Multi-Omics Study of Keystone Species in a Cystic Fibrosis Microbiome
Abstract
1. Introduction
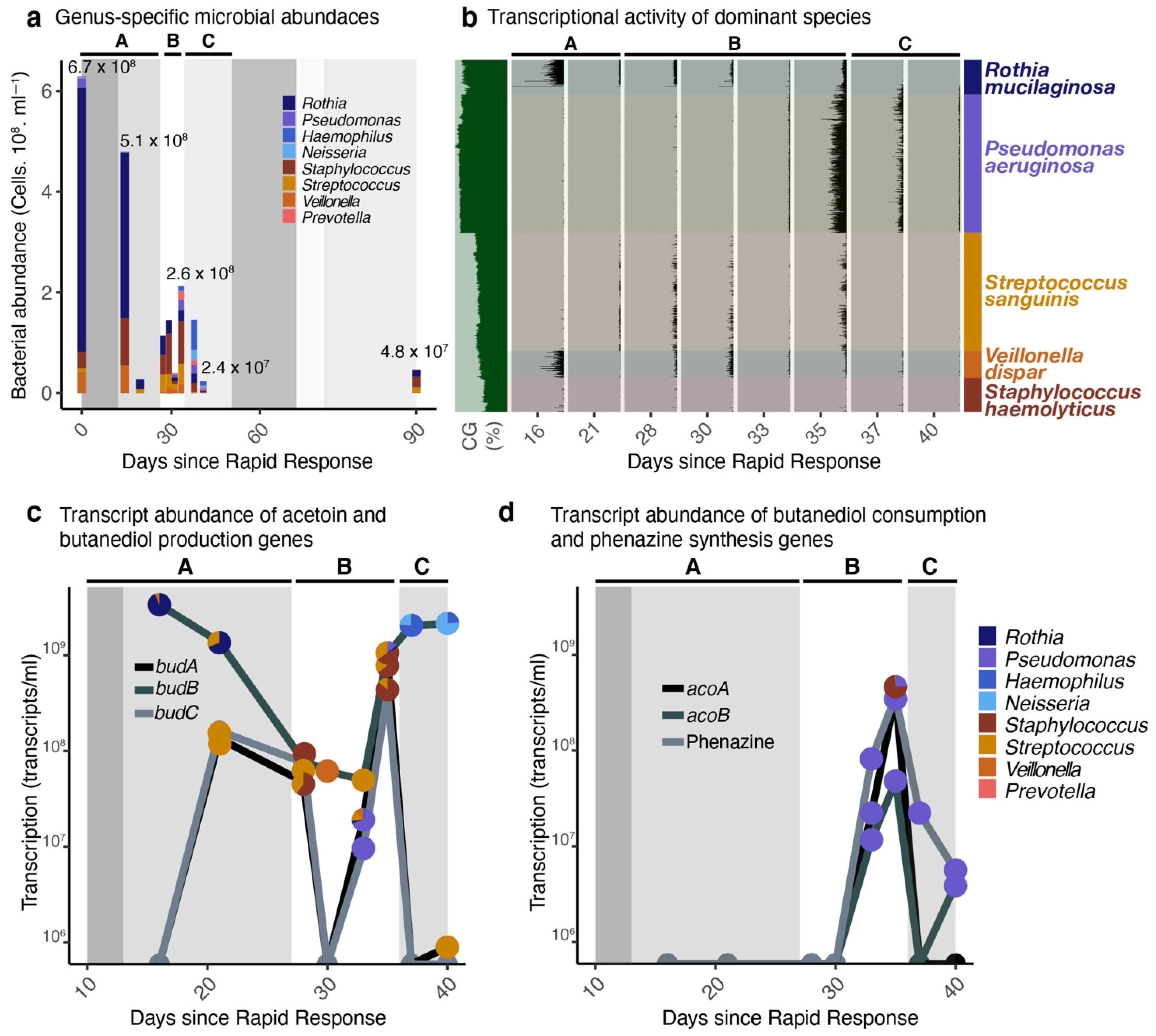
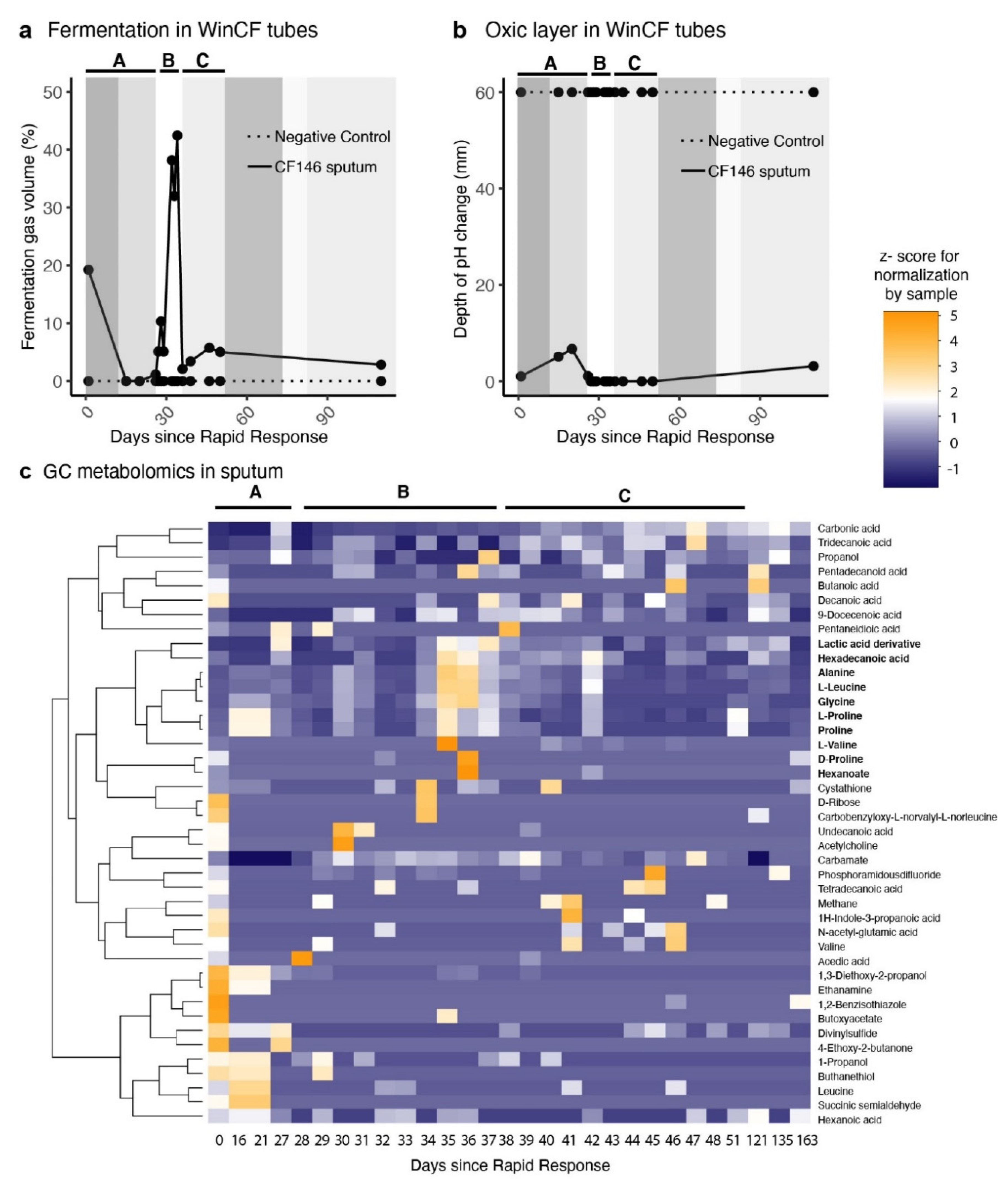
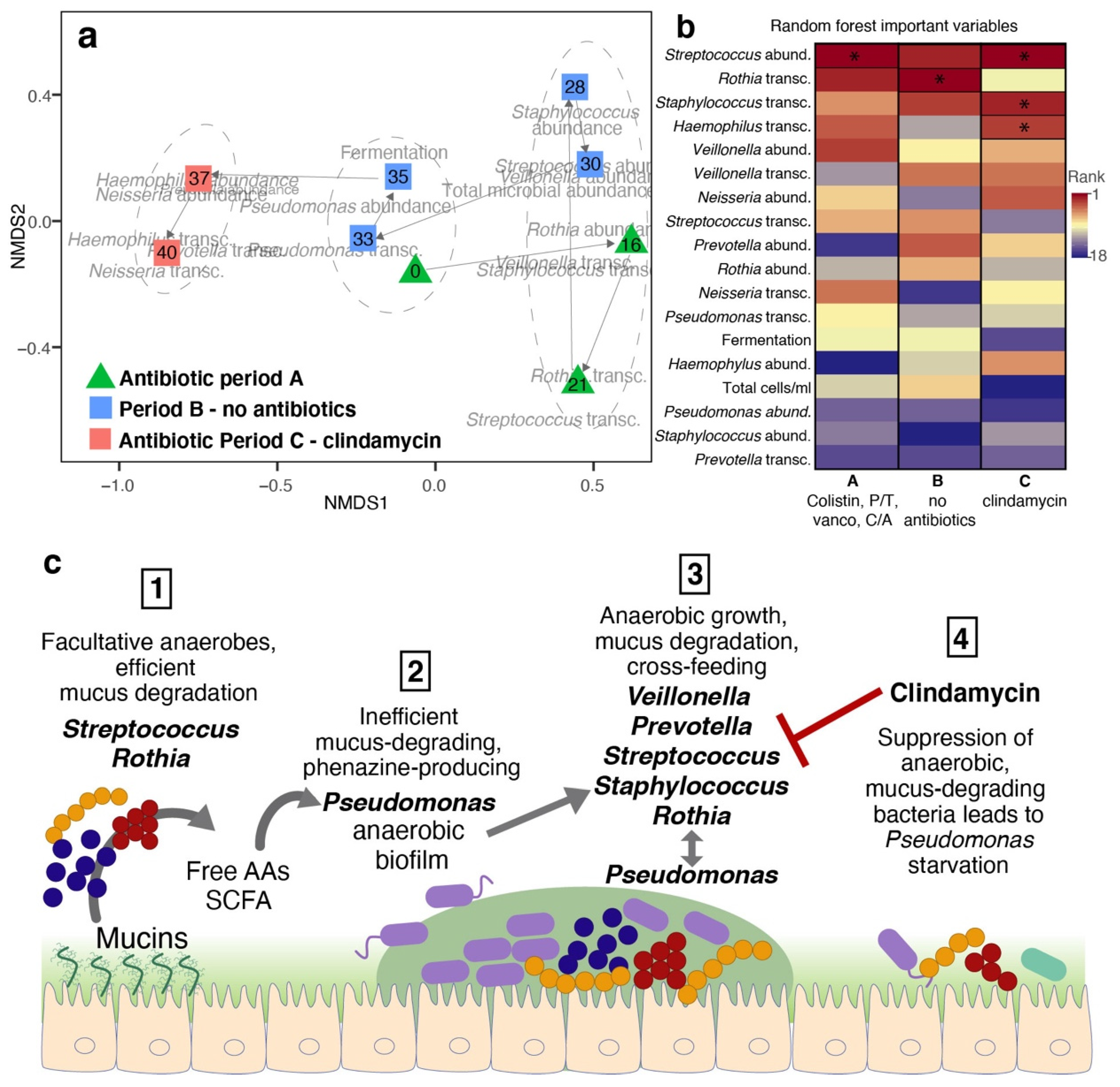
2. Results
3. Discussion
4. Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Power, M.E.; Tilman, D.; Estes, J.A.; Menge, B.A.; Bond, W.J.; Mills, L.S.; Daily, G.; Castilla, J.C.; Lubchenco, J.; Paine, R.T.; et al. Challenges in the quest for keystones. Bioscience 1996, 46, 609–620. [Google Scholar] [CrossRef]
- Hajishengallis, G.; Darveau, R.P.; Curtis, M.A. The keystone-pathogen hypothesis. Nat. Rev. Microbiol. 2012, 10, 717–725. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Schlaeppi, K.; Heijden, M.G.A. Keystone taxa as drivers of microbiome structure and functioning. Nat. Rev. Microbiol. 2018, 16, 567–576. [Google Scholar] [CrossRef] [PubMed]
- Trosvik, P.; Jacques De Muinck, E. Ecology of bacteria in the human gastrointestinal tract-identification of keystone and foundation taxa. Microbiome 2015, 3, 44. [Google Scholar] [CrossRef]
- Röttjers, L.; Faust, K. Can we predict keystones? 17, 1500; 7. [Google Scholar] [CrossRef]
- Roume, H.; Heintz-buschart, A.; Muller, E.E.L.; May, P.; Satagopam, V.P.; Laczny, C.C.; Narayanasamy, S.; Lebrun, L.A.; Hoopmann, M.R.; Schupp, J.M.; et al. Comparative integrated omics: Identification of key functionalities in microbial community-wide metabolic networks. NJP Biofilms Microbiomes 2015, 1, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Schlaeppi, K.; van der Heijden, M.G. Reply to ‘Can we predict microbial keystones’? Nat. Rev. Microbiol. 2019, 17, 2019. [Google Scholar] [CrossRef]
- Lawson, C.E.; Harcombe, W.R.; Hatzenpichler, R. Common principles and best practices for engineering microbiomes. Nat. Rev. Microbiol. 2019, 17, 725–741. [Google Scholar] [CrossRef]
- Lee, H.L.; Shen, H.; Hwang, I.Y.; Ling, H.; Yew, W.S.; Lee, Y.S.; Chang, M.W. Targeted approaches for in situ gut microbiome manipulation. Genes 2018, 9, 351. [Google Scholar] [CrossRef]
- Langdon, A.; Crook, N.; Dantas, G. The effects of antibiotics on the microbiome throughout development and alternative approaches for therapeutic modulation. Genome Med. 2016, 8, 1–16. [Google Scholar] [CrossRef]
- Einarsson, G.; Flanagan, E.; Lee, A.; Elborn, J.S.; Tunny, M.; Plant, B.J. WS03. 1 Longitudinal airway microbiota profiling in cystic fibrosis patients enrolled in the CFMATTERS clinical trial. J. Cyst. Fibros. 2017, 16, S4. [Google Scholar] [CrossRef]
- Cobián Güemes, A.G.; Lim, Y.W.; Quinn, R.A.; Conrad, D.J.; Benler, S.; Maughan, H.; Edwards, R.; Brettin, T.; Cantú, V.A.; Cuevas, D.; et al. Cystic Fibrosis Rapid Response: Translating Multi-omics Data into Clinically Relevant Information. mBio 2019, 10, e00431-19. [Google Scholar] [CrossRef]
- Jethwa, H.; Abraham, S. The evidence for microbiome manipulation in inflammatory arthritis. Rheumatology 2017, 56, 1452–1460. [Google Scholar] [CrossRef]
- Markle, J.G.M.; Frank, D.N.; Adeli, K.; Von Bergen, M.; Danska, J.S. Microbiome manipulation modifies sex-specific risk for autoimmunity. Gut Microbes 2014, 5, 485–493. [Google Scholar] [CrossRef][Green Version]
- Harris, J.K.; De Groote, M.A.; Sagel, S.D.; Zemanick, E.T.; Kapsner, R.; Penvari, C.; Kaess, H.; Deterding, R.R.; Accurso, F.J.; Pace, N.R. Molecular identification of bacteria in bronchoalveolar lavage fluid from children with cystic fibrosis. Proc. Natl. Acad. Sci. USA 2007, 104, 20529–20533. [Google Scholar] [CrossRef]
- Quinton, P.M. Cystic fibrosis: Impaired bicarbonate secretion and mucoviscidosis. Lancet 2008, 372, 415–417. [Google Scholar] [CrossRef]
- Alaiwa, M.H.A.; Reznikov, L.R.; Gansemer, N.D.; Sheets, K.A.; Horswill, A.R.; Stoltz, D.A.; Zabner, J.; Welsh, M.J. pH modulates the activity and synergism of the airway surface liquid antimicrobials β-defensin-3 and LL-37. Proc. Natl. Acad. Sci. USA 2014, 111, 18703–18708. [Google Scholar] [CrossRef] [PubMed]
- Kunzelmann, K.; Schreiber, R.; Hadorn, H.B. Bicarbonate in cystic fibrosis. J. Cyst. Fibros. 2017, 16, 653–662. [Google Scholar] [CrossRef] [PubMed]
- Gibson, R.L.; Burns, J.L.; Ramsey, B.W. Pathophysiology and management of pulmonary infections in cystic fibrosis. Am. J. Respir. Crit. Care Med. 2003, 168, 918–951. [Google Scholar] [CrossRef] [PubMed]
- Conrad, D.; Haynes, M.; Salamon, P.; Rainey, P.B.; Youle, M.; Rohwer, F. Cystic fibrosis therapy: A community ecology perspective. Am. J. Respir. Cell Mol. Biol. 2013, 48, 150–156. [Google Scholar] [CrossRef]
- Sibley, C.D.; Parkins, M.D.; Rabin, H.R.; Duan, K.; Norgaard, J.C.; Surette, M.G. A polymicrobial perspective of pulmonary infections exposes an enigmatic pathogen in Cystic Fibrosis patients. Proc. Natl. Acad. Sci. USA 2008, 105, 15070–15075. [Google Scholar] [CrossRef] [PubMed]
- Harrison, F. Microbial ecology of the cystic fibrosis lung. Microbiology 2007, 153, 917–923. [Google Scholar] [CrossRef] [PubMed]
- Tunney, M.M.; Field, T.R.; Moriarty, T.F.; Patrick, S.; Doering, G.; Muhlebach, M.S.; Wolfgang, M.C.; Boucher, R.; Gilpin, D.F.; McDowell, A. Detection of anaerobic bacteria in high numbers in sputum from patients with cystic fibrosis. Am. J. Respir. Crit. Care Med. 2008, 177, 995–1001. [Google Scholar] [CrossRef]
- Lynch, S.V.; Bruce, K.D. The cystic fibrosis airway microbiome. Cold Spring Harb. Perspect. Med. 2013, 3, a009738. [Google Scholar] [CrossRef]
- Rogers, G.B.; Carroll, M.P.; Serisier, D.J.; Hockey, P.M.; Jones, G.; Bruce, K.D. Characterization of bacterial community diversity in cystic fibrosis lung infections by use of 16S ribosomal DNA terminal restriction fragment length polymorphism profiling. J. Clin. Microbiol. 2004, 42, 5176–5183. [Google Scholar] [CrossRef]
- Whiteson, K.L.; Bailey, B.; Bergkessel, M.; Conrad, D.; Delhaes, L.; Felts, B.; Harris, J.K.; Hunter, R.; Lim, Y.W.; Maughan, H.; et al. The upper respiratory tract as a microbial source for pulmonary infections in Cystic Fibrosis: Parallels from island biogeography. Am. J. Respir. Crit. Care Med. 2014, 189, 1309–1315. [Google Scholar] [CrossRef]
- O’Toole, G.A. Cystic fibrosis airway microbiome: Overturning the old, opening the way for the new. J. Bacteriol. 2018, 200, e00561-17. [Google Scholar] [CrossRef]
- Lim, Y.W.; Schmieder, R.; Haynes, M.; Willner, D.; Furlan, M.; Youle, M.; Abbott, K.; Edwards, R.; Evangelista, J.; Conrad, D.; et al. Metagenomics and metatranscriptomics: Windows on CF-associated viral and microbial communities. J. Cyst. Fibros. 2013, 12, 154–164. [Google Scholar] [CrossRef] [PubMed]
- Lim, Y.W.; Evangelista, J.S.; Schmieder, R.; Bailey, B.; Haynes, M.; Furlan, M.; Maughan, H.; Edwards, R.; Rohwer, F.; Conrad, D. Clinical insights from metagenomic analysis of sputum samples from patients with cystic fibrosis. J. Clin. Microbiol. 2014, 52, 425–437. [Google Scholar] [CrossRef] [PubMed]
- Cowley, E.S.; Kopf, S.H.; LaRiviere, A.; Ziebis, W.; Newman, D.K. Pediatric cystic fibrosis sputum can be chemically dynamic, anoxic, and extremely reduced due to hydrogen sulfide formation. mBio 2015, 6, e00767-15. [Google Scholar] [CrossRef]
- Quinn, R.A.; Lim, W.; Maughan, H.; Conrad, D.; Rohwer, F.; Whiteson, L. Biogeochemical Forces Shape the Composition and Physiology of Polymicrobial Communities in the Cystic Fibrosis Lung. mBio 2014, 5, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Lim, Y.W.; Schmieder, R.; Haynes, M.; Furlan, M.; Matthews, T.D.; Whiteson, K.; Poole, S.J.; Hayes, C.S.; Low, D.A.; Maughan, H.; et al. Mechanistic Model of Rothia mucilaginosa Adaptation toward Persistence in the CF Lung, Based on a Genome Reconstructed from Metagenomic Data. PLoS ONE 2013, 8, e64285. [Google Scholar] [CrossRef]
- Quinn, R.A.; Whiteson, K.; Lim, Y.-W.; Salamon, P.; Bailey, B.; Mienardi, S.; Sanchez, S.E.; Blake, D.; Conrad, D.; Rohwer, F. A Winogradsky-based culture system shows an association between microbial fermentation and cystic fibrosis exacerbation. ISME J. 2015, 9, 1024–1038. [Google Scholar] [CrossRef] [PubMed]
- Whiteson, K.L.; Meinardi, S.; Lim, Y.W.; Schmieder, R.; Maughan, H.; Quinn, R.; Blake, D.R.; Conrad, D.; Rohwer, F. Breath gas metabolites and bacterial metagenomes from cystic fibrosis airways indicate active pH neutral 2, 3-butanedione fermentation. ISME J. 2014, 8, 1247. [Google Scholar] [CrossRef] [PubMed]
- Quinn, R.A.; Whiteson, K.; Lim, Y.W.; Zhao, J.; Conrad, D.; LiPuma, J.J.; Rohwer, F.; Widder, S. Ecological networking of cystic fibrosis lung infections. NPJ Biofilms Microbiomes 2016, 2, 4. [Google Scholar] [CrossRef] [PubMed]
- Flynn, J.M.; Niccum, D.; Dunitz, J.M.; Hunter, R.C. Evidence and Role for Bacterial Mucin Degradation in Cystic Fibrosis Airway Disease. PLoS Pathog. 2016, 12, e1005846. [Google Scholar] [CrossRef] [PubMed]
- Valentini, T.D.; Lucas, S.K.; Binder, K.A.; Cameron, L.C.; Motl, J.A.; Dunitz, J.M.; Hunter, R.C. Bioorthogonal non-canonical amino acid tagging reveals translationally active subpopulations of the cystic fibrosis lung microbiota. Nat. Commun. 2020, 11, 1–11. [Google Scholar] [CrossRef]
- Flynn, J.M.; Cameron, L.C.; Wiggen, T.D.; Dunitz, J.M.; Harcombe, W.R.; Hunter, R.C. Disruption of Cross-Feeding Inhibits Pathogen Growth in the Sputa of Patients with Cystic Fibrosis. mSphere 2020, 5, e00343-20. [Google Scholar] [CrossRef]
- Castner, L.M.; Zimbric, M.; Cahalan, S.; Powell, C.; Caverly, L.J. Outcomes of cystic fibrosis pulmonary exacerbations treated with antibiotics with activity against anaerobic bacteria. J. Cystic Fibros. 2021. [Google Scholar] [CrossRef]
- Phan, J.; Meinardi, S.; Barletta, B.; Blake, D.R.; Whiteson, K. Stable isotope profiles reveal active production of VOCs from human-associated microbes. J. Breath Res. 2017, 11, 17101. [Google Scholar] [CrossRef]
- Phan, J.; Gallagher, T.; Oliver, A.; England, W.E.; Whiteson, K. Fermentation products in the cystic fibrosis airways induce aggregation and dormancy-associated expression profiles in a CF clinical isolate of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 2018, 1–11. [Google Scholar] [CrossRef]
- Gao, B.; Gallagher, T.; Zhang, Y.; Elbadawi-Sidhu, M.; Lai, Z.; Fiehn, O.; Whiteson, K.L. Tracking polymicrobial metabolism in cystic fibrosis airways: Pseudomonas aeruginosa metabolism and physiology are influenced by Rothia mucilaginosa-derived metabolites. mSphere 2018, 3, e00151-18. [Google Scholar] [CrossRef] [PubMed]
- Root, R.K.; Waldvogel, F.; Corey, L.; Stamm, W.E. Clinical Infectious Diseases: A Practical Approach; Oxford University Press: USA, 1999; ISBN 019508103X. [Google Scholar]
- Smith, E.E.; Buckley, D.G.; Wu, Z.; Saenphimmachak, C.; Hoffman, L.R.; D’Argenio, D.A.; Miller, S.I.; Ramsey, B.W.; Speert, D.P.; Moskowitz, S.M. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc. Natl. Acad. Sci. USA 2006, 103, 8487–8492. [Google Scholar] [CrossRef]
- Adamowicz, E.M.; Flynn, J.; Hunter, R.C.; Harcombe, W.R. Cross-feeding modulates antibiotic tolerance in bacterial communities. ISME J. 2018, 2723–2735. [Google Scholar] [CrossRef]
- Climo, M.W.; Israel, D.S.; Wong, E.S.; Williams, D.; Coudron, P.; Markowitz, S.M. Hospital-wide restriction of clindamycin: Effect on the incidence of Clostridium difficile-associated diarrhea and cost. Ann. Intern. Med. 1998, 128, 989–995. [Google Scholar] [CrossRef]
- Ledson, M.J.; Gallagher, M.J.; Cowperthwaite, C.; Convery, R.P.; Walshaw, M.J. Four years’ experience of intravenous colomycin in an adult cystic fibrosis unit. Eur. Respir. J. 1998, 12, 592–594. [Google Scholar] [CrossRef] [PubMed]
- Matsui, H.; Wagner, V.E.; Hill, D.B.; Schwab, U.E.; Rogers, T.D.; Button, B.; Ii, R.M.T.; Superfine, R.; Rubinstein, M.; Iglewski, B.H.; et al. A physical linkage between cystic fibrosis airway surface dehydration and Pseudomonas aeruginosa biofilms. Proc. Natl. Acad. Sci. USA 2006, 103, 18131–18136. [Google Scholar] [CrossRef]
- Goddard, A.F.; Staudinger, B.J.; Dowd, S.E.; Joshi-Datar, A.; Wolcott, R.D.; Aitken, M.L.; Fligner, C.L.; Singh, P.K. Direct sampling of cystic fibrosis lungs indicates that DNA-based analyses of upper-airway specimens can misrepresent lung microbiota. Proc. Natl. Acad. Sci. USA 2012, 109, 13769–13774. [Google Scholar] [CrossRef] [PubMed]
- Liwinski, T.; Leshem, A.; Elinav, E. Breakthroughs and Bottlenecks in Microbiome Research. Trends Mol. Med. 2021, 27, 298–301. [Google Scholar] [CrossRef] [PubMed]
- Crossette, E.; Gumm, J.; Langenfeld, K.; Raskin, L.; Duhaime, M.; Wigginton, K. Metagenomic Quantification of Genes with Internal Standards. mBio 2021, 12, e03173-20. [Google Scholar] [CrossRef]
- Satinsky, B.M.; Gifford, S.M.; Crump, B.C.; Moran, M.A. Use of internal standards for quantitative metatranscriptome and metagenome analysis. Methods Enzymol. 2013, 531, 237–250. [Google Scholar]
- Patel, A.; Noble, R.T.; Steele, J.A.; Schwalbach, M.S.; Hewson, I.; Fuhrman, J.A. Virus and prokaryote enumeration from planktonic aquatic environments by epifluorescence microscopy with SYBR Green, I. Nat. Protoc. 2007, 2, 269. [Google Scholar] [CrossRef]
- Gifford, S.M.; Sharma, S.; Rinta-Kanto, J.M.; Moran, M.A. Quantitative analysis of a deeply sequenced marine microbial metatranscriptome. ISME J. 2011, 5, 461–472. [Google Scholar] [CrossRef]
- Jorth, P.; Ehsan, Z.; Rezayat, A.; Caldwell, E.; Pope, C.; Brewington, J.J.; Goss, C.H.; Benscoter, D.; Clancy, J.P.; Singh, P.K. Direct lung sampling indicates that established pathogens dominate early infections in children with cystic fibrosis. Cell Rep. 2019, 27, 1190–1204. [Google Scholar] [CrossRef]
- Acosta, N.; Heirali, A.; Somayaji, R.; Surette, M.G.; Workentine, M.L.; Sibley, C.D.; Rabin, H.R.; Parkins, M.D. Sputum microbiota is predictive of long-term clinical outcomes in young adults with cystic fibrosis. Thorax 2018, 73, 1016–1025. [Google Scholar] [CrossRef]
- Hankinson, J.L.; Odencrantz, J.R.; Fedan, K.B. Spirometric reference values from a sample of the general US population. Am. J. Respir. Crit. Care Med. 1999, 159, 179–187. [Google Scholar] [CrossRef]
- Schluchter, M.D.; Konstan, M.W.; Drumm, M.L.; Yankaskas, J.R.; Knowles, M.R. Classifying Severity of Cystic Fibrosis Lung Disease Using Longitudinal Pulmonary Function Data. Am. J. Respir. Crit. Care Med. 2006, 174, 780–786. [Google Scholar] [CrossRef]
- Conrad, D.J.; Bailey, B.A.; Hardie, J.A.; Bakke, P.S.; Eagan, T.M.L.; Aarli, B.B. Median regression spline modeling of longitudinal FEV1 measurements in cystic fibrosis (CF) and chronic obstructive pulmonary disease (COPD) patients. PLoS ONE 2017, 12, e0190061. [Google Scholar] [CrossRef]
- Sriramulu, D.D.; Lünsdorf, H.; Lam, J.S.; Römling, U. Microcolony formation: A novel biofilm model of Pseudomonas aeruginosa for the cystic fibrosis lung. J. Med. Microbiol. 2005, 54, 667–676. [Google Scholar] [CrossRef] [PubMed]
- Palmer, K.L.; Aye, L.M.; Whiteley, M. Nutritional cues control Pseudomonas aeruginosa multicellular behavior in cystic fibrosis sputum. J. Bacteriol. 2007, 189, 8079–8087. [Google Scholar] [CrossRef] [PubMed]
- Fung, C.; Naughton, S.; Turnbull, L.; Tingpej, P.; Rose, B.; Arthur, J.; Hu, H.; Harmer, C.; Harbour, C.; Hassett, D.J. Gene expression of Pseudomonas aeruginosa in a mucin-containing synthetic growth medium mimicking cystic fibrosis lung sputum. J. Med. Microbiol. 2010, 59, 1089–1100. [Google Scholar] [CrossRef] [PubMed]
- Cantu, V.A.; Sadural, J.; Edwards, R. PRINSEQ++, a multi-threaded tool for fast and efficient quality control and preprocessing of sequencing datasets. PeerJ Prepr. 2019, 7, e27553v1. [Google Scholar]
- Ponstingl, H.; Ning, Z. SMALT-a new mapper for DNA sequencing reads. F1000 Posters 2010, 1, 313. [Google Scholar]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Eren, A.M.; Esen, Ö.C.; Quince, C.; Vineis, J.H.; Morrison, H.G.; Sogin, M.L.; Delmont, T.O. Anvi’o: An advanced analysis and visualization platform for ‘omics data. PeerJ 2015, 3, e1319. [Google Scholar] [CrossRef] [PubMed]
- Ondov, B.D.; Treangen, T.J.; Melsted, P.; Mallonee, A.B.; Bergman, N.H.; Koren, S.; Phillippy, A.M. Mash: Fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016, 17, 132. [Google Scholar] [CrossRef] [PubMed]
- Smart, K.F.; Aggio, R.B.M.; Van Houtte, J.R.; Villas-Bôas, S.G. Analytical platform for metabolome analysis of microbial cells using methyl chloroformate derivatization followed by gas chromatography–mass spectrometry. Nat. Protoc. 2010, 5, 1709. [Google Scholar] [CrossRef] [PubMed]
- Cobián Güemes, A.G.; Youle, M.; Cantú, V.A.; Felts, B.; Nulton, J.; Rohwer, F. Viruses as winners in the game of life. Annu. Rev. Virol. 2016, 3, 197–214. [Google Scholar] [CrossRef]
- Li, H. Microbiome, metagenomics, and high-dimensional compositional data analysis. Annu. Rev. Stat. Its Appl. 2015, 2, 73–94. [Google Scholar] [CrossRef]
- Thompson, J.; Johansen, R.; Dunbar, J.; Munsky, B. Machine learning to predict microbial community functions: An analysis of dissolved organic carbon from litter decomposition. BioRxiv 2019, 599704. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silveira, C.B.; Cobián-Güemes, A.G.; Uranga, C.; Baker, J.L.; Edlund, A.; Rohwer, F.; Conrad, D. Multi-Omics Study of Keystone Species in a Cystic Fibrosis Microbiome. Int. J. Mol. Sci. 2021, 22, 12050. https://doi.org/10.3390/ijms222112050
Silveira CB, Cobián-Güemes AG, Uranga C, Baker JL, Edlund A, Rohwer F, Conrad D. Multi-Omics Study of Keystone Species in a Cystic Fibrosis Microbiome. International Journal of Molecular Sciences. 2021; 22(21):12050. https://doi.org/10.3390/ijms222112050
Chicago/Turabian StyleSilveira, Cynthia B., Ana G. Cobián-Güemes, Carla Uranga, Jonathon L. Baker, Anna Edlund, Forest Rohwer, and Douglas Conrad. 2021. "Multi-Omics Study of Keystone Species in a Cystic Fibrosis Microbiome" International Journal of Molecular Sciences 22, no. 21: 12050. https://doi.org/10.3390/ijms222112050
APA StyleSilveira, C. B., Cobián-Güemes, A. G., Uranga, C., Baker, J. L., Edlund, A., Rohwer, F., & Conrad, D. (2021). Multi-Omics Study of Keystone Species in a Cystic Fibrosis Microbiome. International Journal of Molecular Sciences, 22(21), 12050. https://doi.org/10.3390/ijms222112050





