Abstract
Huntington’s disease (HD) is a rare hereditary autosomal dominant neurodegenerative disorder, which is caused by expression of mutant huntingtin protein (mHTT) with an abnormal number of glutamine repeats in its N terminus, and characterized by intracellular mHTT aggregates (inclusions) in the brain. Exosomes are small extracellular vesicles that are secreted generally by all cell types and can be isolated from almost all body fluids such as blood, urine, saliva, and cerebrospinal fluid. Exosomes may participate in the spreading of toxic misfolded proteins across the central nervous system in neurodegenerative diseases. In HD, such propagation of mHTT was observed both in vitro and in vivo. On the other hand, exosomes might carry molecules with neuroprotective effects. In addition, due to their capability to cross blood-brain barrier, exosomes hold great potential as sources of biomarkers available from periphery or carriers of therapeutics into the central nervous system. In this review, we discuss the emerging roles of exosomes in HD pathogenesis, diagnosis, and therapy.
Keywords:
extracellular vesicle; exosome; neurodegeneration; Huntington’s disease; huntingtin; polyQ; biomarker; therapy 1. Introduction
Extracellular vesicles (EVs) are phospholipid bilayer membrane enveloped particles released from cells into extracellular environments and body fluids. According to the mechanism of biogenesis, three main EV subtypes can be distinguished, i.e., exosomes, microvesicles, and apoptotic bodies (Figure 1) [1,2]. This review is focused mainly on exosomes, as they are the most studied vesicle subtype in human diseases, including Huntington’s disease. However, the original terminology of EV labeling by the authors of cited literature is respected in our review.
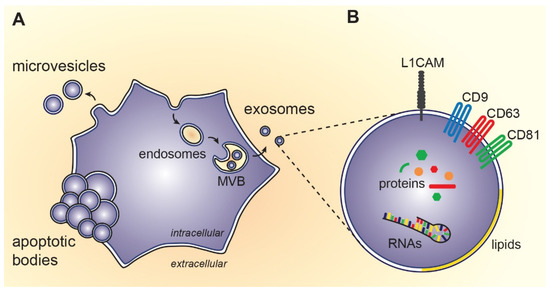
Figure 1.
Biosynthesis and composition of extracellular vesicles: (A) Exosomes originate from endosomal compartment, i.e., intraluminal vesicles of multivesicular bodies (MVB). On the other hand, microvesicles arise by outward budding of cytoplasmic membrane and apoptotic bodies are formed during programmed cell death; (B) Exosomes contain proteins, RNAs, lipids and small molecules of the source cell. Some of the molecules specifically incorporated during exosome biosynthesis can be used as markers for exosome detection (e.g., transmembrane tetraspanins CD9, CD63, CD81) or as a marker of neuronal origin (e.g., L1 cell adhesion molecule, L1CAM).
Exosomes are nano sized EVs (40–120 nm) that were initially identified as a part of reticulocyte maturation in sheep [3]. Exosomes were later observed to be released by almost all cell types [4]. Exosomes play an important role in cell-cell communication and intercellular signaling by transferring molecules into recipient cells [5,6,7]. Due to their roles in the physiological and pathological conditions in the brain, their ability to cross the blood-brain barrier and their cargo, exosomes are a potential source of biomarkers or therapy carriers in neurodegenerative diseases (NDs).
2. Exosome Biogenesis
Exosomes are formed by endocytosis and are released by exocytosis (Figure 1). Inward budding of the plasma membrane results in the formation of small intracellular vesicles which then fuse together and form early endosomes [8]. The invagination of early endosome membranes results in the formation of intraluminal vesicles (ILVs) within large multivesicular bodies (MVBs). During the maturation of early endosomes to late endosomes, some proteins are directly integrated into the invaginating membrane, whilst the cytoplasmic molecules such as proteins, messenger RNAs (mRNAs), and microRNAs (miRNAs) are engulfed and enclosed in the ILV lumen. Some MVBs fuse with the plasma membrane and release their contents, the exosomes, into the extracellular space. Other MVBs are delivered to lysosomes for degradation [9,10,11]. Exosome biogenesis depends on the endosomal sorting complexes required for transport (ESCRTs), which recognize ubiquitinated proteins [12]. ESCRTs consist of four protein complexes ESCRT-0, ESCRT-I and ESCRT-II, and ESCRT-III that work cooperatively to facilitate the MVB formation, vesicle budding, and protein cargo sorting [9,11,13,14].
3. Content of Exosomes
The biological cargo of exosomes varies widely depending on their cell type of origin and their activation sites [15]. They mainly consist of lipids, proteins, nucleic acids, specifically mRNAs and miRNAs, and small molecules [16,17]. More than 4000 different proteins and over 2400 different RNAs have been identified and characterized in exosomes [18].
Owing the endosomal origin, exosomes are enriched in several proteins that are engaged in the biogenesis of MVBs, such as clathrin, ALIX (ALG-2 interacting protein X), TSG101 (tumor susceptibility gene 101) and ubiquitin [19]. In addition, exosomes contain CD9, CD63, CD81, CD82, CD54 and CD11b tetraspanins that serve as distinctive markers [20,21]. Moreover, heat shock proteins (HSP90, HSP70, HSP60) are common exosomal proteins that act as chaperones and play an essential role in cellular responses related to the environmental stress [19]. There are other exosome enriched molecules, including signal transduction molecules (i.e., ADP-ribosylation factor, cell division control protein 42, epidermal growth factor receptor, etc.) and lipid raft-related molecules, such as flotillin, phosphatidylserine, and sphingomyelin [22]. Exosomes contain cytoplasmic proteins (i.e., annexins), enzymes (i.e., protein kinase C, glyceraldehyde 3-phosphate dehydrogenase, etc.), and, depending on exosome origin, immunoregulatory molecules such as the major histocompatibility molecules (MHC) class I and II that have a significant role in immunoregulation by presenting antigenic peptides [19,23].
Exosomes carry functional RNA and miRNA that can be transferred into recipient cells and induce cellular response [7]. In contrast to cells, exosomes contain very little or no ribosomal RNA [24]. RNAs and miRNAs enveloped inside the extracellular vesicles are protected from nucleases, therefore they may have long lasting effects on disease related gene expression. In addition, selected exosomal miRNAs have specific profiles in different central nervous system (CNS) disorders even in the presymptomatic stages and their expression significantly changes under the neuropathological conditions, which make them promising biomarkers [25,26,27].
4. Huntington’s Disease
Huntington’s disease (HD) is a rare, progressive, and incurable neurodegenerative disorder with age of onset between 35 and 45 years and death occurring 15 to 17 years after onset [28,29,30]. The prevalence of HD is 5.7 persons per 100,000 worldwide with an average of 10.6 to 13.7 individuals per 100,000 in the European population [30,31,32]. In certain regions (i.e., Australia, North America, and Western Europe including the United Kingdom), the prevalence of the disease has increased over the past 50 years [33]. HD is characterized by progressive movement disorder (chorea, saccadic eye movement abnormalities, ataxia of speech, dysphagia, etc.), cognitive dysfunction (dementia), and psychiatric disturbances (depression, anxiety, apathy, etc.) [29,30]. Progressive movement failure is the main reason for life-ending complications.
HD has autosomal dominant heredity and is caused by an expanded Cytosine-Adenine-Guanine (CAG) repeat (≥36) in the huntingtin coding gene (IT15) on chromosome 4, resulting in the expression of mutant huntingtin protein (mHTT) with abnormal number of glutamine repeats (polyQ) in its N terminus [32,34]. The CAG repeats below 26 (common range in humans: 17–25) are considered normal and do not cause the disease. The intermediate range repeats (27 to 35) mostly do not cause HD with a few reported exceptions [35]. The CAG repeat range of 36–39 might be found in affected individuals as well as asymptomatic individuals (reduced penetrance alleles), whereas individuals with ≥40 repeats always develop the disorder (fully penetrant alleles) [32]. The number of CAG repeats is inversely correlated with the age of onset, longer repeats predict earlier onset of the disease and vice versa [36,37]. Predictive and diagnostic genetic testing is available to detect the expanded CAG repeats for the affected individuals as well as for individuals at risk [38]. Pretest counselling is essential to consider the test result impact on the patient and family [39].
There is no therapy available to slow down the disease progression and the HD treatment is largely focused on management of symptoms to control the motor and psychiatric disturbances [40,41]. Recently, new therapies targeting either DNA or RNA of mHTT to reduce the mHTT expression are being developed, using antisense oligonucleotides (ASOs), RNA interference (RNAi), zinc finger proteins (ZFPs), and the CRISPR-Cas9. Such huntingtin lowering strategies are considered the most promising treatments as they are targeting the proximate cause of the disease (Table 1) [32,41,42,43,44,45]. Over the past decades, clinical trials targeting the disease modification and symptomatic treatments in HD have begun, a number of these trials have failed and vast number are currently ongoing [46]. Availability of biomarkers, particularly accessible with minimal invasivity, is essential to estimate the treatment efficiency [47].

Table 1.
DNA- and RNA-based strategies targeting mHTT production pathway.
4.1. Huntingtin Protein
Huntingtin (HTT) is a large soluble protein (350 kDa), consisting of 3114 amino acids. The HTT protein is characterized by the presence of polyglutamine (polyQ) region, a proline rich region (PRR), and HEAT repeats (Huntingtin, Elongation factor 3, protein phosphatase 2A, Target of rapamycin 1) that are important for protein interactions, and caspase and calpain cleavage sites in higher vertebrates (Figure 2) [48,49]. The N-terminal 17 amino acids (N17) have been identified as a critical region that has a role in HTT localization, aggregation, and toxicity, and it is the subject of several post translational modifications including acetylation, SUMOylation, phosphorylation, and ubiquitination [49,50].
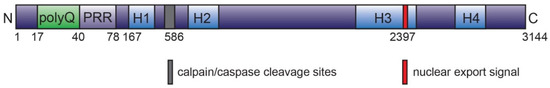
Figure 2.
Overview of human huntingtin protein. Full-length HTT consists of 3144 amino acids, with a total molecular mass of 350 kDa. The N-terminal 17 amino acid region carries several post translational modification sites (such as acetylation, SUMOylation, phosphorylation, ubiquitination, etc.) and is followed by polyglutamine tract (polyQ; expanded in Huntington’s disease), proline rich region (PRR), HEAT repeat domains (H1-H4; the HEAT corresponds to proteins, where they were first described: huntingtin, elongation factor 3 [EF3], protein phosphatase 2A [PP2A], and the yeast kinase TOR). Leucine-rich nuclear export signal is located at the C-terminal region. The protein may undergo post translational modifications and/or protease cleavage on several sites, e.g., caspase-6 cleavage at aspartate 586 [50,72,75,76,77].
PRR is found only in mammals, is variable in the non-HD population, and is essential for HTT interactions with proteins that contain tryptophanes or Src homology 3 domains. PRR deletion has no extreme effects on mouse behavior [67]. The PRR has a proline-proline helix which might be important for stability of the polyQ structure and the tendency of mHTT to aggregate [67]. HEAT repeats consist of around 50 amino acids and contain two antiparallel α-helices forming a hairpin that normally acts as scaffold for various protein complexes and mediates inter and intra molecular interactions [67,68]. A total of 16 HEAT repeats has been identified in the HTT protein and they are organized into 4 clusters [68] (Figure 2). Several proteolytic cleavage sites including proline, glutamic acid, serine, and threonine domains have been identified in HTT. These domains are found on both wild type and mHTT [67,69]. Proteases that are responsible for HTT cleavage include: caspases, calpain, cathepsins, aspartic endopeptidases, and the metalloproteinases [70,71]. HTT is cleaved by caspase 3 at amino acids 513 and 552, caspase 1 at position 572, caspase 2 at amino acid 552, and caspase 6 at amino acid 586 [72,73,74]. The two calpain cleavage sites are located at amino acids 469 and 536, and the MMP-10 metalloproteinase cleaves HTT at amino acid 402 [67,68].
HTT is ubiquitously expressed in the human tissues and organs with higher expression in the CNS and testes. HTT is essential for normal embryonic development [75], as HTT gene knockout is lethal in mice by 8.5 embryonic day [78,79]. HTT is engaged in many cellular and biological functions such as transcription, transport, vesicular trafficking, coordination of cell division, energy metabolism and antiapoptotic activity [42,49], and co-localizes with many organelles such as the nucleus, endoplasmic reticulum, Golgi complex, endosomes, mitochondria, and synaptic vesicles [48,80]. This might reflect its role as a scaffold protein that is engaged in many protein-protein interactions and the formation of multi-protein complexes [81]. Despite decades of research, the HTT roles in cells are not yet fully understood.
4.2. Pathogenesis of Huntington’s Disease
In general, the aggregation of misfolded proteins is the distinctive characteristic of neurodegenerative diseases (NDs) including Alzheimer’s disease (AD), Parkinson’s disease (PD), Amyotrophic lateral sclerosis (ALS), HD, and others. These protein aggregates form different types of inclusions, such as amyloid β and tau in AD, α-synuclein in PD, superoxide dismutase 1 in ALS or mHTT aggregates in HD.
Despite that the gene mutation causing HD was discovered in 1993 [76], the HD pathogenesis is not fully elucidated yet. The expanded polyQ sequence in mHTT protein has tendency to undergo conformational changes to form β-pleated sheet prone to aggregation [77,82]. The early phases of aggregate formation appear to be accelerated by hydrophobic interactions with an amphipathic α-helical structure of N17 [83,84]. Molecular chaperones play a major role in mHTT protein folding and re-folding [85]. Misfolded mHTT or its fragments and oligomers may undergo degradation in ubiquitin-proteasome system (UPS) or by autophagy (Figure 3).
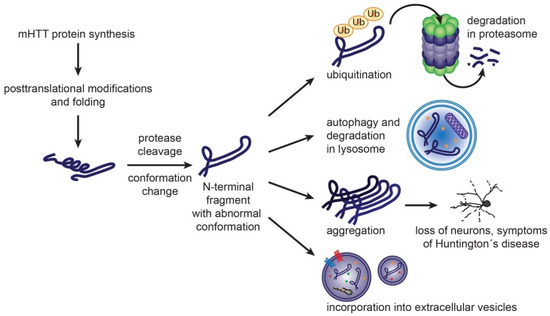
Figure 3.
Formation of mHTT toxic species and cellular mechanisms to handle them. Mutant huntingtin protein carrying expanded polyglutamine sequence is prone to adopt abnormal conformation. Post translational modifications and protease cleavage may contribute to formation of misfolded N-terminal fragments. Molecular chaperones, such as Heat Shock Proteins, participate in re-folding of misfolded proteins. Once unsuccessful, the resulting misfolded proteins can be degraded by ubiquitin-proteasome system or autophagy. Accumulation of misfolded mHTT rich in β-sheet structure leads to aggregate formation, loss of neurons and Huntington’s disease symptoms. Misfolded proteins may be eliminated from neurons also by exosomal secretion [86,87,88].
Under physiological conditions, proteostasis ensures balance between protein synthesis, folding, trafficking and degradation. The impairment of the proteostasis systems aggravates the aggregation of the misfolded huntingtin. Efficacy of proteostasis mechanisms declines with age. Experimental manipulation of distinct proteostasis nodes, such as molecular chaperones, UPS or autophagy may reduce toxic mHTT aggregate formation [81,85].
Post translational modifications also influence the mHTT toxicity, aggregation propensity and intracellular localization. Proteolytic cleavage of mHTT generates N-terminal fragments with increased tendency to aggregate. Such shorter fragments may be also formed by aberrant splicing of HTT coding mRNA [45]. Nuclear localization of mHTT increases its toxicity. On the other hand, HTT phosphorylations at Thr3 and Ser13 and/or Ser16 are potentially protective against aggregation [77]. Ubiquitination, acetylation and SUMoylation target the mHTT protein for degradation via ubiquitin proteasome system or autophagy [34].
The mHTT inclusions can induce a physical block of axonal transport between the cell body and the synaptic cleft and recruit other polyQ-containing proteins (i.e., transcription factors), which then interact with mHTT and might lose their physiological functions, leading to cell death [89,90]. The mHTT inclusions might thus function as sinks where vital proteins are sequestrated, compromising cell survival [91].
5. Exosomes in Huntington’s Disease
In the CNS, exosomes play essential physiological roles in the cell to cell communication and homeostasis maintenance required for normal brain function [92], particularly in neurogenesis [92,93,94,95], in synaptic activity and plasticity [96], myelination [88], and protection and regeneration of neuron after injury and disease [97,98,99] (Figure 4). EVs are released by neural cells including neurons, astrocytes, microglia, and oligodendrocytes [92,93,100] under both normal and pathological conditions. In human CNS, EVs have been isolated from the cerebrospinal fluid (CSF) and from adult brain [101,102]. The cellular origin of the neuronal secreted exosomes can be assessed by the presence of the GPI-anchored prion protein, cell adhesion molecule L1, and subunits of glutamate receptors [103,104].
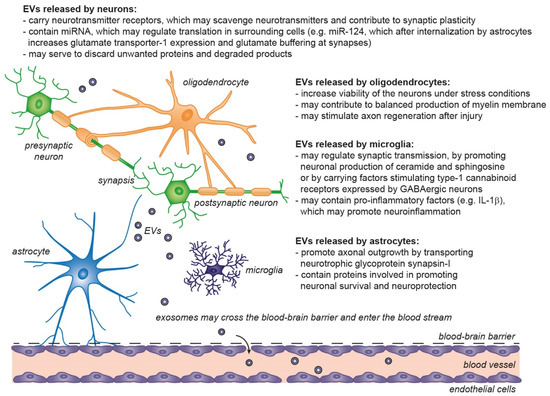
Figure 4.
Physiological roles of extracellular vesicles in the central nervous system. EVs released from neurons and glial cells show neurotrophic and neuroprotective effects, by regulation of synaptic transmission/plasticity, protection against stress and excitotoxicity and may support axon regeneration after injury. Astrocytes and microglia participate in removal of EVs produced by other cells and may contribute to degradation of unwanted proteins (including unfolded proteins) released through EVs [86,87,88,105,106,107].
5.1. Exosomes in Misfolded Protein Spreading
Recently, exosomes have emerged as a common player being able to mediate the pathogenesis of neurodegenerative disorders [108]. It has become clear that proteins related to NDs and prion diseases can be selectively integrated into ILVs of MVBs and subsequently released into the extracellular environment within exosomes [95].
Several studies showed that the neurotoxic misfolded proteins in NDs such as prion protein, α-synuclein, amyloid precursor protein, and tau, are transferred across the CNS by exosomes contributing to the spread and progression of prion disease and amyloidogenesis [16].
In HD, spreading of mHTT between cells is evident mainly from in vitro and animal experiments that revealed internalization of synthetic peptide (44Q) into cells and formation of cytoplasmic aggregates [109], as well as mHTT aggregate transmission between cells [110,111,112,113] and mHTT propagation between neurons [114]. Interestingly, the possibility of the mHTT spreading into genetically unrelated tissue was confirmed in vivo in 3 HD patients undergoing fetal striatal tissue transplantation, where mHTT aggregates were formed in the transplanted tissue [115].
Exosomes are supposed to be involved in such mHTT protein propagation between cells, both on protein and RNA levels. The injection of exosomes released from fibroblasts of HD patients into a newborn mouse brain’s ventricles triggered the manifestation of HD-related behavior and pathology [116]. However, this finding needs further confirmation, as other factors could co-isolate with exosomes from fibroblast conditioned medium by ExoQuick precipitation. At the RNA level, the ability of EVs to deliver toxic expanded trinucleotide repeat RNAs from one cell to another was reported in human 293T cell culture [117]. On the other hand, neuroprotective roles of exosomes secreted from astrocytes and adipose tissue-derived stem cells have been reported. Injection of astrocytic exosomes into the striatum of HD 140Q knock-in (KI) mice reduced the density of mHTT aggregates. Interestingly, presence of mHTT protein in exosomes released by primary astrocytes from 140Q KI mice was not detected, suggesting a potential use of astrocyte-derived exosomes in HD therapy [15]. Similarly, exosomes secreted from adipose-derived stem cells, that are known to release neurotrophic factors, were shown to reduce the mHTT aggregates, mitochondrial dysfunction, and cell apoptosis of in vitro HD model [118]. Interestingly, neuroprotective synaptic chaperone cysteine string protein α (CSPα) was observed to support the export of polyQ expanded (72Q) huntingtin exon 1 from cells via EVs. The CSPα may thus participate in protein homeostasis maintenance at synapses and mHTT clearance from cells [119].
5.2. Diagnostic Potential of Exosomes
Exosomes have large potential as non-invasive diagnostic biomarkers’ carriers for NDs as well as many other diseases. There are several reasons that make them attractive targets for clinical diagnostics and biomarker discovery. First, exosomal contents (lipids, proteins, nucleic acids, etc.) are changing during disease and might reflect the disease progress. Second, exosomes can be isolated non-invasively from easily accessible biological fluids including blood, urine, and saliva. The non-invasive availability is very important for early disease diagnosis especially in NDs. Third, exosomes have a double layer membrane, which protects the potential biomarkers from degradation. Fourth, exosomes are highly stable, making their clinical use practical, as samples can be kept for a long period before analysis. Fifth, exosomes carry markers related to their cellular origin, therefore, the origin can be traced. Finally, exosomes can pass through the blood-brain barrier from blood to brain and vice versa, providing information about nervous cells that is hard to obtain without the use of invasive techniques [12,98,120]. Many studies reported exosomes being biomarker sources for various NDs including AD [104,121,122,123,124], PD [125,126,127,128,129], ALS [118], and prion diseases [95].
Reliable non-invasive biomarkers reflecting disease progression are highly important for patients to provide early diagnostic proof or enable monitoring of currently emerging therapies [26]. Misfolded proteins or protein aggregates/inclusions are the pathological hallmark of many neurodegenerative disorders including HD, therefore the study of these proteins might be essential in developing new biomarkers [118]. Exosomes might contain the mHTT, its fragments, or other proteins reflecting the conditions of exosomes producing CNS cells. Such proteins might serve as potential biomarkers of HD. To date, there are only few studies that analyzed exosomes or their composition in the search for HD biomarkers, and more investigations are in need.
Blood, as the biological fluid easily collectable with minimal invasiveness, contains extracellular vesicles from the whole organism, with the majority (70–90%) of vesicles originating from platelets [130,131]. Thus, the platelets and platelet-derived EVs were searched as potential HD biomarker carriers. However, no differences were found in the number of EVs released by platelets between HD patients and controls and no correlations of platelet-derived EV numbers with age, CAG repeat number and disease stage were observed [132]. Platelets from HD patients contain highest amounts of mHTT among all blood cells [133]. Surprisingly, in EVs derived from platelets, the mHTT protein was undetectable, even after platelet activation [132]. In this study, the platelet-derived EVs were pelleted by centrifugation at 20,000× g for 90 min and the mHTT was detected in EV lysates by highly sensitive 2B7-MW1 Singulex detection assay (sensitivity 4 pg/mL) [132]. Altogether, platelet-derived EVs do not seem to be valuable biomarkers of HD.
Application of omics techniques enabled to map the molecular composition of EV in NDs. While EV protein and miRNA alterations have been studied in more frequent Alzheimer’s and Parkinson’s diseases [25,134], only little is known in HD.
Proteomic analysis of urinary EVs in more than 100 participants was performed by Wang et al., and revealed the enrichment of endolysosomal proteins linked to PD, AD, and HD, which makes the urinary EVs a highly accessible resource for biomarker discovery with particular promise for NDs [135].
Roles of miRNAs in disease pathophysiology are extensively studied as miRNA are significant regulators of gene expression. Several exosomal miRNAs are candidates for ND biomarkers, such as decreased miR-342-3p, miR-125a-5p, miR-125b-5p, miR451a, miR-23a-3p, and miR-126-3p, in AD [136,137,138] or increased miR-331-5p, miR-22*, miR-23a, miR-24, in PD [138,139]. However, in Huntington’s disease, the miRNA content in exosomes was not studied yet. Reed et al. identified 6 significantly increased miRNAs (miR-135b-3p, miR-520f-3p, miR-4317, miR-3928-5p, miR-140-5p, and miR-8082) in CSF in the prodromal HD gene polyQ expansion carriers compared to controls [140]. In blood plasma of symptomatic HD patients, 13 miRNAs (miR-877-5p, miR-223-3p, miR-223-5p, miR-30d-5p, miR-128, miR-22-5p, miR-222-3p, miR-338-3p, miR-130b-3p, miR-425-5p, miR-628-3p, miR-361-5p, miR-942) were upregulated [141]. Several studies reported a decrease of miR-124 expression in HD patients’ brains [142,143], STHdhQ111/HdhQ111 HD cell line, and R6/2 HD mouse [144]. MiR-124 plays a key role in neurogenesis and is the most abundant miRNA in the adult brain [145]. Detection of miR-124 in body fluids, whether in free-form or packed into exosomes, and its possible association with HD, deserve further studies.
5.3. Exosomes in Delivery of Therapeutics
EVs, due to their ability to cross blood-brain barrier [146] and biocompatibility, are promising therapeutic drug carriers into the CNS in NDs with few ongoing preclinical trials in AD and PD [147,148,149]. In HD, exosomes are particularly efficient in delivery of oligonucleotide therapeutics (miRNA and siRNA) (Table 1). Oligonucleotide therapeutics are a novel class of drugs targeting RNA or DNA to prevent expression of the protein responsible for the disease phenotype [54] and primarily include miRNAs, siRNAs and ASOs.
Recently, miRNAs-based therapy has emerged as a promising technique in NDs’ treatments [150]. miR-124 was selected in the majority of exosome-based miRNA delivery studies for ND treatment because it is highly and specifically expressed in all brain regions except for the pituitary gland, and at 100 times lower expression in other tissues, and has a regulatory role in CNS development and diseases [151]. miR-124 supports adult neurogenesis and expression of brain-derived neurotrophic factor by downregulating the expression of repressor RE1-Silencing Transcription Factor (REST) [57,152] and is one of the most downregulated miRNAs in HD. It has been reported that miR-124 slows down the progression of HD in R6/2 HD transgenic mouse through promoting neuronal differentiation and survival [153]. In 2017, Lee et al. developed an exosome based delivery to mouse HD model by generating miR-124 expressing HEK293 cell line to produce the exosomal miR-124 [57]. Such exosomes were injected into the striatum of R6/2 transgenic HD mice which resulted in reduction of REST protein expression. Unfortunately, no significant improvement in rotarod performance was observed in this study one week after miR-124 delivery, and longer time intervals have not been studied [57]. Due to the mild therapeutic efficiency of exosomal miR-124, Lee et al. suggested the increment of miRNAs’ dose packed in the exosomes, and also suggested the use of other miRNAs (i.e., miR-9, miR-22, miR-125b, miR-146a, miR-150, and miR-214) to be delivered by exosomes as they might have greater therapeutic impacts than miR-124 [57].
A small interfering RNA (siRNA) binds a target mRNA, guiding mRNA cleavage through RNA-induced silencing complex (RISC) which can provide effective long-term gene silencing [53,54]. The therapeutic potential of exosome-mediated siRNA delivery in BACHD and N171-82Q mice models was tested by Wu et al. [56]. Modified exosomes expressing the neuron-specific rabies viral glycoprotein (RVG) peptide loaded with siRNA targeting human huntingtin exon 1 (HuHtt) transcript was used. Then, HuHtt-siRNA RVG exosomes were injected intravenously to normal mice, and BACHD and N171-82Q transgenic mice at 10 mg/kg every two days for two weeks. siRNA was efficiently delivered into the mouse brain by RVG-modified exosomes and HuHtt-siRNA RVG exosomes significantly reduced Htt expression up to 46% and 54%, respectively, in transgenic mouse lines. N17-82Q mice receiving RVG exosomes showed improvement on the Rotarod performance. The study indicated the therapeutic potential of HuHtt-siRNA RVG exosomes in HD [56].
Hydrophobically-modified small interfering RNA (hsiRNA) are asymmetric oligonucleotides with chemical modifications that enhance stability and promote cellular internalization. They efficiently bind cellular membranes, enter cells, and stimulate gene silencing [53,54,154]. Exosomes are promising natural nano-devices for therapeutic RNA delivery but loading of the sufficient dose of RNAs remains a challenge [55]. To increase the efficiency, Bicans et al. synthesized a group of cholesterol-conjugated hsiRNAs to be loaded into exosomes. Cholesterol-conjugated hsiRNAs delivered by exosomes (100,000 g fraction) were more effective at inducing HTT mRNA silencing in neurons compared to cholesterol hsiRNA alone. In addition, cholesterol hsiRNAs did not induce the silencing of the target mRNA when delivered by large EVs (10,000 g fraction) [55].
In another study with hsiRNA targeting huntingtin RNA, exosomes were loaded by hsiRNA by simple co-incubation. In in vitro experiments, such exosomes mediated internalization of hsiRNAHTT into primary cortical neurons leading to dose-dependent silencing of Htt mRNA and protein in these cells. Unilateral infusion of such hsiRNAHTT- loaded exosomes into mouse striatum resulted in bilateral hsiRNAHTT distribution in striatal and cortical regions and statistically significant bilateral silencing of up to 35% Htt mRNA [54]. hsiRNAHTT alone, without packing into exosomes, did not show such an effect [54]. This can be a trajectory to use exosomes for delivery of the therapeutic oligonucleotides to treat NDs.
6. Conclusions
HD is a hereditary autosomal dominant neurodegenerative disorder. HD is incurable which represents a great challenge for patients and their families. Currently, there are several therapeutic interventions being developed to prevent or delay the onset of the symptoms and slow down the disease progression. However, the lack of powerful biomarkers that might be used to evaluate the efficiency of the applied therapies is still challenging.
Recently, there has been growing interest in extracellular vesicles (EVs), mainly exosomes, as a potential source for novel non-invasive biomarkers for NDs. Exosomes play significant roles in HDs and many other NDs by acting as a carrier of the misfolded proteins, and other molecules (RNAs and miRNAs) all over tissues and body fluids. Exosomes could be used both as a source of biomarkers to indicate the early stage of the disease and help understanding its pathogenicity, and as a therapeutic moiety, e.g., for delivery of gene silencing therapies. Due to their ability to cross the BBB and their availability in almost all body fluids, exosomes can be a potential source of novel, noninvasive biomarkers for HD, as well as many other diseases. Nonetheless, the use of exosomes as biomarkers in HD is still in the early stages and more studies need to be performed in this field. On the other hand, exosomes as possible carriers of novel therapies for HD and many other NDs are currently being investigated.
Author Contributions
H.A. and H.K.S. performed literature search, drafted and edited the text and prepared figures. P.V. participated in manuscript drafting and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Grantová Agentura České Republiky, project 19-01747S, and by Ministerstvo Školství, Mládeže a Tělovýchovy, project InterCOST LTC18079 under CellFit COST Action CA16119.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the international society for extracellular vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [PubMed]
- Yáñez-Mó, M.; Siljander, P.R.-M.; Andreu, Z.; Zavec, A.B.; Borràs, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef]
- Johnstone, R.M.; Adam, M.; Hammond, J.R.; Orr, L.; Turbide, C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J. Biol. Chem. 1987, 262, 9412–9420. [Google Scholar] [CrossRef]
- Van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef]
- Lakhal, S.; Wood, M.J.A. Exosome nanotechnology: An emerging paradigm shift in drug delivery: Exploitation of exosome nanovesicles for systemic in vivo delivery of RNAi heralds new horizons for drug delivery across biological barriers. BioEssays 2011, 33, 737–741. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.G.; Chopp, M. Exosomes in stroke pathogenesis and therapy. J. Clin. Investig. 2016, 126, 1190–1197. [Google Scholar] [CrossRef]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of MRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Malm, T.; Loppi, S.; Kanninen, K.M. Exosomes in Alzheimer’s disease. Neurochem. Int. 2016, 97, 193–199. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, Y.; Liu, H.; Tang, W.H. Exosomes: Biogenesis, biologic function and clinical potential. Cell Biosci. 2019, 9, 19. [Google Scholar] [CrossRef]
- Jan, A.; Rahman, S.; Khan, S.; Tasduq, S.; Choi, I. Biology, pathophysiological role, and clinical implications of exosomes: A critical appraisal. Cells 2019, 8, 99. [Google Scholar] [CrossRef]
- Yue, B.; Yang, H.; Wang, J.; Ru, W.; Wu, J.; Huang, Y.; Lan, X.; Lei, C.; Chen, H. Exosome biogenesis, secretion and function of exosomal MiRNAs in skeletal muscle myogenesis. Cell Prolif. 2020, 53, e12857. [Google Scholar] [CrossRef]
- Lin, J.; Li, J.; Huang, B.; Liu, J.; Chen, X.; Chen, X.-M.; Xu, Y.-M.; Huang, L.-F.; Wang, X.-Z. Exosomes: Novel biomarkers for clinical diagnosis. Sci. World J. 2015, 2015, 657086. [Google Scholar] [CrossRef]
- Stuffers, S.; Sem Wegner, C.; Stenmark, H.; Brech, A. Multivesicular endosome biogenesis in the absence of ESCRTs. Traffic Cph. Den. 2009, 10, 925–937. [Google Scholar] [CrossRef] [PubMed]
- Katzmann, D.J.; Babst, M.; Emr, S.D. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 2001, 106, 145–155. [Google Scholar] [CrossRef]
- Hong, Y.; Zhao, T.; Li, X.-J.; Li, S. Mutant huntingtin inhibits AB-crystallin expression and impairs exosome secretion from astrocytes. J. Neurosci. 2017, 37, 9550–9563. [Google Scholar] [CrossRef]
- Properzi, F.; Ferroni, E.; Poleggi, A.; Vinci, R. The regulation of exosome function in the CNS: Implications for neurodegeneration. Swiss Med. Wkly. 2015, 145, w14204. [Google Scholar] [CrossRef]
- Zebrowska, A.; Skowronek, A.; Wojakowska, A.; Widlak, P.; Pietrowska, M. Metabolome of exosomes: Focus on vesicles released by cancer cells and present in human body fluids. Int. J. Mol. Sci. 2019, 20, 3461. [Google Scholar] [CrossRef]
- Van Dommelen, S.M.; Vader, P.; Lakhal, S.; Kooijmans, S.A.A.; van Solinge, W.W.; Wood, M.J.A.; Schiffelers, R.M. Microvesicles and exosomes: Opportunities for cell-derived membrane vesicles in drug delivery. J. Control. Release 2012, 161, 635–644. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Zitvogel, L.; Amigorena, S. Exosomes: Composition, biogenesis and function. Nat. Rev. Immunol. 2002, 2, 569–579. [Google Scholar] [CrossRef]
- Beach, A.; Zhang, H.-G.; Ratajczak, M.Z.; Kakar, S.S. Exosomes: An overview of biogenesis, composition and role in ovarian cancer. J. Ovarian Res. 2014, 7, 14. [Google Scholar] [CrossRef]
- Babst, M. MVB vesicle formation: ESCRT-dependent, ESCRT-independent and everything in between. Curr. Opin. Cell Biol. 2011, 23, 452–457. [Google Scholar] [CrossRef] [PubMed]
- Fevrier, B.; Vilette, D.; Archer, F.; Loew, D.; Faigle, W.; Vidal, M.; Laude, H.; Raposo, G. Cells release prions in association with exosomes. Proc. Natl. Acad. Sci. USA 2004, 101, 9683–9688. [Google Scholar] [CrossRef]
- Kalluri, R. The biology and function of exosomes in cancer. J. Clin. Investig. 2016, 126, 1208–1215. [Google Scholar] [CrossRef] [PubMed]
- Statello, L.; Maugeri, M.; Garre, E.; Nawaz, M.; Wahlgren, J.; Papadimitriou, A.; Lundqvist, C.; Lindfors, L.; Collén, A.; Sunnerhagen, P.; et al. Identification of RNA-binding proteins in exosomes capable of interacting with different types of RNA: RBP-facilitated transport of RNAs into exosomes. PLoS ONE 2018, 13, e0195969. [Google Scholar] [CrossRef]
- Xia, X.; Wang, Y.; Huang, Y.; Zhang, H.; Lu, H.; Zheng, J.C. Exosomal MiRNAs in central nervous system diseases: Biomarkers, pathological mediators, protective factors and therapeutic agents. Prog. Neurobiol. 2019, 183, 101694. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, L. Circulating exosomal MiRNA as diagnostic biomarkers of neurodegenerative diseases. Front. Mol. Neurosci. 2020, 13, 53. [Google Scholar] [CrossRef]
- Manna, I.; De Benedittis, S.; Quattrone, A.; Maisano, D.; Iaccino, E.; Quattrone, A. Exosomal MiRNAs as potential diagnostic biomarkers in Alzheimer’s disease. Pharmaceuticals 2020, 13, 243. [Google Scholar] [CrossRef]
- Helder, D.I.; Kaptein, A.A.; van Kempen, G.M.J.; van Houwelingen, J.C.; Roos, R.A.C. Impact of Huntington’s disease on quality of life. Mov. Disord. 2001, 16, 325–330. [Google Scholar] [CrossRef]
- Coulson, N.S.; Buchanan, H.; Aubeeluck, A. Social support in cyberspace: A content analysis of communication within a Huntington’s disease online support group. Patient Educ. Couns. 2007, 68, 173–178. [Google Scholar] [CrossRef]
- Dayalu, P.; Albin, R.L. Huntington disease. Neurol. Clin. 2015, 33, 101–114. [Google Scholar] [CrossRef]
- Wynford-Thomas, R.; Robertson, N.P. The economic burden of chronic neurological disease. J. Neurol. 2017, 264, 2345–2347. [Google Scholar] [CrossRef] [PubMed]
- McColgan, P.; Tabrizi, S.J. Huntington’s disease: A clinical review. Eur. J. Neurol. 2018, 25, 24–34. [Google Scholar] [CrossRef] [PubMed]
- Rawlins, M.D.; Wexler, N.S.; Wexler, A.R.; Tabrizi, S.J.; Douglas, I.; Evans, S.J.W.; Smeeth, L. The prevalence of Huntington’s disease. Neuroepidemiology 2016, 46, 144–153. [Google Scholar] [CrossRef] [PubMed]
- Arrasate, M.; Finkbeiner, S. Protein aggregates in Huntington’s disease. Exp. Neurol. 2012, 238, 1–11. [Google Scholar] [CrossRef]
- Ha, A.D.; Jankovic, J. Exploring the correlates of intermediate CAG repeats in huntington disease. Postgrad. Med. 2011, 123, 116–121. [Google Scholar] [CrossRef]
- Schneider, S.A.; Bird, T. Huntington’s disease, Huntington’s disease look-alikes, and benign hereditary chorea: What’s new? Mov. Disord. Clin. Pract. 2016, 3, 342–354. [Google Scholar] [CrossRef]
- Capiluppi, E.; Romano, L.; Rebora, P.; Nanetti, L.; Castaldo, A.; Gellera, C.; Mariotti, C.; Macerollo, A.; Cislaghi, M.G. Late-onset Huntington’s disease with 40–42 CAG expansion. Neurol. Sci. 2020, 41, 869–876. [Google Scholar] [CrossRef]
- Testa, C.M.; Jankovic, J. Huntington disease: A quarter century of progress since the gene discovery. J. Neurol. Sci. 2019, 396, 52–68. [Google Scholar] [CrossRef]
- Craufurd, D.; MacLeod, R.; Frontali, M.; Quarrell, O.; Bijlsma, E.K.; Davis, M.; Hjermind, L.E.; Lahiri, N.; Mandich, P.; Martinez, A.; et al. Diagnostic genetic testing for Huntington’s disease. Pract. Neurol. 2015, 15, 80–84. [Google Scholar] [CrossRef] [PubMed]
- Dickey, A.S.; La Spada, A.R. Therapy development in Huntington disease: From current strategies to emerging opportunities. Am. J. Med. Genet. A 2018, 176, 842–861. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Feigin, A. Huntington’s disease: New frontiers in therapeutics. Curr. Neurol. Neurosci. Rep. 2021, 21, 10. [Google Scholar] [CrossRef]
- Wild, E.J.; Tabrizi, S. Therapies targeting DNA and RNA in Huntington’s disease. Lancet Neurol. 2017, 16, 837–847. [Google Scholar] [CrossRef]
- Tabrizi, S.J.; Ghosh, R.; Leavitt, B.R. Huntingtin lowering strategies for disease modification in Huntington’s disease. Neuron 2019, 101, 801–819. [Google Scholar] [CrossRef]
- Shannon, K.M. Recent advances in the treatment of Huntington’s disease: Targeting DNA and RNA. CNS Drugs 2020, 34, 219–228. [Google Scholar] [CrossRef] [PubMed]
- Tabrizi, S.J.; Flower, M.D.; Ross, C.A.; Wild, E.J. Huntington disease: New insights into molecular pathogenesis and therapeutic opportunities. Nat. Rev. Neurol. 2020, 16, 529–546. [Google Scholar] [CrossRef]
- Bashir, H. Emerging therapies in Huntington’s disease. Expert Rev. Neurother. 2019, 19, 983–995. [Google Scholar] [CrossRef]
- Przybyl, L.; Wozna-Wysocka, M.; Kozlowska, E.; Fiszer, A. What, when and how to measure—Peripheral biomarkers in therapy of Huntington’s disease. Int. J. Mol. Sci. 2021, 22, 1561. [Google Scholar] [CrossRef] [PubMed]
- Schulte, J.; Littleton, J.T. The biological function of the Huntingtin protein and its relevance to Huntington’s disease pathology. Curr. Trends Neurol. 2011, 5, 65–78. [Google Scholar]
- Tourette, C.; Li, B.; Bell, R.; O’Hare, S.; Kaltenbach, L.S.; Mooney, S.D.; Hughes, R.E. A large scale Huntingtin protein interaction network implicates Rho GTPase signaling pathways in Huntington disease. J. Biol. Chem. 2014, 289, 6709–6726. [Google Scholar] [CrossRef] [PubMed]
- Arndt, J.R.; Chaibva, M.; Legleiter, J. The emerging role of the first 17 amino acids of huntingtin in Huntington’s disease. Biomol. Concepts 2015, 6, 33–46. [Google Scholar] [CrossRef] [PubMed]
- Tabrizi, S.J.; Leavitt, B.R.; Landwehrmeyer, G.B.; Wild, E.J.; Saft, C.; Barker, R.A.; Blair, N.F.; Craufurd, D.; Priller, J.; Rickards, H.; et al. Targeting huntingtin expression in patients with Huntington’s disease. N. Engl. J. Med. 2019, 380, 2307–2316. [Google Scholar] [CrossRef]
- Datson, N.A.; González-Barriga, A.; Kourkouta, E.; Weij, R.; van de Giessen, J.; Mulders, S.; Kontkanen, O.; Heikkinen, T.; Lehtimäki, K.; van Deutekom, J.C.T. The expanded CAG repeat in the huntingtin gene as target for therapeutic RNA modulation throughout the HD mouse brain. PLoS ONE 2017, 12, e0171127. [Google Scholar] [CrossRef]
- Alterman, J.F.; Hall, L.M.; Coles, A.H.; Hassler, M.R.; Didiot, M.-C.; Chase, K.; Abraham, J.; Sottosanti, E.; Johnson, E.; Sapp, E.; et al. Hydrophobically modified SiRNAs silence huntingtin MRNA in primary neurons and mouse brain. Mol. Ther. Nucleic Acids 2015, 4, e266. [Google Scholar] [CrossRef]
- Didiot, M.-C.; Hall, L.M.; Coles, A.H.; Haraszti, R.A.; Godinho, B.M.; Chase, K.; Sapp, E.; Ly, S.; Alterman, J.F.; Hassler, M.R.; et al. Exosome-mediated delivery of hydrophobically modified SiRNA for huntingtin MRNA silencing. Mol. Ther. 2016, 24, 1836–1847. [Google Scholar] [CrossRef] [PubMed]
- Biscans, A.; Haraszti, R.A.; Echeverria, D.; Miller, R.; Didiot, M.-C.; Nikan, M.; Roux, L.; Aronin, N.; Khvorova, A. Hydrophobicity of lipid-conjugated SiRNAs predicts productive loading to small extracellular vesicles. Mol. Ther. 2018, 26, 1520–1528. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Yu, M.; Zhang, L.; Chen, X.; Pei, Z. I02 Systemic injection of exosomal sirna significantly reduced huntingtin expression in transgenic mice of Huntington’s disease. J. Neurol. Neurosurg. Psychiatry 2018, 89, A88–A89. [Google Scholar] [CrossRef]
- Lee, S.-T.; Im, W.; Ban, J.-J.; Lee, M.; Jung, K.-H.; Lee, S.K.; Chu, K.; Kim, M. Exosome-based delivery of MiR-124 in a Huntington’s disease model. J. Mov. Disord. 2017, 10, 45–52. [Google Scholar] [CrossRef]
- Miniarikova, J.; Zanella, I.; Huseinovic, A.; van der Zon, T.; Hanemaaijer, E.; Martier, R.; Koornneef, A.; Southwell, A.L.; Hayden, M.R.; van Deventer, S.J.; et al. Design, characterization, and lead selection of therapeutic MiRNAs targeting huntingtin for development of gene therapy for Huntington’s disease. Mol. Ther. Nucleic Acids 2016, 5, e297. [Google Scholar] [CrossRef] [PubMed]
- Evers, M.M.; Miniarikova, J.; Juhas, S.; Vallès, A.; Bohuslavova, B.; Juhasova, J.; Skalnikova, H.K.; Vodicka, P.; Valekova, I.; Brouwers, C.; et al. AAV5-MiHTT gene therapy demonstrates broad distribution and strong human mutant huntingtin lowering in a Huntington’s disease minipig model. Mol. Ther. 2018, 26, 2163–2177. [Google Scholar] [CrossRef]
- Miniarikova, J.; Zimmer, V.; Martier, R.; Brouwers, C.C.; Pythoud, C.; Richetin, K.; Rey, M.; Lubelski, J.; Evers, M.M.; van Deventer, S.J.; et al. AAV5-MiHTT gene therapy demonstrates suppression of mutant huntingtin aggregation and neuronal dysfunction in a rat model of Huntington’s disease. Gene Ther. 2017, 24, 630–639. [Google Scholar] [CrossRef]
- Pfister, E.; Dinardo, N.; Mondo, E.; Borel, F.; Conroy, F.; Fraser, C.; Gernoux, G.; Han, X.; Hu, D.; Johnson, E.; et al. Artificial MiRNAs reduce human mutant huntingtin throughout the striatum in a transgenic sheep model of Huntington’s disease. Hum. Gene Ther. 2017, 29, 663–673. [Google Scholar] [CrossRef]
- Agustín-Pavón, C.; Mielcarek, M.; Garriga-Canut, M.; Isalan, M. Deimmunization for gene therapy: Host matching of synthetic zinc finger constructs enables long-term mutant huntingtin repression in mice. Mol. Neurodegener. 2016, 11, 64. [Google Scholar] [CrossRef]
- Zeitler, B.; Froelich, S.; Marlen, K.; Shivak, D.A.; Yu, Q.; Li, D.; Pearl, J.R.; Miller, J.C.; Zhang, L.; Paschon, D.E.; et al. Allele-selective transcriptional repression of mutant HTT for the treatment of Huntington’s disease. Nat. Med. 2019, 25, 1131–1142. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.W.; Kim, K.-H.; Chao, M.J.; Atwal, R.S.; Gillis, T.; MacDonald, M.E.; Gusella, J.F.; Lee, J.-M. Permanent inactivation of Huntington’s disease mutation by personalized allele-specific CRISPR/Cas9. Hum. Mol. Genet. 2016, 25, 4566–4576. [Google Scholar] [CrossRef]
- Yang, S.; Chang, R.; Yang, H.; Zhao, T.; Hong, Y.; Kong, H.E.; Sun, X.; Qin, Z.; Jin, P.; Li, S.; et al. CRISPR/Cas9-mediated gene editing ameliorates neurotoxicity in mouse model of Huntington’s disease. J. Clin. Investig. 2017, 127, 2719–2724. [Google Scholar] [CrossRef]
- Roche Provides Update on Tominersen Programme in Manifest Huntington’s Disease. Available online: https://www.roche.com/media/releases/med-cor-2021-03-22b.htm (accessed on 29 March 2021).
- Saudou, F.; Humbert, S. The biology of huntingtin. Neuron 2016, 89, 910–926. [Google Scholar] [CrossRef]
- Zheng, Z.; Diamond, M.I. Huntington disease and the huntingtin protein. Prog. Mol. Biol. Transl. Sci. 2012, 107, 189–214. [Google Scholar] [CrossRef]
- Warby, S.C.; Doty, C.N.; Graham, R.K.; Carroll, J.B.; Yang, Y.-Z.; Singaraja, R.R.; Overall, C.M.; Hayden, M.R. Activated Caspase-6 and Caspase-6-Cleaved Fragments of Huntingtin Specifically Colocalize in the Nucleus. Hum. Mol. Genet. 2008, 17, 2390–2404. [Google Scholar] [CrossRef] [PubMed]
- Tebbenkamp, A.T.N.; Crosby, K.W.; Siemienski, Z.B.; Brown, H.H.; Golde, T.E.; Borchelt, D.R. Analysis of proteolytic processes and enzymatic activities in the generation of huntingtin N-terminal fragments in an HEK293 cell model. PLoS ONE 2012, 7, e50750. [Google Scholar] [CrossRef] [PubMed]
- El-Daher, M.-T.; Hangen, E.; Bruyère, J.; Poizat, G.; Al-Ramahi, I.; Pardo, R.; Bourg, N.; Souquere, S.; Mayet, C.; Pierron, G.; et al. Huntingtin proteolysis releases non-PolyQ fragments that cause toxicity through dynamin 1 dysregulation. EMBO J. 2015, 34, 2255–2271. [Google Scholar] [CrossRef]
- Ehrnhoefer, D.E.; Sutton, L.; Hayden, M.R. Small changes, big impact: Posttranslational modifications and function of huntingtin in huntington disease. Neurosci. Rev. J. Bringing Neurobiol. Neurol. Psychiatry 2011, 17, 475–492. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.D.O.; Schmidt, M.E.; Nguyen, Y.T.; Lazic, N.; Hayden, M.R. Identification of a novel caspase cleavage site in huntingtin that regulates mutant huntingtin clearance. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2019, 33, 3190–3197. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-J.; Cao, Q.; Zhang, Y.; Su, X.-D. Activation and regulation of caspase-6 and its role in neurodegenerative diseases. Annu. Rev. Pharmacol. Toxicol. 2015, 55, 553–572. [Google Scholar] [CrossRef] [PubMed]
- Ross, C.A.; Tabrizi, S.J. Huntington’s disease: From molecular pathogenesis to clinical treatment. Lancet Neurol. 2011, 10, 83–98. [Google Scholar] [CrossRef]
- MacDonald, M.E.; Ambrose, C.M.; Duyao, M.P.; Myers, R.H.; Lin, C.; Srinidhi, L.; Barnes, G.; Taylor, S.A.; James, M.; Groot, N.; et al. A Novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell 1993, 72, 971–983. [Google Scholar] [CrossRef]
- Schaffert, L.-N.; Carter, W.G. Do post-translational modifications influence protein aggregation in neurodegenerative diseases: A systematic review. Brain Sci. 2020, 10, 232. [Google Scholar] [CrossRef]
- Nasir, J.; Floresco, S.B.; O’Kusky, J.R.; Diewert, V.M.; Richman, J.M.; Zeisler, J.; Borowski, A.; Marth, J.D.; Phillips, A.G.; Hayden, M.R. Targeted disruption of the Huntington’s disease gene results in embryonic lethality and behavioral and morphological changes in heterozygotes. Cell 1995, 81, 811–823. [Google Scholar] [CrossRef]
- Duyao, M.P.; Auerbach, A.B.; Ryan, A.; Persichetti, F.; Barnes, G.T.; McNeil, S.M.; Ge, P.; Vonsattel, J.P.; Gusella, J.F.; Joyner, A.L.; et al. Inactivation of the mouse Huntington’s disease gene homolog Hdh. Science 1995, 269, 407–410. [Google Scholar] [CrossRef] [PubMed]
- Parsons, M.P.; Raymond, L.A. Huntington disease. In Neurobiology of Brain Disorders; Elsevier: Amsterdam, The Netherlands, 2015; pp. 303–320. ISBN 978-0-12-398270-4. [Google Scholar]
- Harding, R.J.; Tong, Y.-F. Proteostasis in Huntington’s disease: Disease mechanisms and therapeutic opportunities. Acta Pharmacol. Sin. 2018, 39, 754–769. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.P. Toxic proteins in neurodegenerative disease. Science 2002, 296, 1991–1995. [Google Scholar] [CrossRef]
- Thakur, A.K.; Jayaraman, M.; Mishra, R.; Thakur, M.; Chellgren, V.M.; Byeon, I.-J.L.; Anjum, D.H.; Kodali, R.; Creamer, T.P.; Conway, J.F.; et al. Polyglutamine disruption of the huntingtin exon 1 N terminus triggers a complex aggregation mechanism. Nat. Struct. Mol. Biol. 2009, 16, 380–389. [Google Scholar] [CrossRef]
- Li, S.; Li, X.-J. Multiple pathways contribute to the pathogenesis of huntington disease found. Mol. Neurodegener. 2006, 1, 19. [Google Scholar] [CrossRef][Green Version]
- Koyuncu, S.; Fatima, A.; Gutierrez-Garcia, R.; Vilchez, D. Proteostasis of huntingtin in health and disease. Int. J. Mol. Sci. 2017, 18, 1568. [Google Scholar] [CrossRef]
- Li, H.; Luo, Y.; Zhu, L.; Hua, W.; Zhang, Y.; Zhang, H.; Zhang, L.; Li, Z.; Xing, P.; Zhang, Y.; et al. Glia-derived exosomes: Promising therapeutic targets. Life Sci. 2019, 239, 116951. [Google Scholar] [CrossRef]
- Zhang, G.; Yang, P. A novel cell-cell communication mechanism in the nervous system: Exosomes. J. Neurosci. Res. 2018, 96, 45–52. [Google Scholar] [CrossRef]
- Caruso Bavisotto, C.; Scalia, F.; Marino Gammazza, A.; Carlisi, D.; Bucchieri, F.; Conway de Macario, E.; Macario, A.J.L.; Cappello, F.; Campanella, C. Extracellular vesicle-mediated cell−cell communication in the nervous system: Focus on neurological diseases. Int. J. Mol. Sci. 2019, 20, 434. [Google Scholar] [CrossRef]
- Gunawardena, S.; Her, L.-S.; Brusch, R.G.; Laymon, R.A.; Niesman, I.R.; Gordesky-Gold, B.; Sintasath, L.; Bonini, N.M.; Goldstein, L.S.B. Disruption of axonal transport by loss of huntingtin or expression of pathogenic PolyQ proteins in drosophila. Neuron 2003, 40, 25–40. [Google Scholar] [CrossRef]
- Rossetti, G.; Magistrato, A. Molecular mechanism of Huntington’s disease—A computational perspective. In Huntington’s Disease—Core Concepts and Current Advances; Ersoy Tunal, N., Ed.; InTech: London, UK, 2012; ISBN 978-953-307-953-0. [Google Scholar]
- Wyttenbach, A.; Carmichael, J.; Swartz, J.; Furlong, R.A.; Narain, Y.; Rankin, J.; Rubinsztein, D.C. Effects of heat shock, heat shock protein 40 (HDJ-2), and proteasome inhibition on protein aggregation in cellular models of Huntington’s disease. Proc. Natl. Acad. Sci. USA 2000, 97, 2898–2903. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, L.; Bali, J.; Barr, M.M.; Court, F.A.; Krämer-Albers, E.-M.; Picou, F.; Raposo, G.; van der Vos, K.E.; van Niel, G.; Wang, J.; et al. Emerging roles of extracellular vesicles in the nervous system. J. Neurosci. 2014, 34, 15482–15489. [Google Scholar] [CrossRef] [PubMed]
- Yuyama, K.; Igarashi, Y. Physiological and pathological roles of exosomes in the nervous system. Biomol. Concepts 2016, 7, 53–68. [Google Scholar] [CrossRef]
- Schneider, A.; Simons, M. Exosomes: Vesicular carriers for intercellular communication in neurodegenerative disorders. Cell Tissue Res. 2013, 352, 33–47. [Google Scholar] [CrossRef] [PubMed]
- Vella, L.J.; Sharples, R.A.; Nisbet, R.M.; Cappai, R.; Hill, A.F. The role of exosomes in the processing of proteins associated with neurodegenerative diseases. Eur. Biophys. J. EBJ 2008, 37, 323–332. [Google Scholar] [CrossRef]
- Antonucci, F.; Turola, E.; Riganti, L.; Caleo, M.; Gabrielli, M.; Perrotta, C.; Novellino, L.; Clementi, E.; Giussani, P.; Viani, P.; et al. Microvesicles released from microglia stimulate synaptic activity via enhanced sphingolipid metabolism: Microglial MVs increase sphingolipid metabolism in neurons. EMBO J. 2012, 31, 1231–1240. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Cesca, F.; Loers, G.; Schweizer, M.; Buck, F.; Benfenati, F.; Schachner, M.; Kleene, R. Synapsin I Is an oligomannose-carrying glycoprotein, acts as an oligomannose-binding lectin, and promotes neurite outgrowth and neuronal survival when released via glia-derived exosomes. J. Neurosci. 2011, 31, 7275–7290. [Google Scholar] [CrossRef]
- Kanninen, K.M.; Bister, N.; Koistinaho, J.; Malm, T. Exosomes as new diagnostic tools in CNS diseases. Biochim. Biophys. Acta BBA Mol. Basis Dis. 2016, 1862, 403–410. [Google Scholar] [CrossRef]
- Gharbi, T.; Zhang, Z.; Yang, G.-Y. The function of astrocyte mediated extracellular vesicles in central nervous system diseases. Front. Cell Dev. Biol. 2020, 8, 568889. [Google Scholar] [CrossRef] [PubMed]
- Mrowczynski, O.D.; Zacharia, B.E.; Connor, J.R. Exosomes and their implications in central nervous system tumor biology. Prog. Neurobiol. 2019, 172, 71–83. [Google Scholar] [CrossRef]
- Street, J.M.; Barran, P.E.; Mackay, C.L.; Weidt, S.; Balmforth, C.; Walsh, T.S.; Chalmers, R.T.A.; Webb, D.J.; Dear, J.W. Identification and proteomic profiling of exosomes in human cerebrospinal fluid. J. Transl. Med. 2012, 10, 5. [Google Scholar] [CrossRef] [PubMed]
- Banigan, M.G.; Kao, P.F.; Kozubek, J.A.; Winslow, A.R.; Medina, J.; Costa, J.; Schmitt, A.; Schneider, A.; Cabral, H.; Cagsal-Getkin, O.; et al. Differential expression of exosomal MicroRNAs in prefrontal cortices of schizophrenia and bipolar disorder patients. PLoS ONE 2013, 8, e48814. [Google Scholar] [CrossRef]
- Lachenal, G.; Pernet-Gallay, K.; Chivet, M.; Hemming, F.J.; Belly, A.; Bodon, G.; Blot, B.; Haase, G.; Goldberg, Y.; Sadoul, R. Release of exosomes from differentiated neurons and its regulation by synaptic glutamatergic activity. Mol. Cell. Neurosci. 2011, 46, 409–418. [Google Scholar] [CrossRef]
- Fiandaca, M.S.; Kapogiannis, D.; Mapstone, M.; Boxer, A.; Eitan, E.; Schwartz, J.B.; Abner, E.L.; Petersen, R.C.; Federoff, H.J.; Miller, B.L.; et al. Identification of pre-clinical Alzheimer’s disease by a profile of pathogenic proteins in neurally-derived blood exosomes: A case-control study. Alzheimers Dement. J. Alzheimers Assoc. 2015, 11, 600–607.e1. [Google Scholar] [CrossRef] [PubMed]
- Gassama, Y.; Favereaux, A. Emerging roles of extracellular vesicles in the central nervous system: Physiology, pathology, and therapeutic perspectives. Front. Cell. Neurosci. 2021, 15, 626043. [Google Scholar] [CrossRef] [PubMed]
- Blandford, S.N.; Galloway, D.A.; Moore, C.S. The roles of extracellular vesicle MicroRNAs in the central nervous system. Glia 2018, 66, 2267–2278. [Google Scholar] [CrossRef]
- Paolicelli, R.C.; Bergamini, G.; Rajendran, L. Cell-to-cell communication by extracellular vesicles: Focus on microglia. Neuroscience 2019, 405, 148–157. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Pulliam, L. Exosomes as mediators of neuroinflammation. J. Neuroinflam. 2014, 11, 68. [Google Scholar] [CrossRef] [PubMed]
- Ren, P.-H.; Lauckner, J.E.; Kachirskaia, I.; Heuser, J.E.; Melki, R.; Kopito, R.R. Cytoplasmic penetration and persistent infection of mammalian cells by polyglutamine aggregates. Nat. Cell Biol. 2009, 11, 219–225. [Google Scholar] [CrossRef] [PubMed]
- Costanzo, M.; Abounit, S.; Marzo, L.; Danckaert, A.; Chamoun, Z.; Roux, P.; Zurzolo, C. Transfer of polyglutamine aggregates in neuronal cells occurs in tunneling nanotubes. J. Cell Sci. 2013, 126, 3678–3685. [Google Scholar] [CrossRef]
- Herrera, F.; Tenreiro, S.; Miller-Fleming, L.; Outeiro, T.F. Visualization of cell-to-cell transmission of mutant huntingtin oligomers. PLoS Curr. 2011, 3, RRN1210. [Google Scholar] [CrossRef]
- Yang, W.; Dunlap, J.R.; Andrews, R.B.; Wetzel, R. Aggregated polyglutamine peptides delivered to nuclei are toxic to mammalian cells. Hum. Mol. Genet. 2002, 11, 2905–2917. [Google Scholar] [CrossRef]
- Pearce, M.M.P.; Spartz, E.J.; Hong, W.; Luo, L.; Kopito, R.R. Prion-like transmission of neuronal huntingtin aggregates to phagocytic glia in the drosophila brain. Nat. Commun. 2015, 6, 6768. [Google Scholar] [CrossRef]
- Pecho-Vrieseling, E.; Rieker, C.; Fuchs, S.; Bleckmann, D.; Esposito, M.S.; Botta, P.; Goldstein, C.; Bernhard, M.; Galimberti, I.; Müller, M.; et al. Transneuronal propagation of mutant huntingtin contributes to non-cell autonomous pathology in neurons. Nat. Neurosci. 2014, 17, 1064–1072. [Google Scholar] [CrossRef] [PubMed]
- Cicchetti, F.; Lacroix, S.; Cisbani, G.; Vallières, N.; Saint-Pierre, M.; St-Amour, I.; Tolouei, R.; Skepper, J.N.; Hauser, R.A.; Mantovani, D.; et al. Mutant huntingtin is present in neuronal grafts in huntington disease patients: Transfer of mutant huntingtin to normal tissue. Ann. Neurol. 2014, 76, 31–42. [Google Scholar] [CrossRef]
- Jeon, I.; Cicchetti, F.; Cisbani, G.; Lee, S.; Li, E.; Bae, J.; Lee, N.; Li, L.; Im, W.; Kim, M.; et al. Human-to-mouse prion-like propagation of mutant huntingtin protein. Acta Neuropathol. 2016, 132, 577–592. [Google Scholar] [CrossRef]
- Zhang, X.; Abels, E.R.; Redzic, J.S.; Margulis, J.; Finkbeiner, S.; Breakefield, X.O. Potential transfer of polyglutamine and CAG-repeat RNA in extracellular vesicles in Huntington’s disease: Background and evaluation in cell culture. Cell. Mol. Neurobiol. 2016, 36, 459–470. [Google Scholar] [CrossRef]
- Lee, M.; Liu, T.; Im, W.; Kim, M. Exosomes from adipose-derived stem cells ameliorate phenotype of Huntington’s disease in vitro model. Eur. J. Neurosci. 2016, 44, 2114–2119. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Koutras, C.; Donnelier, J.; Alshehri, M.; Fotouhi, M.; Girard, M.; Casha, S.; McPherson, P.S.; Robbins, S.M.; Braun, J.E.A. Neurons export extracellular vesicles enriched in cysteine string protein and misfolded protein cargo. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef]
- Boukouris, S.; Mathivanan, S. Exosomes in bodily fluids are a highly stable resource of disease biomarkers. Proteom. Clin. Appl. 2015, 9, 358–367. [Google Scholar] [CrossRef]
- Manterola, L.; Guruceaga, E.; Pérez-Larraya, J.G.; González-Huarriz, M.; Jauregui, P.; Tejada, S.; Diez-Valle, R.; Segura, V.; Samprón, N.; Barrena, C.; et al. A small noncoding RNA signature found in exosomes of GBM patient serum as a diagnostic tool. Neuro-oncology 2014, 16, 520–527. [Google Scholar] [CrossRef]
- Vingtdeux, V.; Sergeant, N.; Buée, L. Potential contribution of exosomes to the prion-like propagation of lesions in Alzheimer’s disease. Front. Physiol. 2012, 3. [Google Scholar] [CrossRef]
- Winston, C.N.; Goetzl, E.J.; Akers, J.C.; Carter, B.S.; Rockenstein, E.M.; Galasko, D.; Masliah, E.; Rissman, R.A. Prediction of conversion from mild cognitive impairment to dementia with neuronally derived blood exosome protein profile. Alzheimers Dement. Diagn. Assess. Dis. Monit. 2016, 3, 63–72. [Google Scholar] [CrossRef]
- Goetzl, E.J.; Boxer, A.; Schwartz, J.B.; Abner, E.L.; Petersen, R.C.; Miller, B.L.; Kapogiannis, D. Altered lysosomal proteins in neural-derived plasma exosomes in preclinical Alzheimer disease. Neurology 2015, 85, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Tomlinson, P.R.; Zheng, Y.; Fischer, R.; Heidasch, R.; Gardiner, C.; Evetts, S.; Hu, M.; Wade-Martins, R.; Turner, M.R.; Morris, J.; et al. Identification of distinct circulating exosomes in Parkinson’s disease. Ann. Clin. Transl. Neurol. 2015, 2, 353–361. [Google Scholar] [CrossRef] [PubMed]
- Vella, L.J.; Hill, A.F.; Cheng, L. Focus on extracellular vesicles: Exosomes and their role in protein trafficking and biomarker potential in Alzheimer’s and Parkinson’s disease. Int. J. Mol. Sci. 2016, 17, 173. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.-Y.; Lu, J.-M.; Zhao, Z.-Q.; Li, M.-C.; Lu, T.; An, X.-S.; Xue, L.-J. MicroRNA biomarkers of Parkinson’s disease in serum exosome-like microvesicles. Neurosci. Lett. 2017, 644, 94–99. [Google Scholar] [CrossRef] [PubMed]
- You, Y.; Ikezu, T. Emerging roles of extracellular vesicles in neurodegenerative disorders. Neurobiol. Dis. 2019, 130, 104512. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Liu, C.; Cook, T.J.; Bullock, K.M.; Zhao, Y.; Ginghina, C.; Li, Y.; Aro, P.; Dator, R.; He, C.; et al. Plasma exosomal α-synuclein is likely CNS-derived and increased in Parkinson’s disease. Acta Neuropathol. 2014, 128, 639–650. [Google Scholar] [CrossRef]
- Foster, B.P.; Balassa, T.; Benen, T.D.; Dominovic, M.; Elmadjian, G.K.; Florova, V.; Fransolet, M.D.; Kestlerova, A.; Kmiecik, G.; Kostadinova, I.A.; et al. Extracellular vesicles in blood, milk and body fluids of the female and male urogenital tract and with special regard to reproduction. Crit. Rev. Clin. Lab. Sci. 2016, 53, 379–395. [Google Scholar] [CrossRef]
- Espinosa-Parrilla, Y.; Gonzalez-Billault, C.; Fuentes, E.; Palomo, I.; Alarcón, M. Decoding the role of platelets and related MicroRNAs in aging and neurodegenerative disorders. Front. Aging Neurosci. 2019, 11. [Google Scholar] [CrossRef]
- Denis, H.L.; Lamontagne-Proulx, J.; St-Amour, I.; Mason, S.L.; Weiss, A.; Chouinard, S.; Barker, R.A.; Boilard, E.; Cicchetti, F. Platelet-derived extracellular vesicles in Huntington’s disease. J. Neurol. 2018, 265, 2704–2712. [Google Scholar] [CrossRef]
- Denis, H.L.; Lamontagne-Proulx, J.; St-Amour, I.; Mason, S.L.; Rowley, J.W.; Cloutier, N.; Tremblay, M.-È.; Vincent, A.T.; Gould, P.V.; Chouinard, S.; et al. Platelet abnormalities in Huntington’s disease. J. Neurol. Neurosurg. Psychiatry 2019, 90, 272–283. [Google Scholar] [CrossRef] [PubMed]
- Quiroz-Baez, R.; Hernández-Ortega, K.; Martínez-Martínez, E. Insights into the proteomic profiling of extracellular vesicles for the identification of early biomarkers of neurodegeneration. Front. Neurol. 2020, 11, 580030. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Kojima, K.; Mobley, J.A.; West, A.B. Proteomic analysis of urinary extracellular vesicles reveal biomarkers for neurologic disease. EBioMedicine 2019, 45, 351–361. [Google Scholar] [CrossRef] [PubMed]
- Lugli, G.; Cohen, A.M.; Bennett, D.A.; Shah, R.C.; Fields, C.J.; Hernandez, A.G.; Smalheiser, N.R. Plasma exosomal MiRNAs in persons with and without Alzheimer disease: Altered expression and prospects for biomarkers. PLoS ONE 2015, 10, e0139233. [Google Scholar] [CrossRef] [PubMed]
- Palaniswamy, R.; Sevugan, K.; Sampathkumar Srisharnitha, A. Molecular signatures in exosomes as diagnostic markers for neurodegenerative disorders. Ann. Alzheimers Dement. Care 2020, 4, 12–17. [Google Scholar] [CrossRef]
- Barbagallo, C.; Mostile, G.; Baglieri, G.; Giunta, F.; Luca, A.; Raciti, L.; Zappia, M.; Purrello, M.; Ragusa, M.; Nicoletti, A. Specific signatures of serum MiRNAs as potential biomarkers to discriminate clinically similar neurodegenerative and vascular-related diseases. Cell. Mol. Neurobiol. 2020, 40, 531–546. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.-F.; Qu, M.-W.; Li, G.-C.; Zhang, F.-B.; Rui, H.-C. Circulating exosomal MiRNAs as diagnostic biomarkers in Parkinson’s disease. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 5278–5283. [Google Scholar] [CrossRef]
- Reed, E.R.; Latourelle, J.C.; Bockholt, J.H.; Bregu, J.; Smock, J.; Paulsen, J.S.; Myers, R.H. MicroRNAs in CSF as prodromal biomarkers for huntington disease in the PREDICT-HD study. Neurology 2018, 90, e264–e272. [Google Scholar] [CrossRef]
- Díez-Planelles, C.; Sánchez-Lozano, P.; Crespo, M.C.; Gil-Zamorano, J.; Ribacoba, R.; González, N.; Suárez, E.; Martínez-Descals, A.; Martínez-Camblor, P.; Álvarez, V.; et al. Circulating MicroRNAs in Huntington’s disease: Emerging mediators in metabolic impairment. Pharmacol. Res. 2016, 108, 102–110. [Google Scholar] [CrossRef]
- Johnson, R.; Zuccato, C.; Belyaev, N.D.; Guest, D.J.; Cattaneo, E.; Buckley, N.J. A MicroRNA-based gene dysregulation pathway in Huntington’s disease. Neurobiol. Dis. 2008, 29, 438–445. [Google Scholar] [CrossRef]
- Packer, A.N.; Xing, Y.; Harper, S.Q.; Jones, L.; Davidson, B.L. The bifunctional MicroRNA MiR-9/MiR-9* regulates REST and CoREST and is downregulated in Huntington’s disease. J. Neurosci. Off. J. Soc. Neurosci. 2008, 28, 14341–14346. [Google Scholar] [CrossRef]
- Das, E.; Jana, N.R.; Bhattacharyya, N.P. MicroRNA-124 targets CCNA2 and regulates cell cycle in STHdhQ111/HdhQ111 cells. Biochem. Biophys. Res. Commun. 2013, 437, 217–224. [Google Scholar] [CrossRef]
- Cao, X.; Pfaff, S.L.; Gage, F.H. A functional study of MiR-124 in the developing neural tube. Genes Dev. 2007, 21, 531–536. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, J.; Stewart, T.; Banks, W.A.; Zhang, J. The transport mechanism of extracellular vesicles at the blood-brain barrier. Curr. Pharm. Des. 2017, 23, 6206–6214. [Google Scholar] [CrossRef]
- Kumar, A.; Zhou, L.; Zhi, K.; Raji, B.; Pernell, S.; Tadrous, E.; Kodidela, S.; Nookala, A.; Kochat, H.; Kumar, S. Challenges in biomaterial-based drug delivery approach for the treatment of neurodegenerative diseases: Opportunities for extracellular vesicles. Int. J. Mol. Sci. 2020, 22, 138. [Google Scholar] [CrossRef] [PubMed]
- Muhammad, S.A. Are extracellular vesicles new hope in clinical drug delivery for neurological disorders? Neurochem. Int. 2021, 144, 104955. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Hou, K.; Ji, T.; Wang, X.; Liu, Y.; Zheng, Y.; Xu, J.; Hou, Y.; Chi, G. The role of exosomal MicroRNAs in central nervous system diseases. Mol. Cell. Biochem. 2021. [Google Scholar] [CrossRef]
- Pereira, P.; Queiroz, J.A.; Figueiras, A.; Sousa, F. Current progress on MicroRNAs-based therapeutics in neurodegenerative diseases. Wiley Interdiscip. Rev. RNA 2017, 8. [Google Scholar] [CrossRef]
- Sun, Y.; Luo, Z.-M.; Guo, X.-M.; Su, D.-F.; Liu, X. An updated role of MicroRNA-124 in central nervous system disorders: A review. Front. Cell. Neurosci. 2015, 9, 193. [Google Scholar] [CrossRef]
- Ridolfi, B.; Abdel-Haq, H. Neurodegenerative disorders treatment: The MicroRNA role. Curr. Gene Ther. 2017, 17, 327–363. [Google Scholar] [CrossRef]
- Liu, T.; Im, W.; Mook-Jung, I.; Kim, M. MicroRNA-124 slows down the progression of Huntington’s disease by promoting neurogenesis in the striatum. Neural Regen. Res. 2015, 10, 786–791. [Google Scholar] [CrossRef]
- Didiot, M.-C.; Haraszti, R.A.; Aronin, N.; Khvorova, A. Loading of extracellular vesicles with hydrophobically modified SiRNAs. Methods Mol. Biol. Clifton NJ 2018, 1740, 199–214. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).