Abstract
Nanoparticles (NPs) enhance soybean growth; however, their precise mechanism is not clearly understood. To develop a more effective method using NPs for the enhancement of soybean growth, fiber crosslinked with zinc oxide (ZnO) NPs was prepared. The solution of ZnO NPs with 200 nm promoted soybean growth at the concentration of 10 ppm, while fibers crosslinked with ZnO NPs promoted growth at a 1 ppm concentration. Soybeans grown on fiber cross-linked with ZnO NPs had higher Zn content in their roots than those grown in ZnO NPs solution. To study the positive mechanism of fiber crosslinked with ZnO NPs on soybean growth, a proteomic technique was used. Proteins categorized in photosynthesis and secondary metabolism accumulated more in soybeans grown on fiber crosslinked with ZnO NPs than in those grown in ZnO NPs solution. Furthermore, significantly accumulated proteins, which were NADPH oxidoreductase and tubulins, were confirmed using immunoblot analysis. The abundance of NADPH oxidoreductase increased in soybean by ZnO NPs application. These results suggest that fiber crosslinked with ZnO NPs enhances soybean growth through the increase of photosynthesis and secondary metabolism. Additionally, the accumulation of NADPH oxidoreductase might relate to the effect of auxin with fiber crosslinked with ZnO NPs on soybean growth.
1. Introduction
Metal oxide nanoparticles (NPs) are added to different products due to their unique properties such as catalytic capacity, carcinogenic activity, and drug delivery capabilities [1]. On the other hand, zinc (Zn) is an essential micronutrient for different kinds of living organisms, including plants. Zn deficiency in crops not only restricts their productivity but also influences human health [2]. Zn oxide (ZnO) NPs are extensively utilized due to their optoelectric characteristics [3]; and in products such as plastics, cement, rubber, batteries, and foods [4]. Extensive utilization of ZnO NPs in different products intensified their discharge into the environment. ZnO NPs are added to cosmetic and sunscreen products due to their efficient ultraviolet absorption and reflective properties [5]. Although the incorporation of NPs into the environment creates serious concerns, ZnO NPs are considered to be safe compared with other kinds of NPs.
ZnO NPs suppressed the growth of Salicornia persica [6], Arabidopsis thaliana [7], tomato, and wheat [8]. ZnO NPs decreased chlorophyll content, which leads to reduced photosynthesis [9]. Although ZnO NPs caused negative effects on plants, they increased the fresh/dry weight of leaves/roots in coffee [10], the growth/biomass of lettuce [11], the germination/root growth of maize/cabbage [12], as well as the plant growth of wheat [13] and tobacco [14]. These findings indicate that ZnO NPs have both negative and positive effects on plant growth. ZnO NPs and bulk ZnO treatments in a substance type- and method-dependent manner improved soybean growth and yield [15]. Furthermore, ZnO NPs influenced seed yield and antioxidant-defense system in soil-grown soybean [16]. These reports indicated that the application of ZnO NPs improved the growth and productivity of soybean.
The application of ZnO NPs at low concentrations provides a safe low-risk approach to improving growth, physiology, immunity, and productivity in crops. In this study, to develop a more effective method using NPs for the enhancement of soybean growth, fibers crosslinked with a low concentration of ZnO NPs were prepared. Additionally, ZnO NPs mitigated the risk associated with diverse abiotic [17,18,19,20] and biotic [21] stresses. These reports indicated that ZnO NPs might potentially confer stress tolerance in plants. Because soybean is sensitive against flooding stress [22], it was analyzed whether soybean could survive against flooding stress on fiber crosslinked with ZnO NPs in this study. To investigate the outcome of fiber crosslinked with ZnO NPs on soybean growth, morphological changes were measured among different fibers crosslinked with different sizes and concentrations of ZnO NPs. Morphological results were further studied by proteomic analysis to determine its response mechanisms. Moreover, proteomic results were upheld by immunoblot analysis.
2. Results
2.1. Growth Changes of Soybean Treated by Fibers Crosslinked with ZnO NPs
To develop a more effective method using NPs for the enhancement of soybean growth, three kinds of fibers crosslinked with ZnO NPs were prepared. As fibers, a cupro-long fiber non-woven fabric (BL), a cupro-short fiber spun lace non-woven fabric (BFSL), and a viscose-rayon short fiber spun lace non-woven fabric (RA) were used (Figure S1). Morphological changes of soybean with a variety of fibers crosslinked with ZnO NPs were analyzed to investigate their effects (Figure S1). Silica sand was treated in solution with or without 1 and 10 ppm ZnO NPs of 20 and 200 nm before sowing. When fibers crosslinked with or without 0.15 and 1.5 mg of ZnO NPs of 20 and 200 nm were used, 150 mL of water was supplied in the silica sand, which become 1 and 10 ppm as the final concentration. As control plants, two kinds of control were prepared in this research, which are (i) as control of 1 ppm and 10 ppm ZnO NPs crosslinked with BL, BFSL, and RA, 1 ppm and 10 ppm ZnO NPs solution were used; and (ii) as control for each treatment, BL, BFSL, RA, and solution without ZnO NPs were used. After sowing, four-day-old soybeans were collected and the main-root length, total root-fresh weight, hypocotyl length, and hypocotyl-fresh weight were measured as morphological parameters (Figure 1).
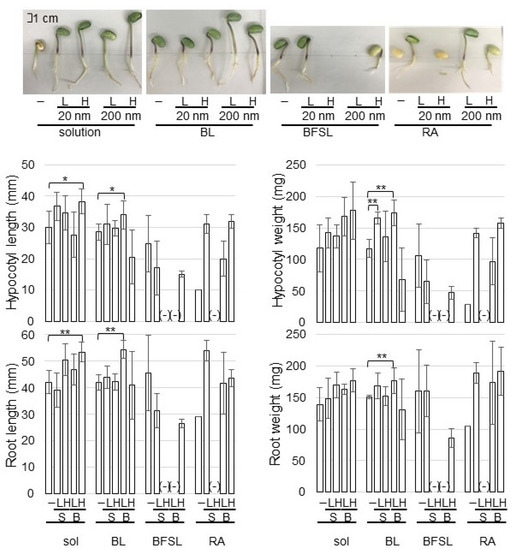
Figure 1.
Effects of ZnO NPs on the morphology of soybean seedlings. Silica sand was treated with or without 1 ppm (L) and 10 ppm (H) ZnO NPs with 20 nm (S) and 200 nm (B) before sowing. When fibers of BL, BFSL, and RA crosslinked with or without 0.15 mg and 1.5 mg of ZnO NPs of 20 nm (S) and 200 nm (B) were used, 150 mL of water was supplied in the silica sand, which become 1 ppm (L) and 10 ppm (H) as final concentration. After sowing, four-day-old soybeans were collected and main-root length, total root-fresh weight, hypocotyl length, and hypocotyl-fresh weight were measured as morphological parameters. The scale bar in the photograph indicates 1 cm. “-” means nontreated soybean for each group as control. Data are presented as the mean ± S.D. from three independent biological replicates. Asterisks indicate significant changes between two treatments according to Student’s t-test (** p < 0.01; * p < 0.05).
Among BL, BFSL, and RA fibers crosslinked with ZnO NPs, the germination of soybean grown on BFSL and RA fibers was very low, resulting in the morphological parameters not being able to be analyzed (Figure 1). The length of root and hypocotyl of soybean grown in a solution of 10 ppm ZnO NPs with 200 nm increased compared with that of nontreatment; however, their weight did not change (Figure 1). On the other hand, the length of root and hypocotyl of soybean grown on BL-fiber crosslinked with ZnO NPs was promoted at the application of 1 ppm concentration compared with that of nontreatment; additionally, their weight also increased compared with nontreatment (Figure 1). Based on morphological results, BL-fiber crosslinked with 1 ppm ZnO NPs of 200 nm was used for Zn-content measurement and proteomic analysis.
2.2. Organ Specificity of Zn Content in Soybean Grown on Fiber Crosslinked with ZnO NPs
To investigate the actions of Zn with organ specificity in soybean, Zn content was measured. Silica sand was treated with 1 ppm ZnO NPs with 200 nm and 1 ppm BL-fiber crosslinked with ZnO NPs with 200 nm. After sowing, root, hypocotyl, and cotyledon were collected from four-day-old soybeans and were measured for Zn content (Figure 2). Soybeans grown on fiber crosslinked with ZnO NPs had higher Zn content in root and hypocotyl than those grown in ZnO NPs solution. Additionally, Zn content in the root was higher than that in the hypocotyl. However, Zn content in cotyledon was the same level between fiber and solution (Figure 2).
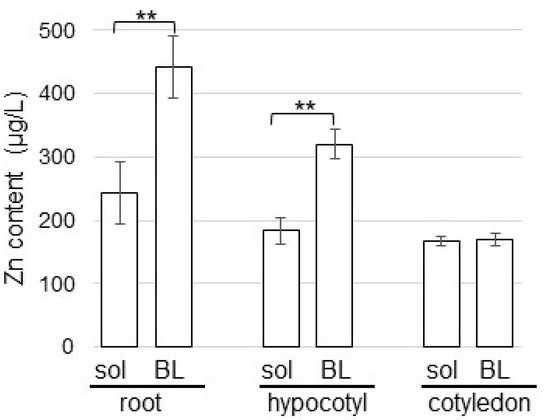
Figure 2.
Effects of ZnO NPs on the Zn content in soybean seedlings. Silica sand was treated with 1 ppm ZnO NPs with 200 nm (sol) and 1 ppm BL-fiber crosslinked with ZnO NPs with 200 nm (BL). After sowing, root, hypocotyl, and cotyledon were collected from four-day-old soybeans and the Zn content was measured. Data are presented as the mean ± S.D. from three independent biological replicates. Asterisks indicate significant changes between the two treatments according to Student’s t-test (** p < 0.01).
2.3. Protein Changes in Soybean Grown on Fiber Crosslinked with ZnO NPs
A proteomic analysis with liquid chromatography (LC)-mass spectrometry (MS) analysis was conducted to investigate the cellular mechanism in soybean growth on fiber crosslinked with ZnO NPs. As for the treatments, four kinds of treatments, which are solution/fiber and nontreatment/ZnO NPs treatment, were performed. Proteins extracted from the soybean root, including hypocotyl after treatment, were analyzed. In total, 7948 proteins and 1647 proteins were identified in the protein database of soybean and A. thaliana, respectively. The proteomic results of all samples from different groups were compared by principal component analysis (PCA), which showed different accumulation patterns of proteins from different treatments (Figure 3). These results indicated that ZnO NPs largely affected soybean protein contents; additionally, this effect was different between fiber and solution (Figure 3). The relative abundance of proteins from soybean treated with ZnO NPs compared to nontreated soybean (Table S1), from soybean grown on fiber crosslinked with ZnO NPs compared to nontreated fiber (Table S2), from soybean grown on fiber compared to a solution (Table S3), and from soybean grown on fiber crosslinked with ZnO NPs compared to ZnO NPs solution (Table S4) were listed.
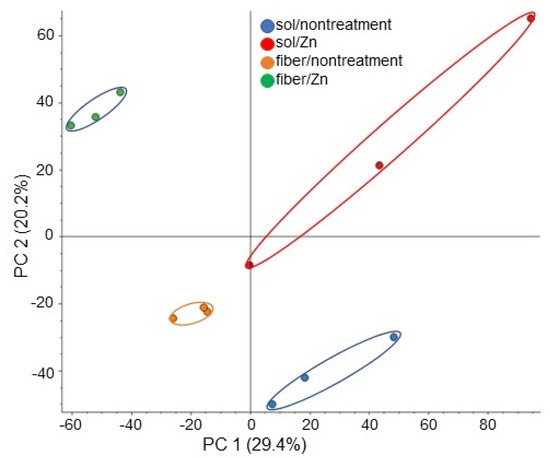
Figure 3.
Overview of total proteomic data from 12 samples based on PCA. Proteins from soybean roots including hypocotyl treated with or without 1 ppm ZnO NPs with 200 nm in solution or BL-fiber were extracted and analyzed using a LC-MS. Blue, red, orange, and green colors indicate nontreated solution, ZnO NPs solution, nontreated fiber, and fiber crosslinked with ZnO NPs sample groups, respectively. Three biological replicates were performed in each group.
The abundance of 471 proteins differentially changed with fold change >1.5 and <2/3 in the soybean treated with ZnO NPs solution compared with nontreated soybean. Among 471 proteins, 273 proteins increased and 198 proteins decreased in the soybean treated with ZnO NPs solution compared with nontreated soybean (Table S3). The functional category of identified proteins was obtained using MapMan Bin codes. The increased proteins were categorized in protein metabolism, stress, photosynthesis, transport, and amino acid metabolism in the functional category, while decreased proteins were mainly involved in RNA metabolism, cell wall, and lipid metabolism (Figure 4A).
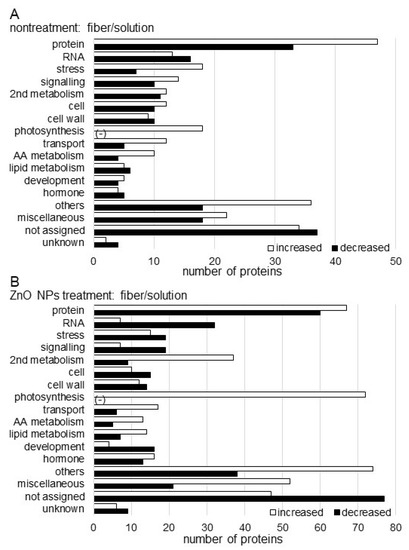
Figure 4.
Functional categorization of proteins identified in the root including hypocotyl of soybean treated with ZnO NPs. Proteins extracted from roots including hypocotyl were analyzed using a LC-MS and significantly changed proteins were identified (p < 0.05). The significantly changed proteins, 471 and 830 in nontreatment (A) and ZnO NPs treatment (B), respectively, between solution and BL-fiber, were functionally categorized using MapMan bin codes. The x-axis indicates the number of identified proteins. Abbreviations: AA, amino acid; protein, protein synthesis/degradation/post-translational modification/targeting/folding/AA activation; cell, cell division/organization/vesicle transport; RNA, RNA metabolism; hormone, hormone metabolism; 2nd metabolism, secondary metabolism; and others, nitrogen metabolism/metal handling/tetrapyrrole synthesis/oxidative pentose pathway/minor carbohydrate/cofactor and vitamin metabolism/gluconeogenesis. The negative sign shows that there is no identified protein in the respective functional category.
Furthermore, the abundance of 830 proteins differentially changed with fold change >1.5 and <2/3 in the soybean grown on a ZnO NPs BL-fiber compared with a nontreated BL-fiber. Among 830 proteins, 470 proteins increased and 360 proteins decreased in the soybean grown on a ZnO NPs BL-fiber compared with nontreated BL-fiber (Table S4). The functional category of identified proteins was obtained using MapMan Bin codes. The increased proteins were categorized in protein metabolism, secondary metabolism, photosynthesis, transport, and amino acid metabolism in the functional category, while decreased proteins were mainly involved in RNA metabolism, stress, signaling, cell construction, and cell wall (Figure 4B).
2.4. Immunoblot Analysis of NADPH Oxidoreductase and Tubulins in Soybean Grown on Fiber Crosslinked with ZnO NPs
Immunoblot analysis of NADPH oxidoreductase and alpha/beta tubulins was carried out to better reveal the change of accumulation of proteins from various treatments. Proteins were extracted from the root and hypocotyl of soybeans, which were treated with or without ZnO NPs on the BL-fiber or solution. Extracted proteins (10 µg) were separated on SDS-polyacrylamide gel. The staining pattern of Coomassie brilliant blue was used as a loading control (Figure S2). To confirm changes of significantly accumulated proteins identified by proteomic analysis (Tables S3 and S4), the accumulation of NADPH oxidoreductase (Figure S3), alpha-tubulin (Figure S4), and beta-tubulin (Figure S5) was analyzed using immunoblot analysis. The abundance of the NADPH oxidoreductase in root and hypocotyl increased with the treatment of ZnO NPs compared to nontreatment in solution and on fiber (Figure 5A). The abundance of alpha-tubulin in hypocotyl increased with or without the treatment of ZnO NPs on the BL-fiber compared to the solution; however, that in root did not change (Figure 5B). On the other hand, the abundance of beta-tubulin did not show any changes (Figure 5C).
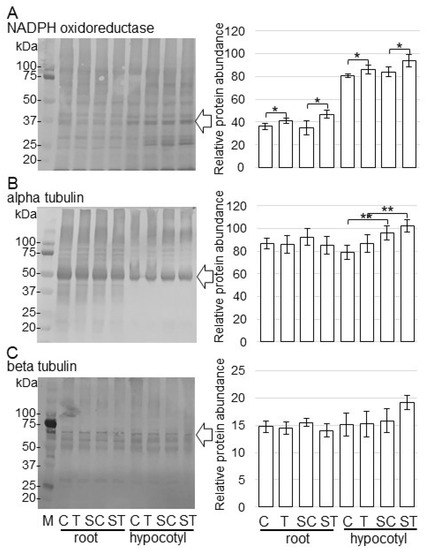
Figure 5.
Accumulation of NADPH oxidoreductase and tubulins in root and hypocotyl of soybean treated with ZnO NPs. Proteins (10 µg) extracted from root and hypocotyl of soybean treated with or without ZnO NPs in solution and BL-fiber, separated on SDS-polyacrylamide gel, and transferred onto membranes. The membranes were cross-reacted with anti-NADPH oxidoreductase (A), anti-alpha tubulin (B), and anti-beta tubulin (C) antibodies. Coomassie brilliant blue pattern was used as a loading control (Figure S2). The integrated densities of bands were calculated using ImageJ software. Data are presented as the mean ± S.D. from three independent biological replicates (Figures S3–S5). Asterisks indicate significant changes between the two treatments according to Student’s t-test (** p < 0.01; * p < 0.05).
2.5. Growth Changes under Flooding Stress of Soybean Grown on Fibers Crosslinked with ZnO NPs
Because ZnO NPs mitigated the risk associated with abiotic and biotic stresses, flooding-stress tolerance was investigated. Soybean seeds were sowed on BL-fiber or in a solution with 1 ppm ZnO NPs with 200 nm. For the non-flooded group, samples were collected at four days after sowing. For the flooded group, soybean seedlings were exposed to two-day-flooding after two-day-germination, and samples were collected. The weight and length of roots including hypocotyl were measured as morphological parameters (Figure 6). Without flooding stress, the weight and length of roots including hypocotyl increased with the treatment of ZnO NPs; however, they did not change under flooding stress, even if they were treated by ZnO NPs (Figure 6).
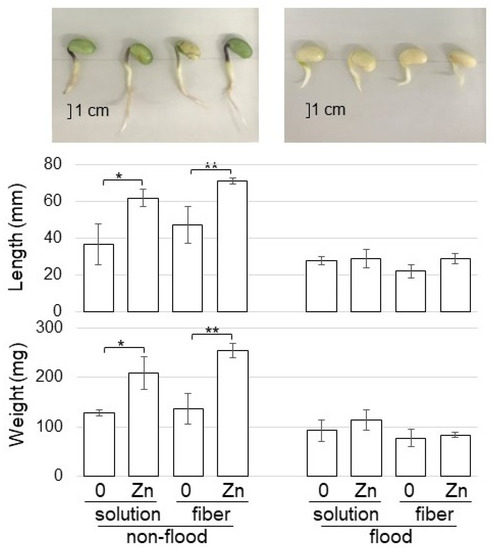
Figure 6.
Morphological effect of fiber crosslinked with ZnO on soybean growth under flooding stress. Soybean seeds were sowed on the BL-fiber or solution with 1 ppm ZnO NPs with 200 nm. For the non-flooded group, samples were collected at four days after sowing. For the flooded group, soybean seedlings were exposed to two-day-flooding after two-day-germination, and samples were collected. The fresh weight and length of hypocotyl and root were measured as morphological parameters. The bar in the photograph indicates one cm. The data are given as the mean ± S.D. from three independent biological replicates. Asterisks indicate significant changes between the two treatments according to Student’s t-test (** p < 0.01; * p < 0.05).
3. Discussions
3.1. Fiber Crosslinked with ZnO NPs Improves Soybean Growth through Zn-Content Increase
Biodegradable fibers are useful materials that are widely used in many applications such as biomedicine, filtration, and food packaging [23]. As fabricating fibers, fibers from cellulose, protein, polylactic acid, polyvinyl alcohol, and polycaprolactone were chosen as the focus [24]. Among these, micro/nanofibers expressed superior properties such as mechanical properties, cell proliferation efficiency, and filtration efficiency [25]. However, their effects on plant growth were limited because of chemical fiber spinning. In this study, the effect of three kinds of non-woven fabrics on plant growth was examined.
ZnO NPs increased the plant growth of coffee [10], lettuce [11], maize/cabbage [12], wheat [13], and tobacco [14]. In soybean, ZnO NPs also increased plant growth and seed yield [15,16]. These reports indicated that the application of ZnO NPs improved soybean growth. To develop a more effective method using ZnO NPs for the enhancement of soybean growth, fiber crosslinked with ZnO NPs was produced. In this study, the solution of ZnO NPs with 200 nm promoted soybean growth at the concentration of 10 ppm, while fibers cross-linked with ZnO NPs promoted growth at a 1 ppm concentration (Figure 1). On the other hand, the length of the root and shoot was reduced with ZnO NPs in radish [26] and spinach [27]. Furthermore, a toxic effect on the growth and development of A. thaliana was also revealed when the plant was exposed to ZnO NPs [28]. In another study, both positive and negative effects were shown in Stevia rebaudiana, depending on the concentration of applied ZnO NPs [29]. These reports with the present study indicate that the response of plants to ZnO NPs depends on the plant species, type, and size/concentration of the NPs.
Zn efficiency and Zn uptake are very important for plant growth and its total content in soil is influenced by several soil properties such as pH, CaCO3, organic matter content, and type of crop, as well as cultivars and nutrient interactions in the soil environment [30,31]. A study on the uptake pathways of ZnO NPs in maize root revealed that the majority of the total ZnO NPs undergo dissolution in the exposure medium, and the released Zn ions are only taken up by the root [32]. Only a small fraction of ZnO NPs absorbed on the root surface can cross the root cortex as a result of speedy cell division and root tip elongation, apart from their entry into the vascular system through the gap of the Casparian strip at the sites of the primary-lateral root junction [33]. Based on these reports, ZnO NPs with 20 nm and 200 nm were used in this study.
Soybeans grown on fiber crosslinked with ZnO NPs have higher Zn content in their roots than those grown in ZnO NPs solution (Figure 2). Once NPs enter the plant cells, such as the vascular bundle and stele, they reach the aerial parts [34] and cellular pores [35] through apoplast or symplast pathways and might interact with cellular and sub-cellular organelles [36]. During this penetration and translocation process, they could damage the sub-cellular organization [36]. However, this direct effect of NPs on sub-cellular organelles was caused by small-sized NPs [37,38]. In the present study, because Zn content in soybean was mainly detected in the root (Figure 2), ZnO with 200 nm might not be able to enter the leaf. Furthermore, because Zn content in soybean grown on fiber crosslinked with ZnO NPs at 200 nm was higher than that of soybean grown in solution (Figure 2), ZnO NPs might be effectively held by fiber. However, confirmatory experiments which directly investigate the plant with analytical technique using a microscope will be a challenge in the future.
3.2. Fiber Crosslinked with ZnO NPs Enhances Photosynthesis and Secondary Metabolism in Soybeans
Because Zn application results in an appreciable increase in leaf area, the content of chlorophyll, other photosynthetic pigments, and stomatal conductance, plant growth, and seed yield are improved [39,40]. In wheat, the content of photosynthetic pigments, such as chlorophyll a, chlorophyll b, and total chlorophyll content, was significantly enhanced by the application of ZnO NPs [41]. Additionally, in this study, photosynthesis-related proteins increased with the application of ZnO NPs solution and further increased in soybean grown on fiber crosslinked with ZnO NPs (Figure 4). These results indicate that ZnO NPs positively affect plant growth through photosynthesis in soybeans.
NPs can also increase the content of plant secondary metabolites in the plant. The levels of secondary metabolites such as anthocyanins, chlorophylls [42], and others were reduced in plants treated with excessive concentrations of NPs; and multiple physiological functions such as photosynthesis/transpiration [43], trace element uptake, nitrogen assimilation, and growth were also inhibited [44,45]. On the other hand, the contents of plant-secondary metabolites were significantly increased when the concentrations were suitable [46]. In the present study, proteins categorized in secondary metabolism significantly increased in soybean grown on fiber crosslinked ZnO NPs (Figure 4). This suggests that the concentration of ZnO NPs might become suitable for soybeans when ZnO NPs crosslinked with fiber, resulting in enhanced soybean growth.
3.3. NADPH Oxidoreductase Increases in Soybean Treated with ZnO NPs
The abundance of NADPH oxidoreductase, which was identified as one of the significantly accumulated proteins using proteomic analysis (Table S4), was confirmed using immunoblot analysis. The abundance of NADPH oxidoreductase increased in soybean by ZnO NPs application (Figure 5). Zn is required for the synthesis of tryptophan, which is a precursor of indole-3-acetic acid, and it also has an active role in the production of the essential growth-hormone auxin [47]. NADPH oxidoreductase increased by the application of auxin and was further raised in rice root and callus by the application of Zn [48]. Hypocotyl elongation was shown to be defective in both light and dark conditions in shy2 and axr2 mutants [49,50], which were later revealed as gain-of-function mutants of AUX/IAAs IAA3 and IAA7, respectively [51,52]. Additionally, in A. thaliana, auxin patterning and cellular growth were linked through a correlated pattern of auxin efflux carrier localization and cortical microtubule orientation [53]. In this study, there was a similar in tubulin (Figure 5), and the length of root and hypocotyl was improved by the application of ZnO NPs (Figure 1). Furthermore, the abundance of NADPH oxidoreductase increased under the same condition (Figure 5). The present results with the previous finding suggest that soybean growth with ZnO NPs might be caused by increasing auxin through NADPH reductase.
3.4. ZnO NPs Do Not Enhance Soybean Growth under Flooding Stress but Enhances It without Stress
ZnO NPs mitigated the risk associated with chilling stress in rice [17], salt stress in tomato [18], drought stress in cucumber [54], arsenic stress in soybean [20], and tobacco mosaic virus [21]. In soybean, Ag NPs with 15 nm improved plant growth under flooding stress by increasing the proteins related to amino acid synthesis and wax formation [37]; and Al2O3 NPs improved plant growth under flooding stress through mitochondrial proteins by regulating membrane permeability and tricarboxylic acid cycle activity [38]. Furthermore, the secondary metabolism of plants has great research value in the quality, flavor, growth/development, and disease therapy [55,56]; and/or tolerance to drought, salt, cold, diseases, and insect pests [57]. These reports indicate that the application of NPs improves stress tolerance; therefore, flooding stress tolerance was investigated for soybean using fiber crosslinked with ZnO NPs (Figure 6). Without flooding stress, soybean growth increased with the treatment of ZnO NPs; unfortunately, however, they did not change under flooding stress, even if they were treated with ZnO NPs (Figure 6). To understand the effect of flooding stress on soybean treated with ZnO NPs, further study is needed to make the process clear.
4. Materials and Methods
4.1. The Preparation of Fiber Crosslinked with ZnO NPs
To prepare fiber crosslinked with NPs, 0.15 and 1.5 mg of ZnO NPs with 20 and 200 nm (Skyspring Nanomaterials, Huston, TX, USA) were used. They become 1 and 10 ppm ZnO NPs as the final concentration with 150 mL of water. For the fibers, a cupro-long fiber non-woven fabric (BL; Asahi Kasei, Osaka, Japan), a cupro-short fiber spun lace non-woven fabric (BFSL), and a viscose-rayon short fiber spun lace non-woven fabric (RA) were used. Each fiber was dissolved with 2 g/L of an anionic active agent (Score Roll FC-250; Hokko Chemical, Tokyo, Japan) and 1 g/L of sodium carbonate at 90 °C, subjected to scouring treatment/dehydration treatment for 20 min, and dried. Next, 50 g/L of a polyfunctional cationizing agent (Cationone KCN; Lion Specialty Chemicals, Tokyo, Japan) and 7 g/L of sodium hydroxide were dissolved at 90 °C, subjected to cationization treatment/ dehydration treatment for 20 min, and dried. In order to obtain a non-woven fabric crosslinked with 0.15 or 1.5 mg of ZnO NPs, the cationized non-woven fabric was cut into a vertical 15 cm/a horizontal 10 cm and submerged in solution at 90 °C for 20 min, and then subjected to a dehydration treatment to naturally dry. The concentration of ZnO NPs was calculated from the exhaustion rate and the notation, after which the length of the non-woven fabric was cut to adjust the supported amount.
4.2. Plant Material
Soybean (Glycine max L. cultivar Enrei) was used in this study. Seeds were sterilized in 2% sodium hypochlorite solution and sown on silica sand. Silica sand was treated in 150 mL solution with or without 1 and 10 ppm ZnO NPs with 20 and 200 nm before sowing. When fibers crosslinked with or without 0.15 mg and 1.5 mg of ZnO NPs with 20 and 200 nm were used, 150 mL of water was supplied in the silica sand, which become 1 ppm and 10 ppm as the final concentration. After sowing, seedlings were maintained at 25 °C in a growth chamber illuminated with white fluorescent light (600 μmol m−2 s−1, 16 h light period/day) and 70% relative humidity. For morphological analysis, 4-day-old soybeans were collected and main-root length, total root-fresh weight, hypocotyl length, and hypocotyl-fresh weight were measured. For proteomic analysis, roots including hypocotyl from 4-day-old soybeans were used. Roots and hypocotyl were collected for the other biochemical assays at 4-day-old soybeans. Three independent experiments were performed as biological triplicates for all experiments. Independent biological replicates were sown on different days.
4.3. Protein Extraction
A portion (300 mg) of samples was ground with a mortar and pestle in 500 µL of lysis buffer, which contains 7 M urea, 2 M thiourea, 5% CHAPS, and 2 mM tributylphosphine. The suspension was centrifuged twice with 16,000× g at 4 °C for 10 min. The detergents in the supernatant were removed using the Pierce Detergent Removal Spin Column (Pierce Biotechnology, Rockford, IL, USA). The method of Bradford [58] was used to determine the protein concentration and bovine serum albumin was used as the standard.
4.4. Protein Enrichment, Reduction, Alkylation, and Digestion
Extracted proteins (100 µg) were adjusted to a final volume of 100 µL, added 400 µL of methanol, and mixed before the addition of 100 µL of chloroform and 300 µL of water. After mixing and centrifugation at 16,000× g for 10 min to achieve phase separation, the upper phase was discarded and 300 µL of methanol was added to the lower phase. After centrifugation at 16,000× g for 10 min, the pellet was collected as the soluble fraction [59]. Proteins were resuspended in 50 mM ammonium bicarbonate, reduced with 50 mM dithiothreitol for 30 min at 56 °C, and alkylated with 50 mM iodoacetamide for 30 min at 37 °C. Alkylated proteins were digested with trypsin (Wako, Osaka, Japan) at a 1:100 enzyme/protein ratio for 18 h at 37 °C. Peptides were desalted with MonoSpin C18 Column (GL Sciences, Tokyo, Japan) and acidified with 1% trifluoroacetic acid.
4.5. Protein Identification Using Nano-Liquid Chromatography Mass Spectrometry
The nano-liquid chromatography (LC) conditions as well as the mass spectrometry (MS) acquisition conditions are described in the previous study [60]. The peptides were loaded onto the LC system (EASY-nLC 1000; Thermo Fisher Scientific, San Jose, CA, USA) equipped with a trap column (Acclaim PepMap 100 C18 LC column, 3 µm, 75 µm ID × 20 mm; Thermo Fisher Scientific), equilibrated with 0.1% formic acid, and eluted with a linear acetonitrile gradient (0–35%) in 0.1% formic acid at a flow rate of 300 nL min−1. The eluted peptides were loaded and separated on the column (EASY-Spray C18 LC column, 3 µm, 75 µm ID × 150 mm; Thermo Fisher Scientific) with a spray voltage of 2 kV (Ion Transfer Tube temperature: 275 °C). The peptide ions were detected using MS (Orbitrap Fusion ETD MS; Thermo Fisher Scientific) in the data-dependent acquisition mode with the installed Xcalibur software (version 4.0; Thermo Fisher Scientific). Full-scan mass spectra were acquired in the MS over 375–1500 m/z with a resolution of 120,000. The most intense precursor ions were selected for collision-induced fragmentation in the linear ion trap at a normalized collision energy of 35%. Dynamic exclusion was employed within 60 s to prevent repetitive selection of peptides.
4.6. Mass-Spectrometry Data Analysis
The MS/MS searches were carried out using MASCOT (version 2.6.2, Matrix Science, London, UK) and SEQUEST HT search algorithms against the UniProtKB Glycine max (Soybean) protein database (version 2021-02; 84,894 proteins) and A. thaliana protein database (version 2021-02; 119,669 proteins) using Proteome Discoverer (version 2.4; Thermo Fisher Scientific). The workflow for both algorithms included spectrum files RC, spectrum selector, MASCOT, SEQUEST HT search nodes, percolator, ptmRS, and minor feature detector nodes. Oxidation of methionine was set as a variable modification and carbamidomethylation of cysteine was set as a fixed modification. Mass tolerances in MS and MS/MS were set at 10 ppm and 0.6 Da, respectively. Trypsin was specified as protease and a maximum of two missed cleavage was allowed. Target-decoy database searches were used for the calculation of the false discovery rate, which was set at 1% for peptide identification.
4.7. Differential Analysis of Proteins Using Mass Spectrometry Data
Label-free quantification was also performed with Proteome Discoverer using precursor ions quantifier nodes. For differential analysis of the relative abundance of peptides and proteins between samples, the freely available software Perseus (version 1.6.15.0) [61] was used. Proteins and peptides abundances were transferred into the log2 scale. Three biological replicates of each sample were grouped and a minimum of three valid values were required in at least one group. Normalization of the abundances was performed to subtract the median of each sample. Missing values were imputed based on a normal distribution (width = 0.3, down-shift = 1.8). Significance was assessed using Student’s t-test analysis. Principal component analysis (PCA) was performed with Proteome Discoverer. Protein functions were categorized using MapMan bin codes [62].
4.8. Immunoblot Analysis
SDS-sample buffer consisting of 60 mM Tris-HCl (pH 6.8), 2% SDS, 10% glycerol, and 5% dithiothreitol was added to protein samples [63]. Proteins (10 µg) quantified by Bradford method [58] were separated by electrophoresis on a 10% SDS-polyacrylamide gel and transferred onto a polyvinylidene difluoride membrane using a semidry transfer blotter (Nippon Eido, Tokyo, Japan). The blotted membrane was blocked for 5 min in Bullet Blocking One regent (Nacalai Tesque, Kyoto, Japan). After blocking, the membrane was cross-reacted with a 1: 1000 dilution of the primary antibodies for 1 h at room temperature. As the primary antibodies, the followings were used: alpha-tubulin (Abcam, Cambridge, UK), beta-tubulin (Proteintech, Rosemont, IL, USA), and NADPH-dependent oxidoreductase [48]. Anti-rabbit IgG conjugated with horseradish peroxidase (Bio-Rad, Hercules, CA, USA) was used as the secondary antibody. After 1 h incubation, signals were detected using the TMB Membrane Peroxidase Substrate kit (Seracare, Milford, MA, USA) following the manufacturer’s protocol. Coomassie brilliant blue staining was used as a loading control. The integrated densities of bands were calculated using ImageJ software (version 1.8, National Institutes of Health, Bethesda, MD, USA).
4.9. Measurement of Zn Content
A portion (100 mg) of samples, which were root, hypocotyl, and cotyledon, was ground with a mortar and pestle in 250 µL of phosphate-buffered saline containing 0.01 M HCl. The suspension was stirred for 20 min and centrifuged with 16,000× g at 4 °C for 10 min. After precipitation of proteins, the supernatant was used for Zn-content assay. Zn content was analyzed using Zn Concentrate Assay Kit (Funakoshi, Tokyo, Japan). Sample (50 µL) was added to 50 µL of chelate color and mixed for 10 min. The absorbance of the mixture was measured at 560 nm.
4.10. Statistical Analysis
The statistical significance of the 2 groups was evaluated by the Student’s t-test. A p-value of less than 0.05 was considered statistically significant.
5. Conclusions
To develop a more effective method using NPs for enhancement of soybean growth, fiber crosslinked with ZnO NPs was used. The solution of ZnO NPs with 200 nm promoted soybean growth at the concentration of 10 ppm, while fibers crosslinked with ZnO NPs promoted growth at a 1 ppm concentration. Among three kinds of fibers, BL was the most effective for soybean growth compared to the other fibers. To study the positive mechanism of BL fiber crosslinked with ZnO NPs on soybean growth, a proteomic technique was used. The main findings of this study are as follows: (i) soybeans grown on fiber crosslinked with ZnO NPs had higher Zn content in their root than those grown in ZnO NPs solution; (ii) proteins categorized in photosynthesis and secondary metabolism accumulated more in soybean grown on fiber crosslinked with ZnO NPs than in those grown in ZnO NPs solution; and (iii) the abundance of NADPH oxidoreductase increased in soybean by ZnO NPs application. These findings suggest that BL-fiber crosslinked with ZnO NPs enhances soybean growth through the increase of photosynthesis and secondary metabolism. Furthermore, the accumulation of NADPH oxidoreductase might relate to the effect of auxin with fiber crosslinked with ZnO NPs on soybean growth. Taken together, ZnO NPs at low doses can be considered a potent substance for potential utilization in plant sciences.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms23137415/s1.
Author Contributions
K.M., S.Y., K.S., R.O., S.A. and R.F. prepared fiber crosslinked with nanoparticles; S.K. performed morphological measurement and protein preparation; H.Y., K.H. and K.T. performed MS analysis; S.K. performed the immunoblot analysis and other biochemical analysis; S.K. analyzed the data and wrote the paper. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Research Grants from Fukui University of Technology (FY 2021) to S.K.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
For MS data, RAW data, peak lists, and result files have been deposited in the ProteomeXchange Consortium [64] via the jPOST [65] partner repository under data-set identifiers PXD031670.
Acknowledgments
Authors are grateful to Nanae Oka of Fukui University of Technology for helping with experiments.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Dubey, P.; Matai, I.; Kumar, S.U.; Sachdey, A.; Bhusan, B.; Gopinath, P. Perturbation of mechanistic cellular system by silver nanoparticle toxicity: Cytotoxic, genotoxic and epigenetic potentials. Adv. Colloid. Interface Sci. 2015, 221, 4–21. [Google Scholar] [CrossRef] [PubMed]
- Pejam, F.; Ardebili, Z.O.; Ladan-Moghadam, A.; Danaee, E. Zinc oxide nanoparticles mediated substantial physiological and molecular changes in tomato. PLoS ONE 2021, 16, e0248778. [Google Scholar] [CrossRef]
- Wang, W.; Vinocur, B.; Shoseyov, O.; Altman, A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 2004, 9, 244–252. [Google Scholar] [CrossRef] [PubMed]
- Sabir, S.; Arshad, M.; Chaudhari, S.K. Zinc oxide nanoparticles for revolutionizing agriculture: Synthesis and applications. Sci. World J. 2014, 2014, 925494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borm, P.; Klaessig, F.C.; Landry, T.D.; Moudgil, B.; Pauluhn, J.; Thomas, K.; Trottier, R.; Wood, S. Research strategies for safety evaluation of nanomaterials, Part V: Role of dissolution in biological fate and effects of nanoscale particles. Toxicol. Sci. 2006, 90, 23–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balážová, Ľ.; Babula, P.; Baláž, M.; Bačkorová, M.; Bujňáková, Z.; Briančin, J.; Kurmanbayeva, A.; Sagi, M. Zinc oxide nanoparticles phytotoxicity on halophyte from genus Salicornia. Plant Physiol. Biochem. 2018, 130, 30–42. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yang, X.; Chen, S.; Li, Q.; Wang, W.; Hou, C.; Gao, X.; Wang, L.; Wang, S. Zinc oxide nanoparticles affect biomass accumulation and photosynthesis in Arabidopsis. Front. Plant Sci. 2016, 6, 1243. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.P.; Li, Q.Q.; Pei, Z.M.; Wang, S.C. Effects of zinc oxide nps on the growth, photosynthetic traits, and antioxidative enzymes in tomato plants. Biol. Plant. 2018, 62, 801–808. [Google Scholar] [CrossRef]
- Prakash, M.G.; Chung, I.M. Determination of zinc oxide nanoparticles toxicity in root growth in wheat (Triticum aestivum L.) seedlings. Acta Biol. Hung. 2016, 67, 286–296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rossi, L.; Fedenia, L.N.; Sharifan, H.; Ma, X.; Lombardini, L. Effects of foliar application of zinc sulfate and zinc nanoparticles in coffee (Coffea arabica L.) plants. Plant Physiol. Biochem. 2019, 135, 160–166. [Google Scholar] [CrossRef]
- Xu, J.; Luo, X.; Wang, Y.; Feng, Y. Evaluation of zinc oxide nanoparticles on lettuce (Lactuca sativa L.) growth and soil bacterial community. Environ. Sci. Pollut. Res. Int. 2018, 25, 6026–6035. [Google Scholar] [CrossRef] [PubMed]
- Pokhrel, L.R.; Dubey, B. Evaluation of developmental responses of two crop plants exposed to silver and zinc oxide nanoparticles. Sci. Total Environ. 2013, 452, 321–332. [Google Scholar] [CrossRef] [PubMed]
- Rizwan, M.; Ali, S.; Ali, B.; Adrees, M.; Arshad, M.; Hussain, A.; Zia-Ur-Rehman, M.; Waris, A.A. Zinc and iron oxide nanoparticles improved the plant growth and reduced the oxidative stress and cadmium concentration in wheat. Chemosphere 2019, 214, 269–277. [Google Scholar] [CrossRef] [PubMed]
- Mazaheri-Tirani, M.; Madadkar-Haghjou, M.; Ismaili, A. Hydroponic grown tobacco plants respond to zinc oxide nanoparticles and bulk exposures by morphological, physiological and anatomical adjustments. Funct. Plant Biol. 2019, 46, 360–375. [Google Scholar] [CrossRef] [PubMed]
- Mirakhorli, T.; Ardebili, Z.O.; Ladan-Moghadam, A.; Danaee, E. Bulk and nanoparticles of zinc oxide exerted their beneficial effects by conferring modifications in transcription factors, histone deacetylase, carbon and nitrogen assimilation, antioxidant biomarkers, and secondary metabolism in soybean. PLoS ONE 2021, 16, e0256905. [Google Scholar] [CrossRef]
- Yusefi-Tanha, E.; Fallah, S.; Rostamnejadi, A.; Pokhrel, L.R. Zinc oxide nanoparticles (ZnO NPs) as a novel nanofertilizer: Influence on seed yield and antioxidant defense system in soil grown soybean (Glycine max cv. Kowsar). Sci. Total Environ. 2020, 738, 140240. [Google Scholar] [CrossRef]
- Song, Y.; Jiang, M.; Zhang, H.; Li, R. Zinc oxide nanoparticles alleviate chilling stress in rice (Oryza Sativa L.) by regulating antioxidative system and chilling response transcription factors. Molecules 2021, 26, 2196. [Google Scholar] [CrossRef]
- Faizan, M.; Bhat, J.; Chen, C.; Alyemeni, M.N.; Wijaya, L.; Ahmad, P.; Yu, F. Zinc oxide nanoparticles (ZnO-NPs) induce salt tolerance by improving the antioxidant system and photosynthetic machinery in tomato. Plant Physiol. Biochem. 2021, 161, 122–130. [Google Scholar] [CrossRef]
- Khan, M.I.R.; Jahan, B.; Alajmi, M.F.; Rehman, M.T.; Khan, N.A. Exogenously-sourced ethylene modulates defense mechanisms and promotes tolerance to zinc stress in mustard (Brassica juncea L.). Plants 2019, 8, 540. [Google Scholar] [CrossRef] [Green Version]
- Bhat, J.A.; Faizan, M.; Bhat, M.A.; Huang, F.; Yu, D.; Ahmad, A.; Bajguz, A.; Ahmad, P. Defense interplay of the zinc-oxide nanoparticles and melatonin in alleviating the arsenic stress in soybean (Glycine max L.). Chemosphere 2022, 288, 132471. [Google Scholar] [CrossRef]
- Abdelkhalek, A.; Al-Askar, A.A. Green synthesized ZnO nanoparticles mediated by Mentha Spicata extract induce plant systemic resistance against Tobacco mosaic virus. Appl. Sci. 2020, 10, 5054. [Google Scholar] [CrossRef]
- Hashimoto, T.; Mustafa, G.; Nishiuchi, T.; Komatsu, S. Comparative analysis of the effect of inorganic and organic chemicals with silver nanoparticles on soybean under flooding stress. Int. J. Mol. Sci. 2020, 21, 1300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, Y.; Lu, T.; Cui, J.; Keshari Samal, S.; Xiong, R.; Huang, C. Bio-based electrospun nanofiber as building blocks for a novel eco-friendly air filtration membrane: A review. Sep. Purif. Technol. 2021, 277, 119623. [Google Scholar] [CrossRef]
- Soares, R.M.D.; Siqueira, N.M.; Prabhakaram, M.P.; Ramakrishna, S. Electrospinning and electrospray of bio-based and natural polymers for biomaterials development. Mater. Sci. Eng. C 2018, 92, 969–982. [Google Scholar] [CrossRef] [PubMed]
- Kai, D.; Liow, S.S.; Loh, X.J. Biodegradable polymers for electrospinning: Towards biomedical applications. Mater. Sci. Eng. C 2014, 45, 659–670. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.; Kumar, A. Investigating long-term effect of nanoparticles on growth of Raphanus sativus plants: A trans-generational study. Ecotoxicology 2018, 27, 23–31. [Google Scholar] [CrossRef]
- Singh, D.; Kumar, A. Impact of irrigation using water containing CuO and ZnO nanoparticles on Spinach oleracea grown in soil media. Bull. Environ. Contam. Toxicol. 2016, 97, 548–553. [Google Scholar] [CrossRef]
- Wan, J.; Wang, R.; Wang, R.; Ju, Q.; Wang, Y.; Xu, J. Comparative physiological and transcriptomic analyses reveal the toxic effects of zno nanoparticles on plant growth. Environ. Sci. Technol. 2019, 53, 4235–4244. [Google Scholar] [CrossRef]
- Javed, R.; Usman, M.; Yücesan, B.; Zia, M.; Gürel, E. Effect of zinc oxide (ZnO) nanoparticles on physiology and steviol glycosides production in micropropagated shoots of Stevia rebaudiana Bertoni. Plant Physiol. Biochem. 2017, 110, 94–99. [Google Scholar] [CrossRef]
- Murakami, M.; Ae, N. Potential for phytoextraction of copper, lead, and zinc by rice (Oryza sativa L.), soybean (Glycine max [L.] Merr.), and maize (Zea mays L.). J. Hazard. Mater. 2009, 162, 1185–1192. [Google Scholar] [CrossRef]
- Khan, A.R.; Wakeel, A.; Muhammad, N.; Liu, B.; Wu, M.; Liu, Y.; Ali, I.; Zaidi, S.H.R.; Azhar, W.; Somg, G.; et al. Involvement of ethylene signaling in zinc oxide nanoparticle-mediated biochemical changes in Arabidopsis thaliana leaves. Environ. Sci. Nano 2019, 6, 341–355. [Google Scholar] [CrossRef]
- Lv, J.; Zhang, S.; Luo, L.; Zhang, J.; Yang, K.; Christie, P. Accumulation, speciation and uptake pathway of ZnO nanoparticles in maize. Environ. Sci. Nano 2015, 2, 68–77. [Google Scholar] [CrossRef]
- Hossain, Z.; Yasmeen, F.; Komatsu, S. Nanoparticles: Synthesis, morphophysiological effects, and proteomic responses of crop plants. Int. J. Mol. Sci. 2020, 21, 3056. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, D.K.; Shweta; Singh, S.; Pandey, R.; Singh, V.P.; Sharma, N.C.; Prasad, S.M.; Dubey, N.K.; Chauhan, D.K. An overview on manufactured nanoparticles in plants: Uptake, translocation, accumulation and phytotoxicity. Plant Physiol. Biochem. 2017, 110, 2–12. [Google Scholar] [CrossRef] [PubMed]
- Rico, C.M.; Majumdar, S.; Duarte-Gardea, M.; Peralta-Videa, J.R.; Gardea-Torresdey, J.L. Interaction of nanoparticles with edible plants and their possible implications in the food chain. J. Agric. Food Chem. 2011, 59, 3485–3498. [Google Scholar] [CrossRef] [Green Version]
- Rajput, V.D.; Minkina, T.; Sushkova, S.; Mandzhieva, S.; Fedorenko, A.; Lysenko, V.; Bederska-Błaszczyk, M.; Olchowik, J.; Tsitsuashvili, V.; Chaplygin, V. Structural and ultrastructural changes in nanoparticle exposed plants. In Nanoscience for Sustainable Agriculture; Pudake, R.N., Chauhan, N., Kole, C., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 281–295. [Google Scholar]
- Mustafa, G.; Sakata, K.; Komatsu, S. Proteomic analysis of soybean root exposed to varying sizes of silver nanoparticles under flooding stress. J. Proteom. 2016, 148, 113–125. [Google Scholar] [CrossRef]
- Mustafa, G.; Komatsu, S. Insights into the response of soybean mitochondrial proteins to various sizes of aluminum oxide nanoparticles under flooding stress. J. Proteome Res. 2016, 15, 4464–4475. [Google Scholar] [CrossRef]
- Yang, W.-D.; Wang, Y.-Y.; Zhao, F.-L.; Ding, Z.-L.; Zhang, X.-C.; Zhu, Z.-Q.; Yang, X.-E. Variation in copper and zinc tolerance and accumulation in 12 willow clones: Implications for phytoextraction. J. Zhejiang Univ.-SCI. B 2014, 15, 788–800. [Google Scholar] [CrossRef] [Green Version]
- Anwaar, S.A.; Ali, S.; Ali, S.; Ishaque, W.; Farid, M.; Farooq, M.A.; Najeeb, U.; Abbas, F.; Sharif, M. Silicon (Si) alleviates cotton (Gossypium hirsutum L.) from zinc (Zn) toxicity stress by limiting Zn uptake and oxidative damage. Environ. Sci. Pollut. Res. 2015, 22, 3441–3450. [Google Scholar] [CrossRef]
- Rai-Kalal, P.; Jajoo, A. Priming with zinc oxide nanoparticles improve germination and photosynthetic performance in wheat. Plant Physiol. Biochem. 2021, 160, 341–351. [Google Scholar] [CrossRef]
- Shankar, S.S.; Rai, A.; Ahmad, A.; Sastry, M. Rapid synthesis of Au, Ag, and bimetallic Au core–Ag shell nanoparticles using Neem (Azadirachta indica) leaf broth. J. Colloid Interface Sci. 2004, 275, 496–502. [Google Scholar] [CrossRef]
- Nair, P.M.G.; Chung, I.M. Assessment of silver nanoparticle-induced physiological and molecular changes in Arabidopsis thaliana. Environ. Sci. Pollut. Res. 2014, 21, 8858–8869. [Google Scholar] [CrossRef]
- Salehi, H.; Miras-Moreno, B.; Chehregani Rad, A.; Pii, Y.; Mimmo, T.; Cesco, S.; Lucini, L. Relatively Low Dosages of CeO2 Nanoparticles in the Solid Medium Induce Adjustments in the Secondary Metabolism and Ionomic Balance of Bean (Phaseolus vulgaris L.) Roots and Leaves. J. Agric. Food Chem. 2020, 68, 67–76. [Google Scholar] [CrossRef]
- Zhang, H.; Lu, L.; Zhao, X.; Zhao, S.; Gu, X.; Du, W.; Wei, H.; Ji, R.; Zhao, L. Metabolomics reveals the “invisible” responses of spinach plants exposed to CeO2 nanoparticles. Environ. Sci. Technol. 2019, 53, 6007–6017. [Google Scholar] [CrossRef]
- Zhang, Y.; Qi, G.; Yao, L.; Huang, L.; Wang, J.; Gao, W. Effects of metal nanoparticles and other preparative materials in the environment on plants: From the perspective of improving secondary metabolites. J. Agric. Food. Chem. 2022, 70, 916–933. [Google Scholar] [CrossRef] [PubMed]
- Ugwu, E.I.; Agunwamba, J.C. A review on the applicability of activated carbon derived from plant biomass in adsorption of chromium, copper, and zinc from industrial wastewater. Environ. Monit. Assess. 2020, 192, 240. [Google Scholar] [CrossRef]
- Oguchi, K.; Tanaka, N.; Komatsu, S.; Akao, S. Characterization of NADPH-dependent oxidoreductase induced by auxin in rice. Physiol. Plant. 2004, 121, 124–131. [Google Scholar] [CrossRef]
- Timpte, C.S.; Wilson, A.K.; Estelle, M. Effects of the axr2 mutation of Arabidopsis on cell shape in hypocotyl and inflorescence. Planta 1992, 188, 271–278. [Google Scholar] [CrossRef]
- Reed, J.W.; Elumalai, R.P.; Chory, J. Suppressors of an Arabidopsis thaliana phyB mutation identify genes that control light signaling and hypocotyl elongation. Genetics 1998, 148, 1295–1310. [Google Scholar] [CrossRef]
- Tian, Q.; Reed, J.W. Control of auxin-regulated root development by the Arabidopsis thaliana SHY2/IAA3 gene. Development 1999, 126, 711–721. [Google Scholar] [CrossRef] [PubMed]
- Nagpal, P.; Walker, L.M.; Young, J.C.; Sonawala, A.; Timpte, C.; Estelle, M.; Reed, J.W. AXR2 encodes a member of the Aux/IAA protein family. Plant Physiol. 2000, 123, 563–574. [Google Scholar] [CrossRef] [Green Version]
- Heisler, M.G.; Hamant, O.; Krupinski, P.; Uyttewaal, M.; Ohno, C.; Jönsson, H.; Traas, J.; Meyerowitz, E.M. Alignment between PIN1 polarity and microtubule orientation in the shoot apical meristem reveals a tight coupling between morphogenesis and auxin transport. PLoS Biol. 2010, 8, e1000516. [Google Scholar] [CrossRef] [Green Version]
- Ghani, M.I.; Saleem, S.; Rather, S.A.; Rehmani, M.S.; Alamri, S.; Rajput, V.D.; Kalaji, H.M.; Saleem, N.; Sial, T.A.; Liu, M. Foliar application of zinc oxide nanoparticles: An effective strategy to mitigate drought stress in cucumber seedling by modulating antioxidant defense system and osmolytes accumulation. Chemosphere 2022, 289, 133202. [Google Scholar] [CrossRef]
- Majnooni, M.B.; Fakhri, S.; Smeriglio, A.; Trombetta, D.; Croley, C.R.; Bhattacharyya, P.; Sobarzo-Sánchez, E.; Farzaei, M.H.; Bishayee, A. Antiangiogenic effects of coumarins against cancer: From chemistry to medicine. Molecules 2019, 24, 4278. [Google Scholar] [CrossRef] [Green Version]
- Naseri, R.; Farzaei, F.; Fakhri, S.; El-Senduny, F.F.; Altouhamy, M.; Bahramsoltani, R.; Ebrahimi, F.; Rahimi, R.; Farzaei, M.H. Polyphenols for diabetes associated neuropathy: Pharmacological targets and clinical perspective. DARU J. Pharm. Sci. 2019, 27, 781–798. [Google Scholar] [CrossRef]
- Osbourn, A.E. Plant secondary metabolites—A primary resource: Biochemistry of plant secondary metabolism and functions of plant secondary metabolites and their exploitation in biotechnology. Trends Biotechnol. 2000, 18, 321–322. [Google Scholar] [CrossRef]
- Bradford, M.M. A Rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Komatsu, S.; Nanjo, Y.; Nishimura, M. Proteomic analysis of the flooding tolerance mechanism in mutant soybean. J. Proteom. 2013, 79, 231–250. [Google Scholar] [CrossRef]
- Li, X.; Rehman, S.U.; Yamaguchi, H.; Hitachi, K.; Tsuchida, K.; Yamaguchi, T.; Sunohara, Y.; Matsumoto, H.; Komatsu, S. Proteomic analysis of the effect of plant-derived smoke on soybean during recovery from flooding stress. J. Proteom. 2018, 181, 238–248. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The perseus computational platform for comprehensive analysis of omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
- Usadel, B.; Nagel, A.; Thimm, O.; Redestig, H.; Blaesing, O.E.; Rofas, N.P.; Selbig, J.; Hannemann, J.; Piques, M.C.; Steinhauser, D.; et al. Extension of the visualization tool mapman to allow statistical analysis of arrays, display of corresponding genes and comparison with known responses. Plant Physiol. 2005, 138, 1195–1204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Vizcaíno, J.A.; Côté, R.G.; Csordas, A.; Dianes, J.A.; Fabregat, A.; Foster, J.M.; Griss, J.; Alpi, E.; Birim, M.; Contell, J.; et al. The Proteomics IDEntifications (PRIDE) Database and associated tools: Status in 2013. Nucleic Acids Res. 2013, 41, 1063–1069. [Google Scholar] [CrossRef] [PubMed]
- Okuda, S.; Watanabe, Y.; Moriya, Y.; Kawano, S.; Yamamoto, T.; Matsumoto, M.; Takami, T.; Kobayashi, D.; Araki, N.; Yoshizawa, A.C.; et al. jPOSTTrepo: An international standard data repository for proteomes. Nucleic Acids Res. 2017, 45, 1107–1111. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).