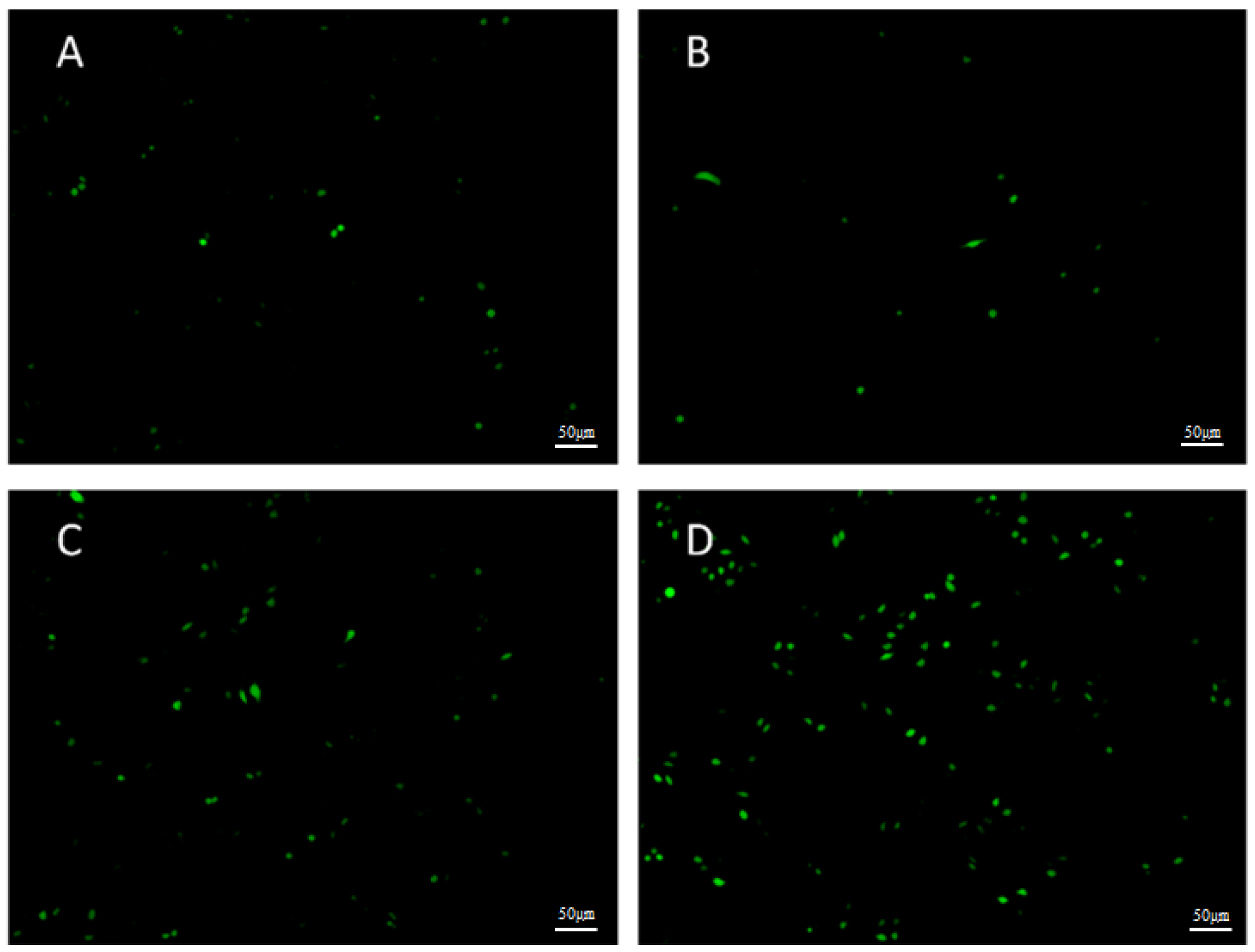
2.5. Mitochondrial Membrane Potential (MMP) Analysis and ROS Analysis
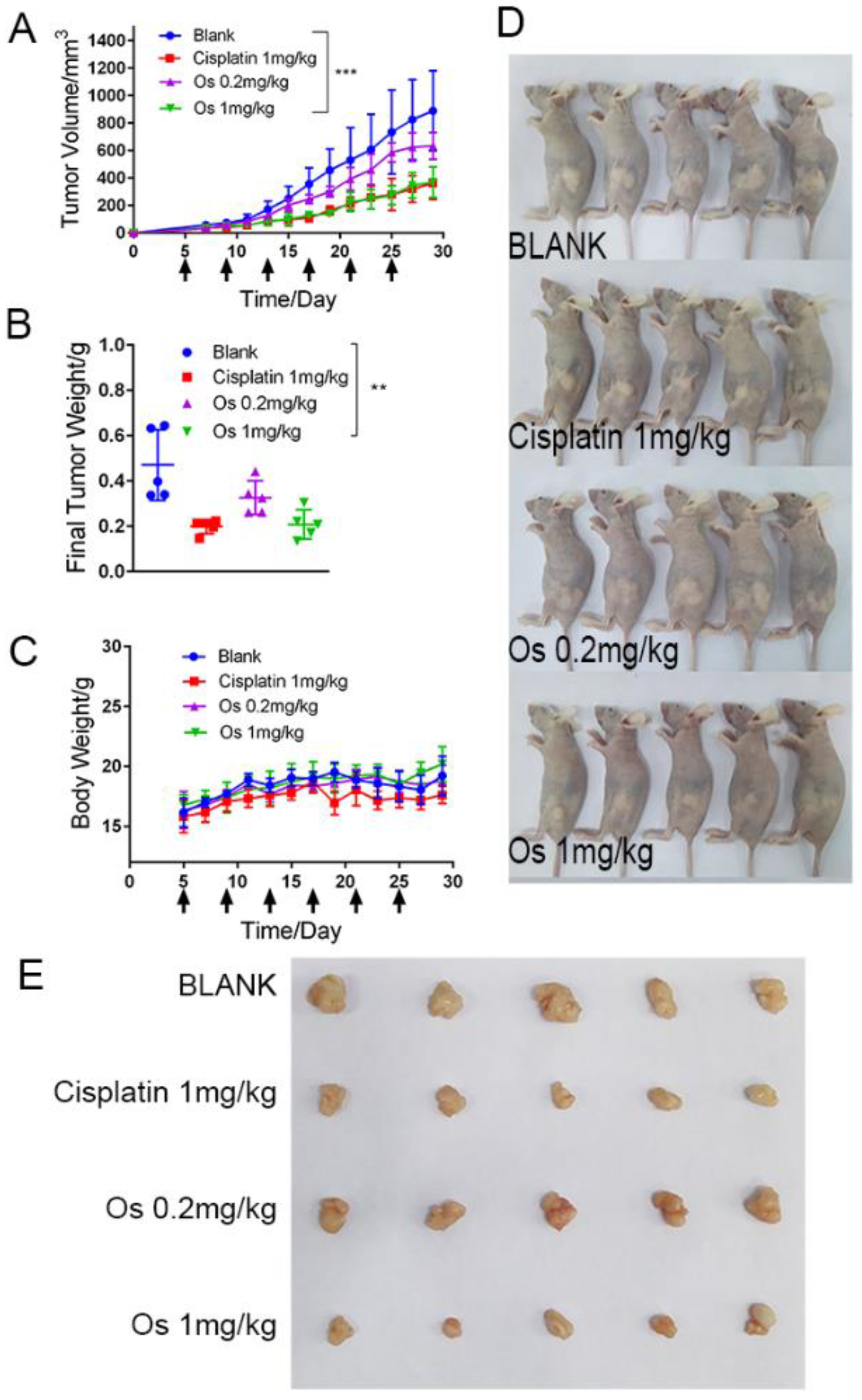
2.6. Anticancer Activity of Na[OsVI(N)(tpm)2] In Vivo
3. Materials and Methods
3.1. Instrumentation
3.2. Materials
3.3. Synthesis of Complex
3.4. X-ray Crystallography
3.5. Stability of Na[OsVI(N)(tpm)2] in DMSO and in Aqueous Solutions in the Absence or Presence of GSH
3.6. Cell Culture Conditions
3.7. MTT Assay
3.8. Proteomic Analysis
3.9. Cell Cycle Analysis
3.10. Apoptosis Evaluation
3.11. Western Blot Analysis
3.12. Analysis of Mitochondrial Membrane Potential (MMP)
3.12.1. Flow Cytometry
3.12.2. Confocal Microscopy
3.13. Detection of Intracellular Reactive Oxygen Species (ROS) by DCFH-DA
3.14. Cell Morphology Observed by an Inverted Microscope
3.15. Xenograft in Nude Mice
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Deo, K.M.; Ang, D.L.; McGhie, B.; Rajamanickam, A.; Dhiman, A.; Khoury, A.; Holland, J.; Bjelosevic, A.; Pages, B.; Gordon, C.; et al. Platinum coordination compounds with potent anticancer activity. Coord. Chem. Rev. 2018, 375, 148–163. [Google Scholar] [CrossRef]
- Meier-Menches, S.M.; Gerner, C.; Berger, W.; Hartinger, C.G.; Keppler, B.K. Structure-activity relationships for ruthenium and osmium anticancer agents—Towards clinical development. Chem. Soc. Rev. 2018, 47, 909–928. [Google Scholar] [CrossRef] [PubMed]
- Xiong, K.; Qian, C.; Yuan, Y.X.; Wei, L.; Liao, X.X.; He, L.T.; Rees, T.W.; Chen, Y.; Wan, J.; Ji, L.N.; et al. Necroptosis Induced by Ruthenium(II) Complexes as Dual Catalytic Inhibitors of Topoisomerase I/II. Angew. Chem.-Int. Ed. 2020, 59, 16631–16637. [Google Scholar] [CrossRef]
- Ye, M.; Huang, W.Q.; Li, Z.X.; Wang, C.X.; Liu, T.; Chen, Y.; Hor, C.H.; Man, W.L.; Ni, W.X. Osmium(VI) nitride triggers mitochondria-induced oncosis and apoptosis. Chem. Commun. 2022, 58, 2468–2471. [Google Scholar] [CrossRef] [PubMed]
- Xiong, X.; Liu, L.-Y.; Mao, Z.-W.; Zou, T. Approaches towards understanding the mechanism-of-action of metallodrugs. Coord. Chem. Rev. 2022, 453, 214311. [Google Scholar] [CrossRef]
- Thota, S.; Rodrigues, D.A.; Crans, D.C.; Barreiro, E.J. Ru(II) Compounds: Next-Generation Anticancer Metallotherapeutics? J. Med. Chem. 2018, 61, 5805–5821. [Google Scholar] [CrossRef]
- Zeng, L.L.; Gupta, P.; Chen, Y.L.; Wang, E.J.; Ji, L.N.; Chao, H.; Chen, Z.S. The development of anticancer ruthenium(II) complexes: From single molecule compounds to nanomaterials. Chem. Soc. Rev. 2017, 46, 5771–5804. [Google Scholar] [CrossRef]
- Needham, R.J.; Sanchez-Cano, C.; Zhang, X.; Romero-Canelon, I.; Habtemariam, A.; Cooper, M.S.; Meszaros, L.; Clarkson, G.J.; Blower, P.J.; Sadler, P.J. In-Cell Activation of Organo-Osmium(II) Anticancer Complexes. Angew. Chem. 2017, 56, 1017–1020. [Google Scholar] [CrossRef]
- Ni, W.X.; Man, W.L.; Cheung, M.T.; Sun, R.W.; Shu, Y.L.; Lam, Y.W.; Che, C.M.; Lau, T.C. Osmium(VI) complexes as a new class of potential anti-cancer agents. Chem. Commun. 2011, 47, 2140–2142. [Google Scholar] [CrossRef]
- Huang, W.Q.; Wang, C.X.; Liu, T.; Li, Z.X.; Pan, C.; Chen, Y.Z.; Lian, X.; Man, W.L.; Ni, W.X. A cytotoxic nitrido-osmium(VI) complex induces caspase-mediated apoptosis in HepG2 cancer cells. Dalton Trans. 2020, 49, 17173–17182. [Google Scholar] [CrossRef] [PubMed]
- Ni, W.-X.; Man, W.-L.; Yiu, S.-M.; Ho, M.; Cheung, M.T.-W.; Ko, C.-C.; Che, C.-M.; Lam, Y.-W.; Lau, T.-C. Osmium(vi) nitrido complexes bearing azole heterocycles: A new class of antitumor agents. Chem. Sci. 2012, 3, 1582. [Google Scholar] [CrossRef]
- Suntharalingam, K.; Johnstone, T.C.; Bruno, P.M.; Lin, W.; Hemann, M.T.; Lippard, S.J. Bidentate ligands on osmium(VI) nitrido complexes control intracellular targeting and cell death pathways. J. Am. Chem. Soc. 2013, 135, 14060–14063. [Google Scholar] [CrossRef] [PubMed]
- Suntharalingam, K.; Lin, W.; Johnstone, T.C.; Bruno, P.M.; Zheng, Y.R.; Hemann, M.T.; Lippard, S.J. A breast cancer stem cell-selective, mammospheres-potent osmium(VI) nitrido complex. J. Am. Chem Soc. 2014, 136, 14413–14416. [Google Scholar] [CrossRef] [PubMed]
- Laws, K.; Bineva-Todd, G.; Eskandari, A.; Lu, C.; O’Reilly, N.; Suntharalingam, K. A Copper(II) Phenanthroline Metallopeptide That Targets and Disrupts Mitochondrial Function in Breast Cancer Stem Cells. Angew. Chem. Int. Ed. 2018, 57, 287–291. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, X.; Ren, Y.; Zhang, J.; Chen, J.; Zhou, W.; Guo, W.; Wang, X.; Chen, H.; Li, M.; et al. Cisplatin-enriching cancer stem cells confer multidrug resistance in non-small cell lung cancer via enhancing TRIB1/HDAC activity. Cell Death Dis. 2017, 8, e2746. [Google Scholar] [CrossRef]
- Thakur, B.; Ray, P. Cisplatin triggers cancer stem cell enrichment in platinum-resistant cells through NF-kappaB-TNFalpha-PIK3CA loop. J. Exp. Clin. Cancer Res. 2017, 36, 164. [Google Scholar] [CrossRef]
- Eskandari, A.; Kundu, A.; Ghosh, S.; Suntharalingam, K. A Triangular Platinum(II) Multi-nuclear Complex with Impressive Cytotoxicity Towards Breast Cancer Stem Cells. Angew. Chem. 2019, 28, 12059–12064. [Google Scholar] [CrossRef]
- Liu, J.-J.; Zhu, H.-L.; Zhao, M.-Y.; Zhao, X.; Zhang, X. Pyrazole derivatives as antitumor, anti-inflammatory and antibacterial agents. Mini Rev. Med. Chem. 2013, 13, 1957–1966. [Google Scholar] [CrossRef]
- Wilson, W.L.; Bottiglieri, N.G. Phase I studies with pyrazole. Cancer Chemother. Rep. 1962, 21, 137–141. [Google Scholar]
- Leung, C.-F.; Yiu, D.T.Y.; Wong, W.-T.; Peng, S.-M.; Lau, T.-C. Synthesis and reactivity of osmium (VI) nitrido complexes containing pyridine-carboxylato ligands. Inorg. Chim. Acta 2009, 362, 3576–3582. [Google Scholar] [CrossRef]
- Che, C.-M.; Lau, T.-C. Ruthenium and Osmium: High Oxidation States. In Comprehensive Coordination Chemistry II; McCleverty, J.A., Meyer, T.J., Eds.; Pergamon: Oxford, UK, 2003; pp. 733–847. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Liang, X.J.; Meng, H.; Wang, Y.; He, H.; Meng, J.; Lu, J.; Wang, P.C.; Zhao, Y.; Gao, X.; Sun, B.; et al. Metallofullerene nanoparticles circumvent tumor resistance to cisplatin by reactivating endocytosis. Proc. Natl. Acad. Sci. USA 2010, 107, 7449–7454. [Google Scholar] [CrossRef] [PubMed]
- Olson, K.A.; Schell, J.C.; Rutter, J. Pyruvate and Metabolic Flexibility: Illuminating a Path Toward Selective Cancer Therapies. Trends Biochem. Sci. 2016, 41, 219–230. [Google Scholar] [CrossRef]
- Gray, L.R.; Tompkins, S.C.; Taylor, E.B. Regulation of pyruvate metabolism and human disease. Cell. Mol. Life Sci. CMLS 2014, 71, 2577–2604. [Google Scholar] [CrossRef] [PubMed]
- Roos, W.P.; Kaina, B. DNA damage-induced cell death by apoptosis. Trends Mol. Med. 2006, 12, 440–450. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Wu, Y.; Chen, Q.; Cao, J.; Hu, K.; Tang, J.; Sang, Y.; Lai, F.; Wang, L.; Zhang, R.; et al. hSSB1 regulates both the stability and the transcriptional activity of p53. Cell Res. 2013, 23, 423–435. [Google Scholar] [CrossRef]
- Guichard, C.; Amaddeo, G.; Imbeaud, S.; Ladeiro, Y.; Pelletier, L.; Maad, I.B.; Calderaro, J.; Bioulac-Sage, P.; Letexier, M.; Degos, F.; et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat. Genet. 2012, 44, 694–698. [Google Scholar] [CrossRef]
- Wong, N.; Yeo, W.; Wong, W.L.; Wong, N.L.; Chan, K.Y.; Mo, F.K.; Koh, J.; Chan, S.L.; Chan, A.T.; Lai, P.B.; et al. TOP2A overexpression in hepatocellular carcinoma correlates with early age onset, shorter patients survival and chemoresistance. Int. J. Cancer 2009, 124, 644–652. [Google Scholar] [CrossRef]
- Wang, J.; Chan, J.Y.; Fong, C.C.; Tzang, C.H.; Fung, K.P.; Yang, M. Transcriptional analysis of doxorubicin-induced cytotoxicity and resistance in human hepatocellular carcinoma cell lines. Liver Int. Off. J. Int. Assoc. Study Liver 2009, 29, 1338–1347. [Google Scholar] [CrossRef] [PubMed]
- Merrill, C.L.; Ni, H.; Yoon, L.W.; Tirmenstein, M.A.; Narayanan, P.; Benavides, G.R.; Easton, M.J.; Creech, D.R.; Hu, C.X.; McFarland, D.C.; et al. Etomoxir-induced oxidative stress in HepG2 cells detected by differential gene expression is confirmed biochemically. Toxicol. Sci. Off. J. Soc. Toxicol. 2002, 68, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Chu, G. Xeroderma pigmentosum complementation group E and UV-damaged DNA-binding protein. DNA Repair 2002, 1, 601–616. [Google Scholar] [CrossRef]
- Qiao, S.; Guo, W.; Liao, L.; Wang, L.; Wang, Z.; Zhang, R.; Xu, D.; Zhang, Y.; Pan, Y.; Wang, Z.; et al. DDB2 is involved in ubiquitination and degradation of PAQR3 and regulates tumorigenesis of gastric cancer cells. Biochem. J. 2015, 469, 469–480. [Google Scholar] [CrossRef]
- Lai, T.C.; Hu, M.C. Regulation of liver receptor homologue-1 by DDB2 E3 ligase activity is critical for hepatic glucose metabolism. Sci. Rep. 2019, 9, 5304. [Google Scholar] [CrossRef] [PubMed]
- Anderson, D.D.; Quintero, C.M.; Stover, P.J. Identification of a de novo thymidylate biosynthesis pathway in mammalian mitochondria. Proc. Natl. Acad. Sci. USA 2011, 108, 15163–15168. [Google Scholar] [CrossRef]
- Santhekadur, P.K.; Rajasekaran, D.; Siddiq, A.; Gredler, R.; Chen, D.; Schaus, S.E.; Hansen, U.; Fisher, P.B.; Sarkar, D. The transcription factor LSF: A novel oncogene for hepatocellular carcinoma. Am. J. Cancer Res. 2012, 2, 269–285. [Google Scholar]
- Xu, Y.; Zhang, C.; Liang, H.; Hu, S.; Li, P.; Liu, L.; Duan, X.; Chen, C.; Zhang, Y.; Dai, P. Dishevelled 1, a pivotal positive regulator of the Wnt signalling pathway, mediates 5-fluorouracil resistance in HepG2 cells. Artif. Cells Nanomed. Biotechnol. 2018, 46, 192–200. [Google Scholar] [CrossRef]
- Scartozzi, M.; Maccaroni, E.; Giampieri, R.; Pistelli, M.; Bittoni, A.; Del Prete, M.; Berardi, R.; Cascinu, S. 5-Fluorouracil pharmacogenomics: Still rocking after all these years? Pharmacogenomics 2011, 12, 251–265. [Google Scholar] [CrossRef]
- Moinova, H.R.; Mulcahy, R.T. An electrophile responsive element (EpRE) regulates beta-naphthoflavone induction of the human gamma-glutamylcysteine synthetase regulatory subunit gene. Constitutive expression is mediated by an adjacent AP-1 site. J. Biol. Chem. 1998, 273, 14683–14689. [Google Scholar] [CrossRef]
- Zipper, L.M.; Mulcahy, R.T. Erk activation is required for Nrf2 nuclear localization during pyrrolidine dithiocarbamate induction of glutamate cysteine ligase modulatory gene expression in HepG2 cells. Toxicol. Sci. Off. J. Soc. Toxicol. 2003, 73, 124–134. [Google Scholar] [CrossRef]
- Okuda, S.; Nishiyama, N.; Saito, H.; Katsuki, H. Hydrogen peroxide-mediated neuronal cell death induced by an endogenous neurotoxin, 3-hydroxykynurenine. Proc. Natl. Acad. Sci. USA 1996, 93, 12553–12558. [Google Scholar] [CrossRef] [PubMed]
- Aboud, O.A.; Weiss, R.H. New opportunities from the cancer metabolome. Clin. Chem. 2013, 59, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Lauvrak, S.U.; Munthe, E.; Kresse, S.H.; Stratford, E.W.; Namlos, H.M.; Meza-Zepeda, L.A.; Myklebost, O. Functional characterisation of osteosarcoma cell lines and identification of mRNAs and miRNAs associated with aggressive cancer phenotypes. Br. J. Cancer 2013, 109, 2228–2236. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Hu, M.; Zhao, R.; Li, P.; Li, M. Dihydromyricetin suppresses the proliferation of hepatocellular carcinoma cells by inducing G2/M arrest through the Chk1/Chk2/Cdc25C pathway. Oncol. Rep. 2013, 30, 2467–2475. [Google Scholar] [CrossRef]
- Santos, S.D.; Wollman, R.; Meyer, T.; Ferrell, J.E., Jr. Spatial positive feedback at the onset of mitosis. Cell 2012, 149, 1500–1513. [Google Scholar] [CrossRef]
- Jung, Y.; Lippard, S.J. Direct cellular responses to platinum-induced DNA damage. Chem. Rev. 2007, 107, 1387–1407. [Google Scholar] [CrossRef]
- Betanzos-Lara, S.; Liu, Z.; Habtemariam, A.; Pizarro, A.M.; Qamar, B.; Sadler, P.J. Organometallic ruthenium and iridium transfer-hydrogenation catalysts using coenzyme NADH as a cofactor. Angew. Chem. 2012, 51, 3897–3900. [Google Scholar] [CrossRef]
- Pierroz, V.; Joshi, T.; Leonidova, A.; Mari, C.; Schur, J.; Ott, I.; Spiccia, L.; Ferrari, S.; Gasser, G. Molecular and cellular characterization of the biological effects of ruthenium(II) complexes incorporating 2-pyridyl-2-pyrimidine-4-carboxylic acid. J. Am. Chem. Soc. 2012, 134, 20376–20387. [Google Scholar] [CrossRef]
- Fischer, U.; Schulze-Osthoff, K. Apoptosis-based therapies and drug targets. Cell Death Differ. 2005, 12 (Suppl. 1), 942–961. [Google Scholar] [CrossRef]
- Enari, M.; Talanian, R.V.; Wong, W.W.; Nagata, S. Sequential activation of ICE-like and CPP32-like proteases during Fas-mediated apoptosis. Nature 1996, 380, 723–726. [Google Scholar] [CrossRef]
- Salvesen, G.S. Caspases: Opening the boxes and interpreting the arrows. Cell Death Differ. 2002, 9, 3–5. [Google Scholar] [CrossRef] [PubMed]
- Smiley, S.T.; Reers, M.; Mottola-Hartshorn, C.; Lin, M.; Chen, A.; Smith, T.W.; Steele, G.D., Jr.; Chen, L.B. Intracellular heterogeneity in mitochondrial membrane potentials revealed by a J-aggregate-forming lipophilic cation JC-1. Proc. Natl. Acad. Sci. USA 1991, 88, 3671–3675. [Google Scholar] [CrossRef]
- Sheldrick, G.M. SHELXT—Integrated space-group and crystal-structure determination. Acta Cryst. Sect. A Found. Adv. 2015, 71, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. Sect. C Struct. Chem. 2015, 71, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Dolomanov, O.V.; Bourhis, L.J.; Gildea, R.J.; Howard, J.A.K.; Puschmann, H. OLEX2: A complete structure solution, refinement and analysis program. J. Appl. Crystallogr. 2009, 42, 339–341. [Google Scholar] [CrossRef]
- Rappsilber, J.; Mann, M.; Ishihama, Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2007, 2, 1896–1906. [Google Scholar] [CrossRef]
- Rappsilber, J.; Ishihama, Y.; Mann, M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 2003, 75, 663–670. [Google Scholar] [CrossRef]
- Huang, C.; Su, T.; Xue, Y.; Cheng, C.; Lay, F.D.; McKee, R.A.; Li, M.; Vashisht, A.; Wohlschlegel, J.; Novitch, B.G.; et al. Cbx3 maintains lineage specificity during neural differentiation. Genes Dev. 2017, 31, 241–246. [Google Scholar] [CrossRef]


| Cell Lines | Na[OsⅥ(N)(tpm)2] | Cisplatin |
|---|---|---|
| HeLa | 10.8 ± 1.0 | 13.4 ± 0.7 |
| A2780 | 5.6 ± 0.4 | 15.9 ± 0.9 |
| MDAMB231 | 9.1 ± 0.6 | 11.3 ± 1.6 |
| HepG2 | 6.5 ± 1.1 | 4.9 ± 0.3 |
| HepG2-stem | 8.2 ± 0.8 | 3.3 ± 0.2 |
| LO-2 | 28.5 ± 5.1 | 5.0 ± 0.3 |
| A549 | 11.5 ± 0.8 | 12.3 ± 1.5 |
| A549CIS | 15.3 ± 2.0 | 149.6 ± 10.6 |
| RI 1 | 1.3 | 12.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, C.; Huang, W.; Ji, P.; Song, F.; Liu, T.; Li, M.; Guo, H.; Huang, Y.; Yu, C.; Wang, C.; et al. A Pyrazolate Osmium(VI) Nitride Exhibits Anticancer Activity through Modulating Protein Homeostasis in HepG2 Cells. Int. J. Mol. Sci. 2022, 23, 12779. https://doi.org/10.3390/ijms232112779
Huang C, Huang W, Ji P, Song F, Liu T, Li M, Guo H, Huang Y, Yu C, Wang C, et al. A Pyrazolate Osmium(VI) Nitride Exhibits Anticancer Activity through Modulating Protein Homeostasis in HepG2 Cells. International Journal of Molecular Sciences. 2022; 23(21):12779. https://doi.org/10.3390/ijms232112779
Chicago/Turabian StyleHuang, Chengyang, Wanqiong Huang, Pengchao Ji, Fuling Song, Tao Liu, Meiyang Li, Hongzhi Guo, Yongliang Huang, Cuicui Yu, Chuanxian Wang, and et al. 2022. "A Pyrazolate Osmium(VI) Nitride Exhibits Anticancer Activity through Modulating Protein Homeostasis in HepG2 Cells" International Journal of Molecular Sciences 23, no. 21: 12779. https://doi.org/10.3390/ijms232112779
APA StyleHuang, C., Huang, W., Ji, P., Song, F., Liu, T., Li, M., Guo, H., Huang, Y., Yu, C., Wang, C., & Ni, W. (2022). A Pyrazolate Osmium(VI) Nitride Exhibits Anticancer Activity through Modulating Protein Homeostasis in HepG2 Cells. International Journal of Molecular Sciences, 23(21), 12779. https://doi.org/10.3390/ijms232112779