GsRSS3L, a Candidate Gene Underlying Soybean Resistance to Seedcoat Mottling Derived from Wild Soybean (Glycine soja Sieb. and Zucc)
Abstract
1. Introduction
2. Results
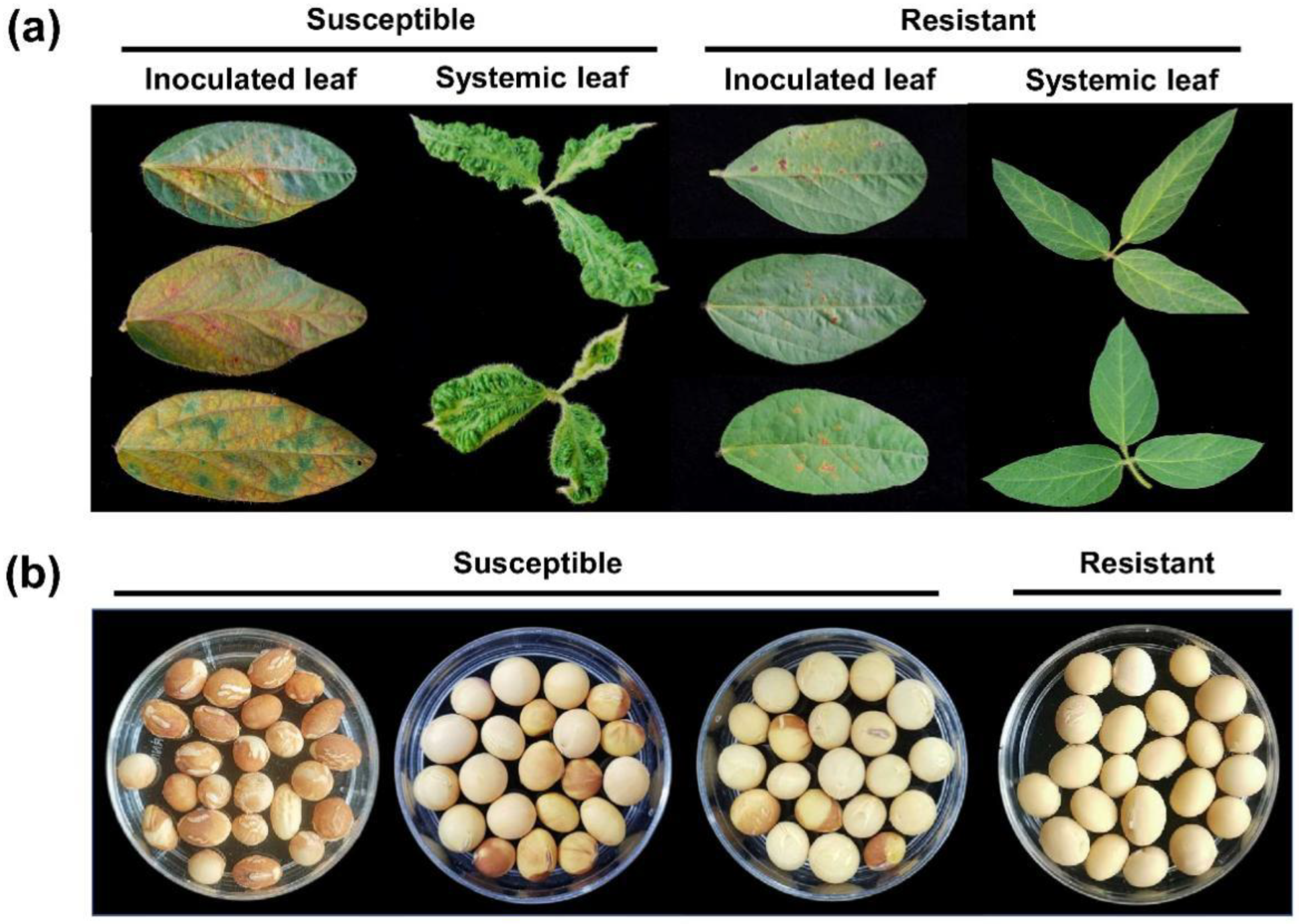
2.1. Response of CSSL Population to SMV
2.2. Identification of Genomic Regions Associated with Resistance to Seed Coat Mottling
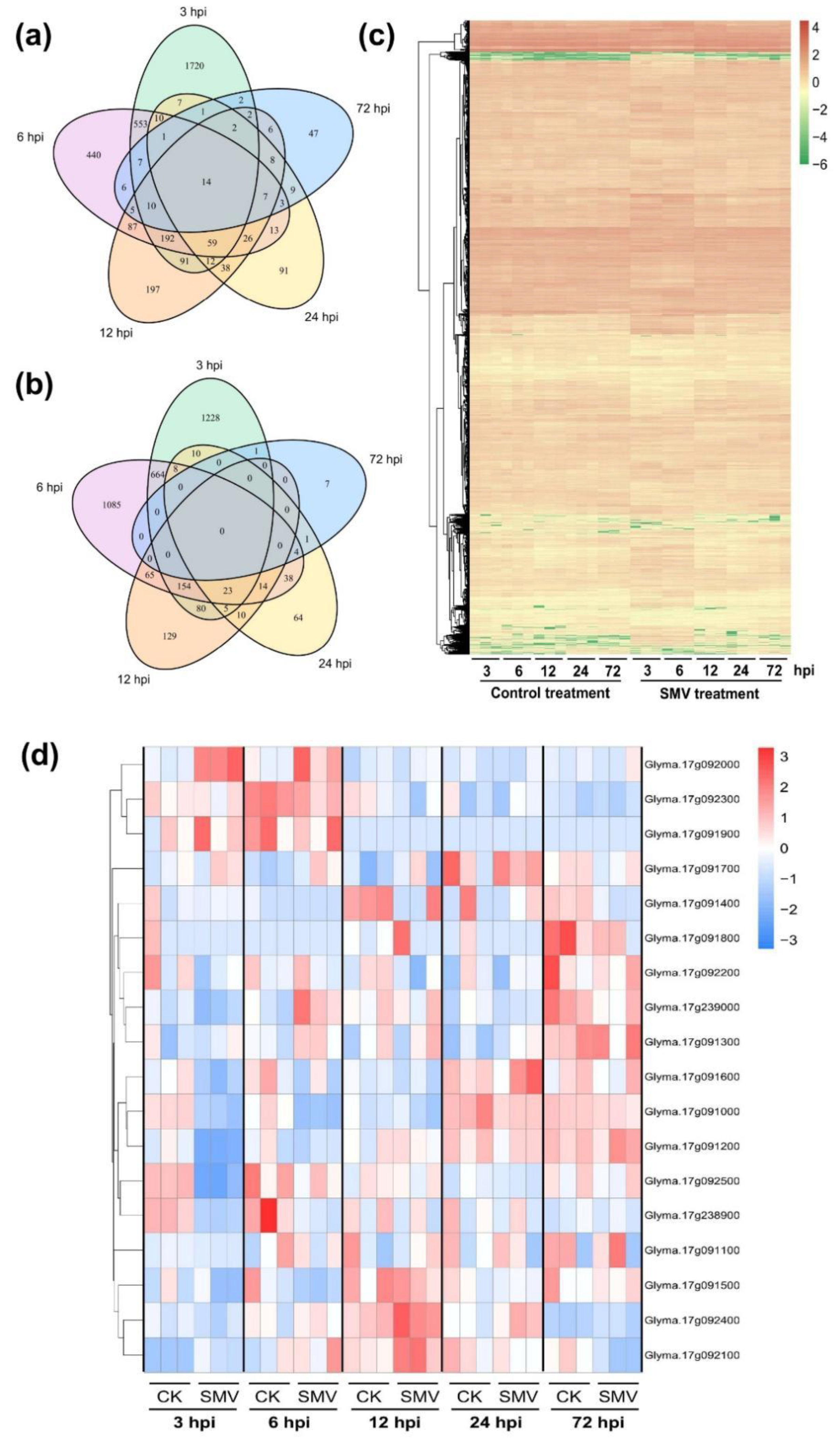
2.3. RNA-Seq Analysis of SN14 Response to SMV Infection
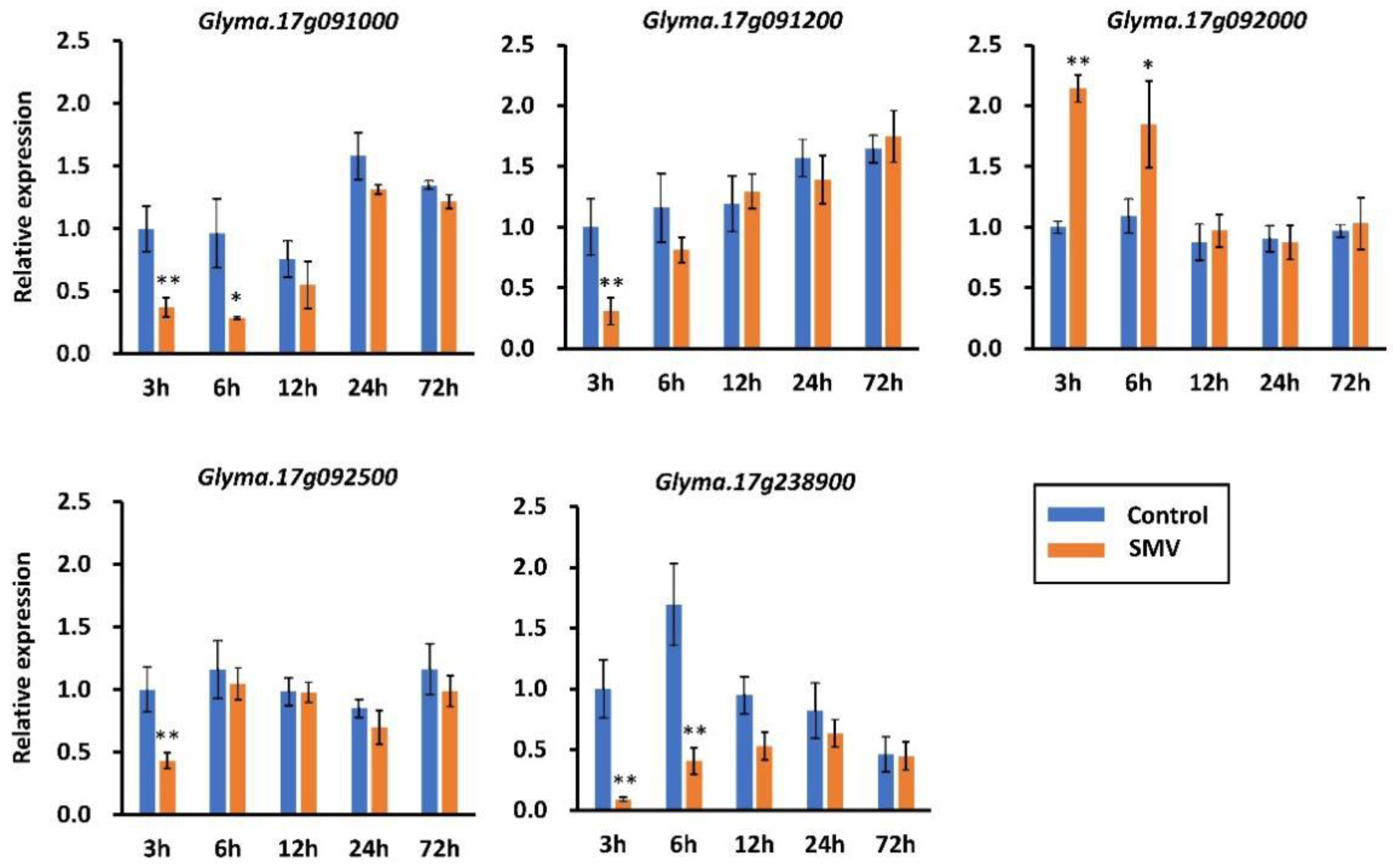
2.4. Prediction and qRT-PCR Validation of Candidate Genes
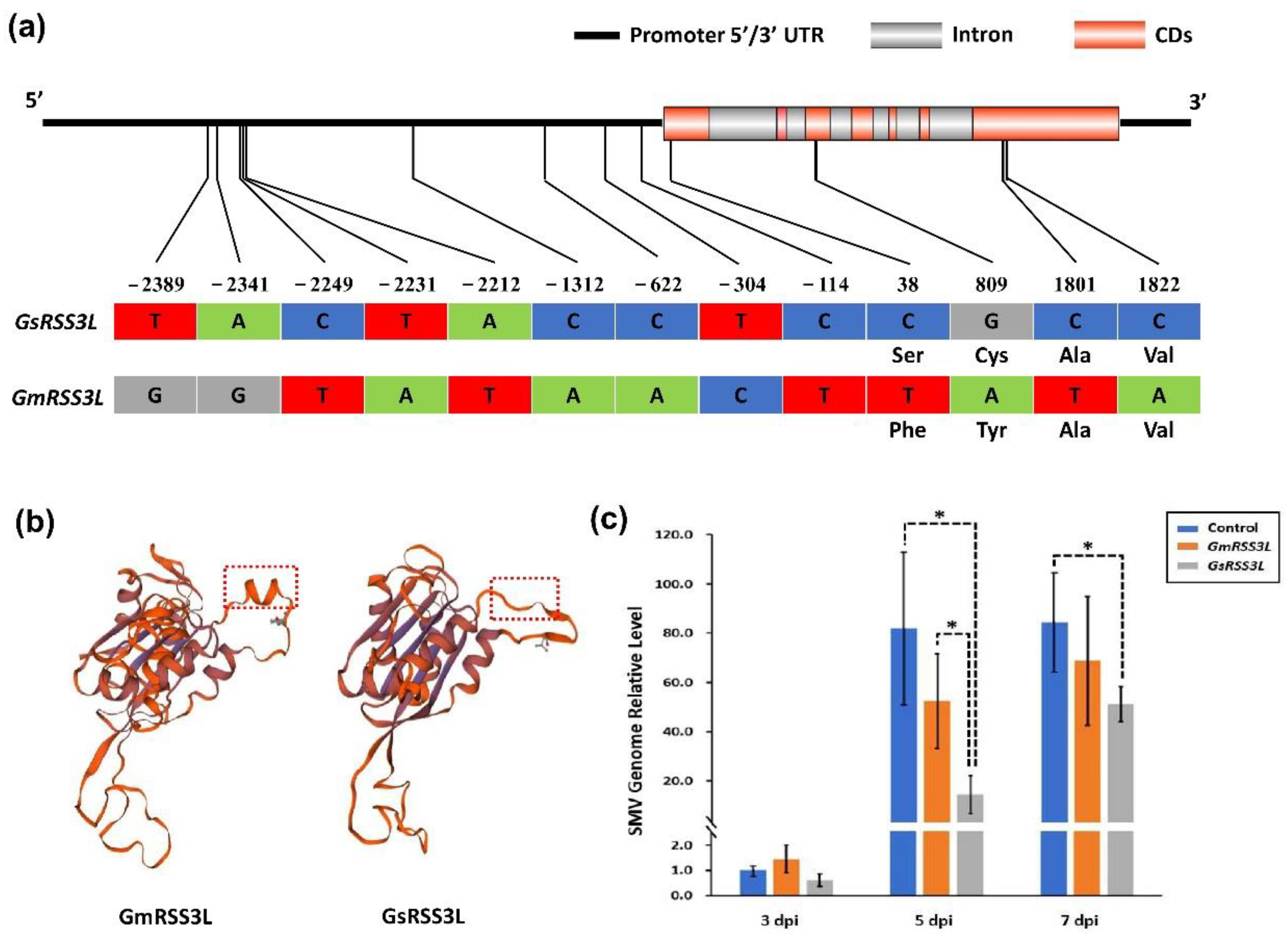
2.5. Agrobacterium-Mediated Transient Expression of Candidate Genes
3. Discussion
4. Materials and Methods
4.1. Genetic Materials
4.2. Virus Culture, Inoculation, and RT-PCR Detection
4.3. QTL Detection
4.4. Transcriptome Analysis
4.5. Prediction and qRT-PCR Validation of Candidate Genes
4.6. Agrobacterium-Mediated Transient Expression of Candidate Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hoeck, J.A.; Fehr, W.R.; Shoemaker, R.C.; Welke, G.A.; Johnson, S.L.; Cianzio, S.R. Molecular marker analysis of seed size in soybean. Crop Sci. 2003, 43, 68–74. [Google Scholar] [CrossRef]
- Widyasari, K.; Alazem, M.; Kim, K. Soybean resistance to soybean mosaic virus. Plants 2020, 9, 219. [Google Scholar] [CrossRef] [PubMed]
- Hill, J.H.; Whitham, S.A. Control of virus disease in soybean. Adv. Virus Res. 2014, 90, 355–390. [Google Scholar] [PubMed]
- Ross, J.P. Effect of aphid-transmitted soybean mosaic virus on yields of closely related resistant and susceptible soybean lines. Crop Sci. 1997, 17, 869–872. [Google Scholar] [CrossRef]
- Rui, R.; Liu, S.; Karthikeyan, A.; Wang, T.; Niu, H.; Yin, J.; Yang, Y.; Wang, L.; Yang, Q.; Zhi, H.; et al. Fine-mapping and identification of a novel locus Rsc15 underlying soybean resistance to Soybean mosaic virus. Theor. Appl. Genet. 2017, 130, 2395–2410. [Google Scholar] [CrossRef]
- Hajimorad, M.R.; Domier, L.L.; Tolin, S.A.; Whitham, S.A.; Saghai Maroof, M.A. Soybean mosaic virus: A successful potyvirus with a wide distribution but restricted natural host range. Mol. Plant Pathol. 2018, 19, 1563–1579. [Google Scholar] [CrossRef]
- Li, K.; Yang, Q.H.; Zhi, H.J.; Gai, J.Y. Identification and distribution of Soybean mosaic virus strains in Southern China. Plant Dis. 2010, 94, 351–357. [Google Scholar] [CrossRef]
- Gao, L.; Wu, Y.; An, J.; Huang, W.; Liu, X.; Xue, Y.; Luan, X.; Lin, F.; Sun, L. Pathogenicity and genome-wide sequence analysis reveals relationships between soybean mosaic virus strains. Arch. Virol. 2022, 167, 517–529. [Google Scholar] [CrossRef]
- Koning, G.; TeKrony, D.M.; Ghabrial, S.A. Soybean seedcoat mottling: Association with Soybean mosaic virus and Phomopsis spp. seed infection. Plant Dis. 2003, 87, 413–417. [Google Scholar] [CrossRef][Green Version]
- Senda, M. Patterning of virus-infected Glycine max seedcoat is associated with suppression of endogenous silencing of chalcone synthase genes. Plant Cell 2004, 16, 807–818. [Google Scholar] [CrossRef]
- Karthikeyan, A.; Li, K.; Jiang, H.; Ren, R.; Li, C.; Zhi, H.; Chen, S.; Gai, J. Inheritance, fine-mapping, and candidate gene analyses of resistance to soybean mosaic virus strain SC5 in soybean. Mol. Genet. Genom. 2017, 292, 811–822. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Li, W.; Zhang, L.; Liu, C.Y.; Zhu, X.S.; Chen, Q.S.; Hu, G. Genetic analysis and molecular mapping of two SMV-resistance traits in soybean: Adult-plant resistance and resistance to seedcoat mottling. J. Integr. Agric. 2010, 9, 11–18. [Google Scholar] [CrossRef]
- Cooper, R.L. A major gene for resistance to seedcoat mottling in soybean. Crop Sci. 1966, 6, 290–292. [Google Scholar] [CrossRef]
- Hu, G.; Gao, F.; Wu, Z. Studies for linkage of resistance to seedcoat mottling on soybean. Acta Agron. Sin. 1996, 22, 555–559. [Google Scholar]
- Capistrano-Gossmann, G.G.; Ries, D.; Holtgrawe, D.; Minoche, A.; Kraft, T.; Frerichmann, S.L.M.; Soerensen, T.R.; Dohm, J.C.; Gonzalez, I.; Schilhabel, M.; et al. Crop wild relative populations of Beta vulgaris allow direct mapping of agronomically important genes. Nat. Commun. 2017, 8, 15708. [Google Scholar] [CrossRef]
- Zhou, Z.; Jiang, Y.; Wang, Z.; Gou, Z.; Lyu, J.; Li, W.; Yu, Y.; Shu, L.; Zhao, Y.; Ma, Y.; et al. Resequencing 302 wild and cultivated accessions identifies genes related to domestication and improvement in soybean. Nat. Biotechnol. 2015, 33, 408–414. [Google Scholar] [CrossRef]
- Xin, D.; Qi, Z.; Jiang, H.; Hu, Z.; Zhu, R.; Hu, J.; Han, H.; Hu, G.; Liu, C.; Chen, Q. QTL location and epistatic effect analysis of 100-seed weight using wild soybean (Glycine soja Sieb. & Zucc.) chromosome segment substitution lines. PLoS ONE 2016, 11, e0149380. [Google Scholar] [CrossRef]
- Toda, Y.; Yoshida, M.; Hattori, T.; Takeda, S. RICE SALT SENSITIVE3 binding to bHLH and JAZ factors mediates control of cell wall plasticity in the root apex. Plant Signal. Behav. 2013, 8, e26256. [Google Scholar] [CrossRef]
- Ma, F.-F.; Wu, X.-Y.; Chen, Y.-X.; Liu, Y.-N.; Shao, Z.-Q.; Wu, P.; Wu, M.; Liu, C.-C.; Wu, W.-P.; Yang, J.-Y.; et al. Fine mapping of the Rsv1-h gene in the soybean cultivar Suweon 97 that confers resistance to two Chinese strains of the soybean mosaic virus. Theor. Appl. Genet. 2016, 129, 2227–2236. [Google Scholar] [CrossRef]
- Tran, P.T.; Widyasari, K.; Seo, J.K.; Kim, K.H. Isolation and validation of a candidate Rsv3 gene from a soybean genotype that confers strain-specific resistance to soybean mosaic virus. Virology 2018, 53, 153–159. [Google Scholar] [CrossRef]
- Ishibashi, K.; Saruta, M.; Shimizu, T.; Shu, M.; Anai, T.; Komatsu, K.; Yamada, N.; Katayose, Y.; Ishikawa, M.; Ishimoto, M.; et al. Soybean antiviral immunity conferred by dsRNase targets the viral replication complex. Nat. Commun. 2019, 10, 4033. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.; Chen, P.; Gergerich, R. Characterization of resistance to Soybean mosaic virus in diverse soybean germplasm. Crop Sci. 2005, 45, 2503–2509. [Google Scholar] [CrossRef]
- Li, C.; Adhimoolam, K.; Yuan, Y.; Yin, J.; Ren, R.; Yang, Y.; Zhi, H. Identification of candidate genes for resistance to Soybean mosaic virus strain SC3 by using fine mapping and transcriptome analyses. Crop Pasture Sci. 2017, 68, 156–166. [Google Scholar] [CrossRef]
- Fu, S.; Zhan, Y.; Zhi, H.; Gai, J.; Yu, D. Mapping of SMV resistance gene Rsc-7 by SSR markers in soybean. Genetica 2006, 128, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Ma, Y.; Yang, Y.; Liu, N.; Li, C.; Song, Y.; Zhi, H. Fine mapping and analyses of RSC8 resistance candidate genes to soybean mosaic virus in soybean. Theor. Appl. Genet. 2011, 122, 555–565. [Google Scholar] [CrossRef]
- Ma, Y.; Li, H.; Wang, D.; Liu, N.; Zhi, H. Molecular mapping and marker assisted selection of soybean mosaic virus resistance gene RSC12 in soybean. Legume Genom. Genet. 2010, 1, 41–46. [Google Scholar]
- Li, K.; Ren, R.; Adhimoolam, K.; Gao, L.; Yuan, Y.; Liu, Z.; Zhong, Y.; Zhi, H. Genetic analysis and identification of two soybean mosaic virus resistance genes in soybean [Glycine max (L.) Merr]. Plant Breed. 2015, 134, 684–695. [Google Scholar] [CrossRef]
- Karthikeyan, A.; Li, K.; Li, C.; Yin, J.; Li, N.; Yang, Y.; Song, Y.; Ren, R.; Zhi, H.; Gai, J.; et al. Fine-mapping and identifying candidate genes conferring resistance to Soybean mosaic virus strain SC20 in soybean. Theor. Appl. Genet. 2018, 131, 461–476. [Google Scholar] [CrossRef]
- Tuyen, D.D.; Lal, S.K.; Xu, D.H. Identification of a major QTL allele from wild soybean (Glycine soja Sieb. & Zucc.) for increasing alkaline salt tolerance in soybean. Theor. Appl. Genet. 2010, 121, 229–236. [Google Scholar] [CrossRef]
- Cao, P.; Zhao, Y.; Wu, F.; Xin, D.; Liu, C.; Wu, X.; Lv, J.; Chen, Q.; Qi, Z. Multi-omics techniques for soybean molecular breeding. Int. J. Mol. Sci. 2022, 23, 4994. [Google Scholar] [CrossRef]
- Guo, X.; Jiang, J.; Liu, Y.; Yu, L.; Chang, R.; Guan, R.; Qiu, L. Identification of a novel salt tolerance-related locus in wild soybean (Glycine soja Sieb. & Zucc.). Front. Plant Sci. 2021, 12, 791175. [Google Scholar] [CrossRef] [PubMed]
- Toda, Y.; Tanaka, M.; Ogawa, D.; Kurata, K.; Kurotani, K.; Habu, Y.; Ando, T.; Sugimoto, K.; Mitsuda, N.; Katoh, E.; et al. RICE SALT SENSITIVE3 forms a ternary complex with JAZ and Class-C bHLH factors and regulates jasmonate-induced gene expression and root cell elongation. Plant Cell 2013, 25, 1709–1725. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Ye, J. Manipulation of jasmonate signaling by plant viruses and their insect vectors. Viruses 2020, 12, 148. [Google Scholar] [CrossRef] [PubMed]
- Lorenzo, O.; Solano, R. Molecular players regulating the jasmonate signalling network. Curr. Opin. Plant Biol. 2005, 8, 532–540. [Google Scholar] [CrossRef]
- Kazan, K.; Manners, J.M. Jasmonate signaling: Toward and integrated view. Plant Physiol. 2008, 146, 1459–1468. [Google Scholar] [CrossRef]
- Zhao, S.; Li, Y. Current understanding of the interplays between host hormones and plant viral infections. PLoS Pathog. 2021, 17, e1009242. [Google Scholar] [CrossRef]
- Laurie-Berry, N.; Joardar, V.; Street, I.H.; Kunkel, B.N. The Arabidopsis thaliana JASMONATE INSENSITIVE 1 gene is required for suppression of salicylic acid-dependent defense during infection by Pseudomonas syringae. Mol. Plant Microbe Int. 2006, 19, 789–800. [Google Scholar] [CrossRef]
- He, Y.; Zhang, H.; Sun, Z.; Li, J.; Hong, G.; Zhu, Q.; Zhou, X.; MacFarlane, S.; Yan, F.; Chen, J. Jasmonic acid-mediated defense suppresses brassinosteroid-mediated susceptibility to Rice black streaked dwarf virus infection in rice. New Phytol. 2017, 214, 388–399. [Google Scholar] [CrossRef]
- Yang, Z.; Huang, Y.; Yang, J.; Yao, S.; Zhao, K.; Wang, D.; Qin, Q.; Bian, Z.; Li, Y.; Lan, Y.; et al. Jasmonate signaling enhances RNA silencing and antiviral defense in rice. Cell Host Microbe 2020, 28, 89–103. [Google Scholar] [CrossRef]
- Yamada, T.; Takagi, K.; Ishimoto, M. Recent advances in soybean transformation and their application to molecular breeding and genomic analysis. Breed. Sci. 2012, 61, 480–494. [Google Scholar] [CrossRef]
- Gao, L.; Zhai, R.; Zhong, Y.K.; Karthikeyan, A.; Ren, R.; Zhang, K.; Li, K.; Zhi, H.J. Screening isolates of Soybean mosaic virus for infectivity in a model plant, Nicotiana benthamiana. Plant Dis. 2015, 99, 442–446. [Google Scholar] [CrossRef] [PubMed]
- Goodin, M.M.; Zaitlin, D.; Naidu, R.A.; Lommel, S.A. Nicotiana benthamiana: Its history and future as a model for plant-pathology interactions. Mol. Plant Microbe Int. 2008, 21, 1015–1026. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarty, R.; Banerjee, R.; Chung, S.M.; Farman, M.; Citovsky, V.; Hogenhout, S.A.; Tzfira, T.; Goodin, M.M. pSITE vectors for stable integration or transient expression of autofluorescent protein fusions in plants: Probing Nicotiana benthamiana-virus interactions. Mol. Plant Microbe Int. 2007, 20, 740–750. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Xi, D.; Yuan, S.; Xu, F.; Zhang, D.; Lin, H. Salicylic acid and jasmonic acid are essential for systemic resistance against Tobacco mosaic virus in Nicotiana benthamiana. Mol. Plant Microbe Int. 2014, 27, 567–577. [Google Scholar] [CrossRef] [PubMed]
- Bao, W.; Yan, T.; Deng, X.; Wuriyanghan, H. Synthesis of full-length cDNA infectious clones of Soybean mosaic virus and functional identification of a key amino acid in the silencing suppressor Hc-Pro. Viruses 2020, 12, 886. [Google Scholar] [CrossRef] [PubMed]
- Hyten, D.L.; Choi, I.; Song, Q.; Specht, J.E.; Carter, T.E.; Shoemaker, R.C.; Hwang, E.; Matukumalli, L.K.; Cregan, P.B. A high density integrated genetic linkage map of soybean and the development of a 1536 universal soy linkage panel for quantitative trait locus mapping. Crop Sci. 2010, 50, 960–968. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550–570. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]

| Block | Chromosome | Physical location 1 | LOD 2 | PVE (%) 3 | Add 4 | |
|---|---|---|---|---|---|---|
| Start | End | |||||
| block9566 | Chr.17 | 7,086,191 | 7,227,595 | 2.50 | 4.57 | −0.02 |
| block9910 | Chr.17 | 39,435,988 | 39,459,788 | 7.90 | 14.65 | −0.05 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, S.; Wang, J.; Yang, X.; Zhang, X.; Xin, X.; Liu, C.; Zou, J.; Cheng, X.; Zhang, N.; Hu, Y.; et al. GsRSS3L, a Candidate Gene Underlying Soybean Resistance to Seedcoat Mottling Derived from Wild Soybean (Glycine soja Sieb. and Zucc). Int. J. Mol. Sci. 2022, 23, 7577. https://doi.org/10.3390/ijms23147577
Song S, Wang J, Yang X, Zhang X, Xin X, Liu C, Zou J, Cheng X, Zhang N, Hu Y, et al. GsRSS3L, a Candidate Gene Underlying Soybean Resistance to Seedcoat Mottling Derived from Wild Soybean (Glycine soja Sieb. and Zucc). International Journal of Molecular Sciences. 2022; 23(14):7577. https://doi.org/10.3390/ijms23147577
Chicago/Turabian StyleSong, Shuang, Jing Wang, Xingqi Yang, Xuan Zhang, Xiuli Xin, Chunyan Liu, Jianan Zou, Xiaofei Cheng, Ning Zhang, Yuxi Hu, and et al. 2022. "GsRSS3L, a Candidate Gene Underlying Soybean Resistance to Seedcoat Mottling Derived from Wild Soybean (Glycine soja Sieb. and Zucc)" International Journal of Molecular Sciences 23, no. 14: 7577. https://doi.org/10.3390/ijms23147577
APA StyleSong, S., Wang, J., Yang, X., Zhang, X., Xin, X., Liu, C., Zou, J., Cheng, X., Zhang, N., Hu, Y., Wang, J., Chen, Q., & Xin, D. (2022). GsRSS3L, a Candidate Gene Underlying Soybean Resistance to Seedcoat Mottling Derived from Wild Soybean (Glycine soja Sieb. and Zucc). International Journal of Molecular Sciences, 23(14), 7577. https://doi.org/10.3390/ijms23147577









