Abstract
Tomato spotted wilt virus (TSWV) is one of the most destructive diseases affecting tomato (Solanum lycopersicum) cultivation and production worldwide. As defenses against TSWV, natural resistance genes have been identified in tomato, including Sw-1a, Sw-1b, sw-2, sw-3, sw-4, Sw-5, Sw-6, and Sw-7. However, only Sw-5 exhibits a high level of resistance to the TSWV. Thus, it has been cloned and widely used in the breeding of tomato with resistance to the disease. Due to the global spread of TSWV, resistance induced by Sw-5 decreases over time and can be overcome or broken by a high concentration of TSWV. How to utilize other resistance genes and identify novel resistance resources are key approaches for breeding tomato with resistance to TSWV. In this review, the characteristics of natural resistance genes, natural resistance resources, molecular markers for assisted selection, and methods for evaluating resistance to TSWV are summarized. The aim is to provide a theoretical basis for identifying, utilizing resistance genes, and developing tomato varieties that are resistant to TSWV.
1. Introduction
Tomato (Solanum lycopersicum) is one of the most important economic vegetable crops. As a major producer and exporter of tomato products the worldwide, China has over one million hectares of harvested area and a total tomato production of ~63 million tonnes in 2019. These values are the highest in the world [1]. Disease is the major biotic stress in tomato production and quality. A total of 136 viral species severely harm tomatoes [2], and the Tomato spotted wilt virus (TSWV) is one of the most harmful.
The TSWV belongs to the species Tospovirus, the genus Orthotospovirus, the family Tospovirdae, and the order Bunyaviridae [3]. It is the only species of RNA globular virus species that infects plants [4]. The TSWV is listed as one of the top 10 most important plants viruses worldwide [5]. The TSWV virions are oblate, easily deformed, and have an envelope structures and continuous protrusion layers on their outer membranes. The genome contains three different genomic RNA strands: Large (L) RNA, Medium (M) RNA, and Small (S) RNA, which encode five proteins. The L RNA is a negative-sense RNA that encodes RNA-dependent RNA polymerase (RdRp) [6], has a replication-related protein function, and can work together with the encoding factors of hosts [7,8]. M RNA is a double-sense and antisense RNA encoding protein in the amino and carboxy-terminal positions within the glycoprotein precursors (GnGc), which play crucial roles in virion assembly, maturation, and release in a host organism [9,10]. M RNA is a sense RNA encoding viral non-structural proteins (NSm) and mainly promotes TSWV infection [11]. S RNA is a double-sense and antisense RNA that encodes nucleocapsid proteins (N) and a sense RNA that encodes non-structural proteins (NSs) [12]; both types of proteins play crucial roles in the TSWV infection cycle [13,14,15]. The TSWV has an extremely wide range of susceptible hosts, including many important agricultural and field crops (infecting more than 1090 plant species), especially tomato, pepper, potato, and tobacco [16,17,18,19,20]. TSWV diseases are widely distributed, and the most severe cases are found in temperate and subtropical areas (Figure 1).
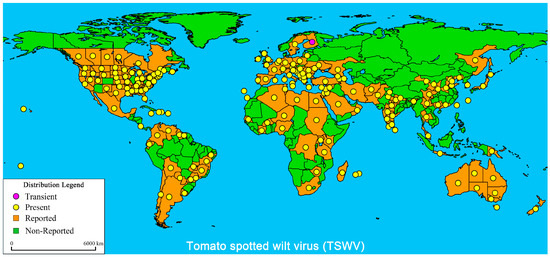
Figure 1.
Global distribution of Tomato spotted wilt virus (TSWV). The global distribution of TSWV disease published by the EPPO Global Database (20 May 2021) and modified about China distribution [21,22,23,24]. The purple marks indicate transient infection, yellow marks indicate present infection, orange marks indicate reported infection, and green marks indicate non-reported infection.
The rapid spread of Frankliniella occidentalis (Western flower thrips) carrying the TSWV has seriously harmed tomato cultivation and production. The TSWV reduces yield in large areas and the marketable value of tomato or even causes the death of tomato plants [25,26]. Numerous tomato plants (such as those that carry the Sw-5 gene) resistant to the TSWV have been screened, and many excellent reviews on the different aspects of TSWV biology, thrips vector-mediated transmission, resistance strategies, and plants’ innate immunity to the TSWV have been published [10,27,28,29,30,31,32,33]. In this study, a systematic overview of global distribution, TSWV symptoms, and inoculation method, organized resistance gene defense against the TSWV, marker-assisted selection markers of resistance breeding, and natural resistant resources is provided, and the focus is innate immunity to TSWV. The aim is to provide a basis for exploring genes involved in resistance against the TSWV and for research on disease resistance breeding in tomato.
2. TSWV Symptoms in Tomato Plants
TSWV infection in field tomatoes is systemic, rendering an entire plant susceptible and resulting in yield losses leading to huge economic losses [34]. Infected tomato plants are usually dwarfed and have necrotic streaks and dark-brown flecks on their leaves, stems, and fruits (Figure 2). The first symptoms in tomato seedlings are inhibited growth points and copper-colored rolls of young leaves. Subsequently, many small dark-brown flecks form, and leaf veins become purple (Figure 2a–c). Growth points appear, leaf necrosis and droop occur, and the stem end forms brown necrotic streaks. The plants show inhibited growth or are completely dwarfed or become deciduous and wilted (Figure 2c,d). In the green fruit period, chlorotic rings appear on the fruit, green fruit is slightly raised, the ring is not obvious, faded necrotic spots appear, which are tumor-like protrusions, and the fruits easily falls off (Figure 2e). During the fruit-ripening period, the fruit has red-yellow or red-white ring spots and bright yellow ring markings, particularly the red ripe fruits (Figure 2f). Identifying resistant resources and genes is the first step to solve these problems.

Figure 2.
Typical symptoms of Tomato spotted wilt virus (TSWV) in different growth stages of field tomato plants. (a,b) dark-brown flecks and necrotic spots; (c,d) turn into copper-colored rolls, purple veins, terminal bud necrosis, and rings on leaves and stems; (e) dark-brown flecks, faded necrotic spots and tumor-like protrusions; (f) chlorotic and bright yellow rings. The white bar represents 25 mm.
3. Methods for Identifying Resistance to TSWV in Tomato
3.1. Mechanical Inoculation
Mechanical inoculation is a simple and effective method for conducting genetic research on resistance to TSWV. A large number of plants can be quickly screened simultaneously with several isolates for the identification of resistant resources [35,36]. An inoculation buffer containing potassium phosphate buffer (0.1 M, pH 7.0), sodium sulphite (0.2% w/v), and polyvinyl pyrolidon (2% w/v) was used. The inoculum was prepared by mixing 1 g of infected leaf tissue with 10 mL of inoculation buffer and 1% carborundum (600 mesh). The fully expanded leaves of the plants at the four-, five, and six-leaf stages were inoculated by rubbing with a brush or cotton swab on the surfaces of the tomato leaves [37]. The inoculated plants were maintained in an environment-controlled greenhouse at 25 °C (day) or 18 °C (night) and at 60% (day) or 95 % (night) of relative humidity for the monitoring of the symptoms. Symptoms were evaluated once a week after inoculation, and six disease grading criteria were used: Asymptomatic, mild, moderate, severe, and whole plant necrosis [38,39]. The incidence rate and disease index were calculated for the identification of resistant tomato plants.
3.2. Thrip Inoculation
Transmission by thrips is difficult to manage, which exposes plants to a high inoculum pressure, and is an extremely effective graft-inoculated method. In general, inoculation needs to be carried out under strictly controlled environmental conditions, which are 25 ± 2 °C temperature, 50% ± 5%/90% ± 5% of relative humidity (day/night), 14 h/10 h (day/night) photoperiod, and cover with anti-thrips mesh (100 mesh). Susceptible plants were inoculated with TSWV isolates at the four-, five, and six-leaf stages and cultivated for symptom development. Then, systemically infected plants were fed to the first instar larvae (0–2 h old) of Frankliniella occidentalis for 2 days to carry the TSWV isolates [26]. The viruliferous larvae were cultivated into adults on healthy plants. Then, the viruliferous adult thrips were used in inoculating plants for 48 h. After the inoculation, the symptoms of the plants were systematically monitored once a week, and resistance and susceptibility were investigated.
Mechanical inoculation and transmission through thrip inoculation are the commonly used methods in screening and identifying resistant germplasm sources in tomato. Mechanical inoculation is easier to implement and is a more effective management method, especially in field natural disease identification. However, it has poor repeatability and depends only on a single test result; it can only identify the resistance of a host to a virus without considering host–vector interactions, resulting in the loss of excellent resistant materials [40]. Furthermore, the method is laborious when a large number of test plants are use. Fortunately, Mandal et al. [41] developed a rapid and efficient pressurized spray inoculation method for the TSWV, but the method seems to be costly and is not widely used. By contrast, transmission by thrips is the closest inoculation method for identifying vector-mediated TSWV resistance components for natural infection. However, this method requires the feeding of viruliferous thrips, and high control on the experimental conditions is required for transmission. However, whether the results pertain to TSWV resistance, thrips resistance, or both is unclear. Mechanical inoculation facilitates the identification of the effects of virus replication and migration, and thrips inoculation facilitates the study of the impact of materials on thrip feeding behavior [42]. Therefore, combining the two methods for identifying plants with different resistance mechanisms according to their complementary information prevents the potential loss of resistance resources [35].
4. Natural Resources Resistant to the TSWV in Tomato
Through the efforts of tomato breeders in the past decades, natural TSWV-resistant germplasm resources have been screened and identified in many tomato lines, genotypes, and cultivars. They are distributed in cultivated and wild tomatoes and mainly distributed in Lycopersicon peruvianum Mill., Lycopersicon chilense Dun., Lycopersicon pimpinellifolium Mill., and other wild tomatoes. Several TSWV resistance genes have been identified in these resources (Table 1). Many germplasm resources of resistance have been discovered from the Porter’s strain of L. pimpinellifolium since 1945. Subsequently, a highly resistant cultivar was detected in the cultivar ‘Stevens’ from S. peruvianum and the LA 1938 from L. chilense [43]. Some plants show high resistance, such as ‘Stevens’, ‘Viradora’, LA0370, LA0445, LA0446, LA2581, LA4445, PE-18/UPV-1, and RDD from L. peruvianum and the LA 1938 from L. chilense. The preferred and safest method for combating TSWV is identifying novel resistant plants. An extensive and in-depth evaluation of tomato plants for which resistance has not been determined is important.

Table 1.
Tomato resources with TSWV resistance genes.
5. Natural Genes Resistant to TSWV in Tomato
Eight loci, namely, Sw-1a, Sw-1b, sw-2, sw-3, sw-4, Sw-5, Sw-6, and Sw-7, for resistance to TSWV have been discovered in the different materials of tomatoes. They originate from cultivated and wild tomatoes, and only Sw-5 has been cloned because of its effective resistance to the TSWV [50,62]. Sw-1a, Sw-1b, sw-2, sw-3, sw-4, and Sw-6 exhibit some degree of resistance to specific TSWV [45,63,64]. As a newly discovered gene in recent years, Sw-7 has a small region range and offers resistance to a wide range of TSWV [65,66,67].
5.1. Sw-1a and Sw-1b
The Sw-1 gene contains two gene clusters, Sw-1a and Sw-1b, and a single dominant and allele pair. Finlay [44] demonstrated that Sw-1a and Sw-1b genes are present in the Pearl Harbor and Porter’s strains of L. pimpinellifolium, and Rey de los Tempranos and Manzana varieties of Lycopersicon esculentum, respectively. The tomato cultivar PI 128657 from L. peruvianum has the Sw-1 gene that resists the isolate TSWV6 [58,68]. However, the isolate specificity and effective resistance of the two gene clusters are limited, particularly in terms of regulating the reactions to the TSWV strains TB2 (tip blight 2), N1 (necrotic), and R1 (ringspot), and TB3 (tip blight 3) [44]. The clusters have been overcome by various TSWV isolates and other tospoviruses [63,64]. Furthermore, the molecular mechanisms and chromosomal locations of the genes are unknown, and thus the genes are rarely used in tomato breeding.
5.2. Sw-2, Sw-3, and Sw-4
Three recessive genes, namely, sw-2, sw-3, and sw-4, for TSWV resistance in tomato have been discovered, which appear to be inherited independently, and come from the Porter’s and Pearl Harbor strains of L. pimpinellifolium, and Rey de los Tempranos and Manzana varieties of L. esculentum [44]. However, these resistance genes, which are isolate specific for TSWV, have been quickly overcome [63,64]. As recessive genes, the genes should show specific resistance in homozygotes. This genetic configuration restricts the development of hybrids [53]. Thus, the genes have not been utilized in commercial breeding.
5.3. Sw-5
Sw-5 is a single dominant quality resistance gene responsible for resistance to a broad range of tospovirus species [64]. Originating from L. peruvianum, this gene has been identified and introgressed in the fresh market tomato cultivar (Lycopersicon esculentum) Stevens [63] and has been mapped near the telomeric region of the long arm in chromosome 9 between the RFLP markers CT71 and CT220 [62]. It is closely linked to the CT220 marker (within 65 kb) [50,51,69]. The Sw-5 locus is a member of a loosely clustered gene family and contains six homologous paralog genes: Sw-5a, Sw-5b, Sw-5c, Sw-5d, Sw-5e, and Sw-5f [31,62]. Sw-5a, Sw-5b, Sw-5c, Sw-5d, and Sw-5e have been cloned in L. peruvianum cv. ‘Stevens’, and Sw-5f has been cloned in L. esculentum ‘Moneymaker’ (Figure 3a,b) [60]. Sw-5* is highly conserved Sw-5a and Sw-5b, and Sw-5aS are highly conserved Sw-5c, Sw-5d, and Sw-5e according to the sequencing and annotation of the tomato genome (S. lycopersicum cv. Heinz 1706) in susceptible tomatoes and have been functionally studied (Figure 3c) [28,70,71].
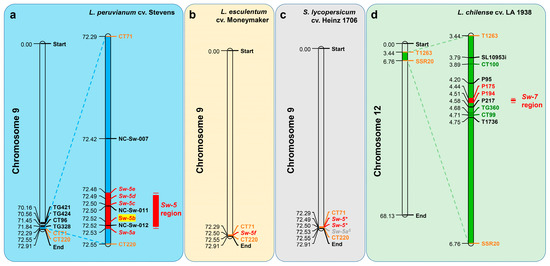
Figure 3.
Mapping of TSWV resistance genes on tomato chromosomes. (a) The site of Sw-5 (Sw-5a, Sw-5b, Sw-5c, Sw-5d, and Sw-5e) on chromosome 9 of L. peruvianum cv. Stevens. The red boxes represent the narrowest area where the Sw-5 cluster region is located [50]; (b) the site of Sw-5f on chromosome 9 of L. esculentum cv. Moneymaker; (c) the site of Sw-5*, Sw-5aS on chromosome 9 of Solanum lycopersicum cv. Heinz 1706; (d) the site of Sw-7 region on chromosome 12 of L. chilense cv. LA 1938. The green boxes represent the narrowest area where the Sw-7 region is currently located. The orange-yellow font markers (T1263 and SSR20) represent the markers mapped by Stevens [72] and Dockter et al. [66]. Green font markers (CT100 and TG360) and red font markers (P175 and P194) represent the markers mapped by Scott et al. [67].
Sw-5a and Sw-5b genes are highly homologous (95%). However, only Sw-5b has broad-spectrum resistance to distinct TSWV isolates [45,63] and is a key gene in resistance to the TSWV [51]. Sw-5b mediates resistance to the related tospovirus species, tomato chlorotic spot virus (TCSV), tomato zonate spot virus (TZSV), and groundnut ring spot virus (GRSV) [51,52,64] and even to the less related impatiens necrotic spot virus (INSV) [73,74]. The roles of the Sw-5c, Sw-5d, and Sw-5e genes in resistance to TSWV and related resistance and molecular mechanisms need further study.
The Sw-5 clustered proteins are members of the resistance (R) gene family and encoded by the amino-terminal Solanaceae domain (SD) and coiled-coil domain (CC) domain; central nucleotide binding-adapter shared by APAF-1, R proteins, and a CED-4 (NB-ARC); and a leucine-rich repeat (LRR) domain (Figure 4b) [50,51,71,75]. Sw-5b is a typical CC-NB-ARC protein. The main reason that Sw-5b has broad-spectrum resistance is that its SD domain can specifically recognize a conserved 21-aa (amino acid) in the TSWV NSm. The NSm region is highly conserved in American-type tospoviruses, but not in Euro-Asian-type tospoviruses [47,76]. The Sw-5b genes have been found in different tomato germplasm materials through the sequencing of the tomato genome [47]. Sequence differences among the genes of different tomato plants are extremely large (Figure 4a,b). Although Sw-5 is widely used in tomato resistance crossbreeding, it is not completely immune to the TSWV. It will be overcome in the presence of a high concentration of TSWV pressure or stable resistance-breaking isolates [77]. The surface of the fruit has ring spots, and the leaves show necrosis [78,79]. Its resistance is limited [80,81].
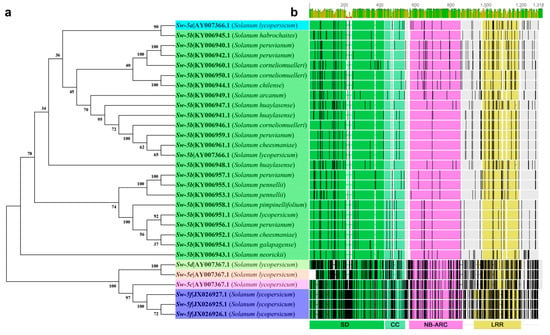
Figure 4.
Cluster and topology analysis of the Sw-5 gene cluster and orthologs from different tomatoes. (a) Cluster analysis based on the Sw-5 gene cluster DNA sequence; (b) alignment and topology analysis based on the Sw-5 protein cluster, an overview of the roles of the amino-terminal Solanaceae domain (SD) and coiled-coil (CC), central nucleotide-binding adaptor shared by apoptotic protease-activating factor-1 (APAF-1), R proteins, and a CED-4 (NB-ARC); and carboxy-terminal leucine-rich repeat domains (LRR) in the Sw-5 protein cluster. The black vertical lines indicate an amino acid difference, and gray blocks indicate the same amino acid sequence.
5.4. Sw-6
A single TSWV resistance gene Sw-6 from L. peruvianum is identified in the L. esculentum introgressed UPV-32 line (Table 1) [53]. The gene is resistant to typical TSWV isolates (e.g., T-941117 and HA-931100 Spanish isolates). However, resistance cannot be identified when screening is performed under greenhouse conditions [82]. The resistance of the Sw-6 locus confers partial resistance, is not as strong that provided by Sw-5, is inherited independent of Sw-5 and the UPV 1 resistance gene (introgressed from L. peruvianum) [45]. It exhibits partial resistance or incomplete dominance when thrip (Frankliniella occidentalis) inoculation is performed by TSWV isolate aggressiveness, and it is effectivity range is narrower than the effectivity ranges of Sw-5 and the UPV 1 resistance gene due to its incomplete penetration and gene dosage effects [45]. Nevertheless, although the TSWV partially conditions Sw-6 to isolate aggressiveness, the resistance of the gene differs from that of the Sw-1a, Sw-1b, sw-2, sw-3, and sw-4 [44,45]. Unfortunately, the molecular mechanism of the Sw-6 resistance gene has not been characterized. Determining whether these genes represent different genes located on distinct chromosomes or on the same chromosomes, and whether they represent different alleles and dosages from a well-known resistance gene cluster, remain unclear [28]. Thus, the resistance levels and persistent resistance in tomato can be improved by facilitating the crossing of the Sw-6 gene with other resistance genes, [68]. The gene plays a positive role in the breeding of disease-resistant tomato.
5.5. Sw-7
Sw-7 is a single dominant quality gene that confers field resistance against various TSWV isolates [43,83,84]. It is derived from the breeding material Y118 (Flag 925-2) selected with S. chilense accession LA 1938 (Table 1) and generally resides between markers T1263 (45.0 cM) and SSR20 (58.2 cM) on chromosome 12 [66,72,82,85]. The region is narrowed between P175 (4.44 Mb) and P194 (4.51 Mb) (Figure 3d) [67]. However, this locus has not been mapped and cloned, and the specific molecular mechanism is unknown.
Sw-7 is not linked to Sw-5 [83], but provides field resistance to the various isolates of the TSWV in Florida, Georgia, Hawaii, and South Africa [86]. Greenhouse utilizing trials are resistant to isolates that overcome tomatoes that are homozygous for Sw-5, and it shows a resistance mechanism different from that of Sw-5 [65,66]. Therefore, Sw-7 was used as an alternative locus conferring resistance to a wide range of TSWV strains. The S. chilense-based germplasm has been promoted in Australia, Thailand, Taiwan, and Italy [86]. Researchers performed a comprehensive transcriptome profiling, functional characterization using an Sw-7 nearly-isogenic line and a TSWV-susceptible parent (Fla.8059) upon inoculation with the TSWV showed the potential involvement of the pathogenesis-related protein 5 (PR-5) in Sw-7 resistance. It is associated with Sw-7 resistance [37,87]. Sw-7 resistance effectively facilitates the breeding of disease-resistant tomatoes and serves as a source of resistance germplasm that provides protection against the TSWV. For the identification of resistance genes for tomato breeding, molecular markers associated with resistance should be developed.
6. Molecular Markers for Resistance to TSWV in Tomato
Due to the geographical specificity of the TSWV isolates distributed with geographical areas, the identification results of natural field inoculation are not reproducible. The disease is limited by many factors, such as environmental influences, which causes great uncertainty for the identification of TSWV-resistant tomato materials. However, in marker-assisted selection (MAS), molecular markers are closely linked to the genes that determine target traits, the desired gene is detected with molecular markers, and target traits are selected. MAS has the advantages of reducing breeding costs and improving breeding selection accuracy and is not affected by environmental conditions. Tomato is considered a model plant for commercial breeding using molecular markers [88]. The development of molecular linkage markers for TSWV resistance genes is mainly focused on the research of Sw-5 and Sw-7 markers. The linkages molecular markers of Sw-1a, Sw-1b, sw-2, sw-3, sw-4, and Sw-6 have not been reported. The linked molecular markers used in resistance breeding against TSWV are summarized in Table 2. The use of these markers has a wide and effective application prospect in the selection and identification of resistant tomato materials, discovery of novel TSWV resistance genes, and acceleration of the breeding processes of tomatoes with TSWV resistance.

Table 2.
Molecular markers associated with TSWV-resistant in tomato.
7. Mechanism of Natural Resistance to the TSWV in Tomato
The viral small interfering RNAs (vsiRNAs) profiles derived from the TSWV genome in an infected tomato were analyzed. The vsiRNAs targeted host genes involved in many pathways, including those related to plant–pathogen interactions [99]. Thus, tomatoes, like other plants, undergo several stages of defense and auto-immunity (Figure 5). Viruses are recognized by the plant pattern-recognition receptors (PRRs), that is, the pathogen–associated molecular patterns (PAMPs) [100]. PAMP-triggered immunity (PTI), which is the first line of defense for immune response in plants when pathogens invade plants [88]. However, rapid pathogen effectors can disrupt PTI response. During virus invasion, nucleotide-binding leucine-rich repeat receptors (NLRs) recognize specific pathogen effectors and trigger effector-triggered immunity (ETI) [100,101,102,103]. Plant NLRs are subdivided into CC-NLRs (CNLs) and Toll/interleukin-1 (TIR)-NLRs (TNLs) according to their N-terminal structures [104]. The NLRs can directly or indirectly identify pathogen effectors and trigger a hypersensitive cell death response (HR) to restrict TSWV to the site of infection [47,105,106].
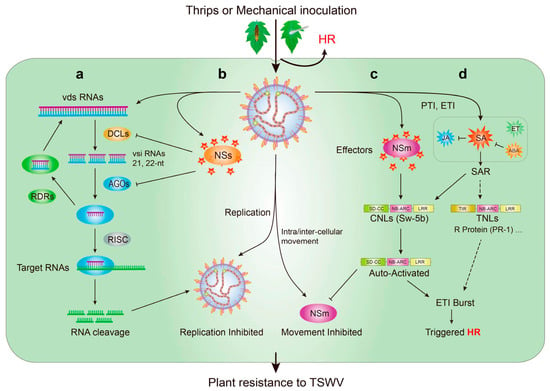
Figure 5.
Mechanism with plant innate immunity against Tomato spotted wilt virus (TSWV). (a) TSWV invaded by thrips or mechanical inoculation, and depend on the plant cellular machineries to complete their life; (b) RNA interference (RNAi) with plant innate immunity is triggered to prevent TSWV invasion, viral dsRNA (vds RNAs) from viral mRNA of TSWV is cleaved into 21, 22-nt viral small interfering RNAs (vsi RNAs) by the RNAse III Dicer-like proteins (DCLs); the vsiRNAs into Argonaute (AGO) activates the RNA-induced silencing complex (RISC) and activates the complementarity and cleavage of viral target RNAs; (c) to further combat TSWV invasion, the CNLs (including Sw-5b protein) from tomato sense the effectors NSm and NSs, robust effector-triggered immunity (ETI); and produce the hypersensitive cell death response (HR); (d) then, trigged PAMP-triggered immunity (PTI) and ETI lead to the accumulation of salicylic acid (SA), jasmonate (JA), ethylene (ET), and abscisic acid (ABA). SA induces the rapid transcriptional activation of a string of resistance (R) genes. Abbreviations: SAR, systemic acquired resistance; RDRs, the RNA-dependent RNA polymerases; SD, Solanaceae domain; CC, coiled-coil domain; NB, nucleotide-binding domain; NB-ARC, Apaf-1, R-protein, and CED-4 domain; LRR, leucine-rich repeat domain; CNLs, coiled-coil nucleotide-binding leucine-rich repeat receptors; TIR, Toll/interleukin-1; TNLs, Toll/interleukin-1 nucleotide-binding leucine-rich repeat receptors; R-protein, resistance protein; NSs, nonstructural protein encoded by the S RNA segment; NSm, nonstructural protein encoded by the M RNA segment.
The tomato Sw-5b belongs to CNLs. NSm from TSWV specifically binds to the extended SD domain of the Sw-5b protein, and the switch activates the receptor. Sw-5b is automatically activated (Figure 5c), and HR is triggered; these processes lead to a robust defense response against tospoviruses [33,47,71,107,108]. The phytohormones salicylic acid (SA), jasmonate (JA), ethylene (ET), and abscisic acid (ABA) play significant roles in PTI and ETI and activate the systemic acquired resistance of plants [109,110,111,112]. The SA signaling pathway has a key role in basal defense against the TSWV in tomato plants [38,113]. TSWV infection robustly up-regulates SA synthesis and increases SA-related defenses [114]. SA accumulates in infected areas and then induces the rapid transcriptional activation of a string of resistance (R) genes (Figure 5d) [115]. It further triggers HR. JA-related response in TSWV-infected plants are repressed by SA, and this process mainly occurs downstream of the JA biosynthesis pathway [116]. The up-regulation of ABA-related genes results in the suppression of SA-mediated defense [114,117]. Pathogenesis-related 1 (PR-1) and PR-5 are associated with Sw-7 resistance and might play a major role in resistance against TSWV infection [37,87].
RNA interference (RNAi), as a conserved regulatory function mechanism, plays pivotal roles in gene regulation and defense against invading viruses. Therefore, TSWV-infected tomatoes and other plants have similar resistance mechanisms against the TSWV [99,118]. The RNAse III Dicer-like proteins (DCLs), Argonautes (AGOs), and RNA-dependent RNA polymerases (RDRs), the three main stages of the RNAi pathway, are triggered after TSWV infection [119,120,121]. The formation of viral dsRNA by TSWV is diced by DCLs culminating in the production of 21and 22 nt vsiRNAs from the three RNA segments of the TSWV [118]. One of these vsiRNAs is recruited onto AGOs and loaded into the RNA-induced silencing complex (RISC) [122]. Through the action of a target mRNA with siRNA-sequence complementarity, this vsiRNA facilitates the cleaving of an RNA target into small fragments or inhibits translation (Figure 5a,b) [123,124]. The RDRs of plants used vsiRNA as a template for synthesizing dsRNA and amplifying of silencing [118,121,125,126]. Thus, plants resist invading viruses though their own RNAi immune mechanisms [120,124,127]. RDR1 has a dual function, is involved in SA resistance pathways, and inhibits the RDR6-mediated antiviral RNAi pathway [128]. The virus does not show weakness. The NSs protein, as a silencing suppressor of TSWV, inhibits RISC activity in plants by binding AGOs, and then the suppressor RNAi mechanism of plants (Figure 5a,b). It also suppresses plant resistance [129,130]. In general, the plant RNAi immune mechanism plays a role in resisting the invasion of defenseless external viruses.
8. Challenge and Prospects
In the breeding of tomato resistant to the TSWV, utilizing existing resistance genes and screening novel resistance genes is a top priority. These methods are environmentally friendly, economical, and effective in alleviating damage due to TSWV infection. Sw-7 has been mapped in the ~70 kb genomic region. We believe it will be cloned soon. Sw-5, Sw-6, and Sw-7 exhibit specific and different resistance mechanisms for TSWV, and complementary resistance is present between them. Thus, we should use these genes as resistance resources and use the MAS technology in aggregating different resistance genes, which will provide stable and lasting resistance. In the plant immune system, the R gene is an essential defense recognition gene [131]. Sw-5 is the only class R identified in tomato, and the Sw-7 locus belongs to the R gene. A detailed R locus physical map was built, and the 368 candidate pathogen recognition genes were found on 12 chromosomes in tomato, including 154 NBS-LRR domain resistance-like genes [70,131,132]. As a marker gene for disease resistance, PR-1 possibly plays a role in resistance to TSWV infection in the Sw-7 line [37]. We infer that the R locus is a readily available resource for screening TSWV resistance genes. TSWV tomato species with high resistance and even completely immune effectiveness have not been found. Therefore, in light of the discovery of novel highly activated TSWV races and increasingly serious spread of TSWV, the collection of resistance resources and the discovery of new resistance genes (including the class of R gene) against TSWV by combining mechanical inoculation with TSWV and MAS are essential for tomato resistance breeding and cultivation production.
The plant innate RNAi mechanism has a significant defense against TSWV invasion, particularly in tomatoes carrying Sw-5 [99,127]. In transgenic plants in tobacco (Nicotiana tabacum) plants, expressed N protein can resist TSWV infection [119,133]; it reflects an RNA-mediated defense mechanism [12,134,135,136]. The constructed RNAi-mediated transgenic plants by targeting the N, NSm and NSs genes of TSWV indicated that enhanced tobacco lines resistance [137,138]. Additionally, the dsRNAs targeting the N gene by the RNAi-based vaccination approach can protect the Nicotiana benthamiana and tomato [139]. As the first layer of defense for immune response, the plant immune system prevents the TSWV invasion through en RNA silencing mechanism [33]. Therefore, the RNAi-mediated technology is feasible for enhancing TSWV resistance and tomato resistance breeding.
The genome-editing technology (GET) has become a common technology in recent years. The clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein-9 nuclease (Cas9) system has been used in improving or mitigating viral disease resistance in plants [140,141]. The CRISPR/Cas9 can efficiently achieve specific site-specific mutations and regulate specific plant immune responses [142]. Strategies to improve plant disease resistance by GET, including, modified R gene, transformed susceptible (S) gene, and targeted degradation of viral genome. The synthetic I2 immune receptor CNL of tomato by CRISPR/Cas9 system demonstrates recognition activity and improves tomato resistance to Phytophthora infestans, Fusarium oxysporum f. sp. lycopersici, and phylogenetically divergent pathogens [143]. Additionally, the integration of NLRs from wild species into cultivated tomato enhances resistance [144]. As a typical S gene from rice, Pi21 has been precisely edited with CRISPR/Cas9 systems, and mutant lines displayed enhanced resistance to blast and bacterial blight in rice [145]. Francisella. novicida Cas9 (FnCas9) targets the cucumber mosaic virus (CMV) RNA and induces resistance to RNA viruses, and resistance is stably inheritable [146]. However, given that GET applications in tomato TSWV resistance breeding have not been reported, we should use tomato-suitable CRISPR/Cas9 technology for tomato resistance breeding [147]. The Sw-5aS from TSWV-susceptible cultivars can be transformed through fixed-site precise editing with CRISPR/Cas9, and Sw-5b-mediated TSWV resistance should be reintroduced for the enhancement of tomato resistance to the TSWV. Additionally, substantial breakthroughs have been achieved in plant engineering of NLRs to defend against pathogens [148]. Huang et al. [149] conducted a stepwise artificial evolution strategy to select Sw-5b mutants, which are effective against resistance-breaking isolates of TSWV, in order to provide a new vision and ideas that resist TSWV by artificial evolution.
9. Conclusions
One of the great challenges faced in the tomato breeding program to TSWV-disease resistance is the achievement of a stable and lasting resistance. Despite the unremitting efforts of researchers for more than 30 years for the identification and introgression of resistance, given the continuous mutation of TSWV and the emergence of new isolates, the detection of new germplasm resistant to TSWV disease has been challenging. In this review, we focused on the TSWV-resistant tomato breeding plants, TSWV resistance genes, and linked molecular markers, especially the systematic overview of TSWV disease resistance mechanisms, and in-depth elucidation of the molecular mechanisms of TSWV resistance. Disease breeding has far-reaching significance. In recent years, new breeding strategies have surpassed the classic breeding methods by systematic summary of the disease-resistance mechanism of plants against TSWV. The use of new breeding strategies, such as RNA silencing mechanism, targeted gene editing, and NLR artificial evolution, to achieve plants resistant to TSWV infection is a new opportunity in tomato breeding, particularly in disease resistance.
Author Contributions
S.Q. and Y.L. conceived the present work; S.Q. and Y.L. drafted the manuscript; Formal analysis, S.Z., M.M.I. and A.H.E.-S.; writing review and editing, M.M.I., A.H.E.-S. and F.Z.; supervision, Y.L.; project administration, Y.L.; funding acquisition, Y.L. All authors have read and agreed to the published version of the manuscript.
Funding
National Key Research and Development Program of China (2016YFD0101703); Key Research and Development Program of Shaanxi Province (2019ZDLNY03-05).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All the data is contained within the article.
Acknowledgments
The authors wish to thank Xiangqiang Zhan from College of Horticulture, Northwest A&F University.
Conflicts of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have influenced the work reported in this paper.
Abbreviations
| TSWV | Tomato spotted wilt virus |
| L RNA | Large RNA |
| M RNA | Medium RNA |
| S RNA | Small RNA |
| RdRp | RNA-dependent RNA polymerase |
| GnGc | Glycoprotein precursors |
| NSm | Viral non-structural proteins |
| NSs | Non-structural proteins |
| N | Nucleocapsid proteins |
| TB2 | TSWV strains tip blight 2 |
| N1 | TSWV strains necrotic |
| R1 | TSWV strains ringspot |
| TB3 | TSWV strains tip blight 3 |
| TCSV | Tomato chlorotic spot virus |
| TZSV | Tomato zonate spot virus |
| GRSV | Groundnut ring spot virus |
| INSV | Impatiens necrotic spot virus |
| R | Resistance gene family |
| SD | Solanaceae domain |
| CC | Coiled-coil domain |
| NB-ARC | Nucleotide binding-adapter shared by APAF-1, R proteins and a CED-4 |
| LRR | Leucine-rich repeat domain |
| APAF-1 | Apoptotic protease-activating factor-1 |
| PR-1 | Pathogenesis-related protein 1 |
| PR-5 | Pathogenesis-related protein 5 |
| MAS | Marker-assisted selection |
| RAPD | Random amplified polymorphic DNA |
| CAPS | Cleaved amplified polymorphic sequence |
| InDel | Insertion-deletion |
| SNP | Single-nucleotide polymorphism |
| SCAR | Sequence characterized amplified regions |
| RFLP | Restriction fragment length polymorphism |
| KASP | Kompetitive Allele-Specific PCR |
| vsiRNAs | Viral small interfering RNAs |
| PRRs | Plant pattern-recognition receptors |
| PAMPs | Pathogen–associated molecular patterns |
| PTI | PAMP-triggered immunity |
| NLRs | Nucleotide-binding leucine-rich repeat receptors |
| ETI | Trigger effector-triggered immunity |
| CNLs | CC-NLRs |
| TNLs | Toll/interleukin-1 (TIR)-NLRs |
| HR | Hypersensitive cell death response |
| RNAi | RNA interference |
| vds RNAs | Viral dsRNA |
| DCLs | RNAse III Dicer-like proteins |
| RDRs | RNA-dependent RNA polymerases |
| AGO | Argonautes |
| RISC | RNA-induced silencing complex |
| SA | Salicylic acid |
| JA | Jasmonate |
| ET | Ethylene |
| ABA | Abscisic acid |
| SAR | Systemic acquired resistance |
| GET | Genome-editing technology |
| CRISPR/Cas9 | Clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein-9 nuclease (Cas9) system |
| S | Susceptible gene |
| CMV | Cucumber mosaic virus |
| FnCas9 | Francisella. novicida Cas9 |
References
- FAO (Food and Agriculture Organization of the United Nations). FAOSTAT—Crops and Livestock Products. Latest Update: 15 September 2021. Available online: http://www.fao.org/faostat/en/#data/QCL (accessed on 28 September 2021).
- Hanssen, I.M.; Lapidot, M.; Thomma, B.P.H.J. Emerging viral diseases of tomato crops. Mol. Plant-Microbe Interact. 2010, 23, 539–548. [Google Scholar] [CrossRef]
- Nilon, A.; Robinson, K.; Pappu, H.R.; Mitter, N. Current status and potential of RNA interference for the management of Tomato spotted wilt virus and thrips vectors. Pathogens 2021, 10, 320. [Google Scholar] [CrossRef] [PubMed]
- Lefkowitz, E.J.; Dempsey, D.M.; Hendrickson, R.C.; Orton, R.J.; Siddell, S.G.; Smith, D.B. Virus taxonomy: The database of the International Committee on Taxonomy of Viruses (ICTV). Nucleic Acids Res. 2018, 46, 708–717. [Google Scholar] [CrossRef] [PubMed]
- Scholthof, K.-B.G.; Adkins, S.; Czosnek, H.; Palukaitis, P.; Jacquot, E.; Hohn, T.; Hohn, B.; Saunders, K.; Candresse, T.; Ahlquist, P.; et al. Top 10 plant viruses in molecular plant pathology. Mol. Plant Pathol. 2011, 12, 938–954. [Google Scholar] [CrossRef] [PubMed]
- De Haan, P.; Kormelink, R.; de Oliveira Resende, R.; van Poelwijk, F.; Peters, D.; Goldbach, R. Tomato spotted wilt virus L RNA encodes a putative RNA polymerase. J. Gen. Virol. 1991, 71, 2207–2216. [Google Scholar] [CrossRef] [PubMed]
- Adkins, S.; Quadt, R.; Choi, T.J.; Ahlquist, P.; German, T. An RNA-dependent RNA-polymerase activity associated with virions of Tomato spotted wilt virus, a plant-infecting and insect-infecting bunyavirus. Virology 1995, 207, 308–311. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-H.; Kim, Y.-S.; Jang, S.-W.; Jeon, Y.-H. Complete genome sequence of Tomato spotted wilt virus from paprika in Korea. Int. J. Plant Pathol. 2013, 2, 121–136. [Google Scholar] [CrossRef]
- Nagata, T.; Inoue-Nagata, A.K.; Prins, M.; Goldbach, R.; Peters, D. Impeded thrips transmission of defective Tomato spotted wilt virus isolates. Phytopathology 2000, 90, 454–459. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Kwon, S.-Y.; Kim, S.T. An insight into the Tomato spotted wilt virus (TSWV), tomato and thrips interaction. Plant Biotechnol. Rep. 2018, 12, 157–163. [Google Scholar] [CrossRef]
- Storms, M.M.H.; Kormelink, R.; Peters, D.; VanLent, J.A.W.M.; Goldbach, R.W. The nonstructural NSm protein of tomato spotted wilt virus induces tubular structures in plant and insect cells. Virology 1995, 214, 485–493. [Google Scholar] [CrossRef]
- Sonoda, S.; Tsumuki, H. Analysis of gene sequences for the nucleocapsid protein from Tomato spotted wilt virus for promoting RNA-mediated cross-protection using the Potato virus X vector system. J. Gen. Plant Pathol. 2004, 70, 239–242. [Google Scholar] [CrossRef]
- Snippe, M.; Borst, J.W.; Goldbach, R.; Kormelink, R. Tomato spotted wilt virus Gc and N proteins interact in vivo. Virology 2007, 357, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Ocampo, T.O.; Peralta, S.M.G.; Bacheller, N.; Uiterwaal, S.; Knapp, A.; Hennen, A.; Ochoa-Martinez, D.L.; Garcia-Ruiz, H. Antiviral RNA silencing suppression activity of Tomato spotted wilt virus NSs protein. Genet. Mol. Res. 2016, 15, 15028625. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Liu, B.; Ding, Z.; Li, G.; Liu, M.; Zhu, D.; Sun, Y.; Dong, S.; Lou, Z. Distinct mechanism for the formation of the ribonucleoprotein complex of Tomato spotted wilt virus. J. Virol. 2017, 91, e00892-17. [Google Scholar] [CrossRef] [PubMed]
- Yeturu, S.; Viera, W.; Garrido, P.; Insuasti, M. First report of Tomato spotted wilt virus infecting tree tomato (Solanum Betaceum cav.) in Ecuador. J. Plant Phytopathol. 2016, 98, 691. [Google Scholar]
- Sivaprasad, Y.; Garrido, P.; Mendez, K.; Pachacama, S.; Garrido, A.; Ramos, L. First report of Tomato spotted wilt virus infecting pepper in Ecuador. J. Phytopathol. 2017, 99, 304. [Google Scholar]
- Oliver, J.E.; Whitfield, A.E. The genus tospovirus: Emerging Bunyaviruses that threaten food security. Ann. Rev. Virol. 2016, 3, 101–124. [Google Scholar] [CrossRef]
- Martinez-Ochoa, N.; Mandal, B.; Csinos, A.S. Evaluation of rhizobacteria to control tomato spotted wilt in tobacco. Phytopathology 2002, 92, S150. [Google Scholar]
- Mandal, B.; Pappu, H.R.; Csinos, A.S.; Culbreath, A.K. Response of peanut, pepper, tobacco, and tomato cultivars to two biologically distinct isolates of Tomato spotted wilt virus. Plant Dis. 2006, 90, 1150–1155. [Google Scholar] [CrossRef]
- Sun, X.H.; Gao, L.L.; Wang, S.L.; Wang, C.L.; Yang, Y.Y.; Wang, X.Y.; Zhu, X.P. First report of Tomato spotted wilt virus infecting pumpkin in China. J. Plant Phytopathol. 2016, 98, 677–697. [Google Scholar]
- Sun, M.; Jing, C.; Chu, C.; Wu, G.; Sun, X.; Xie, Y.; Liu, Y.; Qing, L. Serological detection and molecular identification of Tomato spotted wilt virus in pepper in Chongqing. Acta Hortic. Sin. 2017, 44, 487–494. [Google Scholar]
- Mo, N.; Shi, Y.; Qin, L.; Li, Y.; Liang, Y. Cloning and sequence analysis of Tomato spotted wilt virus coat protein gene in Yangling Region of Shaanxi Province. China Veg. 2019, 3, 36–40. [Google Scholar]
- Yu, M.; Yang, C.; Wang, J.; Hou, Q.; Zhang, S.; Cao, M. First report of Tomato spotted wilt virus (TSWV) isolated from nasturtium (Tropaeolum majus L.) with a serious leaf mosaic disease in China. Plant Dis. 2020, 105, 716. [Google Scholar] [CrossRef] [PubMed]
- Sevik, M.A.; Arli-Sokmen, M. Estimation of the effect of Tomato spotted wilt virus (TSWV) infection on some yield components of tomato. Phytoparasitica 2012, 40, 87–93. [Google Scholar] [CrossRef]
- Chiapello, M.; Bosco, L.; Ciuffo, M.; Ottati, S.; Salem, N.; Rosa, C.; Tavella, L.; Turina, M. Complexity and local specificity of the virome associated with tospovirus-transmitting thrips species. J. Virol. 2021. [Google Scholar] [CrossRef]
- Whitfield, A.E.; Falk, B.W.; Rotenberg, D. Insect vector-mediated transmission of plant viruses. Virology 2015, 479, 278–289. [Google Scholar] [CrossRef]
- Turina, M.; Kormelink, R.; Resende, R.O. Resistance to tospoviruses in vegetable crops: Epidemiological and molecular aspects. Annu. Rev. Phytopathol. 2016, 54, 347–371. [Google Scholar] [CrossRef] [PubMed]
- Khalid, A.; Zhang, Q.; Yasir, M.; Li, F. Small RNA based genetic engineering for plant viral resistance: Application in crop protection. Front. Microbiol. 2017, 8, 43. [Google Scholar] [CrossRef]
- Carbonell, P.; Alonso, A.; Grau, A.; Francisco Salinas, J.; García-Martínez, S.; José Ruiz, J. Twenty years of tomato breeding at EPSO-UMH: Transfer resistance from wild types to local landraces-from the first molecular markers to genotyping by sequencing (GBS). Diversity 2018, 10, 12. [Google Scholar] [CrossRef]
- De Oliveira, A.S.; Boiteux, L.S.; Kormelink, R.; Resende, R.O. The Sw-5 gene cluster: Tomato breeding and research toward orthotospovirus disease control. Front. Plant Sci. 2018, 9, 1055. [Google Scholar] [CrossRef]
- Chen, Y.; Dessau, M.; Rotenberg, D.; Rasmussen, D.A.; Whitfield, A.E. Entry of bunyaviruses into plants and vectors. Adv. Virus Res. 2019, 104, 65–96. [Google Scholar]
- Zhu, M.; Van Grinsven, I.L.; Kormelink, R.; Tao, X. Paving the way to tospovirus infection: Multilined interplays with plant innate immunity. Annu. Rev. Phytopathol. 2019, 57, 41–62. [Google Scholar] [CrossRef]
- Pappu, H.R.; Jones, R.A.C.; Jain, R.K. Global status of tospovirus epidemics in diverse cropping systems: Successes achieved and challenges ahead. Virus Res. 2009, 141, 219–236. [Google Scholar] [CrossRef]
- Roselló, S.; Díez, M.J.; Lacasa, A.; Jordá, C.; Nuez, F. Testing resistance to TSWV introgressed from Lycopersicon peruvianum by artificial transmission techniques. Euphytica 1997, 98, 93–98. [Google Scholar] [CrossRef]
- Kabaş, A.; Fidan, H.; Demirelli, M.B. Identification of new sources of resistance to resistance-breaking isolates of Tomato spotted wilt virus. Saudi J. Biol. Sci. 2021, 28, 3094–3099. [Google Scholar] [CrossRef]
- Padmanabhan, C.; Ma, Q.; Shekasteband, R.; Stewart, K.S.; Hutton, S.F.; Scott, J.W.; Fei, Z.; Ling, K.-S. Comprehensive transcriptome analysis and functional characterization of PR-5 for its involvement in tomato Sw-7 resistance to Tomato spotted wilt tospovirus. Sci. Rep. 2019, 9, 7673. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Gresa, M.P.; Lison, P.; Yenush, L.; Conejero, V.; Rodrigo, I.; Belles, J.M. Salicylic acid is involved in the basal resistance of tomato plants to citrus exocortis viroid and Tomato spotted wilt virus. PLoS ONE 2016, 11, e0166938. [Google Scholar] [CrossRef]
- Shi, Y.; Mo, N.; Qi, S.; Liang, Y. Screening of resistant germplasm of Tomato spotted wilt virus and optimization of artificial identification method. China Veg. 2020, 6, 39–43. [Google Scholar]
- Lacasa, A.; Contreras, J.; Jordá, C.; Díez, M.J.; Roselló, S.; Catalá, M.S.; Costa, J.; Nuez, F. Screening of resistant materials of Lycopersicon spp. to TSWV by means of thrips transmision. Tomato Genet. Coop. Rep. 1994, 44, 16–19. [Google Scholar]
- Mandal, B.; Csinos, A.S.; Martinez-Ochoa, N.; Pappu, H.R. A rapid and efficient inoculation method for Tomato spotted wilt tospovirus. J. Virol. Methods 2008, 149, 195–198. [Google Scholar] [CrossRef] [PubMed]
- Roselló, S.; Soler, S.; José Díez, M.; Rambla, J.L.; Richarte, C.; Nuez, F. New sources for high resistance of tomato to the Tomato spotted wilt virus from Lycopersicon peruvianum. Plant Breed. 1999, 118, 425–429. [Google Scholar] [CrossRef]
- Canady, M.A.; Stevens, M.R.; Barineau, M.S.; Scott, J.W. Tomato spotted wilt virus (TSWV) resistance in tomato derived from Lycopersicon chilense Dun. LA 1938. Euphytica 2001, 117, 19–25. [Google Scholar] [CrossRef]
- Finlay, K.W. Inheritance of spotted wilt resistance in the tomato. II. Five genes controlling spotted wilt resistance in four tomato types. Aust. J. Biol. Sci. 1953, 6, 153–163. [Google Scholar] [CrossRef]
- Roselló, S.; José Díez, M.; Nuez, F. Genetics of Tomato spotted wilt virus resistance coming from Lycopersicon peruvianum. Eur. J. Plant Pathol. 1998, 104, 499–509. [Google Scholar] [CrossRef]
- Maluf, W.R.; Toma-Braghini, M.; Corte, R.D. Progress in breeding tomatoes for resistance to Tomato spotted wilt. Braz. J. Genet. 1991, 14, 509–525. [Google Scholar]
- Zhu, M.; Jiang, L.; Bai, B.H.; Zhao, W.Y.; Chen, X.J.; Li, J.; Liu, Y.; Chen, Z.Q.; Wang, B.T.; Wang, C.L.; et al. The intracellular immune receptor Sw-5b confers broad-spectrum resistance to Tospoviruses through recognition of a conserved 21-amino acid viral effector epitope. Plant Cell 2017, 29, 2214–2232. [Google Scholar] [CrossRef]
- Paterson, R.; Scott, S.; Gergerich, R. Resistance in two Lycopersicon species to an Arkansas isolate of Tomato spotted wilt virus. Euphytica 1989, 43, 173–178. [Google Scholar] [CrossRef]
- Giordano, L.D.; de Avila, A.C.; Charchar, J.M.; Boiteux, L.S.; Ferraz, E. ‘Viradoro’: A tospovirus-resistant processing tomato cultivar adapted to tropical environments. Hortscience 2000, 35, 1368–1370. [Google Scholar] [CrossRef]
- Brommonschenkel, S.H.; Frary, A.; Frary, A.; Tanksley, S.D. The broad-spectrum tospovirus resistance gene Sw-5 of tomato is a homolog of the root-knot nematode resistance gene Mi. Mol. Plant Microbe Interact. 2000, 13, 1130–1138. [Google Scholar] [CrossRef] [PubMed]
- Spassova, M.I.; Prins, T.W.; Folkertsma, R.T.; Klein-Lankhorst, R.M.; Hille, J.; Goldbach, R.W.; Prins, M. The tomato gene Sw5 is a member of the coiled coil, nucleotide binding, leucine-rich repeat class of plant resistance genes and confers resistance to TSWV in tobacco. Mol. Breed. 2001, 7, 151–161. [Google Scholar] [CrossRef]
- Folkertsma, R.T.; Spassova, M.I.; Prins, M.; Stevens, M.R.; Hille, J.; Goldbach, R.W. Construction of a bacterial artificial chromosome (BAC) library of Lycopersicon esculentum cv. Stevens and its application to physically map the Sw-5 locus. Mol. Breed. 1999, 5, 197–207. [Google Scholar] [CrossRef]
- Rosello, S.; Ricarte, B.; Diez, M.J.; Nuez, F. Resistance to Tomato spotted wilt virus introgressed from Lycopersicon peruvianum in line UPV 1 may be allelic to Sw-5 and can be used to enhance the resistance of hybrids cultivars. Euphytica 2001, 119, 357–367. [Google Scholar] [CrossRef]
- Lima, G.S.D.A.; Brommonschenkel, S.H.; Ventura, G.M. Broad-spectrum resistance to tospovirus in accessions of Lycopersicon peruvianum and L. chilense. Summa Phytopathol. 2003, 29, 352–354. [Google Scholar]
- Boiteux, L.S.; Giordano, L.D.B. Screening Lycopersicon germplasm for resistance to a Brazilian isolate of Tomato spotted wilt virus (TSWV). Tomato Genet. Coop. Rep. 1993, 42, 13–14. [Google Scholar]
- Diez, M.; Rosello, S.; Lacasa, A.; Jorda, C.; Costa, J. Agronomic behavior of tomato cultivars and lines resistant to TSWV and influence of inoculation methods. Acta Hortic. 1995, 21, 527–532. [Google Scholar] [CrossRef]
- Holmes, F.O. Resistance to spotted wilt in tomato. Phytopathology 1948, 38, 467–473. [Google Scholar]
- Gilbert, J.C.; Tanaka, J.S. ‘Anahu’, an outstanding hybrid maker. Hawaii Farm Sci. 1971, 20, 6–7. [Google Scholar]
- Gardner, R.G.; Panthee, D.R. Tomato spotted wilt virus-resistant fresh-market tomato breeding lines: NC 58S, NC 123S, NC 127S, and NC 132S. Hortscience 2012, 47, 531–532. [Google Scholar] [CrossRef]
- Rehman, S.; Postma, W.; Tytgat, T.; Prins, P.; Qin, L.; Overmars, H.; Vossen, J.; Spiridon, L.-N.; Petrescu, A.-J.; Goverse, A.; et al. A secreted SPRY domain-containing protein (SPRYSEC) from the plant-parasitic nematode globodera rostochiensis interacts with a CC-NB-LRR protein from a susceptible tomato. Mol. Plant Microbe Interact. 2009, 22, 330–340. [Google Scholar] [CrossRef]
- Kumar, N.K.K.; Ullman, D.E.; Cho, J.J. Evaluation of Lycopersicon germ plasm for Tomato spotted wilt tospovirus resistance by mechanical and thrips transmission. Plant Dis. 1993, 77, 938–941. [Google Scholar] [CrossRef]
- Stevens, M.R.; Lamb, E.M.; Rhoads, D.D. Mapping the Sw-5 locus for Tomato spotted wilt virus resistance in tomatoes using RAPD and RFLP analyses. Theor. Appl. Genet. 1995, 90, 451–456. [Google Scholar] [CrossRef]
- Stevens, M.; Scott, S.; Gergerich, R. Inheritance of a gene for resistance to Tomato spotted wilt virus (TSWV) from Lycopersicon peruvianum Mill. Euphytica 1992, 59, 9–17. [Google Scholar] [CrossRef]
- Boiteux, L.S.; Giordano, L.D. Genetic basis of resistance against two tospovirus species in tomato (Lycopersicon-esculentum). Euphytica 1993, 71, 151–154. [Google Scholar] [CrossRef]
- Price, D.L.; Memmott, F.D.; Hollingsworth, A.; Scott, J.W.; Stevens, M.R. Identification of molecular markers linked to new Tomato spotted wilt virus resistance genes in tomato using AFLP analysis. Hortscience 2007, 42, 855. [Google Scholar]
- Dockter, K.G.; O’Neil, D.S.; Price, D.L.; Scott, J.; Stevens, M.R. Molecular mapping of the Tomato spotted wilt virus resistance gene Sw-7 in tomato. Hortscience 2009, 44, 1123. [Google Scholar]
- Scott, J.; Hutton, S.; Olson, S.; Stevens, M. Spotty Results in Our Sw-7 Tomato Spotted Wilt Virus Research. 2011 Tomato Disease Workshop Meeting Abstract. 2011. Available online: http://vegetablemdonline.ppath.cornell.edu/TDW/Presentations/11%20Scott_TDW_2011.pdf (accessed on 5 October 2021).
- Gordillo, L.F.; Stevens, M.R.; Millard, M.A.; Geary, B. Screening two Lycopersicon peruvianum collections for resistance to Tomato spotted wilt virus. Plant Dis. 2008, 92, 694–704. [Google Scholar] [CrossRef] [PubMed]
- Brommonschenkel, S.H.; Tanksley, S.D. Map-based cloning of the tomato genomic region that spans the Sw-5 tospovirus resistance gene in tomato. Mol. Gen. Genet. 1997, 256, 121–126. [Google Scholar] [CrossRef]
- Andolfo, G.; Sanseverino, W.; Aversano, R.; Frusciante, L.; Ercolano, M.R. Genome-wide identification and analysis of candidate genes for disease resistance in tomato. Mol. Breed. 2014, 33, 227–233. [Google Scholar] [CrossRef]
- De Oliveira, A.S.; Koolhaas, I.; Silva Boiteux, L.; Caldararu, O.F.; Petrescu, A.-J.; Resende, R.O.; Kormelink, R. Cell death triggering and effector recognition by Sw-5 SD-CNL proteins from resistant and susceptible tomato isolines to Tomato spotted wilt virus. Mol. Plant Pathol. 2016, 17, 1442–1454. [Google Scholar] [CrossRef]
- Stevens, M.R. Localization and mapping of Sw-7, a tomato spotted wild virus resistance gene. In Proceedings of the 42nd Tomato Breeders Roundtable, Sacramento, CA, USA, 4–6 April 2009; Available online: http://tgc.ifas.ufl.edu/2009/Stevens%20SW7%20mapping.pdf (accessed on 5 October 2021).
- Dianese, E.C.; Fonseca, M.E.N.; Inoue-Nagata, A.K.; Resende, R.O.; Boiteux, L.S. Search in Solanum (section Lycopersicon) germplasm for sources of broad-spectrum resistance to four Tospovirus species. Euphytica 2011, 180, 307–319. [Google Scholar] [CrossRef]
- Hallwass, M.; de Oliveira, A.S.; de Campos Dianese, E.; Lohuis, D.; Boiteux, L.S.; Inoue-Nagata, A.K.; Resende, R.O.; Kormelink, R. The Tomato spotted wilt virus cell-to-cell movement protein (NSM) triggers a hypersensitive response in Sw-5-containing resistant tomato lines and in Nicotiana benthamiana transformed with the functional Sw-5b resistance gene copy. Mol. Plant Pathol. 2014, 15, 871–880. [Google Scholar] [CrossRef]
- Chen, X.J.; Zhu, M.; Jiang, L.; Zhao, W.Y.; Li, J.; Wu, J.Y.; Li, C.; Bai, B.H.; Lu, G.; Chen, H.Y.; et al. A multilayered regulatory mechanism for the autoinhibition and activation of a plant CC-NB-LRR resistance protein with an extra N-terminal domain. New Phytol. 2016, 212, 161–175. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, A.S.D.; Pallás, V.; Sánchez-Navarro, J.A.; Kormelink, R.; Resende, R.O. The NSm proteins of phylogenetically related tospoviruses trigger Sw-5b–mediated resistance dissociated of their cell-to-cell movement function. Virus Res. 2017, 240, 25–34. [Google Scholar]
- Batuman, O.; Turini, T.A.; Oliveira, P.V.; Rojas, M.R.; Macedo, M.; Mellinger, H.C.; Adkins, S.; Gilbertson, R.L. First report of a resistance-breaking strain of Tomato spotted wilt virus infecting tomatoes with the Sw-5 tospovirus-resistance gene in California. Plant Dis. 2017, 101, 637. [Google Scholar] [CrossRef]
- Aramburu, J.; Marti, M. The occurrence in north-east Spain of a variant of tomato spotted wilt virus (TSWV) that breaks resistance in tomato (Lycopersicon esculentum) containing the Sw-5 gene. Plant Pathol. 2003, 52, 407. [Google Scholar] [CrossRef]
- Ciuffo, M.; Finetti-Sialer, M.M.; Gallitelli, D.; Turina, M. First report in Italy of a resistance-breaking strain of Tomato spotted wilt virus infecting tomato cultivars carrying the Sw5 resistance gene. Plant Pathol. 2005, 54, 564. [Google Scholar] [CrossRef]
- Aramburu, J.; Galipienso, L.; Soler, S.; Lopez, C. Characterization of Tomato spotted wilt virus isolates that overcome the Sw-5 resistance gene in tomato and fitness assays. Phytopathol. Mediterr. 2010, 49, 342–351. [Google Scholar]
- Lopez, C.; Aramburu, J.; Galipienso, L.; Soler, S.; Nuez, F.; Rubio, L. Evolutionary analysis of tomato Sw-5 resistance-breaking isolates of Tomato spotted wilt virus. J. Gen. Virol. 2011, 92, 210–215. [Google Scholar] [CrossRef]
- Price, D.L.; Memmott, F.D.; Scott, J.W.; Olson, S.M.; Stevens, M.R. Identification of molecular markers linked to a new Tomato spotted wilt virus resistance source in tomato. Tomato Genet. Coop. 2007, 57, 35–36. [Google Scholar]
- Scott, J.W.; Stevens, M.R.; Olson, S.M. An alternative source of resistance to Tomato spotted wilt virus. Tomato Genet. Coop. Rep. 2005, 55, 40–41. [Google Scholar]
- Stevens, M.R.; Price, D.L.; Memmott, F.D.; Scott, J.W.; Olson, S.M. Identification of Markers Linked to Sw-7 a New Tomato Spotted Wilt Virus Resistance Gene, Derived from S. chilense. Abstracts from the 2007 Tomato Breeders Roundtable. The Pennsylvania State University, Pennsylvania, USA. Available online: http://tgc.ifas.ufl.edu/2007/2007IndividualAbsPDf/Identification%20of%20Markers%20Linked%20to%20Sw.pdf (accessed on 8 September 2010).
- Stevens, M.R.; Scott, S.J.; Gergerich, R.C. Evaluation of seven Lycopersicon species for resistance to Tomato spotted wilt virus (TSWV). Euphytica 1994, 80, 79–84. [Google Scholar] [CrossRef]
- Stevens, M.R.; Scott, J.W.; Cho, J.J.; Geary, B.D.; Memmott, F.D. A new dominantly inherited source of TSWV resistance in tomato derived from L. chilense, which resists isolates that overcome Sw-5. Hortscience 2006, 41, 991. [Google Scholar] [CrossRef]
- Padmabhan, C.; Zheng, Y.; Shekaste-Band, R.; Stewart, K.; Scott, J.; Fei, Z.; Ling, K. Identification of defense-related genes associated with tomato Sw-7 line against Tomato spotted wilt virus in tomato through transcriptome analysis. Phytopathology 2016, 106, 161. [Google Scholar]
- El-Sappah, A.H.; Islam, M.M.; El-Awady, H.H.; Yan, S.; Qi, S.; Liu, J.; Cheng, G.; Liang, Y. Tomato natural resistance genes in controlling the root-knot nematode. Genes 2019, 10, 925. [Google Scholar] [CrossRef] [PubMed]
- Do Nascimento, I.R.; Maluf, W.R.; Figueira, A.R.; Menezes, C.B.; de Resende, J.T.V.; Faria, M.V.; Nogueira, D.W. Marker assisted identification of tospovirus resistant tomato genotypes in segregating progenies. Sci. Agric. 2009, 66, 298–303. [Google Scholar] [CrossRef]
- Chagué, V.; Mercier, J.C.; Guénard, M.; de Courcel, A.; Vedel, F. Identification and mapping on chromosome 9 of RAPD markers linked to Sw-5 in tomato by bulked segregant analysis. Theor. Appl. Genet. 1996, 92, 1045–1051. [Google Scholar] [CrossRef]
- Śmiech, M.; Rusinowski, Z.; Malepszy, S.; Niemirowicz-Szczytt, K. New RAPD markers of Tomato spotted wilt virus (TSWV) resistance in Lycopersicon esculentum Mill. Acta Physiol. Plant 2000, 22, 299–303. [Google Scholar] [CrossRef]
- Garland, S.; Sharman, M.; Persley, D.; McGrath, D. The development of an improved PCR-based marker system for Sw-5, an important TSWV resistance gene of tomato. Aust. J. Agric. Res. 2005, 56, 285–289. [Google Scholar] [CrossRef]
- Dianese, E.C.; de Fonseca, M.E.N.; Goldbach, R.; Kormelink, R.; Inoue-Nagata, A.K.; Resende, R.O.; Boiteux, L.S. Development of a locus-specific, co-dominant SCAR marker for assisted-selection of the Sw-5 (Tospovirus resistance) gene cluster in a wide range of tomato accessions. Mol. Breed. 2010, 25, 133–142. [Google Scholar] [CrossRef]
- Shi, A.; Vierling, R.; Grazzini, R.; Chen, P.; Caton, H.; Panthee, D. Identification of molecular markers for Sw-5 gene of Tomato spotted wilt virus resistance. Am. J. Biotechnol. Mol. Sci. 2011, 1, 2159–3698. [Google Scholar] [CrossRef]
- Panthee, D.R.; Ibrahem, R. New molecular markers associated with the Sw-5 gene conferring resistance to Tomato spotted wilt virus in tomato. J. Hortic. Sci. Biotechnol. 2013, 88, 129–134. [Google Scholar] [CrossRef]
- Lee, H.J.; Kim, B.; Bae, C.; Kang, W.-H.; Kang, B.-C.; Yeam, I.; Oh, C.-S. Development of a single-nucleotide polymorphism marker for the Sw-5b gene conferring disease resistance to Tomato spotted wilt virus in tomato. Hortic. Sci. Technol. 2015, 33, 730–736. [Google Scholar] [CrossRef]
- Devran, Z.; Kahveci, E. Development and validation of a user-friendly KASP marker for the Sw-5 locus in tomato. Australas. Plant Pathol. 2019, 48, 503–507. [Google Scholar] [CrossRef]
- Tanksley, S.D.; Ganal, M.W.; Prince, J.P.; Devicente, M.C.; Bonierbale, M.W.; Broun, P.; Fulton, T.M.; Giovannoni, J.J.; Grandillo, S.; Martin, G.B.; et al. High-density molecular linkage maps of the tomato and potato genomes. Genetics 1992, 132, 1141–1160. [Google Scholar] [CrossRef] [PubMed]
- Ramesh, S.V.; Williams, S.; Kappagantu, M.; Mitter, N.; Pappu, H.R. Transcriptome-wide identification of host genes targeted by Tomato spotted wilt virus-derived small interfering RNAs. Virus Res. 2017, 238, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.G.; Vance, R.E.; Dangl, J.L. Intracellular innate immune surveillance devices in plants and animals. Science 2016, 354, aaf6395-1–aaf6395-8. [Google Scholar] [CrossRef]
- Caplan, J.; Padmanabhan, M.; Dinesh-Kumar, S.P. Plant NB-LRR immune receptors: From recognition to transcriptional reprogramming. Cell Host Microbe 2008, 3, 126–135. [Google Scholar] [CrossRef] [PubMed]
- Muthamilarasan, M.; Prasad, M. Plant innate immunity: An updated insight into defense mechanism. J. Biosci. 2013, 38, 433–449. [Google Scholar] [CrossRef]
- Ramirez-Prado, J.S.; Abulfaraj, A.A.; Rayapuram, N.; Benhamed, M.; Hirt, H. Plant immunity: From aignaling to epigenetic control of defense. Trends Plant Sci. 2018, 23, 833–844. [Google Scholar] [CrossRef]
- Tian, H.; Wu, Z.; Chen, S.; Ao, K.; Huang, W.; Yaghmaiean, H.; Sun, T.; Xu, F.; Zhang, Y.; Wang, S.; et al. Activation of TIR signalling boosts pattern-triggered immunity. Nature 2021, 1–7. [Google Scholar] [CrossRef]
- Mandadi, K.K.; Scholthof, K.-B.G. Plant immune responses against viruses: How does a virus cause disease? Plant Cell 2013, 25, 1489–1505. [Google Scholar] [CrossRef]
- Chen, H.; Qian, X.; Chen, X.; Yang, T.; Feng, M.; Chen, J.; Cheng, R.; Hong, H.; Zheng, Y.; Mei, Y.; et al. Cytoplasmic and nuclear Sw-5b NLR act both independently and synergistically to confer full host defense against tospovirus infection. New Phytol. 2021, 231, 2262–2281. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Chen, Z.; Wu, Q.; Cai, Y.; Zhang, Y.; Zhao, R.; Yan, J.; Qian, X.; Li, J.; Zhu, M.; et al. The Sw-5b NLR nucleotide-binding domain plays a role in oligomerization and its self-association is important for the activation of cell death signaling. J. Exp. Bot. 2021, 72, 6581–6595. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Huang, H.N.; Zhu, M.; Huang, S.; Zhang, W.H.; Dinesh-Kumar, S.P.; Tao, X.R. A plant immune receptor adopts a two-step recognition mechanism to enhance viral effector perception. Mol. Plant 2019, 12, 248–262. [Google Scholar] [CrossRef] [PubMed]
- Zamora, O.; Schulze, S.; Azoulay-Shemer, T.; Parik, H.; Unt, J.; Brosché, M.; Schroeder, J.I.; Yarmolinsky, D.; Kollist, H. Jasmonic acid and salicylic acid play minor roles in stomatal regulation by CO2, abscisic acid, darkness, vapor pressure deficit, and ozone. Plant J. 2021, 1–44. [Google Scholar] [CrossRef]
- Cao, H.; Glazebrook, J.; Clarke, J.D.; Volko, S.; Dong, X. The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 1997, 88, 57–63. [Google Scholar] [CrossRef]
- Wu, X.; Xu, S.; Zhao, P.; Zhang, X.; Yao, X.; Sun, Y.; Fang, R.; Ye, J. The Orthotospovirus nonstructural protein NSs suppresses plant MYC-regulated jasmonate signaling leading to enhanced vector attraction and performance. PLoS Pathog. 2019, 15, e1007897. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Wu, Q.; Tong, C.; Chen, H.; Miao, D.; Qian, X.; Zhao, X.; Jiang, L.; Tao, X. Characterization of the roles of SGT1/RAR1, EDS1/NDR1, NPR1, and NRC/ADR1/NRG1 in Sw-5b-mediated resistance to Tomato spotted wilt virus. Viruses 2021, 13, 1447. [Google Scholar] [CrossRef] [PubMed]
- Beris, D.; Theologidis, I.; Skandalis, N.; Vassilakos, N. Bacillus amyloliquefaciens strain MBI600 induces salicylic acid dependent resistance in tomato plants against Tomato spotted wilt virus and Potato virus Y. Sci. Rep. 2018, 8, 10320. [Google Scholar] [CrossRef]
- Nachappa, P.; Challacombe, J.; Margolies, D.C.; Nechols, J.R.; Whitfield, A.E.; Rotenberg, D. Tomato spotted wilt virus benefits its thrips vector by modulating metabolic and plant defense pathways in tomato. Front. Plant Sci. 2020, 11, 575564. [Google Scholar] [CrossRef]
- Huang, C. From player to pawn: Viral avirulence factors involved in plant immunity. Viruses 2021, 13, 688. [Google Scholar] [CrossRef]
- Leon-Reyes, A.; Van der Does, D.; De Lange, E.S.; Delker, C.; Wasternack, C.; Van Wees, S.C.M.; Ritsema, T.; Pieterse, C.M.J. Salicylate-mediated suppression of jasmonate-responsive gene expression in Arabidopsis is targeted downstream of the jasmonate biosynthesis pathway. Plant Sci. 2010, 232, 1423–1432. [Google Scholar] [CrossRef]
- Asselbergh, B.; De Vleesschauwer, D.; Höfte, M. Global switches and fine-tuning-ABA modulates plant pathogen defense. Mol. Plant-Microbe Interact. 2008, 21, 709–719. [Google Scholar] [CrossRef]
- Mitter, N.; Koundal, V.; Williams, S.; Pappu, H. Differential expression of Tomato spotted wilt virus-derived viral small RNAs in infected commercial and experimental host plants. PLoS ONE 2013, 8, e76276. [Google Scholar] [CrossRef] [PubMed]
- Konakalla, N.C.; Bag, S.; Deraniyagala, A.S.; Culbreath, A.K.; Pappu, H.R. Induction of plant resistance in tobacco (Nicotiana tabacum) against Tomato spotted wilt orthotospovirus through foliar application of dsRNA. Viruses 2021, 13, 662. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.-W.; Voinnet, O. Antiviral immunity directed by Small RNAs. Cell 2007, 130, 413–426. [Google Scholar] [CrossRef] [PubMed]
- Bai, M.; Yang, G.-S.; Chen, W.-T.; Mao, Z.-C.; Kang, H.-X.; Chen, G.-H.; Yang, Y.-H.; Xie, B.-Y. Genome-wide identification of Dicer-like, Argonaute and RNA-dependent RNA polymerase gene families and their expression analyses in response to viral infection and abiotic stresses in Solanum lycopersicum. Gene 2012, 501, 52–62. [Google Scholar] [CrossRef]
- Kim, Y.J.; Maizel, A.; Chen, X. Traffic into silence: Endomembranes and post-transcriptional RNA silencing. EMBO J. 2014, 33, 968–980. [Google Scholar] [CrossRef]
- Islam, W.; ul Islam, S.; Qasim, M.; Wang, L. Host-Pathogen interactions modulated by small RNAs. RNA Biol. 2017, 14, 891–904. [Google Scholar] [CrossRef]
- Rosa, C.; Kuo, Y.W.; Wuriyanghan, H.; Falk, B.W. RNA interference mechanisms and applications in plant pathology. Annu. Rev. Phytopathol. 2018, 56, 581–610. [Google Scholar] [CrossRef]
- Bernstein, E.; Caudy, A.A.; Hammond, S.M.; Hannon, G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 2001, 409, 363–366. [Google Scholar] [CrossRef]
- Katiyar-Agarwal, S.; Morgan, R.; Dahlbeck, D.; Borsani, O.; Villegas, A.; Zhu, J.-K.; Staskawicz, B.J.; Jin, H. A Pathogen-inducible endogenous siRNA in plant immunity. Proc. Natl. Acad. Sci. USA 2006, 103, 18002–18007. [Google Scholar] [CrossRef]
- Olaya, C.; Fletcher, S.J.; Zhai, Y.; Peters, J.; Margaria, P.; Winter, S.; Mitter, N.; Pappu, H.R. The Tomato spotted wilt virus (TSWV) genome is differentially targeted in TSWV-infected tomato (Solanum lycopersicum) with or without Sw-5 gene. Viruses 2020, 12, 363. [Google Scholar] [CrossRef]
- Ying, X.B.; Dong, L.; Zhu, H.; Duan, C.G.; Du, Q.S.; Lv, D.Q.; Fang, Y.Y.; Garcia, J.A.; Fang, R.X.; Guo, H.S. RNA-dependent RNA polymerase 1 from Nicotiana tabacum suppresses RNA silencing and enhances viral infection in Nicotiana benthamiana. Plant Cell 2010, 22, 1358–1372. [Google Scholar] [CrossRef]
- Giner, A.; Lakatos, L.; García-Chapa, M.; López-Moya, J.J.; Burgyán, J. Viral protein inhibits RISC activity by argonaute binding through conserved WG/GW motifs. PLoS Pathog. 2010, 6, e1000996. [Google Scholar] [CrossRef]
- Hedil, M.; Kormelink, R. Viral RNA silencing suppression: The enigma of bunyavirus NSs proteins. Viruses 2016, 8, 208. [Google Scholar] [CrossRef] [PubMed]
- Andolfo, G.; Sanseverino, W.; Rombauts, S.; Van de Peer, Y.; Bradeen, J.M.; Carputo, D.; Frusciante, L.; Ercolano, M.R. Overview of tomato (Solanum lycopersicum) candidate pathogen recognition genes reveals important Solanum R locus dynamics. New Phytol. 2013, 197, 223–237. [Google Scholar] [CrossRef] [PubMed]
- Ni, X.; Yang, J.; Sun, S.; Yang, W. Identification and analysis of resistance-like genes in the tomato genome. J. Phytopathol. 2014, 162, 137–146. [Google Scholar] [CrossRef]
- Gielen, J.J.L.; de Haan, P.; Kool, A.J.; Peters, D.; van Grinsven, M.Q.J.M.; Goldbach, R.W. Engineered resistance to Tomato spotted wilt virus, a negative-strand RNA virus. Nat. Biotechnol. 1991, 9, 1363–1367. [Google Scholar] [CrossRef]
- Prins, M.; Kikkert, M.; Ismayadi, C.; de Graauw, W.; de Haan, P.; Goldbach, R. Characterization of RNA-mediated resistance to Tomato spotted wilt virus in transgenic tobacco plants expressing NSm gene sequences. Plant Mol. Biol. 1997, 33, 235–243. [Google Scholar] [CrossRef]
- Peng, J.-C.; Chen, T.-C.; Raja, J.A.J.; Yang, C.-F.; Chien, W.-C.; Lin, C.-H.; Liu, F.-L.; Wu, H.-W.; Yeh, S.-D. Broad-spectrum transgenic resistance against distinct tospovirus species at the genus level. PLoS ONE 2014, 9, e96073. [Google Scholar] [CrossRef]
- Yazhisai, U.; Rajagopalan, P.A.; Raja, J.A.J.; Chen, T.-C.; Yeh, S.-D. Untranslatable tospoviral NSs fragment coupled with L conserved region enhances transgenic resistance against the homologous virus and a serologically unrelated tospovirus. Transgenic Res. 2015, 24, 635–649. [Google Scholar] [CrossRef] [PubMed]
- Herrero, S.; Culbreath, A.K.; Csinos, A.S.; Pappu, H.R.; Rufty, R.C.; Daub, M.E. Nucleocapsid gene-mediated transgenic resistance provides protection against Tomato spotted wilt virus epidemics in the field. Phytopathology 2000, 90, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Sonoda, S.; Tsumuki, H. Analysis of RNA-mediated virus resistance by NSs and NSm gene sequences from Tomato spotted wilt virus. Plant Sci. 2004, 166, 771–778. [Google Scholar] [CrossRef]
- Tabein, S.; Jansen, M.; Noris, E.; Vaira, A.M.; Marian, D.; Behjatnia, S.A.A.; Accotto, G.P.; Miozzi, L. The induction of an effective dsRNA-mediated resistance against Tomato spotted wilt virus by exogenous application of double-stranded RNA largely depends on the selection of the viral RNA target region. Front. Plant Sci. 2020, 11, 533338. [Google Scholar] [CrossRef] [PubMed]
- Wright, A.V.; Nuñez, J.K.; Doudna, J.A. Biology and applications of CRISPR systems: Harnessing nature’s toolbox for genome engineering. Cell 2016, 164, 29–44. [Google Scholar] [CrossRef]
- Dong, O.X.; Ronald, P.C. Genetic engineering for disease resistance in plants: Recent progress and future perspectives. Plant Physiol. 2019, 180, 26–38. [Google Scholar] [CrossRef]
- Andolfo, G.; Iovieno, P.; Frusciante, L.; Ercolano, M.R. Genome-editing technologies for enhancing plant disease resistance. Front. Plant Sci. 2016, 7, 1813. [Google Scholar] [CrossRef]
- Giannakopoulou, A.; Steele, J.F.C.; Segretin, M.E.; Bozkurt, T.O.; Zhou, J.; Robatzek, S.; Banfield, M.J.; Pais, M.; Kamoun, S. Tomato I2 immune receptor can be engineered to confer partial resistance to the oomycete Phytophthora infestans in addition to the fungus Fusarium oxysporum. Mol. Plant-Microbe Interact. 2015, 28, 1316–1329. [Google Scholar] [CrossRef]
- Zhang, M.; Coaker, G. Harnessing effector-triggered immunity for durable disease resistance. Phytopathology 2017, 107, 912–919. [Google Scholar] [CrossRef]
- Li, S.; Shen, L.; Hu, P.; Liu, Q.; Zhu, X.; Qian, Q.; Wang, K.; Wang, Y. Developing disease-resistant thermosensitive male sterile rice by multiplex gene editing. J. Integr. Plant Biol. 2019, 61, 1201–1205. [Google Scholar] [CrossRef]
- Zhang, T.; Zheng, Q.; Yi, X.; An, H.; Zhao, Y.; Ma, S.; Zhou, G. Establishing RNA virus resistance in plants by harnessing CRISPR immune system. Plant Biotechnol. J. 2018, 16, 1415–1423. [Google Scholar] [CrossRef]
- Dahan-Meir, T.; Filler-Hayut, S.; Melamed-Bessudo, C.; Bocobza, S.; Czosnek, H.; Aharoni, A.; Levy, A.A. Efficient in planta gene targeting in tomato using geminiviral replicons and the CRISPR/Cas9 system. Plant J. 2018, 95, 5–16. [Google Scholar] [CrossRef] [PubMed]
- Tamborski, J.; Krasileva, K.V. Evolution of plant NLRs: From natural history to precise modifications. Ann. Rev. Virol. 2020, 71, 355–378. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Huang, S.; Li, J.; Wang, H.; Zhao, Y.; Feng, M.; Dai, J.; Wang, T.; Zhu, M.; Tao, X. Stepwise artificial evolution of an Sw-5b immune receptor extends its resistance spectrum against resistance-breaking isolates of Tomato spotted wilt virus. Plant Biotechnol. J. 2021, 1–13. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).