Abstract
Root system architecture (RSA) is an important developmental and agronomic trait that is regulated by various physical factors such as nutrients, water, microbes, gravity, and soil compaction as well as hormone-mediated pathways. Phytohormones act as internal mediators between soil and RSA to influence various events of root development, starting from organogenesis to the formation of higher order lateral roots (LRs) through diverse mechanisms. Apart from interaction with the external cues, root development also relies on the complex web of interaction among phytohormones to exhibit synergistic or antagonistic effects to improve crop performance. However, there are considerable gaps in understanding the interaction of these hormonal networks during various aspects of root development. In this review, we elucidate the role of different hormones to modulate a common phenotypic output, such as RSA in Arabidopsis and crop plants, and discuss future perspectives to channel vast information on root development to modulate RSA components.
1. Introduction
Plants are undeniably the most important source of food, fuel, medicines, and fibres. Apart from the above ground parts of the plants that are actively involved in photosynthesis, the hidden half i.e., the root, controls a wide variety of processes such as nutrients and water acquisition and anchorage in the soil, and also acts as an interface between plant and the earth atmosphere. By sensing and responding to environmental cues and soil nutrition conditions, roots are capable of dramatically altering their architecture and hence enable plants to overcome the challenges posed due to their sessile nature.
Understanding the development and architecture of roots holds potential in improving crop yield and optimizing agricultural land use. The recent development of newer technologies such as advanced digital photography, 3D root imaging, transparent soils, automated rhizotron, X-ray computed tomography, luminescence-fluorescence based imaging systems, and neutron tomography has enabled us to better understand the architecture of complex root systems [1]. Moreover, extensive studies on the Arabidopsis root system have generated crucial insights on molecular mechanisms regulating root architecture development. RSA is influenced by the cross-talk of different hormones as well as by hormone-environment factors that integrate with the plant system via a specific set of downstream regulators that lead to changes in gene expression, signal transduction, and metabolic conversions [2,3]. With the advent of recent technology, the identification of crucial downstream regulators and signalling pathways has now become a reality. For instance, a study by Šimásková and co-workers has shown that cytokinin (CK) induces PIN1 and PIN7 expression via CYTOKININ RESPONSE FACTOR 2 (CRF2) and CRF6 in order to modulate RSA. They showed that domains PCRE7 and PCRE1 of PIN1 and PIN7, respectively, were responsible for their cytokinin-inducibility. A yeast one hybrid (Y1H) screen, ChIP-qPCR, and a protoplast-based luciferase assay demonstrated that the CRF2 and CRF6 transcription factors are positive regulators of PIN7 and PIN1 expression. Additionally, mutant analysis showed that crf236 phenocopies auxin transport defective mutants, confirming that PINs are targets of these transcription factors [4]. Another study shows that FOUR LIPS (FLP) is a transcriptional regulator of PIN3 in the early stages of LR development [5]. However, such interactions have mainly been characterized in model plants such as Arabidopsis, and translating this information into economically important crop plants will open new research possibilities for plant biologists.
This review will briefly outline the root development process from the formation of an embryonic root to the control of root architecture. It also provides insights on various aspects of RSA and how they are regulated by various hormones and their crosstalk in plants.
2. Root System Architecture
The three dimensional structure of the root system is specified as root system architecture (RSA). RSA components include the primary root (PR) length, the number, angle, length, patterning of lateral roots (LR), adventitious roots (AR), and epidermal cell outgrowth called root hair (RH) [6]. There are two types of root systems defined by their branching patterns and developmental origin: taproots and fibrous roots (Figure 1). Taproots that occur in dicots consist of the PR, RH and secondary smaller LRs, and ARs (arising from a root-shoot junction or from the hypocotyl) (Figure 1A). In addition to RH and primary root, a fibrous root system as observed in cereals (Oryza sativa, Zea mays etc.) consists of dense mass of adventitious roots (AR) that occur along with seminal roots, crown roots, and LRs (Figure 1B). However, in cereals, the PR dies as the monocots age, and the adventitious roots form the major portion of the RSA. Depending on the species, RSA displays high variation in morphological traits that help the plant to adapt to a highly competitive environment.
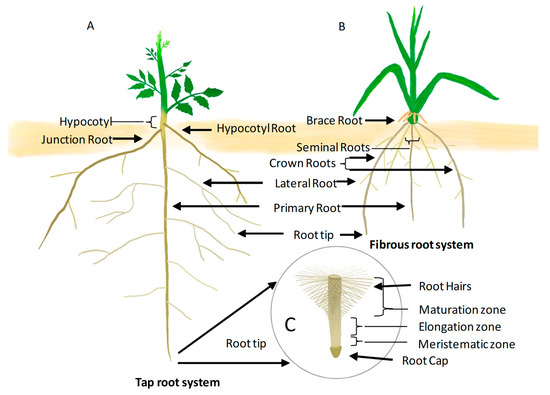
Figure 1.
(A) Tap root system: RSA of a dicot (tomato) showing embryonic primary root, lateral roots, adventitious roots of shoot origin (hypocotyl root), and root-shoot junction (junction root). (B) Fibrous root system: RSA of a monocot (maize) showing embryonic origin primary root and seminal roots, adventitious roots of shoot origin (brace roots) and root-shoot junction (crown root), laterals, and root tip. (C) Root tip of primary root showing root cap, meristematic zone, elongation zone, and maturation zone having root hairs.
2.1. Primary Root Growth and Development
PR is the basic and main component of RSA, is the first to emerge, and is derived from embryonically formed meristematic tissue that forms the hypophysis near the base of the early globular embryo. Asymmetric division takes place, giving rise to upper cell that eventually forms the quiescent centre (QC) and the lower cell that gives rise to the upper cells of the columella. Numerous stem cells surround the QC and, depending on their position, give rise to different tissue types such as columella, vascular tissue, ground tissue, epidermis, and the root cap [7].
The primary root has different developmental zones. In the root apical meristem (RAM), all cells originate from one or more precursor or stem cells at the very tip, called stem cell niche (SCN). The daughter cells divide at different proportions and various rates to generate the division zone (DZ). These cells exit the cell cycle and elongate, generating the elongation zone (EZ). The cells then differentiate and acquire tissue specific features based on their radial position. This zone is called the differentiation zone (Diff zone). The transition zone (TZ) marks the developmental boundary between dividing and differentiating cells [7,8]. Each developmental zone correlates with the graded distribution of hormones, mRNA, peptides, TFs, etc. Auxin and CK play an important role in maintaining the SCN and cell differentiation, respectively. Auxin gradient is expressed as a maximum in SCN and shows a decline towards the TZ [9,10]. The expression of PLETHORA (PLT) TFs correlates with the auxin maxima, declining at the TZ [11], and its expression in SCN is dependent on ARFs such as MONOPTEROS, NON-PHOTOTROPHIC HYPOCOTYL 4 (NPH4) [12]. In turn, PLT contributes to the establishment of auxin maxima by increasing auxin biosynthesis and polar auxin transport (PAT). Cell expansion and differentiation in the root are controlled by CK signalling that promotes the transcription of SHY2 and GH3.17 genes that further shape the auxin gradient, where they position auxin minima at the TZ, leading to differentiation. CKs via ARR1 also promote cell wall remodelling enzyme EXPANSIN1 (EXP1), consequently driving cell differentiation [8].
There are various TFs and genes involved in QC and RAM maintenance. These include WUSCHEL-LIKE HOMEOBOX 5 (WOX5), CYCLIN D3 (CYCD3), CYCLING DOF FACTOR 4 (CDF4), SCARECROW (SCR), SHORTROOT (SHR), HAIRY MERISTEM (HAM), etc. (extensively reviewed in [13]).
There are some substantial differences between dicot and monocot root tissue organization. The cereal tissues are overall larger and more complex with wide cortical cell layers, as compared to the narrow ones in dicots. Additionally, the QC population is drastically larger compared to dicots such as Arabidopsis. Monocots possess polyarch xylem as compared to the tetrarch xylem of dicots. Further, the number of xylems and phloems in dicots vary from 2–8 compared to more than 8 in monocots. In dicots, the pith region is either absent or small and underdeveloped; whereas in monocots the pith is larger and well-developed [14].
2.2. Lateral Root Development
LRs provide better anchorage in soil, help in nutrient assimilation and water uptake, and make symbiotic association with microbes. LR development is primarily governed by auxin activity and auxin response maxima; the downstream signalling cascade regulates LR patterning in angiosperms [15,16,17]. LR formation occurs in the single-layered pericycle tissue at the xylem poles, which are termed as xylem pole pericycle (XPP) cells (extensively reviewed in [15]). After continuous anticlinal division and cell elongation, XPP cells move from meristem to a zone of auxin maximum. However, this auxin maxima is not regular; rather, it appears to behave in an oscillation manner, as high auxin peaks are interrupted by low auxin response [18]. The occurrence of the auxin maxima can be seen by DR5:GUS and DR5:luciferase reporters and can be correlated with LR initiation. In monocots, LR initiation differs, as LRs emerge from phloem poles rather than from XPP. In addition, there can be numerous phloem poles (usually 10 or more); LRs initiate in longitudinal files, with the number of files roughly proportional to the stele diameter [19]. Similar to Arabidopsis, the formation of auxin response maxima is important in LR initiation in cereals such as maize. DR5::RFP lines have indicated the presence of auxin response maxima in differentiating xylem cells and in cells surrounding the protophloem vessels [19]. Additionally, genome-wide transcriptome analysis of LR initiation has highlighted the presence of a common transcriptional regulatory pathway between Arabidopsis and maize, suggesting that genes involved in LR initiation are conserved across angiosperms [20].
2.3. Adventitious Root Development
Postembryonic roots arising from tissues other than roots are termed adventitious. These may be crown roots (from nodes below the ground), brace root (from nodes above the ground), stem roots, or seminal roots [21]. Different root types contribute to structural and absorbance functions. Primary and seminal roots are mostly important in the first stages of plant growth. Brace roots, which take over at later stages of the monocot RSA, play an important role in the structural support of the plants [22].
2.4. Root Hair Development
RH are tip growing extensions from the root epidermis that not only increase the root surface area for better nutrient and water acquisition but also facilitate adhesion of the root to the surrounding rhizosphere and interaction of the root with soil microorganisms such as arbuscular mycorrhizal fungi and nitrogen-fixing bacteria. Several reports cite that overall plant fitness is compromised in several loss-of-function RH mutants when grown in challenging soil conditions [23,24]. Hence, dynamic RH morphogenesis is a trait that can be exploited agriculturally to improve water and nutrient acquisition in heterogeneous soil conditions. RH development starts when a position-dependent signal originates from the underlying cortical cells. Signal perception in epidermal cells that overlie two cortical cells triggers a transcription factor cascade that ultimately inhibits the expression of GLABRA2 (GL2), a core non-RH-determining transcription factor. This causes the expression of ROOT HAIR DEFECTIVE 6 (RHD6) and its homolog RHD6-LIKE1 (RSL1). These proteins induce the expression of downstream basic helix-loop-helix (bHLH) family targets RSL2, RSL4 and homolog of Lotus japonica ROOTHAIRLESS1 (LjRHL1), LRL3 for root hair growth [25]; among them, RSL4 is the key regulator (extensively reviewed in [26,27,28]). Several hormonal cues participate in the development and regulation of RH, and their crosstalk is important to dynamically regulate RH function in changing soil conditions [27].
3. Impact of Hormones in the Regulation of Different Aspects of RSA
3.1. Primary Root
The long standing link between auxin and root development dates back to the time of its discovery, when it was named as root forming hormone [29], and is indispensable in regulating almost every major phase of root growth and development [11,17,30,31]. Low concentrations of auxin have been shown to stimulate PR growth in Arabidopsis and maize, while higher concentrations inhibited root growth through auxin receptor TRANSPORT INHIBITOR RESPONSE 1 (TIR1) mediated signalling via an unknown non-transcriptional mechanism [32,33]. It can be speculated that free Aux/IAA proteins might promote root growth, and ubiquitinated Aux/IAA proteins may lead to inhibition of root growth. Among other signals, auxin is a key instructor of root meristem organogenesis on which multiple phytohormones and other signalling pathways converge to regulate root development. In QC, polar auxin transport (PAT) is an important prerequisite to establish auxin response maxima; however, localized auxin production in the roots also contributes to the establishment of auxin gradient and maxima required for normal root development [34,35]. It was assumed that shoot-derived auxin and its downward transport contributes to a major portion of auxin in the root to create auxin maxima. However, recent reports suggest that auxin is produced locally in the roots via a tryptophan-dependent biosynthesis pathway, which converts tryptophan to indole-3-pyruvic acid (IPyA) to indole-3-acetic acid (auxin) [36,37]. WEAK ETHYLENE INSENSITIVE 8 (WEI8) and its homolog protein TRYPTOPHAN AMINOTRANSFERASE RELATED 2 (TAR2) are the key enzymes that produce and maintain auxin homeostasis in the roots, thereby establishing the root meristem. Plants mutated in wei8tar2 displayed loss of root meristem and reduced root auxin activity. Additionally, through a combination of grafting experiments, Brumos et al. 2018 showed that shoot-derived auxin could not rescue deficiencies of auxin production in roots [36]. Another tryptophan-dependent auxin biosynthesis pathway, which contributes to a lower but significant amount of auxin in roots, was also reported where the root-derived auxin biosynthesis was abolished in cyp79B2cyp79B3, thus suggesting multiple sources of auxin production in roots [34]. Localized auxin production and response machinery along with PAT work together to develop flux separation to maintain auxin maxima in QC [38]. There is also a feedback mechanism of WOX5 and INDOLE-3-ACETIC ACID INDUCIBLE 17 (IAA17) in maintaining auxin maximal response in QC cells that regulate the root patterning in distal stem cell niche. WOX5 activity induces auxin production in QC; however, auxin negatively regulates the WOX5 transcription [39]. IAA17 causes the inhibition of auxin response and therefore promotes WOX5 transcription. IAA17-dependent auxin response suppression restricts the WOX5 expression in QC and therefore establishes the identity of root stem cells. Gain-of-function mutant of IAA17(axr3-1) displayed enhanced auxin maxima in QC and reduced auxin response in distal stem cells, leading to the inhibition of root distal stem cell differentiation [38].
In a recent report, a rice gene named OsFPFL4 modulates root growth by affecting auxin levels. OsFPFL4 overexpressing plants showed shorter primary roots, which were due to more auxin accumulation and altered expression of auxin biosynthesis and transport-related genes [40]. Another gene encodes for NITRILASE 1 enzyme in Arabidopsis, which regulates root growth by modulation of auxin biosynthesis. NIT1 overexpressing plants showed shorter primary roots, which were due to drastic changes of both free IAA and IAN levels [41]. In rice, OsMADS25 regulates root growth through an increased accumulation of auxin in the root. OsMADS25 promoted auxin biosynthesis as well as transport. At the same time, it also reduced auxin degradation to stimulate root growth [42]. IAA also induces cyclic guanosine 3′,5′-monophosphate (cGMP) accumulation in Arabidopsis roots. Exogenous application of cGMP derivative 8-bromo-cGMP increases auxin dependent primary root growth. 8-bromo-cGMP mediates this response through the degradation of AUX/IAA proteins [43]. Phosphatidylinositol-specific phospholipase C2 also governs root growth in Arabidopsis as plc2 mutant showed defects in auxin-mediated multiple root growth phenotypes, including primary root growth [44]. An Arabidopsis exocyst complex subunit SEC6 gene regulates primary root growth through polar auxin transport and PIN protein recycling [45]. WRINKLED1 (WRI1), a key transcriptional regulator of fatty acid biosynthesis, regulates primary root growth through alteration in auxin homeostasis. The wri1-1 loss-of-function mutants showed an increased amount of indole-3-acetic acid (IAA)-Asp conjugates and increased abundance of GH3.3, a gene involved in auxin degradation [46]. In rice, auxin influx carrier OsAUX3 regulates root development as Osaux3-1 and Osaux3-2 mutants showed shorter primary roots (PRs) [47]. In addition, a WUSHEL-related homeobox protein, OsWOX4, regulates primary root elongation in rice. OsWOX4 regulates this response by direct promoter binding and transcriptional activation of OsAUX1 [48]. A multidrug and toxic compound extrusion (MATE) transporter also governs root development through the modulation of auxin homeostasis in the roots [49]. In soybean, GmYUC2a is an important regulator of auxin biosynthesis during root development and nodulation [50]. Beyond transcriptional control, auxin may regulate root growth responses through other, alternate mechanisms such as protein phosphorylation. Through the impact of different auxin concentrations on the root tip, Nikonorova et al. 2021 identified global auxin-mediated changes in protein abundance and phosphorylation regulating root growth promotion and inhibition [51]. This MS-based phosphoproteome approach identified novel growth regulators, such as members of the receptor-like kinases and MAP kinases. This study also suggests that auxin, H+-ATPases, cell wall modifications, and cell wall-sensing receptor-like kinases are tightly embedded in pathways underlying cell elongation. In addition, MKK2 was found to be a potential novel regulator of root growth regulating auxin biosynthesis and signalling [51].
Auxin regulates PR development via interaction with various other phytohormone signalling components. For example, a plant-specific and ethylene-responsive HOMEOBOX PROTEIN52 (HB52) mediates crosstalk between auxin and ethylene signalling in controlling PR elongation. HB52 works downstream to ETHYLENE-INSENSITIVE3 (EIN3) and regulates the transcription of auxin transport-related genes, including PIN2, WAVY ROOT GROWTH1 (WAG1), and WAG2 through binding to their promoter regions (Figure 2A) [52]. In another report, ETHYLENE RESPONSE FACTOR1 (ERF1) mediates the interaction of auxin and ethylene signalling through the transcriptional regulation of ANTHRANILATE SYNTHASE α1 (ASA1) during PR elongation [53].
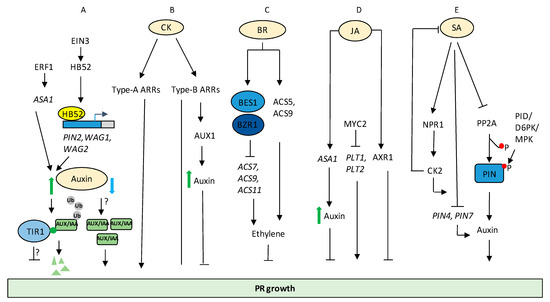
Figure 2.
Regulation of primary root (PR) growth by various hormones and their crosstalk. (A) Auxin effects root growth in a concentration-dependent manner and interacts with ET signalling pathway to control PR elongation. Low concentration of auxin promotes PR growth probably via free AUX/IAAs, while high concentration inhibited PR growth through TIR1-mediated signalling via an unknown non-transcriptional mechanism (B) CK signalling modulates PR growth via AUX-1 mediated auxin translocation. (C) BR induces ET production via stabilizing ACS5, and ACS9 represses ET synthesis through BES1 and BZR1-mediated repression of ACS7, ACS9, and ACS11 to control PR growth. (D) MYC2, a DNA binding bHLH transcription factor in JA signalling, represses the transcriptional expression of PLT1 and PLT2. JA regulates expression of ASA1 and modulates auxin levels to cause root growth inhibition. JA and auxinsignalling occurs via AXR1 to control PR growth. (E) SA via NPR1-mediated signalling establishes an auto regulatory feedback regulation between CK2 and SA to link between SA signalling and auxin transport. SA also led to inhibition of PR elongation in NPR1-independent manner via affecting PP2A, leading to changes in PIN activity and auxin export, resulting in attenuation of root growth. Solid black arrow indicates confirmed pathway. Blue arrow indicates low levels of auxin, green arrow indicates high levels of auxin. Red circle indicates phosphorylation.
Numerous studies have shown how the complex interplay between CK and auxin governs stem cell architecture and meristem size during the early stages of root development. The CK transmembrane receptor ARABIDOPSIS HISTIDINE KINASE 3 (AHK3) together with type-B ARABIDOPSIS RESPONSE REGULATORS (ARR1) and ARR12 modulate auxin redistribution in the root meristem [54]. This change in auxin distribution is brought by modulating the levels of PINs through activation of the auxin repressor SHORT HYPOCOTYL 2 (SHY2)/INDOLE-3-ACETIC ACID INDUCIBLE 3 (IAA3), leading to cell differentiation [4,54]. CK was also shown to regulate the PIN gene expression transcriptionally through CYTOKININ RESPONSE FACTORS (CRFs) [4]. Auxin, on the other hand, mediates the degradation of SHY2, which leads to the upregulation of PIN genes, leading to cell division and expansion [54,55]. Thus, CKs and auxin are involved in the homeostatic feedback regulatory loop that intersects at the common regulatory factor SHY2 to control root meristem size [4,56]. Negative regulators of CK signalling, namely the type-A ARRs, also influence the patterning of root apical meristem [57]. Higher order mutants of type-A ARRs have been shown to have smaller root meristem sizes as compared to the wild type and exhibited enhanced sensitivity towards PAT inhibitor NPA. Following exogenous application of CK, the level of PIN4-GFP was significantly reduced in the root cap. The protein levels, but not the transcript levels, of auxin efflux carriers PIN1 and PIN3 were downregulated in the type-A ARR octuple mutant (arr3,4,5,6,7,8,9,15) roots [57].
External application of CK or increased CK signalling by mutants (higher order type-A ARR) shows negative regulation of PR length both in monocots and dicots [4,58,59,60,61,62,63]. The negative role of CK in root growth is also supported by CK deficient mutants, where the PR grows faster [58]. However, a study on over-expression of cytoplasmic CYTOKININ OXIDASE 7 (CKX7) showed conflicting results of reduced root developmental phenotype from that of other CKXs [64]. Type-B ARR higher order mutants of Arabidopsis displayed increased root length, suggesting a negative role of CK in PR length [65,66,67]. In contrast to this general phenomenon, a recent study showed a novel role of type-B ARRs in positively regulating PR length in rice [68]. Lowering of CK signalling through single and double mutants of CK receptors can increase root meristem size and activity [58,69]; however, CK receptor triple mutant is CK-insensitive and shows decreased root meristem size and activity [69,70,71]. This suggests that a minimum of CK signalling is required for PR development, and inhibitory levels of CK result in lowering of PR growth. CK also controls mitotic divisions in the QC directly by repressing the expression of auxin influx carriers AUXIN RESISTANT 1 (AUX1) and LIKE AUXIN RESISTANT (LAX2) in an ARR1- and ARR12- dependent manner. CK induces mitotic divisions in the QC as evident by the loss-of-function higher order type A ARRs [57]. Consequently, higher endogenous levels of CK in the CK oxidase mutants, ckx3 and ckx5, displayed higher divisions in the QC in comparison to the WT [72], further indicating the positive role of CK in QC divisions. However, the divisions were enhanced in the QC of lax2 roots similarly to CK treated plants, suggesting that CK represses LAX2 expression in the QC. The report also shows that LAX2 is a direct transcriptional target of ARR1. Additionally, the reduction of LAX2 transcript in response to cytokinin was compromised in ahk2ahk4 and arr1-3arr12-1, suggesting that cytokinin receptors and type B ARR are necessary for the regulation of LAX2 [72]. Studies also demonstrate how auxin-CK genetic circuits are modulated by common regulatory genes. AUX1-mediated auxin translocation also regulates CK-dependent root growth inhibition and elongation at the root tip. AUX1 is activated downstream to type B ARR and leads to shootward auxin transport, thereby causing an increase in auxin activity and inhibition of cell elongation at the root tip (Figure 2B) [73]. Another report shows that a rice gene encoding NAC transcription factor 2 (OsNAC2) expressed in the PR tips, crown roots, and LR primordia, affects auxin and CK signalling genes. OsNAC2 physically interacts with the promoters of auxin inactivating GRETCHEN HAGEN (GH3.6 and GH3.8), AUXIN RESPONSE FACTOR 25 (OsARF25), and CYTOKININ OXIDASE 4 (OsCKX4), thus facilitating the integration of auxin and CK signalling to modulate root development in rice [74].
Brassinosteroid (BR) is another key player regulating root growth and development [75,76]. BR regulates root growth in a dose-dependent manner; both high and low doses negatively affect PR growth. Both BR signalling mutants as well as bri1-ems-suppressor 1 (bes1-D) (gain-of-function) mutants have short roots. Additionally, external BR treatment negatively affects PR length [77,78,79]. Contradictory to these reports, in another study, low concentrations of BRs have been shown to promote root growth in WT plants as well as BR-deficient mutants [80], although the changes are small [80]. BR has been shown to affect both cell proliferation and cell elongation in a concentration-dependent manner to modulate root meristem size [81]. All these reports indicate that an optimal level of BR is crucial for root growth and meristem homeostasis. There is a complex cross-talk of BR signalling, BR catabolism, and auxin synthesis culminating in a pattern formation of BRASSINAZOLE-RESISTANT 1 (BZR1) in the meristem and elongation zone. It has been found that low levels of BZR1 are required to maintain the QC, whereas high levels are required in the elongation zone. It has also been shown that a balance between auxin signalling and BR signalling is required to maintain normal root growth [77]. Additionally, BR biosynthetic mutant de-etiolated-2-9 (det2-9), having a short root phenotype, showed increased biosynthesis of ethylene and thereby high accumulation of ethylene. This short root phenotype of det2-9 mutant was partially recovered in mutants det2-9acs9 and det2-9ein3eil1, which are defective in ethylene synthesis and signalling, respectively. Exogenous BR has been shown to regulate ethylene biosynthesis in a dose-dependent manner. BR induces ethylene production via stabilizing ACC SYNTHASE 5 (ACS5) and ACC SYNTHASE 9 (ACS9) (Figure 2C). However, BR signalling represses ethylene synthesis through BES1 and BZR1 mediated repression of ACC SYNTHASE 7 (ACS7), ACS9, and ACC SYNTHASE 11 (ACS11) (Figure 2C). In addition, increased superoxide anions also contribute to the short root phenotype of det2-9 mutants. BR regulates superoxide accumulation via the peroxidase pathway [82]. All these studies indicate that BR regulates root growth through multiple mechanisms. Similarly, BR signalling behaves differently in the epidermal and stele region. BR signalling promotes stem cell proliferation in the epidermal region; however, it induces differentiation of stem cells in the stele region. BR signalling induces target genes in the epidermis particularly related to auxin signalling, but mostly represses genes in the stele [83].
Although involved in mitigating biotic and abiotic stress responses [84,85,86], jasmonic acid (JA) and salicylic acid (SA) are also involved in regulating various parameters of root growth and development. The inhibitory effect of JA on plant growth was one of the first physiological responses reported in the 1980s [87,88]. The first mutant insensitive to JA-mediated inhibition of root growth was jasmonate resistant 1 (jar1), which was later cloned and characterised as JA-Ile synthase [89]. Apart from this, various components of JA signalling also participate in this physiological response. A screen for Arabidopsis mutants unresponsive to root growth inhibition by a bacterial toxin and the JA homologue, coronatine (COR), revealed coronatine insensitive 1 (coi1-1), a null mutant insensitive to JAs [90]. COI1 is at the interface of JA and ethylene (ET) signalling in mediating root growth inhibition. Previous reports show that coi1-16 shows unresponsiveness to ET induced root growth inhibition in light, but not in dark. Furthermore, this response did not require any other components of JA biosynthesis and signalling such as jar1-1, jasmonate insensitive 1 (jin1), allene oxide synthase (aos), or oxophytodienoate-reductase3 (opr3). Thus, the inhibition of Arabidopsis root growth to 1-aminocyclopropane-1-carboxylate (ACC) is light, COI1 dependent, but JA independent and occurs due to inhibition in cell elongation [91,92]. MYC2, a DNA binding bHLH transcription factor in JA signalling, acts as an activator of JA-induced root growth inhibition [93]. Previous reports suggest that MYC2 directly represses the transcriptional expression of PLT1 and PLT2 (Figure 2D) [94]. Moreover, JA may increase auxin levels by inducing the expression of auxin biosynthetic gene ASA1, thereby leading to root growth inhibition (Figure 2D) [95]. Another example of potential interaction between JA and auxin signalling occurs via AUXIN RESISTANT 1 (AXR1). In addition to its role in auxin signalling, axr1-1 shows JA-mediated root growth inhibition and pathogen susceptibility (Figure 2D) [96]. Taken together, the above reports propose that JA signalling is linked to auxin homeostasis, leading to root growth inhibition. An interesting report claims that both endogenous and exogenous JA have an important role in governing the root architecture of Helianthus annuus seedlings. Micromolar concentrations of JA reduced the PR and LR lengths along with LR number by reducing the cortex cell length and cell divisions. However, when JA biosynthesis inhibitor Ibuprofen (IBU) was applied, an increase in cell elongation was observed, thus suggesting that endogenous JA also affect root phenotype in Helianthus annuus [97]. Interestingly, auxins were shown to be not essential for JA induced root growth inhibition in sunflower [97]. Studies have shown that the auxin-JA resistant double mutant (jar1-1axr1-3) was more resistant than the single mutants to inhibition of root growth by methyl jasmonate (MeJA). In addition, the level of resistance between single mutant axr1-3 and double mutant jar1-1axr1-3 was not different when IAA was applied. Additionally, JA produced its phenotype even in the presence of an auxin transport inhibitor. Besides, auxin produced its phenotype even when ibuprofen was applied. Thus, JA appears to act through two pathways: one linked to auxin, and a second via an auxin independent pathway [97]. A recent report highlights the synergistic role of JA and auxin in stem cell activation and root regeneration. Wound induced JA causes ETHYLENE RESPONSE FACTOR 109 (ERF109) activation, which further stimulates CYCLIN D6; 1(CYCD6;1) and ETHYLENE RESPONSE FACTOR 115 (ERF115). They then modulate RETINOBLASTOMA-RELATED (RBR)-SCARECROW (SCR) module to allow root tissue regeneration. Upon wounding, auxin accumulation also takes place, which then activates several regeneration regulators of this pathway [98].
SA, in a concentration-dependent manner, regulates root growth and development through interaction with the auxin machinery. A report by Zhao and coworkers (2015) suggests that exogenous application of 50 µM of SA promoted root waving along with a reduction in PR elongation in Arabidopsis. This SA-mediated root waving response was regulated via NITRIC OXIDE-ASSOCIATED PROTEIN1 (AtNOA1) and NONEXPRESSER OF PATHOGENESIS-RELATED GENES 1 (NPR1). Atnoa1 mutants displayed a disrupted root waving phenotype as a result of altered auxin distribution at the root tip, suggesting interplay of SA signalling and auxin transporters through AtNOA1 to modulate root waving in Arabidopsis [99]. Another report shows that exogenous application of 100 µM SA inhibited root elongation in Arabidopsis [100]. Armengot et al. 2014 revealed that the mutants of protein kinase CK2, a Ser/Thr kinase accumulated high levels of SA, which further resulted in the impediment of PR growth (Figure 2E). CONSTITUTIVE PATHOGEN RESPONSE (CPR) mutants having high constitutive levels of SA exhibited a similar phenotype as the CK2 loss-of-function mutants. In addition, SA enhanced the transcription of CK2 encoding genes via the NPR1-mediated signalling, thereby implying an auto regulatory feedback regulation between CK2 and SA [101]. Loss of CK2 activity led to reduction in the SA-mediated repression of auxin transporter genes including PIN4 and PIN7, thus establishing a link between SA signalling and auxin transport [101]. Exogenous application of 3 µM SA inhibits PR elongation and causes shortening of cell length. SA led to auxin redistribution and a pronounced auxin response at the root tip in a PATHOGENESIS–RELATED 1 (PR1)-independent manner. At concentrations higher than 50 µM SA, the growth related processes in the root were ceased. Lower concentrations of SA promoted the expression of TRP AMINOTRANSFERASE OF ARABIDOPSIS 1(TAA1) and PIN1 and suppressed PIN2 and PIN7 expression. A high concentration of SA upregulated the expression of TAA1 and inhibited the expression of PIN efflux transporters [100]. Utilizing mathematical modelling of auxin distribution after SA exposure, Pasternak et al. 2019 hypothesized that root tip accumulated auxin after treatment with low concentration of SA, whereas high concentrations of SA led to depletion in auxin levels. Accumulation of auxin after the application of a lower level of SA causes the distal meristem to become enlarged. Endodermal periclinal divisions were higher in Arabidopsis root treated with a lower concentration of SA as a result of enhanced auxin levels in the endodermis. However, lesser vascular cell files were demonstrated in SA-treated roots in comparison to control due to reduced accumulation of auxin in the vascular cells [100]. However, administration of 1.5 µM SA under stress-free conditions increased the total root biomass in Zea mays [102]. SA-deficient mutant abnormal inflorescence meristem1 (aim1) in Oryza sativa exhibits a defect in root growth and showed reduced meristem activity. Reduced SA levels in aim1 led to further decrement in the reactive oxygen species (ROS) levels due to enhanced expression of ROS scavenging-related genes. The authors hypothesize that AIM1 is involved in SA biosynthesis and represses the ROS scavenging genes to accumulate ROS for higher root meristem activity via induction of transcriptional repressors WRKY62 and WRKY76 [103]. Recent observations by Tan et al. 2020 concluded that SA led to inhibition of PR elongation in an NPR1-independent manner (Figure 2E). The reduced PR growth was a result of inhibition of serine/threonine PROTEIN PHOSPHATASE 2A (PP2A) activity by SA, leading to increased phosphorylation of PIN protein by different kinases such as PINOID (PID)/D6PK/MAPKs, thereby affecting PIN activity andauxin export, resulting in attenuation of root growth (Figure 2E) [104].
3.2. Lateral Roots
Auxin is commonly known to induce LR formation as suggested in different auxin biosynthesis and signalling mutants in Arabidopsis and tomato [105,106]; however, there are reports suggesting that exogenous auxin application (25nM) reduced the induction of LRs [107]. In the roots, a regular pattern of root branching depends on the oscillatory gene activity that determines prebranch sites. A recent report suggests that local auxin source in the root cap is derived from the auxin precursor indole-3-butyric acid (IBA), which regulates the oscillation amplitude and therefore determines the origin of the prebranch site. An IBA-regulated gene, MEMBRANE-ASSOCIATED KINASE REGULATOR4 (MAKR4), converts the prebranch site into a regular spacing of lateral organs [108]. LBD16/ASYMMETRIC LEAVES2-LIKE18 (ASL18), which is a direct downstream target of SOLITARY ROOT (SLR)/IAA14-ARF7-ARF19 auxin signalling module, regulates LR development possibly through transcriptional regulation through its related LBD/ASLs, causing nuclear migration and asymmetric cell division for LR initiation and primordium development (Figure 3) [109]. A very recent report shows the role of ABCB transporter in LR emergence [110]. The report suggests that loss of ABCB21 reduces rootward auxin transport and delays LR emergence [110]. These results support a primary role for ABCB21 in regulating auxin distribution supplementary to the primary ABCB auxin transporters ABCB1 and 19 [110]. It is a well-established fact that ARF7 and the LATERAL ORGAN BOUNDARIES DOMAIN (LBD) module are required for Arabidopsis LR development. A recent report suggests that PR1 homolog PRH1 regulates LR development in Arabidopsis via the auxin signalling pathway. It was shown that the expression of PRH1 was induced upon auxin, and mutants of PRH1 had fewer LRs. Additionally, PRH1 was shown to be a direct transcriptional target of ARF7 and LBDs [111]. PRH1 might then regulate the expression of EXPANSIN (EXP) genes, which then affect cell wall loosening (Figure 3).
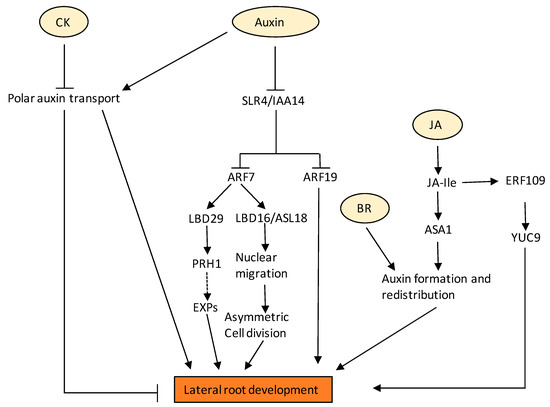
Figure 3.
Regulation of LR development by various hormones and their crosstalk. Auxin via SLR4/ARF7-ARF9 signalling module promotes LR development. Auxin-induced expression of PRH1 is dependent on ARF7 and LBD29. PRH1 may promote LR development by regulating the expression of EXP genes that promote cell wall loosening. SLR4/ARF7-ARF9 signalling module also activates the expression of LBD16/ASL18 that are involved in the symmetry breaking of LR founder cells for LR initiation and primordium development. JA positively regulates lateral root formation by modulating auxin biosynthesis and homeostasis. JA induces the expression of ASA1 both through COI1 and ERF109. ERF109 also induces the expression of another auxin biosynthetic gene, YUC9. BR regulates lateral root development by interacting with auxin. At low concentration, BR modulates auxin transport and promotes lateral root development. CK negatively regulates LR initiation and development by inhibiting auxin carriers. Solid black arrows indicate confirmed pathway. Dotted arrow indicates pathways for which there is little or no evidence.
Although the optimization of LR development is primarily governed by auxin, various studies highlight the role of CKs in LR patterning and growth [60,63,112]. Previous reports demonstrate that change in the CK levels, either through increased endogenous signalling or through external application, inhibits LR production [113,114]. In line with this, a decrease in CK signalling increased LRs [58,65,71,115]. CK insensitive mutants have increased LR formation as a result of increased auxin perception [114], whereas LR formation in ahk triple mutant was severely impaired as a result of a decrease in cell division [70]. The inhibition of LR by CK is brought indirectly by changing auxin transport by disrupting PIN1 localization (Figure 3) [116]. This results in a change of cell division pattern of LRFC in the outer layer from undergoing periclinal division [113]. Upon application of exogenous CK, a rapid modulation in PIN1 dynamics on the plasma membrane showed the role of CK in controlling auxin flow. CK also led to the post-transcriptional degradation of PIN1 proteins through targeted lysis in vacuoles [116]. The endocytic trafficking of PIN proteins is regulated by AHK4 and type-B ARR (ARR2 and ARR12) [117]. Plants perturbed in their perception and/or signalling of CKs have large meristem sizes [71] and a higher number of LRs [65], and exhibit increased root branching phenotype [58], implying the important role of CKs in maintaining homeostasis in regulating RSA [66]. A recent report claims that the impact of CK on LR elongation is tissue- and concentration-specific as low CK levels reduced the length of the laterals in the apical/proximal region of the root system. By contrast, high levels of CK reduced the length along the entire root system, but with a more pronounced effect in the apical zones [63]. Another interesting report introduces TRANSPORTER OF IBA1 (TOB1), as a new player in LR development. Michniewicz and co-workers show that the TOB1 gene is responsible for transport of IBA to the cytoplasm and converts it into active IAA. They also show that it is a direct target of the CK response pathway, and mutants altered in TOB1 show an increased number of emerged laterals. Thus, TOB1 acts as a link between CK signalling and auxin to fine-tune RSA [118].
Apart from their role in inhibiting LR growth, CKs regulate the spacing of LRP along the PR in Arabidopsis, as disruption of CK signalling genes led to irregularities in spacing of consecutive LRP sites [119]. High local endogenous CK levels in pericycle cells neighbouring the pre-existing LRs inhibit LRP initiation at proximity to existing ones. This spacing is regulated via blockage of pericycle founder cells from G2 to M phase transition by CK [60,119,120].
BRs are also one of the major factors regulating LR development. BRs modulate LR development primarily via interacting with auxin. BR signalling mutant brassinosteroid insensitive 1 (bri1) has been reported to have a decreased number of LRs, suggesting the possible involvement of BRs in LR development. Furthermore, a low concentration of BR has a positive effect on LR numbers. BRs promote the initiation of LR primordia by increasing acropetal auxin transport (Figure 3) [121]. At low levels of auxin, BRs and auxin exhibited a synergistic effect on LR formation, which was inhibited by NPA, an auxin transport inhibitor [121]. However, higher concentrations of BRs have a negative effect on LR formation [122]. Higher concentrations of BRs have been shown to induce the expression of several AUX/IAAs, which play a vital role in LR formation [123]. These reports suggest that high concentrations of BRs inhibit LR formation, possibly via inhibiting auxin signalling through these AUX/IAAs [81]. CK has been shown to antagonise the effect of BRs on LR development. CK receptor double mutants ahk2ahk3 showed increased sensitivity to BRs in LR elongation, indicating the antagonistic actions of these two hormones in LR development [113].
Various reports strongly suggest the role of JA in LR development. It was found that the promoters of all four members of ALLENE OXIDECYCLASE (AOC), a JA biosynthesis gene, were highly expressed in LR primordia, suggesting the involvement of JA in LR formation and development [124]. Several reports show the positive effect of JA on LR initiation and growth by affecting auxin biosynthesis and homeostasis. It was found that exogenous application of MeJA represses LRP initiation in asa1-1. Transcriptional studies further validated that MeJA induces the expression of ASA1 in a COI1-dependent manner that ultimately leads to increased local auxin accumulation in the root basal meristem and, as a result, promotes LR formation (Figure 3) [95]. Another mechanism by which JA promotes LR formation is by inducing the expression of ETHYLENE RESPONSE FACTOR 109 (ERF 109), which then binds to the GCC boxes in the promoters of ASA1 and YUCCA 9 (YUC9) (Figure 3) [125]. COI1 is also involved in JA-induced pericycle cell activation and LR formation, positioning, and emergence on bends and requires a canonical auxin signalling pathway [126]. A very recent report highlights the negative role of JA in regulating auxin induced LR formation, but independently of COIreceptor [127]. Higher concentrations of (−) JA and (+) JA were shown to counter the promontory effect of auxin by stabilizing DII-VENUS and suppressing the expression of PUCHI and LATERAL ORGAN BOUNDARIES-DOMAIN 29 (LBD29), involved in LR formation [127].
SA has been shown to enhance root biomass and a number of emerged LRs in Catharanthus roseus [128]. The distance between the root tip and the first emerged LR primordia was also decreased, which could be due to SA-induced reduction in the level of CK in the root cap or lower CK sensitivity in apical cells of the root. Femtomolar (fM) concentrations of SA displayed a larger root cap with a higher number of columella cells [128].SA also enhanced the LR formation in a dose-dependent manner [100]. Armengot and co-workers revealed that the mutants of CK2, a Ser/Thr kinase, accumulated high levels of SA, which further resulted in the impediment of LR growth [101]. Pasternak et al. 2019 reported inhibition of LR number with increasing concentrations of SA [100]. Similarly, the application of 20 µM SA or 40 µM SA to Arabidopsis seedlings inhibited LR organogenesis by changing the auxin homeostasis through the cellular redistribution of PIN proteins [104].
3.3. Adventitious Roots
Lateral and adventitious roots mostly share common regulatory networks except for a few regulatory mechanisms that distinguish lateral and AR pathways. AUXIN RESPONSE FACTOR 7 (ARF7) and AUXIN RESPONSE FACTOR 19(ARF19) are key molecular players in the regulation of LR initiation and transcriptionally activate LATERAL ORGAN BOUNDARIES-DOMAIN (LBD) genes. In the case of ARs, molecular functions of ARFs are overtaken by WUSCHEL RELATED HOMEOBOX 11 (WOX11) transcription factor. WOX11 responds to wound-induced auxin in the procambium and its surrounding cells and works redundantly with its homologous protein WUSCHEL RELATED HOMEOBOX 12 (WOX12) to activate downstream LBD16 and 29, resulting in the fate transition from procambium to root founder cell [129]. However, a few reports suggest that ARF7/19 also govern the formation of adventitious roots from hypocotyls, which precludes the discrimination of molecular pathways of LR and adventitious roots [130]. Auxin transporters are also important for AR development. For example, an ATP-binding cassette efflux family, ABCB19, has been linked to AR formation [131]. Other members of the same transporter are found to transport IBA [132,133], which has been tested to have the highest ability to induce adventitious rooting [134]. Other hormones such as ethylene have been shown to regulate the positioning of AR primordia [135]. In the fern, Ceratopteris richardii, an intermediate clade WOX (IC-WOX) gene, CrWOXA, which is exclusively expressed in root founder cells during LR and AR initiation, regulates AR organogenesis. CrWOXA is a direct target of the auxin signalling pathway, which in turn may directly activate the WUSCHEL-clade WOX (WC-WOX) gene, CrWUL [136]. A similar case is observed in Arabidopsis, where WOX5/7 expression is directly regulated by WOX11/12 required for root founder cell division during AR initiation [137]. AR initiation is mediated by ARF6, 8, and 17 modules, which control the expression of GH3.3, GH3.5, and GH3.6 (Figure 4). ARF 6 and 8 are positive regulators, whereas ARF17 is a negative regulator, thus altering auxin homeostasis. Thus, a complex regulatory network exists that fine-tunes AR initiation (Figure 4) [138,139].
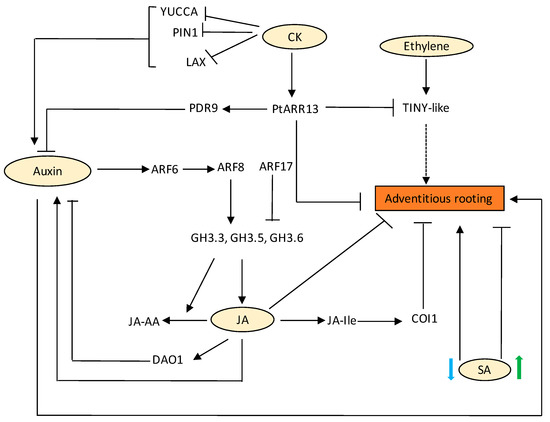
Figure 4.
Regulation of AR development by various hormones and their crosstalk. Auxin controls AR initiation by activating ARF6 and ARF8, leading to downregulation of COI1-mediated JA signalling. GH3.3, GH3.5, and GH3.6 are regulated by ARF6, ARF8, and ARF17. The 3 GH3s control JA homeostasis. JA level controls JA-Ile levels. JA-Ile negatively regulates AR development by activating COI1-dependent signalling. Feedback regulation by DAO1 is activated by JA signalling, which then regulates IAA homeostasis. SA at low concentration promotes AR development, whereas at higher concentrations it inhibits AR development. CK regulates auxin homeostasis by negatively affecting auxin carriers PIN, LAX, and YUCCA1 genes, thereby regulating adventitious rooting through these pathways. In poplar cuttings, CK signalling activates PtARR13, which represses AR formation by promoting the expression of PDR9 and inhibiting the expression of TINY-Like TF. Solid black arrows indicate confirmed pathway. Dotted arrow indicates pathways for which there is little or no evidence. Blue arrow indicates low level of SA and green arrow indicates high level of SA.
Research conducted on crop plants indicates the role of CK in concert with auxin in the regulation of AR induction and development. In apple rootstocks, N6-benzyladenine (BA) was shown to inhibit AR formation via restricting root primordium growth [140]. In tomato, zeatin levels increased during AR induction and extension, which was reduced after IAA treatment [141]. CK also negatively affect PIN, YUCCA6 and LAX3 expression, leading to reduced auxin flow and hence, AR repression (Figure 4) [135]. Transcriptomic studies dealing with poplar softwood cuttings revealed that PtARRs were significantly downregulated during a time course of 6 h that was concomitant with an increase in auxin signalling genes, depicting a general antagonism between auxin and CK during AR induction [142]. The PtARR13 (a positive CK regulator) overexpressing shoots exhibited differential expression of two most important genes; PLEIOTROPIC DRUG RESISTANCE TRANSPORTER9 (PDR9) and TINY, which are induced via auxin and ethylene, respectively, in normal WT excised shoots [142], implying that CK inhibits AR development through interactions with other hormonal networks (Figure 4). In Arabidopsis, the CK receptor ahk4 mutants, also known as wooden leg-3 (wol-3), and the higher order mutant ahk2ahk3ahk4 exhibited enhanced AR formation on the hypocotyl [70,143]. Phenotypic analysis of plants overexpressing the CK-degrading enzymes such as the CYTOKININ OXIDASE/DEHYDROGENASE (AtCKX) gene family also demonstrates the role of CKs in AR formation. The 35S:AtCKX2 and 35S:AtCKX4 lines displayed enhanced AR, which was almost 7 times as much in comparison to the WT [58]. However, in Vigna callus, kinetin alone was able to induce rhizogenic activity of the callus in terms of AR formation [144] pointing out the fact that distinct signals in different plant species determine specific outcomes in terms of AR induction and formation. BR has been reported to induce adventitious rooting in cucumber at lower concentrations through a nitric oxide-dependent pathway, whereas a higher concentration was inhibitory [145].
JA differentially regulates AR formation in different species. In Arabidopsis, JA negatively regulates adventitious rooting via COI1 and MYC2/3/4 dependent JA signalling pathway by altering the JA-Ile homeostasis. jar1-1, coi1-16, myc2, myc3, and myc4 form numerous adventitious roots as compared to the WT, and this response was dependent on ARF 6 and ARF8 (Figure 4) [139]. Additionally, GH3.3, GH 3.5, and GH3.6 catalyse the production of inactive forms of JA, such as JA-Asp, JA-Trp, and JA-Met, thus controlling JA homeostasis and AR initiation (Figure 4) [139]. However, JA enhanced the AR formation in leafy cuttings of Petunia hybrid [146]. Very recently, Druege and co-workers established that early wound-induced JA accumulation leads to enhanced IAA accumulation and hence stimulates AR production in Petunia cuttings [147]. The effect of JA in AR development depends on experimental conditions. At low sub-micromolar concentrations, MeJA has been shown to promote AR development in dark grown hypocotyls of Arabidopsis and thin cell layers of N. tabacum when applied together with IBA and kinetin [148]. Recently Lakehal and co-workers proposed that TIR1 and AUXIN SIGNALING F-BOX 2 (AFB2) interact with IAA6, IAA9, and IAA17 to control JA homeostasis and AR initiation in Arabidopsis [138]. They also showed that a feedback circuit exists between IAA and JA that is mediated by DIOXYGENASE FOR AUXIN OXIDATION (DAO1) and COI1-dependent JA signalling. DAO1 catalyses the conversion of IAA to oxindole-3-acetic acid (oxIAA), hence leading to reduced free IAA levels and AR initiation in Arabidopsis. They also showed that the expression of DAO1 is induced by JA signalling (Figure 4) [149].
SA has also been shown to promote the formation of secondary adventitious roots in Arabidopsis seedlings at a lower concentration (30 µM) and showed inhibition of AR growth at concentrations higher than 30 µM SA [100]. At a concentration of 200 µM, SA initiated AR formation in mung bean (Phaseolus radiatus L.) hypocotyl cuttings, with maximum effect at 400 µM SA. Further increases in SA concentration inhibited the response. Treatment of mung bean hypocotyl cuttings with N, N’-dimethyl thiourea (DMTU), an H2O2 scavenger, led to a decline in the number of ARs. Similarly, diphenyleneiodonium (DPI), a specific inhibitor of membrane-linked NADPH oxidase, also restricted AR formation, suggesting H2O2 functions synergistically with SA to induce adventitious rooting [150].
3.4. Root Hair
Phytohormone auxin promotes the growth of RH [151]. ARF5/7/8/19 bind to the promoter of RSL4 to control its expression in trichoblasts [152]. ARF19 was also shown to regulate the expression of RSL2(Figure 5) [153]; however, overexpression of auxin repressors ARFs in RH (ARF1-4, ARF9-11, and ARF16) suppressed RH growth (Figure 5) [154], suggesting differential activity of ARFs. Among other phytohormones, it was found that auxin and ethylene act synergistically in regulating RH growth. A transcriptome study suggests that almost 90% of the auxin-regulated genes underlying RH growth were synergistic to ethylene [25]. Moreover, other hormones such as JA, BR, and strigolactone pathways may regulate root hair growth possibly through interconnections with the auxin/ethylene signalling pathways [155]. BR inhibits root hair growth. The expression of root hair-related AUX/IAA (such as AXR2, AXR3, and SLR) was increased by the application of EpiBL and repressed in the BR-insensitive bri1 mutant, suggesting a possibility that BR may suppress root hair growth through the suppression of auxin signalling. JA positively regulates root hair growth. The interconnection of JA and auxin was shown as axr2 mutants were found to be resistant to exogenous JA in primary root inhibition, and JA was also shown to promote auxin biosynthesis. However, no direct evidence supports idea that JA regulates root hairs through auxin (extensively reviewed by Lee and Cho, 2013). In RH cells, auxin promotes membrane depolarization in a concentration- and pH-dependent manner that is strongly dependent on AUX1-mediated auxin transport. AUX1-mediated auxin transport involved SCF TIR1/AFB-type Ca2+ signalling in the RH as treatment with SCFTIR1/AFB inhibitor auxinole blocked AUX1 transport and IAA-triggered calcium signals [156]. Using 3D modelling, Jones et al. 2009 showed that AUX1-dependent auxin transport via non-hair cells maintains the auxin flow in the development of RH [157]. In a recent study, auxin was found to govern the transcriptional regulation of a cell wall receptor protein ERULUS (ERU) in an ARF7- and 19- dependent manner (Figure 5). ERU is a member of the Catharanthus roseus RECEPTOR-LIKE KINASE 1-LIKE (CrRLK1L) subfamily of cell wall receptors and regulates RH cell wall composition and modulates pectin dynamics [26]. MEDIATOR18 (MED18) protein, through the modulation of auxin signalling and transport, influences the growth of PR, LR, and RH development [158]. The energy status of plants also affects RH development. In a recent study, glucose-activated TOR was found to phosphorylate and stabilize PIN2 at the plasma membrane, which results in the shootward auxin transport to the RH [159,160].
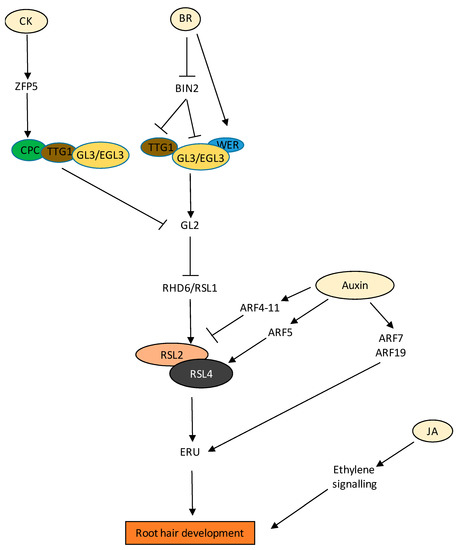
Figure 5.
Regulation of RH development by various hormones and their crosstalk.BR positively regulates the expression of WER, GL2 (negative regulator of RH development) and inhibits BIN2, which is then unable to phosphorylate GL3/EGL3 and TTG1. Presence of BR promotes functional TF complex formation between TTG1, GL3/EGL3, and WER, promoting GL2 expression, leading to inhibition of the RH development pathway. CK induces TF ZFP5, which promotes the expression of CPC (positive regulator of RH development), leading to the formation of functional TF complex of TTG1, GL3/EGL3, and CPC. The complex inhibits the expression of GL2, thus causing activation of the RH development pathway. The presence of auxin liberates the ARFs from AUX/IAAs. The ARFs show different specificity to RH development. ARF4-11 inhibits, whereas ARF6 promotes, the RH development pathway. ARF5 directly interacts with the promotor of RSL4, leading to RH development. Auxin also directly regulates the expression of RSL4 target gene ERU via ARF7/ARF19. JA positively influences RH growth via ET signalling. Solid black arrows indicate confirmed pathways.
There are very few reports citing the development of RH by CK. CK promotes RH formation at the differentiation zone regulating a similar set of RH-specific genes, as observed for auxin and ethylene (Zhang et al. 2016). CK independently regulate RH formation to that of auxin and ethylene (Zhang et al. 2016). CK levels are also modulated in RH of root nodule-forming species, such as in Medicago trancatula via several cytokinin biosynthesis and signalling genes, including CYTOKININ RESPONSE 1(CRE1) and type A ARRs such as MtARRA2, MtARRA8, MtARRA9, and MtARRA10 in response to nodulation factors, indicating that CK signalling negatively impacts root epidermal infections [161,162].
In Arabidopsis, 6-Benzylaminopurine (BAP) treated seedlings showed enhanced RH growth in response to low hormonal concentrations (10 µM) [163]. Apparently, this profound effect was auxin- and ethylene-independent, as similar results were achieved in axr1, ethylene response 1 (etr1), and ethylene inhibitor Aminoethoxyvinylglycine (AVG) treated seedlings, respectively (Zhang et al. 2016), while on the other hand, CKX2 overexpression line (35S:CKX2) displayed abnormal short-hair phenotypes [161]. In addition, several proteins and TFs regulate RH development, and are themselves orchestrated by upstream phytohormonal networks. This is exemplified via molecular characterization of the C2H2 zinc finger protein, ZINC FINGER PROTEIN 5 (ZFP5), whose mutant has fewer RHs than the WT [164]; its expression is transcriptionally increased by BAP application (Figure 5). In effect, ZFP5 itself induces the expression of another RH-localised RB MYB protein, CAPRICE (CPC), involved in RH patterning by binding to its promoter, which then inhibits negative regulators of RH development such as GLABRA2 (GL2) (Figure 5) [165].
In the case of RH, the relative spatial distribution of BRI1 plays a crucial role. Earlier, it was reported that BRs play an essential role in the establishment of epidermal cell fate in Arabidopsis roots. BRs convey the positional information of hair and non-hair cells along the roots via BRI1 [166]. Later, it was found that expression of BRI1 in hair cells has a promontory effect, whereas in the case of non-hair cells, its expression is inhibitory [167]. Additionally, BRASSINOSTEROID-INSENSITIVE 2 (BIN2) also plays a key role in regulating the distribution of hair (H) and non-hair (N) cells in the root epidermis. Root hair formation is determined by transcription factor complex formation and lateral communication between H- and N-cells. At high BR concentrations, BIN2 regulates this distribution primarily via the phosphorylation of two major transcription factors, ENHANCER OF GLABRA3 (EGL3) and TRANSPARENT TESTA GLABRA 1 (TTG1), leading to functional TF complex and subsequent GL2 expression and determination of the non-hair cell fate in all epidermal cells (Figure 5) [168]. It has also been reported that expression of BRI1 under protophloem-specific promoters rescued root meristem phenotypic defects of bri1 brl1 brl3 triple receptor mutants [169]. All of these results suggest that not only optimal concentration of BR but tissue-specific expression of BR signalling components also play a crucial role in root development.
JA and MeJA also positively influence RH growth via ethylene signalling (Figure 5) [170]. It was found that the effects of JAs were abolished in the ethylene-insensitive mutants etr1-1 and etr1-3, or by ethylene biosynthesis inhibitors (AVG) [170].
3.5. Gravitropism
Auxin is the major hormone that drives the growth of an organ (PRs or LRs) towards the gravity vector. The angle at which an organ grows with respect to the gravity vector is called the gravitropic set point angle (GSA) [171]. It is a well-established fact that the distribution of auxin drives the root growth in response to gravity via the starch statolith hypothesis [172]. In response to gravity-stimulation of PRs, polarized localization of PIN3 and PIN7 redirects auxin flux towards the lower side of the PR, thus, causing differential cell elongation and root tip bending [173,174,175]. The dual role of auxin in gravity perception and gravity response was demonstrated by Zhang and co-workers. They reported thatTIR1-dependent auxin signalling module TIR1-AFB-IAA17 mediates starch granule formation and gravitropic perception in root tips by regulating the expression of starch granule synthesis genes PHOSPHOGLUCOMUTASE (PGM), ADENOSINE DIPHOSPHATE GLUCOSE PYROPHOSPHORYLASE (ADG1), and STARCH SYNTHASE 4 (SS4) (Figure 6A) [31]. Downstream signalling of auxin reorients PR growth. Auxin influx and efflux transporters distribute differential auxin gradient in the upper and lower sides of the gravity-stimulated roots. The asymmetric distribution of auxin towards the lower side of the root is modulated by AUX1 and PINs [175,176]. While auxin is the central component of root gravitropic responses, ET is implicated in this process through crosstalk with auxin, which also involves para-aminobenzoic acid (PABA) (Figure 6A) [176]. The interference of ET in root graviresponses is mediated through its interaction with the auxin pathway at the level of auxin biosynthesis and transport. In gravistimulated roots, PABA promotes an asymmetric auxin response, which causes the asymmetric growth responsible for root curvature. This activity requires the auxin response transcription factors AUXIN RESPONSE FACTOR7 (ARF7) and ARF19 as well as ethylene biosynthesis and signalling, indicating that PABA activity requires both auxin and ethylene pathways (Figure 6A) [176].
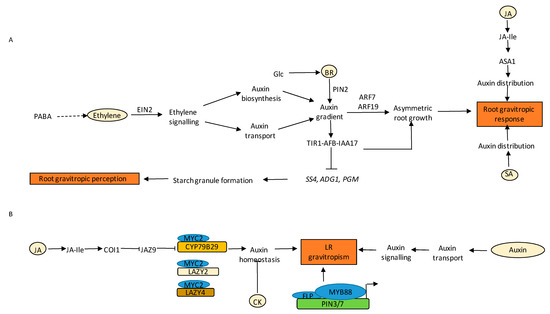
Figure 6.
Regulation of PR and LR gravitropism by various hormones and their crosstalk. (A) PABA works upstream of ET signalling, through which it regulates auxin biosynthesis and transport. This ultimately leads to ARF7/19 mediated auxin activity effects on asymmetric growth promotion followed by root gravitropic response. Glc acts via BR to regulate root gravitropism. BR regulates root gravitropism by interacting with auxin via PIN2 distribution. Auxin signalling module TIR1-AFB-IAA17 mediates starch granule formation and gravitropic perception by inhibiting the expression of PGM, ADG1, and SS4. JA influences gravitropic response by modulating auxin levels through SA1 induction. SA affects auxin transport and redistribution, leading to gravitropic bending. (B) The presence of JA activates COI1-mediated JA signalling that causes proteasomal degradation of JAZ9, leading to the liberation of MYC2, which then binds to the promotors of CYP79B2 and LAZ2 and LAZY4, leading to changes in auxin homeostasis and LR gravitropism. Auxin transport and signalling modules are essential to regulate vertical orientation of LRs. The binding of FLP and MYB88 TFs leads to transcriptional activation of PIN3 and PIN7, which then lead to gravitropic bending of LRs. CK perturbs auxin homeostasis, leading to horizontal branching of LRs. Solid black arrows indicate confirmed pathways.
Biochemical and computational analyses have revealed that besides auxin, CK also influences gravity responses in plants [112,177,178]. The combinatorial effect of auxin-CK on root gravitropic responses is observed at the protein transport and differential expression levels of PINs [178]. Several mutant lines defective in endogenous CK perception and signalling such as ahk3 and higher-order mutants; ahk2 ahk3 ahk4, ahp1 ahp2 ahp3 ahp4 ahp5, arr1 arr10 arr12, and the CK degrading lines 35S:AtCKX2 and 35S:AtCKX3, had higher root gravitropic angles than the WT [178]. The expression of PIN3 was also shown to be significantly lower in these lines as compared to the WT in the root tip plasma membrane of columella cells. However lateral expansion of PIN7 levels was also observed in the columella cells, although the intensity was comparable to the WT [178]. It is known that CK alters auxin sensitivity and transport in root columella cells. AUX1 signals were diminished in 35S:AtCKX2 and 35S:AtCKX3 lines, suggesting endogenous CKs regulate AUX1 in root tips [178]. Root caps cells were also shown for their potential in producing CKs upon gravistimulation in Arabidopsis, indicating the importance of de novo CK synthesis [179,180]. The ARR5 exhibited an asymmetrical pattern and redistribution at the lower side following a 90 degree shift in root orientation [180]. The tips aligned towards BAP-applied sites, causing a discernible bending and triggering an inhibition in root elongation during gravity sensing [180].
There are several reports indicating the involvement of BR signalling in the regulation of root gravitropic responses. Exogenous BR application is known to increase gravitropic curvature of PRs [181,182]. BR signalling mutants showed reduced gravitropic curvature, whereas transgenic plants overexpressing BRI1 were hypersensitive to gravity. BR and auxin show a synergistic effect on gravitropic curvature at low concentrations of auxin primarily via increasing acropetal and basipetal auxin transport [181,182]. BR has also been found to promote the activity of RHO-RELATED PROTEIN FROM PLANTS 2 (ROP2) GTPase, resulting in enhanced polar accumulation of PIN2 protein and thereby increased gravitropic response [182]. However, high concentrations of auxin antagonise BR-induced root gravitropic curvature and vice versa [183]. Exogenous BR could also affect gravitropic responses by regulating the actin cytoskeleton and PIN2 localization pattern, and thereby the auxin gradient in a way similar to auxin [184]. A recent report indicated that BR antagonises PIN2 endocytosis [185]. BR-mediated modulation of endocytic sorting and distribution of PIN2 plays a crucial role in the formation of lateral PIN2 gradient in gravistimulated roots. This lateral gradient regulates auxin signalling in gravistimulated roots and thereby root growth deviation (Figure 6A) [185]. In addition to this, BR plays a key role in glucose (Glc) mediated root directional responses (Figure 6A) [186]. Exogenous BR dramatically increased Glc-mediated directional responses. Additionally, BR signalling mutant bri1-6 showed reduced root deviation in the presence of Glc, whereas bzr1-1D showed increased root deviation in thepresence of Glc. Glc enhances BR signalling by increasing the endocytosis of BRI1. It was found that BR modulates Glc-induced root deviation responses by regulating PAT (Figure 6A) [186]. A recent report has implicated the involvement of BR signalling in root gravitropic response in maize. Treatment with BR resulted in more curvature of PRs of maize [187].
JA is also found to be a crucial regulator of root gravitropism. Studies on rice coleoptiles revealed an increased amount of JA upon gravistimulation. In addition, a JA gradient was formed opposite to the internal auxin gradient across the stimulated organ during a gravitropic response that worked in an IAA manner. Consistent with these observations, a JA-deficient rice mutant, hebiba, bent slowly upon gravitropic stimulus, thus, suggesting that JA might accelerate the bending response [188]. Moreover, Trp conjugates of JA (JA-Trp) that act as IAA antagonists have been reported to cause agravitropism in a dose-dependent fashion. The response is TIR1-dependent but COI1-independent [189]. JA also influences gravitropism via ASA1, which further leads to changes in auxin homeostasis (Figure 6A). Thus, JA might work via auxin to regulate tropic responses in roots. Another report demonstrates that MeJA impairs lateral auxin distribution in WT roots upon gravistimulation and therefore slows root gravitropic response [190].
Administration of SA led to inhibition of gravitropic response in maize roots, which was an indirect result of inhibition of ethylene biosynthesis by SA. In the recent studies by Tan and co-workers, the effect of exogenous SA on various root growth parameters was examined. They concluded from their studies that exposure of 20 µM or 40 µM SA to Arabidopsis seedlings led to partial agravitropic roots in an NPR1-independent manner [104]. SA also affected the auxin redistribution in the PRs and was involved in the gravitropic bending via auxin efflux carrier PIN2. SA leads to the agravitropic response via blocking PP2A activity, causing the cellular SA levels to increase, thereby resulting in phosphorylation of PIN proteins and affecting auxin transport and redistribution in the cell (Figure 6A) [104]. SA also modulates root gravitropism by affecting clathrin-mediated endocytosis, as exposure to 50 µM SA in wild type Arabidopsis seedlings led to a reduction in gravitropic root curvature. However, the clathrin heavy chain (chc2-2) exhibited less sensitivity to SA-induced gravitropic response [191]. A recent study by Ke et al. 2021 reported that high concentrations of SA impaired the root gravitropism in Arabidopsis seedlings via condensation of PIN2 into hyperclusters through REMORIN 1.2 (REM1.2) dependent nanodomain compartmentalization. As a result, the movement of PIN2 by clathrin-mediated endocytosis and lateral diffusion was restricted, thereby interfering with the asymmetric distribution of auxin, leading to the inhibition of gravitropic responses [192].
Not only the main root, but other parts of the RSA such as LRs also show gravitropism [193,194]. In such cases, the LRs partially suppress positive ortho-gravitropic growth and grow radially to cover more area in the soil. Supplying exogenous auxin and an endogenous increase in auxin levels causes LRs to grow at a more vertical GSA. Reports by Roychoudhry and co-workers have shown that the entire TIR1/AFB-Aux/IAA-ARF-dependent auxin signalling module is necessary to establish GSA in LRs (Figure 6B) [194]. Recently emerged LRs (stage I) show strong but transient PIN3 expression, presumably limiting the strength of auxin redistribution in (stage II) LRs that are establishing their GSA [193,195]. At later stages, the expression of PIN3 diminishes, and PIN4 and PIN7 are expressed to maintain vertical GSA [193]. While auxin promotes gravitropic bending of LRs, CK function as an anti-gravitropic component that is shown to suppress cellular elongation in the upper portion of stage II LRs, thus promoting the radial expansion of RSA (Figure 6B) [112]. Apart from this, there are other molecular components, such as LAZY gene family in Arabidopsis, that has also been reported to have a role in maintaining GSA by controlling the distribution of PINs. LAZY (LZY)/DEEPER ROOTING (DRO)/NEGATIVE GRAVITROPIC RESPONSE OF ROOTS (NGR) genes affect auxin flow towards the lower side of the LR in gravistimulated LRs. The lzy1lzy2lzy3 mutant showed an inverse auxin distribution and asymmetry in the PIN3 expression in the columella cells of the LR, resulting in a horizontally placed LR [196,197]. A recent report by Sharma et al. 2020 also showed a promising involvement of JA-auxin machinery in the regulation of LR angle via LAZY genes. JA signalling transcription factor MYC2 directly activates the transcription of CYP79B2, an auxin biosynthetic gene, and LAZY2/4 in the regulation of LR angle. In addition, JA treatment also affects auxin transport machinery and PIN2 localization in the stage II LR, leading to a downward LR orientation (Figure 6B) [198]. The Arabidopsis R2R3-MYB transcription factor FOUR LIPS (FLP) and its paralog, MYB88, act redundantly but differently in the regulation of primary and LR gravity responses. FLP alone is responsible for the transcription regulation of PIN3 and PIN7 in gravity-stimulating cells in the PRs, whereas FLP-MYB88 both act redundantly to control LR gravitropic set point angle (GSA) (Figure 6B) [5]. The Arabidopsis FERONIA (FER) receptor kinase, which integrates multiple hormonal and environmental signals, governs auxin-mediated LR and root gravitropic responses. The fer-4 mutants showed increased LR branching and delayed gravitropism, which was found to be associated with aberrant PIN2 polarity and an altered polar auxin transport [199]. Previous reports revealed that NGR is necessary for the positive gravitropism of roots. A recent study suggests that NGR, a plasma membrane protein exclusively expressed in the columella and LR cap cells, governs positive gravitropism through regulation of auxin efflux carrier PIN3 localization and modulation of lateral auxin flow in response to gravity [200]. A Lotus japonica mutant defective in LAZY3, a functional ortholog of LAZY1, showed negative gravitropism in primary and LRs. This negative gravitropic response in lazy3 mutant was due to reduced polar auxin transport [201]. In Arabidopsis, DEEPER ROOTING 1 (DRO1) mutation causes horizontally placed LRs and altered GSA, while DRO1 overexpression showed steeper LR angles [202]. In a recent report, Atdro1 primary and LRs showed defects in establishing an auxin gradient in response to gravity [203]. Collectively, all of these results indicate a promising involvement of active auxin signalling and transport machinery in the regulation of root gravitropic responses in LRs.
4. Conclusions and Future Prospects
The RSA is continuously modified by abiotic and biotic signals from the soil that interact with various endogenous cues and turn them into different cellular responses. Phytohormones affect several overlapping processes and their actions depend on specific hormone combinations rather than on the action of a single hormone individually. This review has focussed on various examples of crosstalk between auxin-JA, auxin-CK, etc., suggesting that different hormones affect a number of common responses in plants [95,96]. Moreover, these interactions do not occur via a simple linear mechanism; rather, there are many feedback loops and the involvement of various downstream activators and repressors that drive a single development output robustly. For example, binding of MYC2 transcription factor on the promoters of LAZY2, LAZY4, and auxin-responsive gene CYP79B2 leads to gravitropic bending of LRs [198]. A recent review by Semeradova et al. 2020 summarizes that auxin transport machinery is post-translationally modified by multiple hormones that enable the rapid modulation of the auxin flow to control plant growth and development [204]. Additionally, several other reports as mentioned in this review have further clarified how almost all plant hormones influence the rate of auxin homeostasis [95,118,149], thus making it clear that auxin is indispensable in controlling all aspects of the plant life cycle.
It is very essential to map key transcriptional and post translational pathways and their genes that define the distinct nature of PR and LRs, as that might be the key in understanding hormonal and developmental outputs. Another interesting area is to pinpoint DNA polymorphisms that control a particular root trait of interest. This provides a powerful approach to identify new root branching regulatory genes. A challenging step will be to study the regulation of different root traits under more realistic conditions than under highly aseptic growth conditions such as agar plates where we might miss novel adaptive strategies employed by the plants to survive in their microenvironment. Thus, there is a need to integrate systems biology-aided computational methods (SimRoot) [205] and computer technology such as magnetic resonance imaging and microscale computed tomography [206] that will not only enable us to integrate information related to the overall organization of how roots sense various endogenous and environmental signals, and how they turn them into cellular responses, but also assist in devising predictive models that may identify the crucial regulators, hubs, and integrators involved in regulating RSA. Such multiscale mechanistic insights will underpin efforts to develop crops with improved root systems.
Author Contributions
A.L. and M.S. (Manvi Sharma) conceptualized the article. M.S. (Manvi Sharma), D.S., M.S. (Mohan Sharma), A.T., H.B.S., P.A., H.K.B. and B.N.S. wrote the original draft. M.S., H.B.S., and H.K.B. prepared the figures. A.L. and M.S. (Manvi Sharma) reviewed and finalized the article. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Acknowledgments
The research in AL laboratory is supported by a project grant from Department of Biotechnology, Government of India (Grant No. BT/HRD/NWBA/37/01/2015) and Core Grant from the National Institute of Plant Genome Research. M.S., M.S., and D.S. duly acknowledge research fellowships from National Institute of Plant Genome Research, Govt of India. P.A., H.B.S., and H.K.B. duly acknowledge research fellowships from Department of Biotechnology. A.T. duly acknowledges research fellowship from University Grants Commission, Govt. Of India. The authors acknowledge DBT-eLibrary Consortium (DeLCON) for providing access to e-resources.
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Tötzke, C.; Kardjilov, N.; Lenoir, N.; Manke, I.; Oswald, S.E.; Tengattini, A. What comes NeXT?—High-speed neutron tomography at ILL. Opt. Express 2019, 27, 28640. [Google Scholar] [CrossRef]
- Liu, J.; Rowe, J.; Lindsey, K. Hormonal crosstalk for root development: A combined experimental and modeling perspective. Front. Plant Sci. 2014, 5, 116. [Google Scholar] [CrossRef]
- Sengupta, D.; Reddy, A.R. Simplifying the root dynamics: From complex hormone—Environment interactions to specific root architectural modulation. Plant Growth Regul. 2018, 85, 337–349. [Google Scholar] [CrossRef]
- Šimášková, M.; O’Brien, J.A.; Khan, M.; Van Noorden, G.; Ötvös, K.; Vieten, A.; De Clercq, I.; Van Haperen, J.M.A.; Cuesta, C.; Hoyerová, K.; et al. Cytokinin response factors regulate PIN-FORMED auxin transporters. Nat. Commun. 2015, 6, 8717. [Google Scholar] [CrossRef]
- Wang, H.Z.; Yang, K.Z.; Zou, J.J.; Zhu, L.L.; Xie, Z.D.; Morita, M.T.; Tasaka, M.; Friml, J.; Grotewold, E.; Beeckman, T.; et al. Transcriptional regulation of PIN genes by FOUR LIPS and MYB88 during Arabidopsis root gravitropism. Nat. Commun. 2015, 6, 8822. [Google Scholar] [CrossRef]
- Orman-Ligeza, B.; Parizot, B.; Gantet, P.P.; Beeckman, T.; Bennett, M.J.; Draye, X. Post-embryonic root organogenesis in cereals: Branching out from model plants. Trends Plant Sci. 2013, 18, 459–467. [Google Scholar] [CrossRef]
- Dolan, L.; Janmaat, K.; Willemsen, V.; Linstead, P.; Poethig, S.; Roberts, K.; Scheres, B. Cellular organisation of the Arabidopsis thaliana root. Development 1993, 119, 71–84. [Google Scholar] [CrossRef] [PubMed]
- Svolacchia, N.; Salvi, E.; Sabatini, S. Arabidopsis primary root growth: Let it grow, can’t hold it back anymore! Curr. Opin. Plant Biol. 2020, 57, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Mellor, N.L.; Voß, U.; Janes, G.; Bennett, M.J.; Wells, D.M.; Band, L.R. Auxin fluxes through plasmodesmata modify root-tip auxin distribution. Development 2020, 147, dev181669. [Google Scholar] [CrossRef]
- Sabatini, S.; Beis, D.; Wolkenfelt, H.; Murfett, J.; Guilfoyle, T.; Malamy, J.; Benfey, P.; Leyser, O.; Bechtold, N.; Weisbeek, P.; et al. An Auxin-Dependent Distal Organizer of Pattern and Polarity in the Arabidopsis Root Plants Contain Many Different Cell Types in Ordered Spatial Patterns. Cell 1999, 99, 463–472. [Google Scholar] [CrossRef]
- Galinha, C.; Hofhuis, H.; Luijten, M.; Willemsen, V.; Blilou, I.; Heidstra, R.; Scheres, B. PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 2007, 449, 1053–1057. [Google Scholar] [CrossRef] [PubMed]
- Billou, I.; Xu, J.; Wildwater, M.; Willemsen, V.; Paponov, I.; Frimi, J.; Heldstra, R.; Aida, M.; Palme, K.; Scheres, B. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 2005, 433, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Drisch, R.C.; Stahl, Y. Function and regulation of transcription factors involved in root apical meristem and stem cell maintenance. Front. Plant Sci. 2015, 6, 505. [Google Scholar] [CrossRef]
- Hochholdinger, F.; Zimmermann, R. Conserved and diverse mechanisms in root development. Curr. Opin. Plant Biol. 2008, 11, 70–74. [Google Scholar] [CrossRef]
- Motte, H.; Vanneste, S.; Beeckman, T. Molecular and environmental regulation of root development. Annu. Rev. Plant Biol. 2019, 70, 465–488. [Google Scholar] [CrossRef] [PubMed]
- Lavenus, J.; Goh, T.; Roberts, I.; Guyomarc’h, S.; Lucas, M.; De Smet, I.; Fukaki, H.; Beeckman, T.; Bennett, M.; Laplaze, L. Lateral root development in Arabidopsis: Fifty shades of auxin. Trends Plant Sci. 2013, 18, 450–458. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Scheres, B. Lateral root formation and the multiple roles of auxin. J. Exp. Bot. 2018, 69, 155–167. [Google Scholar] [CrossRef]
- Moreno-Risueno, M.A.; Van Norman, J.M.; Moreno, A.; Zhang, J.; Ahnert, S.E.; Benfey, P.N. Oscillating gene expression determines competence for periodic Arabidopsis root branching. Science 2010, 329, 1306–1311. [Google Scholar] [CrossRef] [PubMed]
- Jansen, L.; Roberts, I.; de Rycke, R.; Beeckman, T. Phloem-associated auxin response maxima determine radial positioning of lateral roots in maize. Philos. Trans. R. Soc. B Biol. Sci. 2012. [Google Scholar] [CrossRef] [PubMed]
- Jansen, L.; Hollunder, J.; Roberts, I.; Forestan, C.; Fonteyne, P.; Van Quickenborne, C.; Zhen, R.G.; Mckersie, B.; Parizot, B.; Beeckman, T. Comparative transcriptomics as a tool for the identification of root branching genes in maize. Plant Biotechnol. J. 2013. [Google Scholar] [CrossRef] [PubMed]
- Steffens, B.; Rasmussen, A. The physiology of adventitious roots. Plant Physiol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, J.A.; Rasmussen, A.; Traini, R.; Voß, U.; Sturrock, C.; Mooney, S.J.; Wells, D.M.; Bennett, M.J. Branching out in roots: Uncovering form, function, and regulation. Plant Physiol. 2014, 170, 603–617. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Kurten, E.L.; Monshausen, G.; Hummel, G.M.; Gilroy, S.; Baldwin, I.T. NaRALF, a peptide signal essential for the regulation of root hair tip apoplastic pH in Nicotiana attenuata, is required for root hair development and plant growth in native soils. Plant J. 2007, 52, 877–890. [Google Scholar] [CrossRef] [PubMed]
- Miguel, M.A.; Postma, J.A.; Lynch, J.P. Phene synergism between root hair length and basal root growth angle for phosphorus acquisition. Plant Physiol. 2015, 167. [Google Scholar] [CrossRef]
- Bruex, A.; Kainkaryam, R.M.; Wieckowski, Y.; Kang, Y.H.; Bernhardt, C.; Xia, Y.; Zheng, X.; Wang, J.Y.; Lee, M.M.; Benfey, P.; et al. A gene regulatory network for root epidermis cell differentiation in Arabidopsis. PLoS Genet. 2012, 8, e1002446. [Google Scholar] [CrossRef]
- Schoenaers, S.; Balcerowicz, D.; Breen, G.; Hill, K.; Zdanio, M.; Mouille, G.; Holman, T.J.; Oh, J.; Wilson, M.H.; Nikonorova, N.; et al. The auxin-regulated CrRLK1L kinase ERULUS controls cell wall composition during root hair tip growth. Curr. Biol. 2018, 28, 722–732.e6. [Google Scholar] [CrossRef]
- Vissenberg, K.; Claeijs, N.; Balcerowicz, D.; Schoenaers, S. Hormonal regulation of root hair growth and responses to the environment in Arabidopsis. J. Exp. Bot. 2020, 71, 2412–2427. [Google Scholar] [CrossRef]
- Schoenaers, S.; Balcerowicz, D.; Vissenberg, K. Molecular mechanisms regulating root hair tip growth: A comparison with pollen tubes. In Pollen Tip Growth: From Biophysical Aspects to Systems Biology; Springer: Cham, Switzerland, 2017; ISBN 9783319566450. [Google Scholar]
- Went, F.W. The dual effect of auxin on root formation. Am. J. Bot. 1939, 26, 24. [Google Scholar] [CrossRef]
- Zhu, Q.; Gallemí, M.; Pospíšil, J.; Žádníková, P.; Strnad, M.; Benková, E. Root gravity response module guides differential growth determining both root bending and apical hook formation in Arabidopsis. Development 2019, 146, dev175919. [Google Scholar] [CrossRef]
- Zhang, Y.; He, P.; Ma, X.; Yang, Z.; Pang, C.; Yu, J.; Wang, G.; Friml, J.; Xiao, G. Auxin-mediated statolith production for root gravitropism. New Phytol. 2019, 224, 761–774. [Google Scholar] [CrossRef]
- Barbez, E.; Dünser, K.; Gaidora, A.; Lendl, T.; Busch, W. Auxin steers root cell expansion via apoplastic pH regulation in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2017, 114, 201613499. [Google Scholar] [CrossRef] [PubMed]
- Fendrych, M.; Akhmanova, M.; Merrin, J.; Glanc, M.; Hagihara, S.; Takahashi, K.; Uchida, N.; Torii, K.U.; Friml, J. Rapid and reversible root growth inhibition by TIR1 auxin signalling. Nat. Plants 2018, 4, 453–459. [Google Scholar] [CrossRef] [PubMed]
- Ljung, K.; Hull, A.K.; Celenza, J.; Yamada, M.; Estelle, M.; Normanly, J.; Sandberg, G. Sites and regulation of auxin biosynthesis in Arabidopsis roots. Plant Cell 2005, 17, 1090–1104. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, Y.; Men, S.; Fischer, U.; Stepanova, A.N.; Alonso, J.M.; Ljung, K.; Grebe, M. Local auxin biosynthesis modulates gradient-directed planar polarity in Arabidopsis. Nat. Cell Biol. 2009, 11, 731–738. [Google Scholar] [CrossRef] [PubMed]
- Brumos, J.; Robles, L.M.; Yun, J.; Vu, T.C.; Jackson, S.; Alonso, J.M.; Stepanova, A.N. Local auxin biosynthesis is a key regulator of plant development. Dev. Cell 2018, 47, 306–318.e5. [Google Scholar] [CrossRef] [PubMed]
- Morffy, N.J.; Strader, L.C. Locally sourced: Auxin biosynthesis and transport in the root meristem. Dev. Cell 2018, 47, 262–264. [Google Scholar] [CrossRef]
- Tian, H.; Wabnik, K.; Niu, T.; Li, H.; Yu, Q.; Pollmann, S.; Vanneste, S.; Govaerts, W.; Rolčík, J.; Geisler, M.; et al. WOX5-IAA17 feedback circuit-mediated cellular auxin response is crucial for the patterning of root stem cell niches in Arabidopsis. Mol. Plant 2014, 7, 277–289. [Google Scholar] [CrossRef]
- Ding, Z.; Friml, J. Auxin regulates distal stem cell differentiation in Arabidopsis roots. Proc. Natl. Acad. Sci. USA 2010, 107, 12046–12051. [Google Scholar] [CrossRef]
- Guo, Y.; Wu, Q.; Xie, Z.; Yu, B.; Zeng, R.; Min, Q.; Huang, J. OsFPFL4 is involved in the root and flower development by affecting auxin levels and ROS accumulation in rice (Oryza sativa). Rice 2020, 13, 2. [Google Scholar] [CrossRef]
- Lehmann, T.; Janowitz, T.; Sánchez-Parra, B.; Alonso, M.M.P.; Trompetter, I.; Piotrowski, M.; Pollmann, S. Arabidopsis NITRILASE 1 contributes to the regulation of root growth and development through modulation of auxin biosynthesis in seedlings. Front. Plant Sci. 2017, 8, 36. [Google Scholar] [CrossRef]
- Zhang, G.; Xu, N.; Chen, H.; Wang, G.; Huang, J. OsMADS25 regulates root system development via auxin signalling in rice. Plant J. 2018, 95, 1004–1022. [Google Scholar] [CrossRef]
- Nan, W.; Wang, X.; Yang, L.; Hu, Y.; Wei, Y.; Liang, X.; Mao, L.; Bi, Y. Cyclic GMP is involved in auxin signalling during Arabidopsis root growth and development. J. Exp. Bot. 2014, 65, 1571–1583. [Google Scholar] [CrossRef]
- Chen, X.; Li, L.; Xu, B.; Zhao, S.; Lu, P.; He, Y.; Ye, T.; Feng, Y.Q.; Wu, Y. Phosphatidylinositol-specific phospholipase C2 functions in auxin-modulated root development. Plant Cell Environ. 2019, 42, 1441–1457. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Feng, Y.; Liu, Y.; Bao, Y. Mutations in exocyst complex subunit SEC6 gene impaired polar auxin transport and PIN protein recycling in Arabidopsis primary root. Plant Sci. 2016, 250, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Kong, Q.; Ma, W.; Yang, H.; Ma, G.; Mantyla, J.J.; Benning, C. The Arabidopsis WRINKLED1 transcription factor affects auxin homeostasis in roots. J. Exp. Bot. 2017, 68, 4627–4634. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Qiao, J.Y.; Yu, C.L.; Chen, H.; Sun, C.D.; Huang, L.Z.; Li, C.Y.; Geisler, M.; Qian, Q.; Jiang, D.A.; et al. The auxin influx carrier, OsAUX3, regulates rice root development and responses to aluminium stress. Plant Cell Environ. 2019, 42, 1125–1138. [Google Scholar] [CrossRef]
- Chen, R.; Xu, N.; Yu, B.; Wu, Q.; Li, X.; Wang, G.; Huang, J. The WUSCHEL-related homeobox transcription factor OsWOX4 controls the primary root elongation by activating OsAUX1 in rice. Plant Sci. 2020, 298, 110575. [Google Scholar] [CrossRef]
- Upadhyay, N.; Kar, D.; Datta, S. A multidrug and toxic compound extrusion (MATE) transporter modulates auxin levels in root to regulate root development and promotes aluminium tolerance. Plant Cell Environ. 2020, 43, 745–759. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, W.; Zuo, Y.; Zhu, L.; Hastwell, A.H.; Chen, L.; Tian, Y.; Su, C.; Ferguson, B.J.; Li, X. GmYUC2a mediates auxin biosynthesis during root development and nodulation in soybean. J. Exp. Bot. 2019. [Google Scholar] [CrossRef]
- Nikonorova, N.; Murphy, E.; Fonseca de Lima, C.F.; Zhu, S.; van de Cotte, B.; Vu, L.D.; Balcerowicz, D.; Li, L.; Kong, X.; De Rop, G.; et al. Auxin-triggered changes in the Arabidopsis root tip (phospho)proteome reveal novel root growth regulators. bioRxiv 2021. [Google Scholar] [CrossRef]
- Miao, Z.Q.; Zhao, P.X.; Mao, J.L.; Yu, L.H.; Yuan, Y.; Tang, H.; Liu, Z.B.; Xiang, C. Bin HOMEOBOX PROTEIN52 mediates the crosstalk between ethylene and auxin signaling during primary root elongation by modulating auxin transport-related gene expression. Plant Cell 2018, 30, 2761–2778. [Google Scholar] [CrossRef] [PubMed]
- Mao, J.L.; Miao, Z.Q.; Wang, Z.; Yu, L.H.; Cai, X.T.; Xiang, C. Bin Arabidopsis ERF1 mediates cross-talk between ethylene and auxin biosynthesis during primary root elongation by regulating ASA1 expression. PLoS Genet. 2016. [Google Scholar] [CrossRef]
- Dello Ioio, R.; Nakamura, K.; Moubayidin, L.; Perilli, S.; Taniguchi, M.; Morita, M.T.; Aoyama, T.; Costantino, P.; Sabatini, S. A genetic framework for the control of cell division and differentiation in the root meristem. Science 2008, 322, 1380–1384. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.H.; Liu, Y.B.; Zhang, X.S. Auxin-cytokinin interaction regulates meristem development. Mol. Plant 2011, 4, 616–625. [Google Scholar] [CrossRef]
- Chapman, E.J.; Estelle, M. Cytokinin and auxin intersection in root meristems. Genome Biol. 2009, 10, 210. [Google Scholar] [CrossRef]
- Zhang, W.; To, J.P.C.; Cheng, C.Y.; Eric Schaller, G.; Kieber, J.J. Type-A response regulators are required for proper root apical meristem function through post-transcriptional regulation of PIN auxin efflux carriers. Plant J. 2011, 68, 1–10. [Google Scholar] [CrossRef]
- Werner, T.; Motyka, V.; Laucou, V.; Smets, R.; Van Onckelen, H.; Schmülling, T. Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 2003, 15, 2532–2550. [Google Scholar] [CrossRef] [PubMed]
- Werner, T.; Nehnevajova, E.; Köllmer, I.; Novák, O.; Strnad, M.; Krämer, U.; Schmülling, T. Root-specific reduction of cytokinin causes enhanced root growth, drought tolerance, and leaf mineral enrichment in Arabidopsis and tobacco. Plant Cell 2010, 22, 3905–3920. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Mo, X.; Shou, H.; Wu, P. Cytokinin-mediated cell cycling arrest of pericycle founder cells in lateral root initiation of Arabidopsis. Plant Cell Physiol. 2006, 47, 1112–1123. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Fang, J.; Xu, F.; Wang, W.; Sun, X.; Chu, J.; Cai, B.; Feng, Y.; Chu, C. CYTOKININ OXIDASE/DEHYDROGENASE4 integrates cytokinin and auxin signaling to control rice crown root formation. Plant Physiol. 2014, 165, 1035–1046. [Google Scholar] [CrossRef]
- Ding, W.; Tong, H.; Zheng, W.; Ye, J.; Pan, Z.; Zhang, B.; Zhu, S. Isolation, characterization and transcriptome analysis of a cytokinin receptor mutant osckt1 in rice. Front. Plant Sci. 2017, 8, 88. [Google Scholar] [CrossRef] [PubMed]
- Márquez, G.; Alarcón, M.V.; Salguero, J. Cytokinin inhibits lateral root development at the earliest stages of lateral root primordium initiation in maize primary root. J. Plant Growth Regul. 2019, 38, 83–92. [Google Scholar] [CrossRef]
- Köllmer, I.; Novák, O.; Strnad, M.; Schmülling, T.; Werner, T. Overexpression of the cytosolic cytokinin oxidase/dehydrogenase (CKX7) from Arabidopsis causes specific changes in root growth and xylem differentiation. Plant J. 2014, 78, 359–371. [Google Scholar] [CrossRef] [PubMed]
- Mason, M.G.; Mathews, D.E.; Argyros, D.A.; Maxwell, B.B.; Kieber, J.J.; Alonso, J.M.; Ecker, J.R.; Schaller, G.E. Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell 2005, 17, 3007–3018. [Google Scholar] [CrossRef] [PubMed]
- Dello Ioio, R.; Linhares, F.S.; Scacchi, E.; Casamitjana-Martinez, E.; Heidstra, R.; Costantino, P.; Sabatini, S. Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr. Biol. 2007, 17, 678–682. [Google Scholar] [CrossRef] [PubMed]
- Hill, K.; Mathews, D.E.; Kim, H.J.; Street, I.H.; Wildes, S.L.; Chiang, Y.H.; Mason, M.G.; Alonso, J.M.; Ecker, J.R.; Kieber, J.J.; et al. Functional characterization of type-B response regulators in the Arabidopsis cytokinin respons. Plant Physiol. 2013, 162, 212–224. [Google Scholar] [CrossRef] [PubMed]
- Worthen, J.M.; Yamburenko, M.V.; Lim, J.; Nimchuk, Z.L.; Kieber, J.J.; Schaller, G.E. Type-B response regulators of rice play key roles in growth, development and cytokinin signaling. Development 2019, 146, dev174870. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, M.; Pischke, M.S.; Mähönen, A.P.; Miyawaki, K.; Hashimoto, Y.; Seki, M.; Kobayashi, M.; Shinozaki, K.; Kato, T.; Tabata, S.; et al. In planta functions of the Arabidopsis cytokinin receptor family. Proc. Natl. Acad. Sci. USA 2004, 101, 8821–8826. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, C.; Ohashi, Y.; Sato, S.; Kato, T.; Tabata, S.; Ueguchi, C. Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis. Plant Cell 2004, 16, 1365–1377. [Google Scholar] [CrossRef]
- Riefler, M.; Novak, O.; Strnad, M.; Schmülling, T. Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 2006, 18, 40–54. [Google Scholar] [CrossRef]
- Zhang, W.; Swarup, R.; Bennett, M.; Schaller, G.E.; Kieber, J.J. Cytokinin induces cell division in the Quiescent CENTER of the Arabidopsis root apical meristem. Curr. Biol. 2013, 23, 1979–1989. [Google Scholar] [CrossRef]
- Street, I.H.; Mathews, D.E.; Yamburkenko, M.V.; Sorooshzadeh, A.; John, R.T.; Swarup, R.; Bennett, M.J.; Kieber, J.J.; Schaller, G.E. Cytokinin acts through the auxin influx carrier AUX1 to regulate cell elongation in the root. Development 2016, 143, 3982–3993. [Google Scholar] [CrossRef] [PubMed]
- Mao, C.; He, J.; Liu, L.; Deng, Q.; Yao, X.; Liu, C.; Qiao, Y.; Li, P.; Ming, F. OsNAC2 integrates auxin and cytokinin pathways to modulate rice root development. Plant Biotechnol. J. 2020, 18, 429–442. [Google Scholar] [CrossRef] [PubMed]
- Saini, S.; Sharma, I.; Pati, P.K. Versatile roles of brassinosteroid in plants in the context of its homoeostasis, signaling and crosstalks. Front. Plant Sci. 2015, 6, 950. [Google Scholar] [CrossRef]
- Planas-Riverola, A.; Gupta, A.; Betegoń-Putze, I.; Bosch, N.; Ibanḛs, M.; Cano-Delgado, A.I. Brassinosteroid signaling in plant development and adaptation to stress. Development 2019, 146, dev151894. [Google Scholar] [CrossRef] [PubMed]
- Chaiwanon, J.; Wang, Z.Y. Spatiotemporal brassinosteroid signaling and antagonism with auxin pattern stem cell dynamics in Arabidopsis roots. Curr. Biol. 2015, 25, 1031–1042. [Google Scholar] [CrossRef] [PubMed]
- González-García, M.P.; Vilarrasa-Blasi, J.; Zhiponova, M.; Divol, F.; Mora-García, S.; Russinova, E.; Caño-Delgado, A.I. Brassinosteroids control meristem size by promoting cell cycle progression in Arabidopsis roots. Development 2011, 138, 849–859. [Google Scholar] [CrossRef] [PubMed]
- Hacham, Y.; Holland, N.; Butterfield, C.; Ubeda-Tomas, S.; Bennett, M.J.; Chory, J.; Savaldi-Goldstein, S. Brassinosteroid perception in the epidermis controls root meristem size. Development 2011, 138, 839–848. [Google Scholar] [CrossRef]
- Müssig, C.; Shin, G.H.; Altmann, T. Brassinosteroids promote root growth in Arabidopsis. Plant Physiol. 2003, 133, 1261–1271. [Google Scholar] [CrossRef]
- Wei, Z.; Li, J. Brassinosteroids regulate root growth, development, and symbiosis. Mol. Plant 2016, 9, 86–100. [Google Scholar] [CrossRef]
- Lv, B.; Tian, H.; Zhang, F.; Liu, J.; Lu, S.; Bai, M.; Li, C.; Ding, Z. Brassinosteroids regulate root growth by controlling reactive oxygen species homeostasis and dual effect on ethylene synthesis in Arabidopsis. PLoS Genet. 2018, 14, e1007144. [Google Scholar] [CrossRef] [PubMed]
- Vragović, K.; Selaa, A.; Friedlander-Shani, L.; Fridman, Y.; Hacham, Y.; Holland, N.; Bartom, E.; Mockler, T.C.; Savaldi-Goldstein, S. Translatome analyses capture of opposing tissuespecific brassinosteroid signals orchestrating root meristem differentiation. Proc. Natl. Acad. Sci. USA 2015, 112, 923–928. [Google Scholar] [CrossRef]
- Sharma, M.; Laxmi, A. Jasmonates: Emerging players in controlling temperature stress tolerance. Front. Plant Sci. 2016, 6, 1129. [Google Scholar] [CrossRef] [PubMed]
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 2013, 111, 1021–1058. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.I.R.; Fatma, M.; Per, T.S.; Anjum, N.A.; Khan, N.A. Salicylic acid-induced abiotic stress tolerance and underlying mechanisms in plants. Front. Plant Sci. 2015, 6, 462. [Google Scholar] [CrossRef] [PubMed]
- Ueda, J.; Kato, J. Isolation and identification of a senescence-promoting substance from wormwood (Artemisia absinthium L.). Plant Physiol. 1980, 66, 246–249. [Google Scholar] [CrossRef]
- Dathe, W.; Sembdner, G. Endogenous plant hormones of the broad bean, Vicia faba L. III. Distribution of abscisic acid and gibberellins in the pistil at anthesis. Biochem. Physiol. Pflanz. 1981, 176, 590–594. [Google Scholar] [CrossRef]
- Staswick, P.E.; Su, W.; Howell, S.H. Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc. Natl. Acad. Sci. USA 1992, 89, 6837–6840. [Google Scholar] [CrossRef] [PubMed]
- Xie, D.X.; Feys, B.F.; James, S.; Nieto-Rostro, M.; Turner, J.G. COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 1998, 280, 1091–1094. [Google Scholar] [CrossRef]
- Ellis, C.; Turner, J.G. A conditionally fertile coi1 allele indicates cross-talk between plant hormone signalling pathways in Arabidopsis thaliana seeds and young seedlings. Planta 2002, 215, 549–556. [Google Scholar] [CrossRef]
- Adams, E.; Turner, J. COI1, a jasmonate receptor, is involved in ethylene-induced inhibition of Arabidopsis root growth in the light. J. Exp. Bot. 2010, 61, 4373–4386. [Google Scholar] [CrossRef] [PubMed]
- Dombrecht, B.; Xue, G.P.; Sprague, S.J.; Kirkegaard, J.A.; Ross, J.J.; Reid, J.B.; Fitt, G.P.; Sewelam, N.; Schenk, P.M.; Manners, J.M.; et al. MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell Online 2007, 19, 2225–2245. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Sun, J.; Zhai, Q.; Zhou, W.; Qi, L.; Xu, L.; Wang, B.; Chen, R.; Jiang, H.; Qi, J.; et al. The basic helix-loop-helix transcription factor myc2 directly represses plethora expression during jasmonate-mediated modulation of the root stem cell niche in Arabidopsis. Plant Cell 2011, 23, 3335–3352. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Xu, Y.; Ye, S.; Jiang, H.; Chen, Q.; Liu, F.; Zhou, W.; Chen, R.; Li, X.; Tietz, O.; et al. Arabidopsis ASA1 is important for jasmonate-mediated regulation of auxin biosynthesis and transport during lateral root formation. Plant Cell 2009, 21, 1495–1511. [Google Scholar] [CrossRef]
- Tiryaki, I.; Staswick, P.E. An Arabidopsis mutant defective in jasmonate response is allelic to the auxin-signaling mutant axr1. Plant Physiol. 2002, 130, 887–894. [Google Scholar] [CrossRef]
- Corti Monzón, G.; Pinedo, M.; Lamattina, L.; de la Canal, L. Sunflower root growth regulation: The role of jasmonic acid and its relation with auxins. Plant Growth Regul. 2012, 66, 129–136. [Google Scholar] [CrossRef]
- Zhou, W.; Lozano-Torres, J.L.; Blilou, I.; Zhang, X.; Zhai, Q.; Smant, G.; Li, C.; Scheres, B. A jasmonate signaling network activates root stem cells and promotes regeneration. Cell 2019, 177, 942–956.e14. [Google Scholar] [CrossRef]
- Zhao, X.; Wang, J.; Yuan, J.; Wang, X.L.; Zhao, Q.P.; Kong, P.T.; Zhang, X. NITRIC OXIDE-ASSOCIATED PROTEIN1 (AtNOA1) is essential for salicylic acid-induced root waving in Arabidopsis thaliana. New Phytol. 2015, 207, 211–224. [Google Scholar] [CrossRef]
- Pasternak, T.; Groot, E.P.; Kazantsev, F.V.; Teale, W.; Omelyanchuk, N.; Kovrizhnykh, V.; Palme, K.; Mironova, V.V. Salicylic acid affects root meristem patterning via auxin distribution in a concentration-dependent manner. Plant Physiol. 2019, 180, 1725–1739. [Google Scholar] [CrossRef]
- Armengot, L.; Marquès-Bueno, M.M.; Soria-Garcia, A.; Müller, M.; Munné-Bosch, S.; Martínez, M.C. Functional interplay between protein kinase CK2 and salicylic acid sustains PIN transcriptional expression and root development. Plant J. 2014, 78, 411–423. [Google Scholar] [CrossRef]
- Agtuca, B.; Rieger, E.; Hilger, K.; Song, L.; Robert, C.A.M.; Erb, M.; Karve, A.; Ferrieri, R.A. Carbon-11 reveals opposing roles of auxin and salicylic acid in regulating leaf physiology, leaf metabolism, and resource allocation patterns that impact root growth in Zea mays. J. Plant Growth Regul. 2014, 33, 328–339. [Google Scholar] [CrossRef]
- Xu, L.; Zhao, H.; Ruan, W.; Deng, M.; Wang, F.; Peng, J.; Luo, J.; Chen, Z.; Yi, K. ABNORMAL INFLORESCENCE MERISTEM1 functions in salicylic acid biosynthesis to maintain proper reactive oxygen species levels for root meristem activity in rice. Plant Cell 2017, 29, 560–574. [Google Scholar] [CrossRef]
- Tan, S.; Abas, M.; Verstraeten, I.; Glanc, M.; Molnár, G.; Hajný, J.; Lasák, P.; Petřík, I.; Russinova, E.; Petrášek, J.; et al. Salicylic acid targets protein phosphatase 2A to attenuate growth in plants. Curr. Biol. 2020, 30, 381–395.e8. [Google Scholar] [CrossRef] [PubMed]
- Muday, G.K.; Lomax, T.L.; Rayle, D.L. Characterization of the growth and auxin physiology of roots of the tomato mutant, diageotropica. Planta 1995, 195, 548–553. [Google Scholar] [CrossRef] [PubMed]
- Banda, J.; Bellande, K.; von Wangenheim, D.; Goh, T.; Guyomarc’h, S.; Laplaze, L.; Bennett, M.J. Lateral root formation in Arabidopsis: A well-ordered LRexit. Trends Plant Sci. 2019, 24, 826–839. [Google Scholar] [CrossRef]
- Ivanchenko, M.G.; Napsucialy-Mendivil, S.; Dubrovsky, J.G. Auxin-induced inhibition of lateral root initiation contributes to root system shaping in Arabidopsis thaliana. Plant J. 2010, 64, 740–752. [Google Scholar] [CrossRef] [PubMed]
- Xuan, W.; Audenaert, D.; Parizot, B.; Möller, B.K.; Njo, M.F.; De Rybel, B.; De Rop, G.; Van Isterdael, G.; Mähönen, A.P.; Vanneste, S.; et al. Root cap-derived auxin pre-patterns the longitudinal axis of the Arabidopsis root. Curr. Biol. 2015, 25, 1381–1388. [Google Scholar] [CrossRef]
- Goh, T.; Joi, S.; Mimura, T.; Fukaki, H. The establishment of asymmetry in Arabidopsis lateral root founder cells is regulated by LBD16/ASL18 and related LBD/ASL proteins. Development 2012, 139, 883–893. [Google Scholar] [CrossRef]
- Jenness, M.K.; Carraro, N.; Pritchard, C.A.; Murphy, A.S. The Arabidopsis ATP-BINDING CASSETTE transporter ABCB21 regulates auxin levels in cotyledons, the root pericycle, and leaves. Front. Plant Sci. 2019, 10, 806. [Google Scholar] [CrossRef]
- Zhang, F.; Tao, W.; Sun, R.; Wang, J.; Li, C.; Kong, X.; Tian, H.; Ding, Z. PRH1 mediates ARF7-LBD dependent auxin signaling to regulate lateral root development in Arabidopsis thaliana. PLoS Genet. 2020, 16, e1008044. [Google Scholar] [CrossRef] [PubMed]
- Waidmann, S.; Ruiz Rosquete, M.; Schöller, M.; Sarkel, E.; Lindner, H.; LaRue, T.; Petřík, I.; Dünser, K.; Martopawiro, S.; Sasidharan, R.; et al. Cytokinin functions as an asymmetric and anti-gravitropic signal in lateral roots. Nat. Commun. 2019, 10, 3540. [Google Scholar] [CrossRef] [PubMed]
- Laplaze, L.; Benkova, E.; Casimiro, I.; Maes, L.; Vanneste, S.; Swarup, R.; Weijers, D.; Calvo, V.; Parizot, B.; Herrera-Rodriguez, M.B.; et al. Cytokinins act directly on lateral root founder cells to inhibit root initiation. Plant Cell 2007, 19, 3889–3900. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Ramireddy, E.; Schmülling, T. Lateral root formation and growth of Arabidopsis is redundantly regulated by cytokinin metabolism and signalling genes. J. Exp. Bot. 2013, 64, 5021–5032. [Google Scholar] [CrossRef]
- Hutchison, C.E.; Li, J.; Argueso, C.; Gonzalez, M.; Lee, E.; Lewis, M.W.; Maxwell, B.B.; Perdue, T.D.; Schaller, G.E.; Alonso, J.M.; et al. The Arabidopsis histidine phosphotransfer proteins are redundant positive regulators of cytokinin signaling. Plant Cell 2006, 18, 3073–3087. [Google Scholar] [CrossRef] [PubMed]
- Marhavý, P.; Duclercq, J.; Weller, B.; Feraru, E.; Bielach, A.; Offringa, R.; Friml, J.; Schwechheimer, C.; Murphy, A.; Benková, E. Cytokinin controls polarity of PIN1-dependent auxin transport during lateral root organogenesis. Curr. Biol. 2014, 24, 1031–1037. [Google Scholar] [CrossRef] [PubMed]
- Marhavý, P.; Bielach, A.; Abas, L.; Abuzeineh, A.; Duclercq, J.; Tanaka, H.; Pařezová, M.; Petrášek, J.; Friml, J.; Kleine-Vehn, J.; et al. Cytokinin modulates endocytic trafficking of PIN1 auxin efflux carrier to control plant organogenesis. Dev. Cell 2011, 21, 796–804. [Google Scholar] [CrossRef]
- Michniewicz, M.; Ho, C.H.; Enders, T.A.; Floro, E.; Damodaran, S.; Gunther, L.K.; Powers, S.K.; Frick, E.M.; Topp, C.N.; Frommer, W.B.; et al. TRANSPORTER OF IBA1 links auxin and cytokinin to influence root architecture. Dev. Cell 2019, 50, 599–609.e4. [Google Scholar] [CrossRef]
- Chang, L.; Ramireddy, E.; Schmülling, T. Cytokinin as a positional cue regulating lateral root spacing in Arabidopsis. J. Exp. Bot. 2015, 66, 4759–4768. [Google Scholar] [CrossRef]
- Bielach, A.; Podlešáková, K.; Marhavý, P.; Duclercq, J.; Cuesta, C.; Müller, B.; Grunewald, W.; Tarkowski, P.; Benkováa, E. Spatiotemporal regulation of lateral root organogenesis in Arabidopsis by cytokinin. Plant Cell 2012, 24, 3967–3981. [Google Scholar] [CrossRef]
- Bao, F.; Shen, J.; Brady, S.R.; Muday, G.K.; Asami, T.; Yang, Z. Brassinosteroids interact with auxin to promote lateral root development in Arabidopsis. Plant Physiol. 2004, 134, 1624–1631. [Google Scholar] [CrossRef]
- Gupta, A.; Singh, M.; Laxmi, A. Interaction between glucose and brassinosteroid during the regulation of lateral root development in Arabidopsis. Plant Physiol. 2015, 168, 307–320. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Park, P.J.; Hwang, H.J.; Lee, S.Y.; Oh, M.H.; Kim, S.G. Brassinosteroid signals control expression of the AXR3/IAA17 gene in the cross-talk point with auxin in root development. Biosci. Biotechnol. Biochem. 2006, 70, 768–773. [Google Scholar] [CrossRef] [PubMed]
- Stenzel, I.; Otto, M.; Delker, C.; Kirmse, N.; Schmidt, D.; Miersch, O.; Hause, B.; Wasternack, C. ALLENE OXIDE CYCLASE (AOC) gene family members of Arabidopsis thaliana: Tissue-and organ-specific promoter activities and in vivo heteromerization. J. Exp. Bot. 2012, 63, 6125–6138. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.T.; Xu, P.; Zhao, P.X.; Liu, R.; Yu, L.H.; Xiang, C.B. Arabidopsis ERF109 mediates cross-talk between jasmonic acid and auxin biosynthesis during lateral root formation. Nat. Commun. 2014, 5, 5833. [Google Scholar] [CrossRef] [PubMed]
- Raya-González, J.; Pelagio-Flores, R.; López-Bucio, J. The jasmonate receptor COI1 plays a role in jasmonate-induced lateral root formation and lateral root positioning in Arabidopsis thaliana. J. Plant Physiol. 2012, 169, 1348–1358. [Google Scholar] [CrossRef] [PubMed]
- Ishimaru, Y.; Hayashi, K.; Suzuki, T.; Fukaki, H.; Prusinska, J.; Meester, C.; Quareshy, M.; Egoshi, S.; Matsuura, H.; Takahashi, K.; et al. Jasmonic acid inhibits auxin-induced lateral rooting independently of the CORONATINE INSENSITIVE1 receptor. Plant Physiol. 2018, 177, 1704–1716. [Google Scholar] [CrossRef]
- Echevarría-Machado, I.; Rosa, R.M.; Larqué-Saavedra, A. Responses of transformed Catharanthus roseus roots to femtomolar concentrations of salicylic acid. Plant Physiol. Biochem. 2007, 45, 501–507. [Google Scholar] [CrossRef]
- Liu, J.; Sheng, L.; Xu, Y.; Li, J.; Yang, Z.; Huang, H.; Xu, L. WOX11 and 12 are involved in the first-step cell fate transition during de novo root organogenesis in Arabidopsis. Plant Cell 2014, 26, 1081–1093. [Google Scholar] [CrossRef]
- Wilmoth, J.C.; Wang, S.; Tiwari, S.B.; Joshi, A.D.; Hagen, G.; Guilfoyle, T.J.; Alonso, J.M.; Ecker, J.R.; Reed, J.W. NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation. Plant J. 2005, 43, 118–130. [Google Scholar] [CrossRef]
- Sukumar, P.; Maloney, G.S.; Muday, G.K. Localized induction of the ATP-binding cassette B19 auxin transporter enhances adventitious root formation in Arabidopsis. Plant Physiol. 2013, 162, 1392–1405. [Google Scholar] [CrossRef]
- Růžička, K.; Ljung, K.; Vanneste, S.; Podhorská, R.; Beeckman, T.; Friml, J.; Benková, E. Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. Plant Cell 2007, 19, 2197–2212. [Google Scholar] [CrossRef] [PubMed]
- Strader, L.C.; Bartel, B. Transport and metabolism of the endogenous auxin precursor indole-3-butyric acid. Mol. Plant 2011, 4, 477–486. [Google Scholar] [CrossRef] [PubMed]
- Nordström, A.C.; Jacobs, F.A.; Efiasson, L. Effect of exogenous indole-3-acetic acid and indole-3-butyric acid on internal levels of the respective auxins and their conjugation with aspartic acid during adventitious root formation in pea cuttings. Plant Physiol. 1991, 96, 856–861. [Google Scholar] [CrossRef] [PubMed]
- Li, S.-W. Molecular bases for the regulation of adventitious root generation in plants. Front. Plant Sci. 2021, 12, 614072. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Zhang, Y.; Liu, W.; Wang, H.; Wen, S.; Zhang, Y.; Xu, L.; Rowe, H. Molecular evolution of auxin-mediated root initiation in plants. Mol. Biol. Evol. 2020, 37, 1387–1393. [Google Scholar] [CrossRef]
- Hu, X.; Xu, L. Transcription factors WOX11/12 directly activate WOX5/7 to promote root primordia initiation and organogenesis. Plant Physiol. 2016, 172, 2363–2373. [Google Scholar] [CrossRef]
- Lakehal, A.; Chaabouni, S.; Cavel, E.; Le Hir, R.; Ranjan, A.; Raneshan, Z.; Novák, O.; Păcurar, D.I.; Perrone, I.; Jobert, F.; et al. A molecular framework for the control of adventitious rooting by TIR1/AFB2-Aux/IAA-dependent auxin signaling in Arabidopsis. Mol. Plant 2019, 12, 1499–1514. [Google Scholar] [CrossRef]
- Gutierrez, L.; Mongelard, G.; Floková, K.; Pǎcurar, D.I.; Novák, O.; Staswick, P.; Kowalczyk, M.; Pǎcurar, M.; Demailly, H.; Geiss, G.; et al. Auxin controls Arabidopsis adventitious root initiation by regulating jasmonic acid homeostasis. Plant Cell 2012, 24, 2515–2527. [Google Scholar] [CrossRef]
- Mao, J.; Zhang, D.; Meng, Y.; Li, K.; Wang, H.; Han, M. Inhibition of adventitious root development in apple rootstocks by cytokinin is based on its suppression of adventitious root primordia formation. Physiol. Plant. 2019, 166, 663–676. [Google Scholar] [CrossRef]
- Guan, L.; Tayengwa, R.; Cheng, Z.M.; Peer, W.A.; Murphy, A.S.; Zhao, M. Auxin regulates adventitious root formation in tomato cuttings. BMC Plant Biol. 2019, 19, 435. [Google Scholar] [CrossRef]
- Ramirez-Carvajal, G.A.; Morse, A.M.; Dervinis, C.; Davis, J.M. The cytokinin type-B response regulator PtRR13 is a negative regulator of adventitious root development in populus 1[C][W][OA]. Plant Physiol. 2009, 150, 759–771. [Google Scholar] [CrossRef]
- Kuroha, T.; Ueguchi, C.; Sakakibara, H.; Satoh, S. Cytokinin receptors are required for normal development of auxin-transporting vascular tissues in the hypocotyl but not in adventitious roots. Plant Cell Physiol. 2006, 47, 234–243. [Google Scholar] [CrossRef]
- Soh, W.Y.; Choi, P.S.; Cho, D.Y. Effects of cytokinin on adventitious root formation in callus cultures of Vigna unguiculata (L.) walp. Vitr. Cell. Dev. Biol. Plant 1998, 34, 189–195. [Google Scholar] [CrossRef]
- Li, Y.; Wu, Y.; Liao, W.; Hu, L.; Dawuda, M.M.; Jin, X.; Tang, Z.; Yang, J.; Yu, J. Nitric oxide is involved in the brassinolide-induced adventitious root development in cucumber. BMC Plant Biol. 2020, 20, 102. [Google Scholar] [CrossRef]
- Lischweski, S.; Muchow, A.; Guthörl, D.; Hause, B. Jasmonates act positively in adventitious root formation in petunia cuttings. BMC Plant Biol. 2015, 15, 229. [Google Scholar] [CrossRef]
- Druege, U.; Hilo, A.; Pérez-Pérez, J.M.; Klopotek, Y.; Acosta, M.; Shahinnia, F.; Zerche, S.; Franken, P.; Hajirezaei, M.R. Molecular and physiological control of adventitious rooting in cuttings: Phytohormone action meets resource allocation. Ann. Bot. 2019, 123, 929–949. [Google Scholar] [CrossRef] [PubMed]
- Fattorini, L.; Hause, B.; Gutierrez, L.; Veloccia, A.; Della Rovere, F.; Piacentini, D.; Falasca, G.; Altamura, M.M. Jasmonate promotes auxin-induced adventitious rooting in dark-grown Arabidopsis thaliana seedlings and stem thin cell layers by a cross-talk with ethylene signalling and a modulation of xylogenesis. BMC Plant Biol. 2018. [Google Scholar] [CrossRef]
- Lakehal, A.; Dob, A.; Novák, O.; Bellini, C. A DAO1-mediated circuit controls auxin and jasmonate crosstalk robustness during adventitious root initiation in Arabidopsis. Int. J. Mol. Sci. 2019, 20, 4428. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Zhu, C.; Ma, X.; Li, G.; Gan, L.; Ng, D.; Xia, K. Hydrogen peroxide is a second messenger in the salicylic acid-triggered adventitious rooting process in mung bean seedlings. PLoS ONE 2013, 8, e84580. [Google Scholar] [CrossRef] [PubMed]
- Grierson, C.; Nielsen, E.; Ketelaarc, T.; Schiefelbein, J. Root hairs. Arab. Book 2014. [Google Scholar] [CrossRef]
- Mangano, S.; Denita-Juarez, S.P.; Choi, H.S.; Marzol, E.; Hwang, Y.; Ranocha, P.; Velasquez, S.M.; Borassi, C.; Barberini, M.L.; Aptekmann, A.A.; et al. Molecular link between auxin and ROS-mediated polar growth. Proc. Natl. Acad. Sci. USA 2017, 114, 5289–5294. [Google Scholar] [CrossRef] [PubMed]
- Bhosale, R.; Giri, J.; Pandey, B.K.; Giehl, R.F.H.; Hartmann, A.; Traini, R.; Truskina, J.; Leftley, N.; Hanlon, M.; Swarup, K.; et al. A mechanistic framework for auxin dependent Arabidopsis root hair elongation to low external phosphate. Nat. Commun. 2018, 9, 1409. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.-S.; Seo, M.; Cho, H.-T. Two TPL-binding motifs of ARF2 are involved in repression of auxin responses. Front. Plant Sci. 2018, 9, 372. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.D.W.; Cho, H.T. Auxin, the organizer of the hormonal/environmental signals for root hair growth. Front. Plant Sci. 2013, 4, 448. [Google Scholar] [CrossRef]
- Dindas, J.; Scherzer, S.; Roelfsema, M.R.G.; Von Meyer, K.; Müller, H.M.; Al-Rasheid, K.A.S.; Palme, K.; Dietrich, P.; Becker, D.; Bennett, M.J.; et al. AUX1-mediated root hair auxin influx governs SCFTIR1/AFB-type Ca2+ signaling. Nat. Commun. 2018, 9, 1174. [Google Scholar] [CrossRef]
- Jones, A.R.; Kramer, E.M.; Knox, K.; Swarup, R.; Bennett, M.J.; Lazarus, C.M.; Leyser, H.M.O.; Grierson, C.S. Auxin transport through non-hair cells sustains root-hair development. Nat. Cell Biol. 2009, 11, 78–84. [Google Scholar] [CrossRef]
- Raya-González, J.; Oropeza-Aburto, A.; López-Bucio, J.S.; Guevara-García, Á.A.; de Veylder, L.; López-Bucio, J.; Herrera-Estrella, L. MEDIATOR18 influences Arabidopsis root architecture, represses auxin signaling and is a critical factor for cell viability in root meristems. Plant J. 2018, 96, 895–909. [Google Scholar] [CrossRef]
- Yuan, X.; Xu, P.; Yu, Y.; Xiong, Y. Glucose-TOR signaling regulates PIN2 stability to orchestrate auxin gradient and cell expansion in Arabidopsis root. Proc. Natl. Acad. Sci. USA 2020, 117, 32223–32225. [Google Scholar] [CrossRef]
- Retzer, K.; Weckwerth, W. The tor-auxin connection upstream of root hair growth. Plants 2021, 10, 150. [Google Scholar] [CrossRef]
- Boivin, S.; Fonouni-Farde, C.; Frugier, F. How auxin and cytokinin phytohormones modulate root microbe interactions. Front. Plant Sci. 2016, 7, 1240. [Google Scholar] [CrossRef] [PubMed]
- Jardinaud, M.F.; Boivin, S.; Rodde, N.; Catrice, O.; Kisiala, A.; Lepage, A.; Moreau, S.; Roux, B.; Cottret, L.; Sallet, E.; et al. A laser dissection-RNAseq analysis highlights the activation of cytokinin pathways by nod factors in the Medicago truncatula root epidermis. Plant Physiol. 2016, 171, 2256–2276. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Huang, L.; Yan, A.; Liu, Y.; Liu, B.; Yu, C.; Zhang, A.; Schiefelbein, J.; Gan, Y. Multiple phytohormones promote root hair elongation by regulating a similar set of genes in the root epidermis in Arabidopsis. J. Exp. Bot. 2016, 67, 6363–6372. [Google Scholar] [CrossRef] [PubMed]
- An, L.; Zhou, Z.; Sun, L.; Yan, A.; Xi, W.; Yu, N.; Cai, W.; Chen, X.; Yu, H.; Schiefelbein, J.; et al. A zinc finger protein gene ZFP5 integrates phytohormone signaling to control root hair development in Arabidopsis. Plant J. 2012, 72, 474–490. [Google Scholar] [CrossRef] [PubMed]
- Wada, T.; Kurata, T.; Tominaga, R.; Koshino-Kimura, Y.; Tachibana, T.; Goto, K.; Marks, M.D.; Shimura, Y.; Okada, K. Role of a positive regulator of root hair development, CAPRICE, in Arabidopsis root epidermal cell differentiation. Development 2002, 129, 5409–5419. [Google Scholar] [CrossRef]
- Kuppusamy, K.T.; Chen, A.Y.; Nemhauser, J.L. Steroids are required for epidermal cell fate establishment in Arabidopsis roots. Proc. Natl. Acad. Sci. USA 2009, 106, 8073–8076. [Google Scholar] [CrossRef]
- Fridman, Y.; Elkouby, L.; Holland, N.; Vragović, K.; Elbaum, R.; Savaldi-Goldstein, S. Root growth is modulated by differential hormonal sensitivity in neighboring cells. Genes Dev. 2014, 28, 912–920. [Google Scholar] [CrossRef]
- Cheng, Y.; Zhu, W.; Chen, Y.; Ito, S.; Asami, T.; Wang, X. Brassinosteroids control root epidermal cell fate via direct regulation of a MYB-bHLH-WD40 complex by GSK3-like kinases. eLife 2014, 2014. [Google Scholar] [CrossRef]
- Kang, Y.H.; Breda, A.; Hardtke, C.S. Brassinosteroid signaling directs formative cell divisions and protophloem differentiation in Arabidopsis root meristems. Development 2017, 144, 272–280. [Google Scholar] [CrossRef]
- Zhu, C.; Gan, L.; Shen, Z.; Xia, K. Interactions between jasmonates and ethylene in the regulation of root hair development in Arabidopsis. J. Exp. Bot. 2006, 57, 1299–1308. [Google Scholar] [CrossRef]
- Digby, J.; Firn, R.D. The gravitropic set-point angle (GSA): The identification of an important developmentally controlled variable governing plant architecture. Plant Cell Environ. 1995, 18, 1434–1440. [Google Scholar] [CrossRef]
- Sato, E.M.; Hijazi, H.; Bennett, M.J.; Vissenberg, K.; Swarup, R. New insights into root gravitropic signalling. J. Exp. Bot. 2015, 66, 2155–2165. [Google Scholar] [CrossRef] [PubMed]
- Friml, J.; Wiśniewska, J.; Benková, E.; Mendgen, K.; Palme, K. Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 2002, 415, 806–809. [Google Scholar] [CrossRef] [PubMed]
- Vanneste, S.; Friml, J. Auxin: A trigger for change in plant development. Cell 2009, 136, 1005–1016. [Google Scholar] [CrossRef] [PubMed]
- Kleine-Vehn, J.; Ding, Z.; Jones, A.R.; Tasaka, M.; Morita, M.T.; Friml, J. Gravity-induced PIN transcytosis for polarization of auxin fluxes in gravity-sensing root cells. Proc. Natl. Acad. Sci. USA 2010, 107, 22344–22349. [Google Scholar] [CrossRef] [PubMed]
- Nziengui, H.; Lasok, H.; Kochersperger, P.; Ruperti, B.; Rébeillé, F.; Palme, K.; Ditengou, F.A. Root gravitropism is regulated by a crosstalk between para-aminobenzoic acid, ethylene, and auxin. Plant Physiol. 2018, 178, 1370–1389. [Google Scholar] [CrossRef]
- Aloni, R.; Aloni, E.; Langhans, M.; Ullrich, C.I. Role of cytokinin and auxin in shaping root architecture: Regulating vascular differentiation, lateral root initiation, root apical dominance and root gravitropism. Ann. Bot. 2006, 97, 883–893. [Google Scholar] [CrossRef]
- Pernisova, M.; Prat, T.; Grones, P.; Harustiakova, D.; Matonohova, M.; Spichal, L.; Nodzynski, T.; Friml, J.; Hejatko, J. Cytokinins influence root gravitropism via differential regulation of auxin transporter expression and localization in Arabidopsis. New Phytol. 2016, 212, 497–509. [Google Scholar] [CrossRef]
- Miyawaki, K.; Matsumoto-Kitano, M.; Kakimoto, T. Expression of cytokinin biosynthetic isopentenyltransferase genes in Arabidopsis: Tissue specificity and regulation by auxin, cytokinin, and nitrate. Plant J. 2004, 37, 128–138. [Google Scholar] [CrossRef]
- Aloni, R.; Langhans, M.; Aloni, E.; Ullrich, C.I. Role of cytokinin in the regulation of root gravitropism. Planta 2004, 220, 177–182. [Google Scholar] [CrossRef]
- Kim, S.K.; Chang, S.C.; Lee, E.J.; Chung, W.S.; Kim, Y.S.; Hwang, S.; Lee, J.S. Involvement of brassinosteroids in the gravitropic response of primary root of maize. Plant Physiol. 2000, 123, 997–1004. [Google Scholar] [CrossRef]
- Li, L.; Xu, J.; Xu, Z.H.; Xue, H.W. Brassinosteroids stimulate plant tropisms through modulation of polar auxin transport in Brassica and Arabidopsis. Plant Cell 2005, 17, 2738–2753. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.-W.; Lee, S.M.; Joo, S.-H.; Yun, H.S.; Lee, Y.; Kaufman, P.B.; Kirakosyan, A.; Kim, S.-H.; Nam, K.H.; Lee, J.S.; et al. Elongation and gravitropic responses of Arabidopsis roots are regulated by brassinolide and IAA. Plant Cell Environ. 2007, 30, 679–689. [Google Scholar] [CrossRef] [PubMed]
- Lanza, M.; Garcia-Ponce, B.; Castrillo, G.; Catarecha, P.; Sauer, M.; Rodriguez-Serrano, M.; Páez-García, A.; Sánchez-Bermejo, E.; Tc, M.; Leo del Puerto, Y.; et al. Role of actin cytoskeleton in brassinosteroid signaling and in its integration with the auxin response in plants. Dev. Cell 2012, 22, 1275–1285. [Google Scholar] [CrossRef] [PubMed]
- Retzer, K.; Akhmanova, M.; Konstantinova, N.; Malínská, K.; Leitner, J.; Petrášek, J.; Luschnig, C. Brassinosteroid signaling delimits root gravitropism via sorting of the Arabidopsis PIN2 auxin transporter. Nat. Commun. 2019, 10, 5516. [Google Scholar] [CrossRef]
- Singh, M.; Gupta, A.; Laxmi, A. Glucose control of root growth direction in Arabidopsis thaliana. J. Exp. Bot. 2014, 65, 2981–2993. [Google Scholar] [CrossRef]
- Trevisan, S.; Forestan, C.; Brojanigo, S.; Quaggiotti, S.; Varotto, S. Brassinosteroid application affects the growth and gravitropic response of maize by regulating gene expression in the roots, shoots and leaves. Plant Growth Regul. 2020, 92, 117–130. [Google Scholar] [CrossRef]
- Gutjahr, C.; Riemann, M.; Müller, A.; Düchting, P.; Weiler, E.W.; Nick, P. Cholodny–Went revisited: A role for jasmonate in gravitropism of rice coleoptiles. Planta 2005, 222, 575–585. [Google Scholar] [CrossRef]
- Staswick, P.E. The tryptophan conjugates of jasmonic and indole-3-acetic acids are endogenous auxin inhibitors. Plant Physiol. 2009, 150, 1310–1321. [Google Scholar] [CrossRef]
- Sun, J.; Chen, Q.; Qi, L.; Jiang, H.; Li, S.; Xu, Y.; Liu, F.; Zhou, W.; Pan, J.; Li, X.; et al. Jasmonate modulates endocytosis and plasma membrane accumulation of the Arabidopsis pin2 protein. New Phytol. 2011, 191, 360–375. [Google Scholar] [CrossRef]
- Du, Y.; Tejos, R.; Beck, M.; Himschoot, E.; Li, H.; Robatzek, S.; Vanneste, S.; Friml, J. Salicylic acid interferes with clathrin-mediated endocytic protein trafficking. Proc. Natl. Acad. Sci. USA 2013, 110, 7946–7951. [Google Scholar] [CrossRef]
- Ke, M.; Ma, Z.; Wang, D.; Sun, Y.; Wen, C.; Huang, D.; Chen, Z.; Yang, L.; Tan, S.; Li, R.; et al. Salicylic acid regulates PIN2 auxin transporter hyperclustering and root gravitropic growth via Remorin-dependent lipid nanodomain organisation in Arabidopsis thaliana. New Phytol. 2021, 229, 963–978. [Google Scholar] [CrossRef]
- Rosquete, M.R.; Von Wangenheim, D.; Barbez, E.; Stelzer, E.H.K.; Maizel, A.; Benkova, E. An auxin transport mechanism restricts positive orthogravitropism in lateral roots. Curr. Biol. 2013, 23, 817–822. [Google Scholar] [CrossRef] [PubMed]
- Roychoudhry, S.; Del Bianco, M.; Kieffer, M.; Kepinski, S. Auxin controls gravitropic setpoint angle in higher plant lateral branches. Curr. Biol. 2013, 23, 1497–1504. [Google Scholar] [CrossRef] [PubMed]
- Rosquete, M.R.; Waidmann, S.; Kleine-Vehn, J. PIN7 auxin carrier has a preferential role in terminating radial root expansion in Arabidopsis thaliana. Int. J. Mol. Sci. 2018, 19, 1238. [Google Scholar] [CrossRef]
- Taniguchi, M.; Furutani, M.; Nishimura, T.; Nakamura, M.; Fushita, T.; Iijima, K.; Baba, K.; Tanaka, H.; Toyota, M.; Tasaka, M.; et al. The Arabidopsis LAZY1 family plays a key role in gravity signaling within statocytes and in branch angle control of roots and shoots. Plant Cell 2017, 29, 1984–1999. [Google Scholar] [CrossRef]
- Yoshihara, T.; Spalding, E.P. LAZY genes mediate the effects of gravity on auxin gradients and plant architecture. Plant. Physiol. 2017, 175, 959–969. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Sharma, M.; Jamsheer, M.; Laxmi, A. Jasmonic acid coordinates with light to regulate branching angle of Arabidopsis lateral roots. bioRxiv 2020. [Google Scholar] [CrossRef]
- Dong, Q.K.; Zhang, Z.W.; Liu, Y.T.; Tao, L.Z.; Liu, H.L. FERONIA regulates auxin-mediated lateral root development and primary root gravitropism. FEBS Lett. 2019, 593, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Ge, L.; Chen, R. Negative gravitropic response of roots directs auxin flow to control root gravitropism. Plant. Cell Environ. 2019, 42, 2372–2383. [Google Scholar] [CrossRef]
- Chen, Y.; Xu, S.; Tian, L.; Liu, L.; Huang, M.; Xu, X.; Song, G.; Wu, P.; Sato, S.; Jiang, H.; et al. LAZY3 plays a pivotal role in positive root gravitropism in Lotus japonicus. J. Exp. Bot. 2020, 71, 168–177. [Google Scholar] [CrossRef]
- Guseman, J.M.; Webb, K.; Srinivasan, C.; Dardick, C. DRO1 influences root system architecture in Arabidopsis and Prunus species. Plant J. 2017, 89, 1093–1105. [Google Scholar] [CrossRef] [PubMed]
- Waite, J.M.; Collum, T.D.; Dardick, C. AtDRO1 is nuclear localized in root tips under native conditions and impacts auxin localization. Plant Mol. Biol. 2020, 103, 197–210. [Google Scholar] [CrossRef]
- Semeradova, H.; Montesinos, J.C.; Benkova, E. All roads lead to auxin: Post-translational regulation of auxin transport by multiple hormonal pathways. Plant Commun. 2020, 1, 100048. [Google Scholar] [CrossRef] [PubMed]
- Postma, J.A.; Lynch, J.P. Root cortical aerenchyma enhances the growth of maize on soils with suboptimal availability of nitrogen, phosphorus, and potassium. Plant Physiol. 2011, 156, 1190–1201. [Google Scholar] [CrossRef]
- Mooney, S.J.; Pridmore, T.P.; Helliwell, J.; Bennett, M.J. Developing X-ray computed tomography to non-invasively image 3-D root systems architecture in soil. Plant Soil 2012, 352, 1–22. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).