Contribution of Yeast Studies to the Understanding of BCL-2 Family Intracellular Trafficking
Abstract
1. Introduction
2. Intrinsic Molecular Determinants of BCL-2 Family Members Localization
- -
- Anti-apoptotic proteins, which includes BCL-2, BCL-xL, MCL-1, et al.
- -
- Multidomain pro-apoptotic proteins, which includes BAX, BAK, and BOK.
- -
- BH3-only proteins, which includes BID, BIM, BAD, PUMA, NOXA, et al.
2.1. Role of the Hydrophobic C-Terminal α-Helix BCL-2, BCL-xL, and BAX
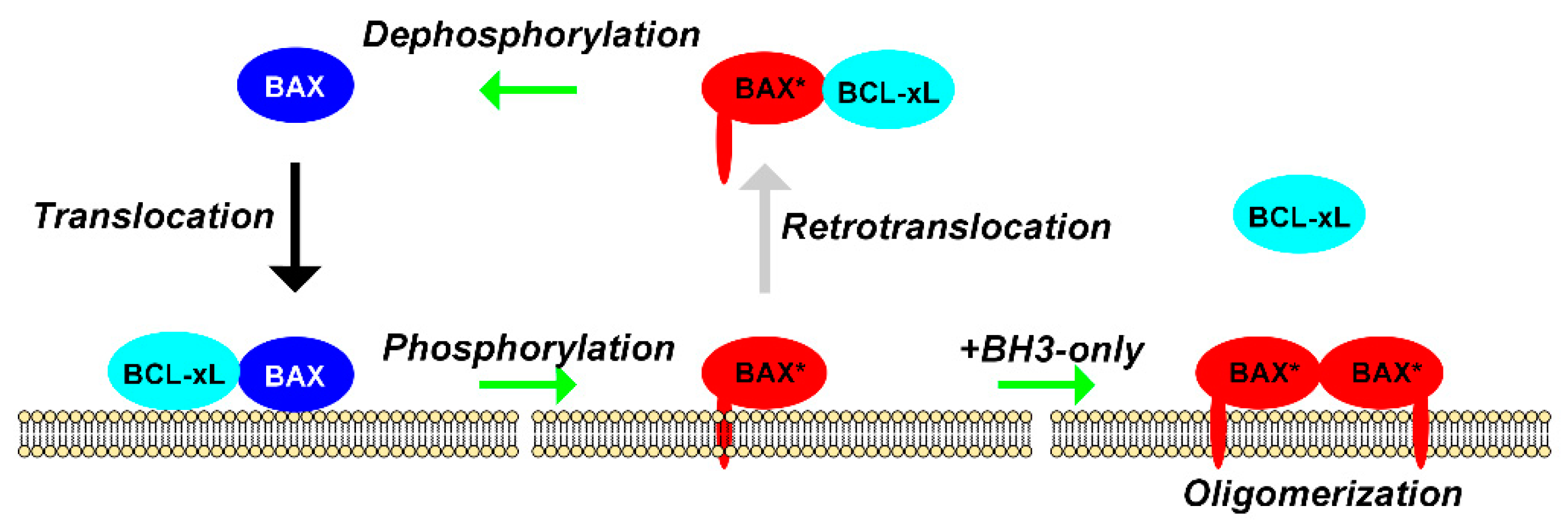
2.2. Translocation and Retrotranslocation of BAX
2.3. BCL-xL-Dependent Stimulation of Translocation/Retrotranslocation v/s BAX Phosphorylation
3. Regulators of BCL-2 Family Members Localization
3.1. Mitochondrial Receptors
3.2. Mitochondria-Associated Membranes (MAM) and Mitochondria-ER Contacts (MERC)
4. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cory, S.; Huang, D.C.S.; Adams, J.M. The Bcl-2 family: Roles in cell survival and oncogenesis. Oncogene 2003, 22, 8590–8607. [Google Scholar] [CrossRef]
- Youle, R.J.; Strasser, A. The BCL-2 protein family: Opposing activities that mediate cell death. Nat. Rev. Mol. Cell Biol. 2008, 9, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Kale, J.; Osterlund, E.J.; Andrews, D.W. BCL-2 family proteins: Changing partners in the dance towards death. Cell Death Differ. 2018, 25, 65–80. [Google Scholar] [CrossRef] [PubMed]
- Lanave, C.; Santamaria, M.; Saccone, C. Comparative genomics: The evolutionary history of the Bcl-2 family. Gene 2004, 333, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Aouacheria, A.; De Laval, V.R.; Combet, C.; Hardwick, J.M. Evolution of Bcl-2 homology motifs: Homology versus homoplasy. Trends Cell Biol. 2013, 23, 103–111. [Google Scholar] [CrossRef]
- Aouacheria, A.; Combet, C.; Tompa, P.; Hardwick, J.M. Redefining the BH3 Death Domain as a ‘Short Linear Motif’. Trends Biochem. Sci. 2015, 40, 736–748. [Google Scholar] [CrossRef]
- Banjara, S.; Suraweera, C.D.; Hinds, M.G.; Kvansakul, M. The Bcl-2 Family: Ancient Origins, Conserved Structures, and Divergent Mechanisms. Biomolecules 2020, 10, 128. [Google Scholar] [CrossRef]
- Manara, A.; Imanifard, Z.; Fracasso, L.; Bellin, D.; Crimi, M. Plants expressing murine pro-apoptotic protein Bid do not have enhanced PCD. BMC Res. Notes 2020, 13, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Carmona-Gutierrez, D.; Bauer, M.A.; Zimmermann, A.; Aguilera, A.; Austriaco, N.; Ayscough, K.; Balzan, R.; Bar-Nun, S.; Barrientos, A.; Belenky, P.; et al. Guidelines and recommendations on yeast cell death nomenclature. Microb. Cell 2018, 5, 4–31. [Google Scholar] [CrossRef]
- Suhaili, S.H.; Karimian, H.; Stellato, M.; Lee, T.-H.; Aguilar, M.-I. Mitochondrial outer membrane permeabilization: A focus on the role of mitochondrial membrane structural organization. Biophys. Rev. 2017, 9, 443–457. [Google Scholar] [CrossRef]
- Kalkavan, H.; Green, D.R. MOMP, cell suicide as a BCL-2 family business. Cell Death Differ. 2018, 25, 46–55. [Google Scholar] [CrossRef]
- Moldoveanu, T.; Czabotar, P.E. BAX, BAK, and BOK: A Coming of Age for the BCL-2 Family Effector Proteins. Cold Spring Harb. Perspect. Biol. 2020, 12, a036319. [Google Scholar] [CrossRef] [PubMed]
- Edlich, F.; Banerjee, S.; Suzuki, M.; Cleland, M.M.; Arnoult, D.; Wang, C.; Neutzner, A.; Tjandra, N.; Youle, R.J. Bcl-xL Retrotranslocates Bax from the Mitochondria into the Cytosol. Cell 2011, 145, 104–116. [Google Scholar] [CrossRef] [PubMed]
- Teijido, O.; Dejean, L. Upregulation of Bcl2 inhibits apoptosis-driven BAX insertion but favors BAX relocalization in mitochondria. FEBS Lett. 2010, 584, 3305–3310. [Google Scholar] [CrossRef] [PubMed]
- Renault, T.T.; Teijido, O.; Missire, F.; Ganesan, Y.T.; Velours, G.; Arokium, H.; Beaumatin, F.; Llanos, R.; Athané, A.; Camougrand, N.; et al. Bcl-xL stimulates Bax relocation to mitochondria and primes cells to ABT-737. Int. J. Biochem. Cell Biol. 2015, 64, 136–146. [Google Scholar] [CrossRef]
- Todt, F.; Cakir, Z.; Reichenbach, F.; Youle, R.J.; Edlich, F. The C-terminal helix of Bcl-xL mediates Bax retrotranslocation from the mitochondria. Cell Death Differ. 2012, 20, 333–342. [Google Scholar] [CrossRef] [PubMed]
- Schellenberg, B.; Wang, P.; Keeble, J.A.; Rodriguez-Enriquez, R.; Walker, S.; Owens, T.W.; Foster, F.; Tanianis-Hughes, J.; Brennan, K.; Streuli, C.H.; et al. Bax Exists in a Dynamic Equilibrium between the Cytosol and Mitochondria to Control Apoptotic Priming. Mol. Cell 2013, 49, 959–971. [Google Scholar] [CrossRef] [PubMed]
- Popgeorgiev, N.; Jabbour, L.; Gillet, G. Subcellular Localization and Dynamics of the Bcl-2 Family of Proteins. Front. Cell Dev. Biol. 2018, 6, 13. [Google Scholar] [CrossRef] [PubMed]
- Ruvolo, P.P.; Deng, X.; May, W.S. Phosphorylation of Bcl2 and regulation of apoptosis. Leukemia 2001, 15, 515–522. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Beauchemin, M.; Bertrand, R. Phospho-Bcl-xL(Ser62) influences spindle assembly and chromosome segregation during mitosis. Cell Cycle 2014, 13, 1313–1326. [Google Scholar] [CrossRef] [PubMed]
- Beaumatin, F.; El Dhaybi, M.; Lasserre, J.-P.; Salin, B.; Moyer, M.P.; Verdier, M.; Manon, S.; Priault, M. N52 monodeamidated Bcl-xL shows impaired oncogenic propertiesin vivoandin vitro. Oncotarget 2016, 7, 17129–17143. [Google Scholar] [CrossRef]
- Giacomello, M.; Pellegrini, L. The coming of age of the mitochondria–ER contact: A matter of thickness. Cell Death Differ. 2016, 23, 1417–1427. [Google Scholar] [CrossRef]
- Kwak, C.; Shin, S.; Park, J.-S.; Jung, M.; Nhung, T.T.M.; Kang, M.-G.; Lee, C.; Kwon, T.-H.; Park, S.K.; Mun, J.Y.; et al. Contact-ID, a tool for profiling organelle contact sites, reveals regulatory proteins of mitochondrial-associated membrane formation. Proc. Natl. Acad. Sci. USA 2020, 117, 12109–12120. [Google Scholar] [CrossRef]
- Légiot, A.; Céré, C.; Dupoiron, T.; Kaabouni, M.; Camougrand, N.; Manon, S. Mitochondria-Associated Membranes (MAMs) are involved in Bax mitochondrial localization and cytochrome c release. Microb. Cell 2019, 6, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Lalier, L.; Mignard, V.; Joalland, M.-P.; Lanoé, D.; Cartron, P.-F.; Manon, S.; Vallette, F.M. TOM20-mediated transfer of Bcl2 from ER to MAM and mitochondria upon induction of apoptosis. Cell Death Dis. 2021, 12, 1–11. [Google Scholar] [CrossRef]
- Motz, C.; Martin, H.; Krimmer, T.; Rassow, J. Bcl-2 and Porin Follow Different Pathways of TOM-dependent Insertion into the Mitochondrial Outer Membrane. J. Mol. Biol. 2002, 323, 729–738. [Google Scholar] [CrossRef]
- Bellot, G.; Cartron, P.-F.; Er, E.; Oliver, L.; Juin, P.; Armstrong, L.C.; Bornstein, P.; Mihara, K.; Manon, S.; Vallette, F.M. TOM22, a core component of the mitochondria outer membrane protein translocation pore, is a mitochondrial receptor for the proapoptotic protein Bax. Cell Death Differ. 2006, 14, 785–794. [Google Scholar] [CrossRef] [PubMed]
- Cartron, P.-F.; Bellot, G.; Oliver, L.; Grandier-Vazeille, X.; Manon, S.; Vallette, F.M. Bax inserts into the mitochondrial outer membrane by different mechanisms. FEBS Lett. 2008, 582, 3045–3051. [Google Scholar] [CrossRef] [PubMed]
- Renault, T.T.; Grandier-Vazeille, X.; Arokium, H.; Velours, G.; Camougrand, N.; Priault, M.; Teijido, O.; Dejean, L.M.; Manon, S. The cytosolic domain of human Tom22 modulates human Bax mitochondrial translocation and conformation in yeast. FEBS Lett. 2012, 586, 116–121. [Google Scholar] [CrossRef] [PubMed]
- Frank, D.O.; Dengjel, J.; Wilfling, F.; Kozjak-Pavlovic, V.; Häcker, G.; Weber, A. The Pro-Apoptotic BH3-Only Protein Bim Interacts with Components of the Translocase of the Outer Mitochondrial Membrane (TOM). PLoS ONE 2015, 10, e0123341. [Google Scholar] [CrossRef] [PubMed]
- Cartron, P.-F.; Petit, E.; Bellot, G.; Oliver, L.; Vallette, F.M. Metaxins 1 and 2, two proteins of the mitochondrial protein sorting and assembly machinery, are essential for Bak activation during TNF alpha triggered apoptosis. Cell. Signal. 2014, 26, 1928–1934. [Google Scholar] [CrossRef]
- Petit, E.; Cartron, P.-F.; Oliver, L.; Vallette, F.M. The phosphorylation of Metaxin 1 controls Bak activation during TNFα induced cell death. Cell. Signal. 2017, 30, 171–178. [Google Scholar] [CrossRef]
- Chou, C.-H.; Lee, R.-S.; Yang-Yen, H.-F. An Internal EELD Domain Facilitates Mitochondrial Targeting of Mcl-1 via a Tom70-dependent Pathway. Mol. Biol. Cell 2006, 17, 3952–3963. [Google Scholar] [CrossRef][Green Version]
- Kreimendahl, S.; Rassow, J. The Mitochondrial Outer Membrane Protein Tom70—Mediator in Protein Traffic, Membrane Contact Sites and Innate Immunity. Int. J. Mol. Sci. 2020, 21, 7262. [Google Scholar] [CrossRef] [PubMed]
- Lutter, M.; Fang, M.; Luo, X.; Nishijima, M.; Xie, X.-S.; Wang, X. Cardiolipin provides specificity for targeting of tBid to mitochondria. Nat. Cell Biol. 2000, 2, 754–756. [Google Scholar] [CrossRef] [PubMed]
- Gonzalvez, F.; Pariselli, F.; Dupaigne, P.; Budihardjo, I.I.; Lutter, M.; Antonsson, B.; Diolez, P.; Manon, S.; Martinou, J.-C.; Goubern, M.; et al. tBid interaction with cardiolipin primarily orchestrates mitochondrial dysfunctions and subsequently activates Bax and Bak. Cell Death Differ. 2005, 12, 614–626. [Google Scholar] [CrossRef] [PubMed]
- Priault, M.; Camougrand, N.; Kinnally, K.W.; Vallette, F.M.; Manon, S. Yeast as a tool to study Bax/mitochondrial interactions in cell death. FEMS Yeast Res. 2003, 4, 15–27. [Google Scholar] [CrossRef][Green Version]
- Renault, T.T.; Dejean, L.M.; Manon, S. A brewing understanding of the regulation of Bax function by Bcl-xL and Bcl-2. Mech. Ageing Dev. 2017, 161, 201–210. [Google Scholar] [CrossRef]
- Manon, S. Investigating BCL-2 Family Protein Interactions in Yeast. Adv. Struct. Saf. Stud. 2018, 1877, 93–109. [Google Scholar] [CrossRef]
- Büttner, S.; Ruli, D.; Vögtle, F.-N.; Galluzzi, L.; Moitzi, B.; Eisenberg, T.; Kepp, O.; Habernig, L.; Carmona-Gutierrez, D.; Rockenfeller, P.; et al. A yeast BH3-only protein mediates the mitochondrial pathway of apoptosis. EMBO J. 2011, 30, 2779–2792. [Google Scholar] [CrossRef]
- Cebulski, J.; Malouin, J.; Pinches, N.; Cascio, V.; Austriaco, N. Yeast Bax Inhibitor, Bxi1p, Is an ER-Localized Protein That Links the Unfolded Protein Response and Programmed Cell Death in Saccharomyces cerevisiae. PLoS ONE 2011, 6, e20882. [Google Scholar] [CrossRef]
- Hanada, M.; Aimé-Sempé, C.; Sato, T.; Reed, J.C. Structure-Function Analysis of Bcl-2 Protein. J. Biol. Chem. 1995, 270, 11962–11969. [Google Scholar] [CrossRef] [PubMed]
- Zha, H.; Aimé-Sempé, C.; Sato, T.; Reed, J.C. Proapoptotic Protein Bax Heterodimerizes with Bcl-2 and Homodimerizes with Bax via a Novel Domain (BH3) Distinct from BH1 and BH2. J. Biol. Chem. 1996, 271, 7440–7444. [Google Scholar] [CrossRef]
- Greenhalf, W.; Stephan, C.; Chaudhuri, B. Role of mitochondria and C-terminal membrane anchor of Bcl-2 in Bax induced growth arrest and mortality in Saccharomyces cerevisiae. FEBS Lett. 1996, 380, 169–175. [Google Scholar] [CrossRef]
- Manon, S.; Chaudhuri, B.; Guérin, M. Release of cytochrome c and decrease of cytochrome c oxidase in Bax-expressing yeast cells, and prevention of these effects by coexpression of Bcl-xL. FEBS Lett. 1997, 415, 29–32. [Google Scholar] [CrossRef]
- Clow, A.; Greenhalf, W.; Chaudhuri, B. Under respiratory growth conditions, Bcl-x(L) and Bcl-2 are unable to overcome yeast cell death triggered by a mutant Bax protein lacking the membrane anchor. JBIC J. Biol. Inorg. Chem. 1998, 258, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Kane, D.; Sarafian, T.; Anton, R.; Hahn, H.; Gralla, E.; Valentine, J.; Örd, T.; Bredesen, D. Bcl-2 inhibition of neural death: Decreased generation of reactive oxygen species. Science 1993, 262, 1274–1277. [Google Scholar] [CrossRef]
- Trancíková, A.; Weisová, P.; Kiššová, I.; Zeman, I.; Kolarov, J. Production of reactive oxygen species and loss of viability in yeast mitochondrial mutants: Protective effect of Bcl-x. FEMS Yeast Res. 2004, 5, 149–156. [Google Scholar] [CrossRef]
- Greenwood, M.T.; Ludovico, P. Expressing and functional analysis of mammalian apoptotic regulators in yeast. Cell Death Differ. 2009, 17, 737–745. [Google Scholar] [CrossRef] [PubMed]
- Henke, N.; Lisak, D.A.; Schneider, L.; Habicht, J.; Pergande, M.; Methner, A.; Nickel, N. The ancient cell death suppressor BAX inhibitor-1. Cell Calcium 2011, 50, 251–260. [Google Scholar] [CrossRef]
- Silva, R.D.; Manon, S.; Goncalves, J.; Saraiva, L.; Corte-Real, M. The Importance of Humanized Yeast to Better Understand the Role of Bcl-2 Family in Apoptosis: Finding of Novel Therapeutic Opportunities. Curr. Pharm. Des. 2011, 17, 246–255. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Polcic, P.; Jaká, P.; Mentel, M. Yeast as a tool for studying proteins of the Bcl-2 family. Microb. Cell 2015, 2, 74–87. [Google Scholar] [CrossRef]
- Polčic, P.; Mentel, M. Reconstituting the Mammalian Apoptotic Switch in Yeast. Genes 2020, 11, 145. [Google Scholar] [CrossRef]
- Gross, A.; McDonnell, J.M.; Korsmeyer, S.J. BCL-2 family members and the mitochondria in apoptosis. Genes Dev. 1999, 13, 1899–1911. [Google Scholar] [CrossRef]
- Petros, A.M.; Olejniczak, E.T.; Fesik, S.W. Structural biology of the Bcl-2 family of proteins. Biochim. Biophys. Acta 2004, 1644, 83–94. [Google Scholar] [CrossRef]
- Hinds, M.G.; Smits, C.; Fredericks-Short, R.; Risk, J.M.; Bailey, M.; Huang, D.C.S.; Day, C.L. Bim, Bad and Bmf: Intrinsically unstructured BH3-only proteins that undergo a localized conformational change upon binding to prosurvival Bcl-2 targets. Cell Death Differ. 2006, 14, 128–136. [Google Scholar] [CrossRef] [PubMed]
- Rogers, J.M.; Wong, C.T.; Clarke, J. Coupled Folding and Binding of the Disordered Protein PUMA Does Not Require Particular Residual Structure. J. Am. Chem. Soc. 2014, 136, 5197–5200. [Google Scholar] [CrossRef]
- Liang, X.H.; Kleeman, L.K.; Jiang, H.H.; Gordon, G.; Goldman, J.E.; Berry, G.; Herman, B.; Levine, B. Protection against Fatal Sindbis Virus Encephalitis by Beclin, a Novel Bcl-2-Interacting Protein. J. Virol. 1998, 72, 8586–8596. [Google Scholar] [CrossRef]
- Liang, X.H.; Jackson, S.; Seaman, M.; Brown, K.; Kempkes, B.; Hibshoosh, H.; Levine, B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nat. Cell Biol. 1999, 402, 672–676. [Google Scholar] [CrossRef]
- Meléndez, A.; Tallóczy, Z.; Seaman, M.N.J.; Eskelinen, E.-L.; Hall, D.H.; Levine, B. Autophagy Genes Are Essential for Dauer Development and Life-Span Extension in C. elegans. Science 2003, 301, 1387–1391. [Google Scholar] [CrossRef] [PubMed]
- Schweers, R.L.; Zhang, J.; Randall, M.S.; Loyd, M.R.; Li, W.; Dorsey, F.C.; Kundu, M.; Opferman, J.T.; Cleveland, J.L.; Miller, J.L.; et al. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc. Natl. Acad. Sci. USA 2007, 104, 19500–19505. [Google Scholar] [CrossRef] [PubMed]
- Sandoval, H.; Thiagarajan, P.; Dasgupta, S.K.; Schumacher, A.; Prchal, J.T.; Chen, M.; Wang, J. Essential role for Nix in autophagic maturation of erythroid cells. Nat. Cell Biol. 2008, 454, 232–235. [Google Scholar] [CrossRef] [PubMed]
- Kataoka, T.; Holler, N.; Micheau, O.; Martinon, F.; Tinel, A.; Hofmann, K.; Tschopp, J. Bcl-rambo, a Novel Bcl-2 Homologue That Induces Apoptosis via Its Unique C-terminal Extension. J. Biol. Chem. 2001, 276, 19548–19554. [Google Scholar] [CrossRef]
- Murakawa, T.; Yamaguchi, O.; Hashimoto, A.; Hikoso, S.; Takeda, T.O.; Oka, T.; Yasui, H.; Ueda, H.; Akazawa, Y.; Nakayama, H.; et al. Bcl-2-like protein 13 is a mammalian Atg32 homologue that mediates mitophagy and mitochondrial fragmentation. Nat. Commun. 2015, 6, 7527. [Google Scholar] [CrossRef]
- García-Sáez, A.J.; Mingarro, I.; Pérez-Payá, E.; Salgado, J. Membrane-Insertion Fragments of Bcl-xL, Bax, and Bid. Biochemistry 2004, 43, 10930–10943. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, T.; Schlipf, S.; Sanz, J.; Neubert, K.; Stein, R.; Borner, C. Characterization of the signal that directs Bcl-xL, but not Bcl-2, to the mitochondrial outer membrane. J. Cell Biol. 2003, 160, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Priault, M.; Camougrand, N.; Chaudhuri, B.; Manon, S. Role of the C-terminal domain of Bax and Bcl-xL in their localization and function in yeast cells. FEBS Lett. 1999, 443, 225–228. [Google Scholar] [CrossRef]
- Garenne, D.; Renault, T.T.; Manon, S. Bax mitochondrial relocation is linked to its phosphorylation and its interaction with Bcl-xL. Microb. Cell 2016, 3, 597–605. [Google Scholar] [CrossRef]
- Pécot, J.; Maillet, L.; Le Pen, J.; Vuillier, C.; Trécesson, S.D.C.; Fétiveau, A.; Sarosiek, K.A.; Bock, F.J.; Braun, F.; Letai, A.; et al. Tight Sequestration of BH3 Proteins by BCL-xL at Subcellular Membranes Contributes to Apoptotic Resistance. Cell Rep. 2016, 17, 3347–3358. [Google Scholar] [CrossRef] [PubMed]
- Warren, C.F.A.; Wong-Brown, M.W.; Bowden, N.A. BCL-2 family isoforms in apoptosis and cancer. Cell Death Dis. 2019, 10, 1–12. [Google Scholar] [CrossRef]
- Hsu, Y.-T.; Wolter, K.G.; Youle, R.J. Cytosol-to-membrane redistribution of Bax and Bcl-XL during apoptosis. Proc. Natl. Acad. Sci. USA 1997, 94, 3668–3672. [Google Scholar] [CrossRef]
- Wolter, K.G.; Hsu, Y.-T.; Smith, C.L.; Nechushtan, A.; Xi, X.-G.; Youle, R.J. Movement of Bax from the Cytosol to Mitochondria during Apoptosis. J. Cell Biol. 1997, 139, 1281–1292. [Google Scholar] [CrossRef]
- Horie, C.; Suzuki, H.; Sakaguchi, M.; Mihara, K. Characterization of Signal That Directs C-Tail–anchored Proteins to Mammalian Mitochondrial Outer Membrane. Mol. Biol. Cell 2002, 13, 1615–1625. [Google Scholar] [CrossRef]
- Gómez-Fernández, J.C. Functions of the C-terminal domains of apoptosis-related proteins of the Bcl-2 family. Chem. Phys. Lipids 2014, 183, 77–90. [Google Scholar] [CrossRef]
- Tremblais, K.; Oliver, L.; Juin, P.; Le Cabellec, M.T.; Meflah, K.; Vallette, F.M. The C-Terminus of bax Is Not a Membrane Addressing/Anchoring Signal. Biochem. Biophys. Res. Commun. 1999, 260, 582–591. [Google Scholar] [CrossRef] [PubMed]
- Oliver, L.; Priault, M.; Tremblais, K.; LeCabellec, M.-T.; Meflah, K.; Manon, S.; Vallette, F.M. The substitution of the C-terminus of bax by that of bcl-xL does not affect its subcellular localization but abrogates its pro-apoptotic properties. FEBS Lett. 2000, 487, 161–165. [Google Scholar] [CrossRef]
- Suzuki, M.; Youle, R.J.; Tjandra, N. Structure of Bax. Cell 2000, 103, 645–654. [Google Scholar] [CrossRef]
- Martínez-Senac, M.D.M.; Corbalán-García, S.; Gómez-Fernández, J.C. Conformation of the C-Terminal Domain of the Pro-Apoptotic Protein Bax and Mutants and Its Interaction with Membranes. Biochemistry 2001, 40, 9983–9992. [Google Scholar] [CrossRef] [PubMed]
- Peng, R.; Tong, J.-S.; Li, H.; Yue, B.; Zou, F.; Yu, J.; Zhang, L. Targeting Bax interaction sites reveals that only homo-oligomerization sites are essential for its activation. Cell Death Differ. 2013, 20, 744–754. [Google Scholar] [CrossRef][Green Version]
- Wang, Q.; Sun, S.-Y.; Khuri, F.; Curran, W.J.; Deng, X. Mono- or Double-Site Phosphorylation Distinctly Regulates the Proapoptotic Function of Bax. PLoS ONE 2010, 5, e13393. [Google Scholar] [CrossRef]
- Arokium, H.; Ouerfelli, H.; Velours, G.; Camougrand, N.; Vallette, F.M.; Manon, S. Substitutions of Potentially Phosphorylatable Serine Residues of Bax Reveal How They May Regulate Its Interaction with Mitochondria. J. Biol. Chem. 2007, 282, 35104–35112. [Google Scholar] [CrossRef]
- Simonyan, L.; Renault, T.T.; Novais, M.J.D.C.; Sousa, M.J.; Real, M.C.; Camougrand, N.; Gonzalez, C.; Manon, S. Regulation of Bax/mitochondria interaction by AKT. FEBS Lett. 2016, 590, 13–21. [Google Scholar] [CrossRef]
- Kuwana, T.; King, L.E.; Cosentino, K.; Suess, J.; Garcia-Saez, A.J.; Gilmore, A.P.; Newmeyer, D.D. Mitochondrial residence of the apoptosis inducer BAX is more important than BAX oligomerization in promoting membrane permeabilization. J. Biol. Chem. 2020, 295, 1623–1636. [Google Scholar] [CrossRef]
- Gardai, S.J.; Hildeman, D.A.; Frankel, S.K.; Whitlock, B.B.; Frasch, S.C.; Borregaard, N.; Marrack, P.; Bratton, D.L.; Henson, P.M. Phosphorylation of Bax Ser184 by Akt Regulates Its Activity and Apoptosis in Neutrophils. J. Biol. Chem. 2004, 279, 21085–21095. [Google Scholar] [CrossRef]
- Xin, M.; Deng, X. Nicotine Inactivation of the Proapoptotic Function of Bax through Phosphorylation. J. Biol. Chem. 2005, 280, 10781–10789. [Google Scholar] [CrossRef]
- Xin, M.; Gao, F.; May, W.S.; Flagg, T.; Deng, X. Protein Kinase Cζ Abrogates the Proapoptotic Function of Bax through Phosphorylation. J. Biol. Chem. 2007, 282, 21268–21277. [Google Scholar] [CrossRef]
- Xin, M.; Li, R.; Xie, M.; Park, D.; Owonikoko, T.K.; Sica, G.L.; Corsino, P.E.; Zhou, J.; Ding, C.; White, M.A.; et al. Small-molecule Bax agonists for cancer therapy. Nat. Commun. 2014, 5, 1–11. [Google Scholar] [CrossRef]
- Morano, K.A.; Thiele, D.J. The Sch9 protein kinase regulates Hsp90 chaperone complex signal transduction activity in vivo. EMBO J. 1999, 18, 5953–5962. [Google Scholar] [CrossRef] [PubMed]
- Cartron, P.-F.; Arokium, H.; Oliver, L.; Meflah, K.; Manon, S.; Vallette, F.M. Distinct Domains Control the Addressing and the Insertion of Bax into Mitochondria. J. Biol. Chem. 2005, 280, 10587–10598. [Google Scholar] [CrossRef] [PubMed]
- Arokium, H.; Camougrand, N.; Vallette, F.M.; Manon, S.; He, Q.-M.; Wei, Y.-Q.; Tian, L.; Zhao, X.; Su, J.-M.; Yang, L.; et al. Studies of the Interaction of Substituted Mutants of BAX with Yeast Mitochondria Reveal That the C-terminal Hydrophobic α-Helix Is a Second ART Sequence and Plays a Role in the Interaction with Anti-apoptotic BCL-xL. J. Biol. Chem. 2004, 279, 52566–52573. [Google Scholar] [CrossRef] [PubMed]
- Simonyan, L.; Légiot, A.; Lascu, I.; Durand, G.; Giraud, M.-F.; Gonzalez, C.; Manon, S. The substitution of Proline 168 favors Bax oligomerization and stimulates its interaction with LUVs and mitochondria. Biochim. Biophys. Acta 2017, 1859, 1144–1155. [Google Scholar] [CrossRef]
- Willis, S.N.; Fletcher, J.I.; Kaufmann, T.; Van Delft, M.F.; Chen, L.; Czabotar, P.E.; Ierino, H.; Lee, E.F.; Fairlie, W.D.; Bouillet, P.; et al. Apoptosis Initiated When BH3 Ligands Engage Multiple Bcl-2 Homologs, Not Bax or Bak. Science 2007, 315, 856–859. [Google Scholar] [CrossRef] [PubMed]
- Marani, M.; Tenev, T.; Hancock, D.; Downward, J.; Lemoine, N.R. Identification of Novel Isoforms of the BH3 Domain Protein Bim Which Directly Activate Bax to Trigger Apoptosis. Mol. Cell. Biol. 2002, 22, 3577–3589. [Google Scholar] [CrossRef] [PubMed]
- Gavathiotis, E.; Suzuki, M.; Davis, M.L.; Pitter, K.; Bird, G.H.; Katz, S.G.; Tu, H.-C.; Kim, H.; Cheng, E.H.-Y.; Tjandra, N.; et al. BAX activation is initiated at a novel interaction site. Nat. Cell Biol. 2008, 455, 1076–1081. [Google Scholar] [CrossRef]
- Gallenne, T.; Gautier, F.; Oliver, L.; Hervouet, E.; Noël, B.; Hickman, J.A.; Geneste, O.; Cartron, P.-F.; Vallette, F.M.; Manon, S.; et al. Bax activation by the BH3-only protein Puma promotes cell dependence on antiapoptotic Bcl-2 family members. J. Cell Biol. 2009, 185, 279–290. [Google Scholar] [CrossRef] [PubMed]
- Gonzalvez, F.; Bessoule, J.-J.; Rocchiccioli, F.; Manon, S.; Petit, P.X. Role of cardiolipin on tBid and tBid/Bax synergistic effects on yeast mitochondria. Cell Death Differ. 2005, 12, 659–667. [Google Scholar] [CrossRef]
- Yao, Y.; Nisan, D.; Fujimoto, L.M.; Antignani, A.; Barnes, A.; Tjandra, N.; Youle, R.J.; Marassi, F.M. Characterization of the membrane-inserted C-terminus of cytoprotective BCL-XL. Protein Expr. Purif. 2016, 122, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Marassi, F.M. Reconstitution and Characterization of BCL-2 Family Proteins in Lipid Bilayer Nanodiscs. Methods Mol. Biol. 2019, 1877, 233–246. [Google Scholar] [CrossRef] [PubMed]
- Dingeldein, A.P.; Lindberg, M.J.; Åden, J.; Zhong, X.; Stoll, R.; Gröbner, G. Bax to the future—A novel, high-yielding approach for purification and expression of full-length Bax protein for structural studies. Protein Expr. Purif. 2019, 158, 20–26. [Google Scholar] [CrossRef]
- Upreti, M.; Galitovskaya, E.N.; Chu, R.; Tackett, A.J.; Terrano, D.T.; Granell, S.; Chambers, T.C. Identification of the Major Phosphorylation Site in Bcl-xL Induced by Microtubule Inhibitors and Analysis of Its Functional Significance. J. Biol. Chem. 2008, 283, 35517–35525. [Google Scholar] [CrossRef]
- Pfanner, N.; Warscheid, B.; Wiedemann, N. Mitochondrial proteins: From biogenesis to functional networks. Nat. Rev. Mol. Cell Biol. 2019, 20, 267–284. [Google Scholar] [CrossRef]
- Colin, J.; Garibal, J.; Mignotte, B.; Guénal, I. The mitochondrial TOM complex modulates bax-induced apoptosis in Drosophila. Biochem. Biophys. Res. Commun. 2009, 379, 939–943. [Google Scholar] [CrossRef] [PubMed]
- Ross, K.; Rudel, T.; Kozjak-Pavlovic, V. TOM-independent complex formation of Bax and Bak in mammalian mitochondria during TNFα-induced apoptosis. Cell Death Differ. 2009, 16, 697–707. [Google Scholar] [CrossRef] [PubMed]
- Vance, J.E. MAM (mitochondria-associated membranes) in mammalian cells: Lipids and beyond. Biochim. Biophys. Acta 2014, 1841, 595–609. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Hildreth, R.L.; Colberg-Poley, A.M. Human Cytomegalovirus Inhibits Apoptosis by Proteasome-Mediated Degradation of Bax at Endoplasmic Reticulum-Mitochondrion Contacts. J. Virol. 2013, 87, 5657–5668. [Google Scholar] [CrossRef] [PubMed]
- Martins, V.M.; Fernandes, T.R.; Lopes, D.; Afonso, C.B.; Domingues, M.R.; Côrte-Real, M.; Sousa, M.J. Contacts in Death: The Role of the ER–Mitochondria Axis in Acetic Acid-Induced Apoptosis in Yeast. J. Mol. Biol. 2019, 431, 273–288. [Google Scholar] [CrossRef]
- Michel, A.H.; Kornmann, B. The ERMES complex and ER–mitochondria connections. Biochem. Soc. Trans. 2012, 40, 445–450. [Google Scholar] [CrossRef]
- Lang, A.; Peter, A.T.J.; Kornmann, B. ER–mitochondria contact sites in yeast: Beyond the myths of ERMES. Curr. Opin. Cell Biol. 2015, 35, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Vance, J.E. Phospholipid Synthesis and Transport in Mammalian Cells. Traffic 2014, 16, 1–18. [Google Scholar] [CrossRef]
- Modi, S.; López-Doménech, G.; Halff, E.F.; Covill-Cooke, C.; Ivankovic, D.; Melandri, D.; Arancibia-Cárcamo, I.L.; Burden, J.J.; Lowe, A.R.; Kittler, J.T. Miro clusters regulate ER-mitochondria contact sites and link cristae organization to the mitochondrial transport machinery. Nat. Commun. 2019, 10, 1–15. [Google Scholar] [CrossRef] [PubMed]
- White, R.R.; Lin, C.; Leaves, I.; Castro, I.G.; Metz, J.; Bateman, B.C.; Botchway, S.W.; Ward, A.D.; Ashwin, P.; Sparkes, I. Miro2 tethers the ER to mitochondria to promote mitochondrial fusion in tobacco leaf epidermal cells. Commun. Biol. 2020, 3, 1–8. [Google Scholar] [CrossRef]
- Kornmann, B.; Osman, C.; Walter, P. The conserved GTPase Gem1 regulates endoplasmic reticulum-mitochondria connections. Proc. Natl. Acad. Sci. USA 2011, 108, 14151–14156. [Google Scholar] [CrossRef] [PubMed]
- Kornmann, B.; Currie, E.; Collins, S.R.; Schuldiner, M.; Nunnari, J.; Weissman, J.S.; Walter, P. An ER-Mitochondria Tethering Complex Revealed by a Synthetic Biology Screen. Science 2009, 325, 477–481. [Google Scholar] [CrossRef]
- Csordás, G.; Várnai, P.; Golenár, T.; Roy, S.; Purkins, G.; Schneider, T.G.; Balla, T.; Hajnóczky, G. Imaging Interorganelle Contacts and Local Calcium Dynamics at the ER-Mitochondrial Interface. Mol. Cell 2010, 39, 121–132. [Google Scholar] [CrossRef]
- Gautier, F.; Guillemin, Y.; Cartron, P.-F.; Gallenne, T.; Cauquil, N.; Le Diguarher, T.; Casara, P.; Vallette, F.M.; Manon, S.; Hickman, J.A.; et al. Bax Activation by Engagement with, Then Release from, the BH3 Binding Site of Bcl-xL. Mol. Cell. Biol. 2010, 31, 832–844. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dimitrakakis, C.; Konstadoulakis, M.; Messaris, E.; Kymionis, G.; Karayannis, M.; Panoussopoulos, D.; Michalas, S.; Androulakis, G. Molecular markers in breast cancer: Can we use c-erbB-2, p53, bcl-2 and bax gene expression as prognostic factors? Breast 2002, 11, 279–285. [Google Scholar] [CrossRef] [PubMed]
- Schniewind, B.; Christgen, M.; Kurdow, R.; Haye, S.; Kremer, B.; Kalthoff, H.; Ungefroren, H. Resistance of pancreatic cancer to gemcitabine treatment is dependent on mitochondria-mediated apoptosis. Int. J. Cancer 2004, 109, 182–188. [Google Scholar] [CrossRef] [PubMed]
- Merino, D.; Kelly, G.L.; Lessene, G.; Wei, A.H.; Roberts, A.W.; Strasser, A. BH3-Mimetic Drugs: Blazing the Trail for New Cancer Medicines. Cancer Cell 2018, 34, 879–891. [Google Scholar] [CrossRef]
- Rudin, C.M.; Hann, C.L.; Garon, E.B.; De Oliveira, M.R.; Bonomi, P.D.; Camidge, D.R.; Chu, Q.; Giaccone, G.; Khaira, D.; Ramalingam, S.S.; et al. Phase II Study of Single-Agent Navitoclax (ABT-263) and Biomarker Correlates in Patients with Relapsed Small Cell Lung Cancer. Clin. Cancer Res. 2012, 18, 3163–3169. [Google Scholar] [CrossRef] [PubMed]
- Souers, A.J.; Leverson, J.D.; Boghaert, E.R.; Ackler, S.L.; Catron, N.D.; Chen, J.; Dayton, B.D.; Ding, H.; Enschede, S.H.; Fairbrother, W.J.; et al. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat. Med. 2013, 19, 202–208. [Google Scholar] [CrossRef]
- Kumar, S.K.; Harrison, S.J.; Cavo, M.; de la Rubia, J.; Popat, R.; Gasparetto, C.; Hungria, V.; Salwender, H.; Suzuki, K.; Kim, I.; et al. Venetoclax or placebo in combination with bortezomib and dexamethasone in patients with relapsed or refractory multiple myeloma (BELLINI): A randomised, double-blind, multicentre, phase 3 trial. Lancet Oncol. 2020, 21, 1630–1642. [Google Scholar] [CrossRef]
- Kandel, E.S.; Hay, N. The Regulation and Activities of the Multifunctional Serine/Threonine Kinase Akt/PKB. Exp. Cell Res. 1999, 253, 210–229. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, K.M.; Anderson, N.G. The protein kinase B/Akt signalling pathway in human malignancy. Cell. Signal. 2002, 14, 381–395. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rouchidane Eyitayo, A.; Gonin, M.; Arokium, H.; Manon, S. Contribution of Yeast Studies to the Understanding of BCL-2 Family Intracellular Trafficking. Int. J. Mol. Sci. 2021, 22, 4086. https://doi.org/10.3390/ijms22084086
Rouchidane Eyitayo A, Gonin M, Arokium H, Manon S. Contribution of Yeast Studies to the Understanding of BCL-2 Family Intracellular Trafficking. International Journal of Molecular Sciences. 2021; 22(8):4086. https://doi.org/10.3390/ijms22084086
Chicago/Turabian StyleRouchidane Eyitayo, Akandé, Mathilde Gonin, Hubert Arokium, and Stéphen Manon. 2021. "Contribution of Yeast Studies to the Understanding of BCL-2 Family Intracellular Trafficking" International Journal of Molecular Sciences 22, no. 8: 4086. https://doi.org/10.3390/ijms22084086
APA StyleRouchidane Eyitayo, A., Gonin, M., Arokium, H., & Manon, S. (2021). Contribution of Yeast Studies to the Understanding of BCL-2 Family Intracellular Trafficking. International Journal of Molecular Sciences, 22(8), 4086. https://doi.org/10.3390/ijms22084086






