Abstract
Casein kinase I (CK1), a ubiquitous serine/threonine (Ser/Thr) protein kinase in eukaryotes, plays pivotal roles in a wide spectrum of cellular functions including metabolism, cell cycle progression, developmental control and stress response. Plant CK1 evolves a lineage expansion, resulting in a unique branch of members exclusive to the kingdom. Among them, Arabidopsis Mut9p-LIKE KINASEs (MLKs) target diverse substrates including histones and the key regulatory proteins involving in physiological processes of light signaling, circadian rhythms, phytohormone and plant defense. Deregulation of the kinase activity by mutating the enzyme or the phosphorylation sites of substrates causes developmental disorders and susceptibility to adverse environmental conditions. Recent findings suggest that MLKs have evolved as a general kinase that modifies transcription factors or primary regulatory proteins in a dynamic way. Here, we summarize the current knowledge of the roles of MLKs and MLK orthologs in several commercially important crops.
1. Introduction
Casein kinase I (CK1), a highly conserved serine/threonine (Ser/Thr) protein kinase, has been identified ubiquitously in eukaryotes ranging from yeast to human since the 1970s [1,2,3,4]. The dual-specificity Ser/Thr kinases evolutionary conserved on both structure and function in eukaryotes have been identified as a family of monomeric kinases [5,6,7,8,9]. In mammals, seven CK1 isoforms (alpha, beta, gamma1-3, delta and epsilon), together with the various splice variants, are involved in a variety of cellular processes, such as chromosome segregation and cellular differentiation, by phosphorylating a wide range of key regulatory proteins [9,10,11,12,13].
Plant CK1 evolved an ancient lineage duplication event, resulting in the division of CK1 into two subsets, i.e., casein kinase 1 like (CKL) and a unique group with members exclusively from plant species [14]. The latter subset has attracted great attention from plant scientists in the last decade, and functional characterization of several members from model plants and crops has revealed their essentiality for plant growth and development. Among them, Arabidopsis Mut9p-LIKE KINASEs (MLKs) appear to act as a general kinase to orchestrate a wide range of developmental and stress response pathways, such as light signaling, circadian rhythms and phytohormone [3,4,15,16,17,18].
The purpose of this review is to provide an overview of the roles of plant CK1, especially the MLK family members in growth and development. We summarize the regulation of CK1 expression and the kinase activity, and the phosphorylation target sites, as far as the available literature permits. In addition, multiple CK1-modulated pathways are discussed with a focus on light signaling, phytohormone, circadian clock and stress response based on the recent research achievements of CK1 in model plants Arabidopsis and rice.
2. Plant-Specific CK1 in Arabidopsis and Major Crops
Plant genomes as compared with other eukaryotic organisms encode a large number of CK1 protein kinases [19]. For example, Arabidopsis and rice, model dicot and monocot, have 17 and 15 CK1 encoding genes, respectively [20,21]. These members are grouped phylogenetically into two main clusters, i.e., CK1-like (CKL) cluster and a plant-specific CK1 cluster containing members exclusively from plants [14,21]. For the latter cluster, the first characterized member was Mut9p in the green alga Chlamydomonas reinhardtii [14]. Four Arabidopsis (MLK1-4) and six rice CK1s belong to the cluster [21]. The MLK homologs in angiosperm including amborellales, chlorophyta, lycopodiophyta, monocot and dicot showed a tendency to increase from lower to higher plants (Table 1), suggesting an expansion during evolution probably due to the sessile life. This review described mainly the recent findings of MLKs and MLK orthologs in rice and several crops.

Table 1.
The number of Ser/Thr kinases and MLK homologs in the angiosperm species.
Phylogenetically apart from CKL protein kinases, the MLK homologs in higher plants are evolutionarily conserved (>60% identity) and the region of homology is largely limited to the catalytic motif containing the canonical domains of eukaryotic protein kinases [14]. The Ser/Thr kinase domain contains putative kinase catalytic loop, substrate recognition region and ATP-binding site [22,23], which is supportive of the finding that the plant CK1 uses ATP as phosphate donor to phosphorylate substrate proteins [24]. Interestingly, the N-terminal extension with varying length (from 20 amino acids to up to 410 amino acids), which is absent in animal CKI isomers, differs from one another [14,16] (Figure 1A), whereas the carboxyl (C)-terminal non-catalytic region (about 280 amino acids) is highly conserved (Figure 1B). In contrast, mammalian CKI family members possess a highly conserved amino (N)-terminal domain, but a variable C-terminal extension [23]. Both MLK homologs and human CKI are positively charged, suggesting that these kinases prefer acidic substrates such as serine and threonine residues.
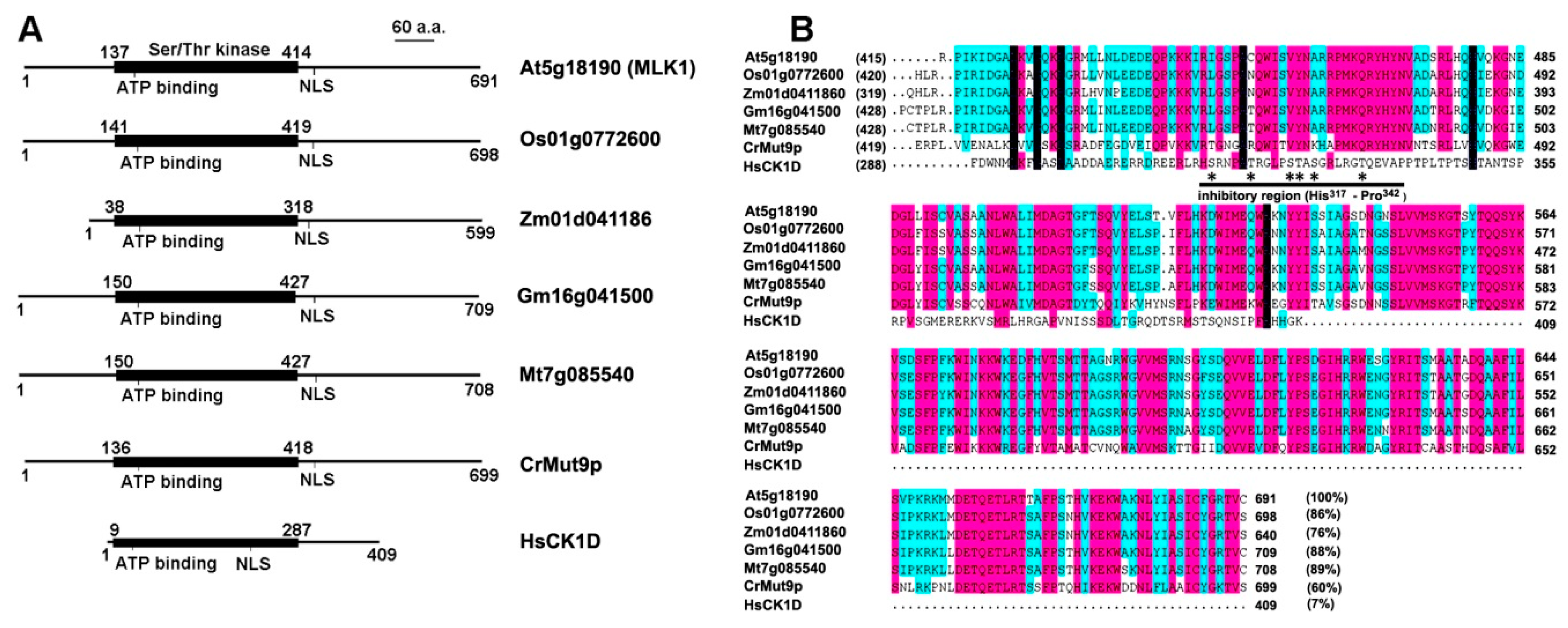
Figure 1.
Functional domains and the C-terminal sequence alignment of MLK homologs with human CK1 delta (CK1D). (A) Functional domains of CK1 in plants and human. The CK1 family members share a highly conserved (63% to 92% identity) catalytic domain (Ser/Thr kinase domain) of about 280 amino acids. In addition to the kinase domain, the representative kinases contain an ATP-binding site and a nuclear localization signal (NLS); (B) Alignment of the C-terminal sequence (the non-catalytic region). The sequence identity of the C-terminal segment to MLK1 (At5g18190) is bracketed. The inhibitory region of human CK1D is underlined (from histone 317 to proline 342), and the potential phosphorylation sites are indicated with star. At, Arabidopsis thaliana; Cr, Chlamydomonas reinhardtii; Gm, Glycine max; Hs, Homo sapiens; Mt, Medicago truncatula; Os, Oryza sativa; Zm, Zea mays.
3. Regulation of CK1s Expression and Activity
Although MLKs split from CKLs [21,25], they appear to share similar catalytic structure and mechanism. Similar to animal CK1 enzymes which have been described as constitutively active [24], the spatial and temporal expression pattern of MLKs and rice homologs showed that these genes were ubiquitously expressed in all tissues at different developmental stages (http://bbc.botany.utoronto.ca/efp) [3,26]. The catalytic activity of CK1 relies on the conserved amino acid residues which are critical for the three-dimensional structure. The kinase domain of MLK homologs is about 60% identical to that of CKL and the predicted three-dimensional structure resembles the typical CK1 [16]. The structural model of Mut9p uncovered a group of residues determining the Mut9p specificity in association with the histone H3 tail. Among them, Asp 266 (D266) at the position equivalent to the substrate-binding pocket of CK1 was associated with phosphorylatable histone residue T3 [14]. Mutation of the conserved Asp, D267 in the case of MLK4/ PPK1 (photoregulatory protein kinases), caused the elimination of in vitro catalytic activity of MLK4/PPK1 [16]. Moreover, an invariant Lys residue (K174 of Mut9p or K175 of MLK4/PPK1), which is implicated in anchoring and orienting the ATP phosphate donor [27], was indispensable for in vitro kinase activity in the green alga and Arabidopsis, respectively [14,16]. It is likely that the abolishment of kinase activity by replacing the conserved amino acids is attributed to the alteration of the enzyme structure or substrate specificity determination.
CK1 activity is modulated by inhibitors and the effectors affecting the kinase localization and compartmentalization. Several ATP-competitive small molecules have been identified and characterized as CK1-specific inhibitors in animals [28]. Among them, CK1-7 (N-2-aminoethyl-5-chloroisoquinoline-8-sulfonamide) is the first ATP-competitive inhibitor with no selectivity towards CK1 isoforms [29]. Application of CKI-7 has effectively eliminated the catalytic activity of CK1 in a wide spectrum of plant species, such as rice, broccoli and Chlamydomonas [14,30,31]. PF670462, a highly CK1-selective inhibitor, and small molecule IC261 have been applied in investigation of CKL-mediated circadian rhythm [7,32]. Recent chemical screening has demonstrated that PHA767491, an animal CDC7 (cell division control protein 7) inhibitor, and analogs, such as AMI-23, -212 and -331, inhibited CK1 activity [20,33]. Interestingly, mammalian CK1 activity was also affected by inhibitory auto-phosphorylation occurring especially in the highly divergent C-terminal domain and occasionally in the kinase domain. C-terminus truncation elevated kinase activity [34], suggesting the inhibitory effect of auto-phosphorylation was overcome. In spite of a long C-terminus with more potential phosphosites than the non-plant species, MLKs lack the inhibitory auto-phosphorylation domain [16], implying that plants have a distinct or more complicated self-regulation mechanism. Regardless of the report that the nuclear plant kinases were also localized in the cytosol [35], Mut9p, MLK1-4 and EL1 (early flowering 1) have been experimentally shown to reside exclusively in the nucleus [3,16,22,36,37]. These findings suggest that MLK homologs modulate phosphorylation-dependent regulations predominantly in the nucleus, where kinase and substrate are in close proximity.
4. Target Proteins Phosphorylated by MLK Homologs
Considerable efforts have been made to uncover plant phosphoproteomes, but the phosphorylation events provided are far from complete due to the fact that phosphoproteomic approaches including in vivo and in vitro methods are limited in precisely localizing the phosphorylation sites on the proteins. Several databases such as PhosPhAt [38] and P3DB [39] provide information of mass spectrometry-based phosphorylation sites identified or predicted from several plant species. Currently, 56, 857 phosphorylation sites accounting for only 2.6% of the predicted ones including serine, threonine and tyrosine sites have been experimentally determined in Arabidopsis using PhosPhAt 4.0 [40] (http://phosphat.uni-hohenheim.de/statistics.html). Further verification of these phosphorylation sites would be beneficial for the functional characterization of the phosphorylation events. With the rapid development of methodology for protein phosphorylation site(s) identification, a new generation MS instrument, such as quadrupole Orbitrap high-resolution Mass spectrometry, has been used to target hundreds of peptides within one single LC-MS/MS experiment and generated full MS/MS spectra containing sufficient fragmentations for precise localization of the phosphorylated residues [16,41].
As plant CK1 is ubiquitously expressed in all tissues and the protein is localized in the nucleus and/or the cytoplasm, it is not surprising that a broad range of proteins including histones and signaling component proteins have been identified as CK1 targets in model plants and crops. Table 2 enlisted an exhaustive list of CK1 substrates identified in the angiosperm during the last decade.

Table 2.
Summary of the CK1 substrates identified in plants.
4.1. Histones Targeted by MLK Famliy Members
Histones are the main protein components of chromatin, a tightly packed higher order structure in the eukaryote nucleus. The basic structure unit of chromatin is the nucleosome that is composed of 147 bp DNA wrapped around an octamer consisting of two copies of each of the four core histones (H3, H4, H2A and H2B) linked by histone H1 [42]. In addition to the globular domain, each of the core histones has a tail protruding from the nucleosome and most of the post-translational modifications including phosphorylation occur at these amino-terminal tails (reviewed by [43,44,45]). As a reversible process, phosphorylation is one of the most important ways of modulating a chromatin structure and accessibility by regulating the switches between condensed and relaxed states of the chromatin higher order structure (reviewed by [46,47,48]).
Identification of upstream kinases and the target site(s) of individual enzyme is complicated due to the fact that there are numerous candidate kinases responsible for histone phosphorylation and that multiple sites of plant histones are potential phosphorylation targets. In plants, histone phosphorylation modulated by the canonical kinases, such as AtAurora3 and AtHaspin, has been recently reviewed [43,46,49]. In vitro assay showed that Chlamydomonus Mut9p and Arabidopsis MLK1 and MLK4 phosphorylated histones including H3, H2A and H4, albeit at different intensity [14,22,37]. Partial depletion of phosphorylated H3 at T3 (H3T3ph) was observed in both mut9 mutant strain and Arabidopsis mlk1mlk2 double mutant [14,22], confirming that Mut9p and MLK1/MLK2 are responsible for in vivo H3T3 phosphorylation in the green alga and Arabidopsis, respectively. The existence of other kinases, especially the paralogos, may explain the incomplete loss of H3T3ph in the mutants. Additionally, MLK4 was reported to phosphorylate H2AS95 residue in vitro, while the global level of phosphorylated H2AS95 did not alter clearly in the loss-of-function mlk4 mutant [37], implying that H2AS95 is not likely the main target of MLK4 in Arabidopsis. Collectively, histones are phosphorylated by MLK homologos in model plants Chlamydomonus and Arabidopsis.
4.2. Signaling Components Targeted by Plant CK1
In addition to the conserved histone residues, plant CK1 including CKL and MLK family members have been predicted, based on structural analysis, to have a broad range of substrates. For example, a rice lipase and an integral negative regulatory domain (NRD) of dehydration-responsive element-binding protein 2A (DREB2A) in Arabidopsis were potential phosphorylation targets of CKL protein kinases [50,51]. Arabidopsis CKL3/CK1.3 and CKL4/CK1.4 phosphorylated photoreceptor CRY2 (CRYPTOCHROME 2) at S587 and T603 in vitro in blue-light dependent manner [21], and CKL2 physically interacted with and phosphorylated ADF4 (actin depolymerizing factor 4) in the regulation of actin filament reorganization and stomatal closure [52]. Recently, CKL4 was found to regulate circadian clock by phosphorylating the transcriptional repressor PRR5 (PSEUDO-RESPONSE REGULATIOR 5) and TOC1 (TIMING OF CAB EXPRESSION 1) in Arabidopsis [20]. In sesame and cotton, CKL protein was involved in phosphorylation of SebHLH (the basic region/helix-loop-helix transcriptional factor) [17] and GhTCP15 (TEOSINTE BRANCHED1/CYCLOIDEA/PCF 15) [53], respectively. In the last decade, extensive studies have focused on the functions and regulatory mechanisms of MLK family members in model plant Arabidopsis and rice, and a wide range of substrates have been identified primarily as transcription factors or regulatory proteins involving in basic physiological processes (Table 2).
5. Biological Functions of Plant CK1
Phosphorylation of an amino acid residue can have multiple consequences, including change of enzyme activity, subcellular localization, or protein stability, which affect the interaction with other molecules (protein, DNA, RNA, etc.) and eventually alter the biological functions. In non-plant eukaryotes, the CK1-dependent phosphorylation has been shown to play diverse biological roles [9,11], but the research of plant CK1 lags far behind. The main reason is that compared with animals, plants have a larger number of CK1 encoding genes, which share a high degree of redundancy in both the catalytic and regulatory domains relative to the animal systems. A steadily growing list of both CK1 substrates and the interacting partners adds another level of complexity in dissecting the kinase functions in plants.
An increasing number of proteins involving in plant growth and stress response have been confirmed as MLK targets in Arabidopsis via mutating the kinase activity site or the phosphosites of the target proteins (reviewed in [57,58,59]). These findings suggest that MLKs act as a general kinase in a myriad of cellular processes of plant growth and development. Indeed, mlk quadruple mutant is lethal [16,60], supporting the vital nature of MLKs in Arabidopsis. The role of the canonical plant CK1 in phosphorylation-dependent regulation of chromatin and cell cycle has been reviewed [43,46,49]. We described, here, the recent findings on the functions of plant CK1, especially MLK family members in light signaling, circadian rhythm, phytohormone and stress response. The information provides a connection among the molecular events mediated by plant CK1 through direct/indirect modification or interaction with the target proteins.
5.1. Light Signaling
Plants perceive and response to light signal, a major developmental cue, using multiple families of photoreceptors including phytochromes and cryptochromes, which absorb red/far-red light and blue light, respectively. It has been documented that the stability of CRY2 was negatively regulated by CKL-mediated phosphorylation [21,61] and light signaling components, including HY5 (long hypocotyle 5), HFR1 (long hypocotyl in far-red 1), COP1 (CONSTITUTIVE PHOTOMORPHOGENIC 1) and PIF1 (PHYTOCHROME-INTERACTING FACTOR 1) were phosphorylation targets of CK2 [5], suggesting that phosphorylation plays critical roles in modulating the photoreceptors as light sensors. Recently, two independent studies have revealed the delicate coaction of phytochrome-cryptochrome by PPK/MLK-mediated phosphorylation of PIF3 and CYR2 [58]. Compared with ppk124 triple mutants, which showed a reduced level of light-induced PIF3 phosphorylation and degradation, more robust reduction was detected in PPKs quadruple-knockdown transgenic lines [15], suggesting that PPKs/MLKs in Arabidopsis collectively promote light-induced degradation of PIF3 by phosphorylation. The triple mutants of ppk123/124/134 also displayed hypersensitivity to red light, which was strongly suppressed by mutating phyB, suggesting the hypersensitivity is attributed to the reduced degradation of phyB [15]. In an accompanying paper, the level of phosphorylated blue-light receptor CRY2 was clearly reduced in the ppk triple mutants with photo-hypersensitivity to blue light. PPKs were also shown to interact with and catalyze the blue light-dependent phosphorylation of CRY2, although individual kinase preferred different phosphosties. Substitution of several Ser/Thr residues at the phosphosites of CRY2 resulted in abolished phosphorylation [16]. Thus, PPKs/MLKs are the major kinases catalyzing the phosphorylation of the two types of photoreceptors in Arabidopsis. Notably, the binding affinity of PPK1 with PIF3 was not affected by deletion of the C-terminal domain of PPK1 [15]. However, the C-terminal of PPKs is indispensable for the interaction with CYR2 in response to blue light [16]. It is tempting to speculate that the C-terminal domain of PPKs is likely to contribute to the receptor discrimination of blue-light from red-light as light spectrum shifts. Expressing phosphor-deficient and phosphor-mimic forms of the substrates in the corresponding mutant backgrounds could help to elucidate the function of CK1-mediated phosphorylation in light signaling.
5.2. Circadian Clock
Like most organisms, plants have evolved an internal timing mechanism, the circadian clock, which is an autoregulatory, endogenous biological cycle with a period of approximately 24 h. A functional and accurate clock-dependent synchronization of physiology with the environment allows for efficient allocation of resources and optimum fitness. CK1 has been known to affect timekeeping across metazoans and fungi [9,62]. Early evidence of the implication of plant CK1 in circadian clock was found in a unicellular marine algal species Ostreococcus tauri [7]. The basal alga of the green lineage contained only one CK1 with diurnal fluctuation. Overexpression O. tauri CK1 caused a long-period phenotype and treatment with chemical inhibitor altered circadian period [7].
In higher plants, the clock output processes, such as flowering time and hypocotyl elongation, are clearly affected in CK1-deficient mutants. For example, mutation of rice EL1/Hd16 (heading date 16) caused earlier flowering [3,4]. Hd16 was found to phosphorylate OsPRR37 (Oryza sativa pseudo-response regulator 37), an ortholog of Arabidopsis circadian clock components PRRs, and act upstream of flowering regulators Ghd7 (Grain number, plant height, and heading date 7) and Hd2 [55]. In contrast, Arabidopsis mlk4 and the higher order of mlk4- combined mutant(s) flowered late with retarded elongation of hypocotyl [60,63]. The proteome-wide analysis revealed physical interaction between the rhythmic oscillators and protein kinases in Arabidopsis [60]. Among them, MLKs were shown to interplay with the evening complex components ELF3 (EARLY FLOWERING 3) and ELF4 in the assay of affinity purification and mass spectrometry (AP-MS) [60]. MLK1 and MLK2 regulated hypocotyl elongation by interacting with CCA1 (CIRCADIAN CLOCK ASSOCIATED1), which bound to the promoter of DWF4 (DWARF4) [63]. MLK4 interplayed with CCA1 to affect the expression of GIGANTEA (GI), a positive flowering regulator [37]. Moreover, CKL4 phosphorylated the clock regulators, PRR5 and TOC1, and the circadian period of Arabidopsis was lengthened by simultaneous knockdown CKLs [20]. In prr5 toc1 mutants the period-lengthening effect was attenuated by inhibition of CK1 [64].
Therefore, plant CK1 plays a critical role in regulating the circadian clock via posttranslational modification of the central clock regulators CCA1 and PRRs (including TOC1), as it does in animal systems. Future studies aiming to decipher the interacting network between the circadian components targeted by plant CK1 should broaden our knowledge of the kinase in the epigenetic regulation of circadian rhythm.
5.3. Phytohormone
Phytohormones are generally small organic molecules that regulate both the physiological and developmental processes of plants through a complex network of signal transduction pathways. This section described the recent findings on the CK1-mediated phosphorylation of signal transduction components and the downstream proteins of phytohormone gibberellin (GA), auxin and abscisic acid (ABA).
An increasing body of experimental findings has demonstrated the involvement of CK1 in GA signaling pathway. Mutation of rice EL1 caused enhanced GA response with taller plant height. EL1 phosphorylated rice DELLA protein SLR1 (SLENDER RICE 1) and mutation at its phosphorylation site altered GA signaling [3]. In contrast, Arabidopsis mlk1mlk2 double mutant with shorter hypocotyl was hyposensitive to GA. Although the phosphorylation target(s) of MLK1/MLK2 are not known, both kinases were found to interact with RGA, a DELLA protein REPRESSOR OF ga1-3, to regulate hypocotyl elongation [63]. These observations suggest that CK1 plays a critical role in regulating the activity or turnover of GA-responsive transcription factors.
CK1 has been implicated to regulate auxin synthesis or signaling pathways in plants. For example, rice OsCKI1 knockdown mutants displayed abnormal root development with altered auxin content [31]. Comparison of two rice cultivars, Asominori and NIL (ltg1), representing dominant LTG1 (Low Temperature Growth 1) allele with functional CKL and the ltg1 allele, respectively, revealed that the latter possessed higher content of IAA (indole-3-acetic acid) than the former. Consistently, the shoot growth of NIL (ltg1) was less sensitive to NAA (1-naphthylacetic acid) treatment and was less affected by the auxin transport inhibitor NPA (N-1-naphthylphthalamic acid) relative to Asominori [65], suggesting that rice CKL encoding gene LTG1 affects plant growth through an auxin-dependent process. Research on cotton CKL homolog showed that compared with Col-0, the IAA content in GhCK1 overexpressing Arabidopsis explants was higher, while lower in GhCKI RNAi line, suggesting a correlation between GhCK1 expression level and IAA content [18]. GhTCP15 was identified as the substrate of GhCK1, and its phosphorylation affected IAA content by modulating the transcription of GhPIF4 [53]. Further identification and functional characterization of CK1 substrates relevant to auxin synthesis or transport should help in understanding the mechanism of CK1-mediated phosphorylation in auxin signaling.
ABA is an important phytohormone in plant response to adverse environmental conditions by regulating stomata opening and closure. CK1-mediated phosphorylation of the transcriptional factors involving in ABA signaling appears to play a role in plant stress response or adaption. Early research found that application of ABA induced SeCK1 transcription in developing sesame seeds [17]. Deletion of Arabidopsis CKL2 caused stomatal closure less sensitive to ABA and phosphorylation of ADF4 (actin depolymerizing factor 4) by CKL2 inhibited actin filament disassembly [52]. Recently, MLKs/AELs (Arabidopsis EL1-like) have been reported to phosphorylate ABA recepetors PYR/PYLs (PYRABACTIN RESISTANCE/PYR1-LIKE). Deduced phosphorylation level of the receptors mimicked the hypersensitivity of mlk triple mutants to ABA treatment [2], indicating that MLK-mediated phosphorylation negatively regulates ABA response. Thus, the altered sensitivity of the CK1-defective mutants to ABA is supportive of the implication of CK1 in ABA signal transduction although how these ABA-involving processes are connected has yet to be explored.
As phytohormones act through a complex network of signal transduction pathways, it is not surprising that mutation of the ubiquitous plant CK1 altered plant response to multiple relevant phytohormones. Rice OsCKI1 knockdown mutant displayed abnormal root development with altered auxin content and was also hyposensitive to brassinosteroid (BR) and ABA treatment during germination. Consequently, in OsCKI1-deficient plants, the expression level of the genes related to hormone metabolism and signal transduction was altered [31]. Supportively, an investigation of the global phosphorylation dynamics regulated by BRs using mass spectrometry (MS)-based phosphoproteomics revealed that most phosphoproteins had strong connection with BR signaling components, and many substrates were the downstream components of auxin and ABA signaling [66]. Further verification of these proteins would help to elucidate the role of CK1-mediated phosphorylation in BR, which may bridge the downstream events among the phytohormone signaling pathways.
5.4. Plant Stress Response
Due to the sessile lifestyle, plant establishes its stress defense at the level of protection and adaption. Chromatin alterations in chromatin composition and histone modification are emerging as integral and complex elements of stress responses in plants [67,68]. Post-translational modifications of histones including histone phosphorylation appear to be an obvious response of plant chromatin to stress signals. For example, when cultured BY-2 cells of tobacco were exposed to sucrose or NaCl stress, the phosphorylation of histone H3 at S10 and T3 was increased [69], and the dynamic changes of histone were found to be correlated with the upregulation of some stress-inducible genes [25,70,71]. In recent years, CK1 has been found to affect chromatin substructures by phosphorylating histone residue(s). Arabidopsis mlk1mlk2 double mutant with diminished level of H3T3 phosphorylation was hypersensitive to osmotic stress including salt and drought. mlk1mlk2 exhibited partial decondensation in pericentromeric/knob heterochromatin, and about one third of MLK1/MLK2-dependent genes were stress-related according to the annotation [22]. These findings suggest that changes of MLK-mediated histone phosphorylation occur globally, which provides a direct link between CK1-mediated phosphorylation of histone H3 and stress response.
In addition to global phosphorylation, CK1 has been reported to be potentially responsible for phosphorylation of the negative regulatory domain (NRD) of DREB2A under normal growth conditions, which in turn facilitates the degradation of DREB2A. Upon heat stress, phosphorylation NRD of DREB2A was decreased, and application of a highly CK1-selective inhibitor caused the accumulation of DREB2A in a dose-dependent manner [50], suggesting that CK1-mediated phosphorylation is likely implicated in response to heat stress by targeting DREB2A. Additionally, Wirthmuelle et al. found that salicylic acid (SA)-induced defense marker genes were strongly upregulated in triple mutant mlk1, 3, 4, implying that MLKs affect plant immunity via transcriptional regulation of SA signaling [72].
Plant CKL genes from several species are involved in response to environmental cues, especially extreme temperature. In a high temperature (HT)-sensitive cotton line, for instance, GhCK1 was induced by HT and overexpression of GhCK1 resulted in elevated ABA levels [18]. At the early anther development stage, AtCKL2 and AtCKL7 were HT-induced due to the binding of AtMYB24 at the promoters [73]. In contrast, a rice dominant allele of LTG1, which encodes a CKL protein, conferred low temperature (LT) tolerance, while the recessive ltg1 allele was hypersensitive to LT [65]. It would be promising to identify the main signaling factors which act as direct targets or functional partners of CK1 in stress response.
6. Concluding Remarks
Plants, owing to the sessile nature of their lifestyle, have a large number of CK1 encoding genes including both the canonical CK1 family members and the CK1s unique to the kingdom. The latter ones play fundamental roles in various cellular, physiological and developmental processes by phosphorylating or interacting with target proteins. During the last decade, research has been mainly focused on the regulatory mechanisms of CK1 in model plants. The findings have considerably advanced our understanding of the genetic regulation network mediated by CK1, however, there are still many questions to answer in order to address how plant CK1 functions. For example, what specifies the plant-specific CK1 from animal CK1? It would be promising to explore the roles of the C-terminus, which is distinct from that of the animals. As some nuclear protein CK1s also reside in the cytoplasm, identification of the non-nuclear targets would be likely to provide insights on CK1 functions in cytoplasm. CK1s, especially the MLK homologs, are implicated in multiple signaling pathways, but how they orchestrate these pathways is poorly understood. Further investigation of CK1 substrates and interacting partners would help to unveil the biological roles of CK1 in plant growth and development.
Author Contributions
J.K. and Z.W. wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financed by the Agricultural Science and Technology Innovation Program (ASTIP-IAS-TS-14) and the National Natural Science Foundation of China (31772663) to Z.W.
Acknowledgments
We thank our lab members for helpful discussion and critical reading of the manuscript. Our apology goes to all authors whose work could not be cited due to the limitation of space.
Conflicts of Interest
The authors declare that they have no competing interests.
Abbreviations
| AEL | Arabidopsis early-flowering 1-like |
| CCA1 | CIRCADIAN CLOCK-ASSOCIATED 1 |
| CDC7 | Cell division control protein 7 |
| CK1 | Casein kinase I |
| CRY2 | CRYPTOCHROME 2 |
| DREB2A | Dehydration-responsive element-binding protein 2A |
| ELF3/4 | EARLY FLOWERING 3/4 |
| Ghd7 | Grain number, plant height, and heading date 7 |
| H3T3ph | Phosphorylation of histone H3 at threonine 3 |
| Hd16 | Heading date 16 |
| LTG1 | Low temperature growth 1 |
| MLK | MUT9-like kinase |
| PIF | PHYTOCHROME INTERACTING FACTOR |
| PPK | Photoregulatory protein kinase |
| PRR | PSEUDO-RESPONSE REGULATOR |
| PYR1/PYL | PYRABACTIN RESISTANCE1/PYR1-LIKE |
| SLR1 | SLENDER RICE 1 |
| TCP15 | TEOSINTE BRANCHED1-CYCLOIDEA-PCF transcription factor 15 |
| TOC1 | TIMING OF CAB EXPRESSION 1 |
References
- Kumar, R.; Tao, M. Multiple forms of casein kinase from rabbit erythrocytes. Biochim. Biophys. Acta 1975, 410, 87–98. [Google Scholar] [CrossRef]
- Chen, H.H.; Qu, L.; Xu, Z.H.; Zhu, J.K.; Xue, H.W. El1-like casein kinases suppress aba signaling and responses by phosphorylating and destabilizing the aba receptors pyr/pyls in arabidopsis. Mol. Plant 2018, 11, 706–719. [Google Scholar] [CrossRef]
- Dai, C.; Xue, H.W. Rice early flowering1, a cki, phosphorylates della protein slr1 to negatively regulate gibberellin signalling. Embo J. 2010, 29, 1916–1927. [Google Scholar] [CrossRef]
- Hori, K.; Ogiso-Tanaka, E.; Matsubara, K.; Yamanouchi, U.; Ebana, K.; Yano, M. Hd16, a gene for casein kinase i, is involved in the control of rice flowering time by modulating the day-length response. Plant. J. 2013, 76, 36–46. [Google Scholar] [CrossRef]
- Mulekar, J.J.; Huq, E. Expanding roles of protein kinase ck2 in regulating plant growth and development. J. Exp. Bot. 2014, 65, 2883–2893. [Google Scholar] [CrossRef]
- Lee, J.Y. Versatile casein kinase 1: Multiple locations and functions. Plant. Signal. Behav. 2009, 4, 652–654. [Google Scholar] [CrossRef]
- van Ooijen, G.; Hindle, M.; Martin, S.F.; Barrios-Llerena, M.; Sanchez, F.; Bouget, F.Y.; O’Neill, J.S.; Le Bihan, T.; Millar, A.J. Functional analysis of casein kinase 1 in a minimal circadian system. PLoS ONE 2013, 8, e70021. [Google Scholar] [CrossRef]
- Schittek, B.; Sinnberg, T. Biological functions of casein kinase 1 isoforms and putative roles in tumorigenesis. Mol. Cancer 2014, 13, 231. [Google Scholar] [CrossRef]
- Knippschild, U.; Gocht, A.; Wolff, S.; Huber, N.; Lohler, J.; Stoter, M. The casein kinase 1 family: Participation in multiple cellular processes in eukaryotes. Cell. Signal. 2005, 17, 675–689. [Google Scholar] [CrossRef]
- Wang, X.; Hoekstra, M.F.; DeMaggio, A.J.; Dhillon, N.; Vancura, A.; Kuret, J.; Johnston, G.C.; Singer, R.A. Prenylated isoforms of yeast casein kinase i, including the novel yck3p, suppress the gcs1 blockage of cell proliferation from stationary phase. Mol. Cell. Biol. 1996, 16, 5375–5385. [Google Scholar] [CrossRef]
- Gross, S.D.; Anderson, R.A. Casein kinase i: Spatial organization and positioning of a multifunctional protein kinase family. Cell. Signal. 1998, 10, 699–711. [Google Scholar] [CrossRef]
- Zhai, L.; Graves, P.R.; Robinson, L.C.; Italiano, M.; Culbertson, M.R.; Rowles, J.; Cobb, M.H.; DePaoli-Roach, A.A.; Roach, P.J. Casein kinase i gamma subfamily. Molecular cloning, expression, and characterization of three mammalian isoforms and complementation of defects in the saccharomyces cerevisiae yck genes. J. Biol. Chem. 1271, 270, 12717–12724. [Google Scholar] [CrossRef]
- Green, C.L.; Bennett, G.S. Identification of four alternatively spliced isoforms of chicken casein kinase i alpha that are all expressed in diverse cell types. Gene 1998, 216, 189–195. [Google Scholar] [CrossRef]
- Casas-Mollano, J.A.; Jeong, B.R.; Xu, J.; Moriyama, H.; Cerutti, H. The mut9p kinase phosphorylates histone h3 threonine 3 and is necessary for heritable epigenetic silencing in chlamydomonas. Proc. Natl. Acad. Sci. USA 2008, 105, 6486–6491. [Google Scholar] [CrossRef]
- Ni, W.; Xu, S.L.; Gonzalez-Grandio, E.; Chalkley, R.J.; Huhmer, A.F.R.; Burlingame, A.L.; Wang, Z.Y.; Quail, P.H. Ppks mediate direct signal transfer from phytochrome photoreceptors to transcription factor pif3. Nat. Commun. 2017, 8, 15236. [Google Scholar] [CrossRef]
- Liu, Q.; Wang, Q.; Deng, W.; Wang, X.; Piao, M.; Cai, D.; Li, Y.; Barshop, W.D.; Yu, X.; Zhou, T.; et al. Molecular basis for blue light-dependent phosphorylation of arabidopsis cryptochrome 2. Nat. Commun. 2017, 8, 15234. [Google Scholar] [CrossRef]
- Kim, M.J.; Go, Y.S.; Lee, S.B.; Kim, Y.S.; Shin, J.S.; Min, M.K.; Hwang, I.; Suh, M.C. Seed-expressed casein kinase i acts as a positive regulator of the sefad2 promoter via phosphorylation of the sebhlh transcription factor. Plant. Mol. Biol. 2010, 73, 425–437. [Google Scholar] [CrossRef]
- Min, L.; Zhu, L.; Tu, L.; Deng, F.; Yuan, D.; Zhang, X. Cotton ghcki disrupts normal male reproduction by delaying tapetum programmed cell death via inactivating starch synthase. Plant. J. 2013, 75, 823–835. [Google Scholar] [CrossRef]
- Lehti-Shiu, M.D.; Shiu, S.H. Diversity, classification and function of the plant protein kinase superfamily. Philos. Trans. R. Soc. London. Ser. Biol. Sci. 2012, 367, 2619–2639. [Google Scholar] [CrossRef]
- Uehara, T.N.; Mizutani, Y.; Kuwata, K.; Hirota, T.; Sato, A.; Mizoi, J.; Takao, S.; Matsuo, H.; Suzuki, T.; Ito, S.; et al. Casein kinase 1 family regulates prr5 and toc1 in the arabidopsis circadian clock. Proc. Natl. Acad. Sci. USA 2019, 116, 11528–11536. [Google Scholar] [CrossRef]
- Tan, S.T.; Dai, C.; Liu, H.T.; Xue, H.W. Arabidopsis casein kinase1 proteins ck1.3 and ck1.4 phosphorylate cryptochrome2 to regulate blue light signaling. Plant. Cell 2013, 25, 2618–2632. [Google Scholar] [CrossRef]
- Wang, Z.; Casas-Mollano, J.A.; Xu, J.; Riethoven, J.J.; Zhang, C.; Cerutti, H. Osmotic stress induces phosphorylation of histone h3 at threonine 3 in pericentromeric regions of arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2015, 112, 8487–8492. [Google Scholar] [CrossRef]
- Cheong, J.K.; Virshup, D.M. Casein kinase 1: Complexity in the family. Int. J. Biochem. Cell Biol. 2011, 43, 465–469. [Google Scholar] [CrossRef]
- Tuazon, P.T.; Traugh, J.A. Casein kinase i and ii--multipotential serine protein kinases: Structure, function, and regulation. Adv. Second Messenger Phosphoprot. Res. 1991, 23, 123–164. [Google Scholar]
- Cerutti, H.; Casas-Mollano, J.A. Histone h3 phosphorylation: Universal code or lineage specific dialects? Epigenetics 2009, 4, 71–75. [Google Scholar] [CrossRef]
- Winter, D.; Vinegar, B.; Nahal, H.; Ammar, R.; Wilson, G.V.; Provart, N.J. An "electronic fluorescent pictograph" browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2007, 2, e718. [Google Scholar] [CrossRef]
- Hanks, S.K.; Hunter, T. The eukaryotic protein kinase superfamily: Kinase (catalytic) domain structure and classification. Faseb J. 1995, 9, 576–596. [Google Scholar] [CrossRef]
- Knippschild, U.; Kruger, M.; Richter, J.; Xu, P.; Garcia-Reyes, B.; Peifer, C.; Halekotte, J.; Bakulev, V.; Bischof, J. The ck1 family: Contribution to cellular stress response and its role in carcinogenesis. Front. Oncol. 2014, 4, 96. [Google Scholar] [CrossRef]
- Chijiwa, T.; Hagiwara, M.; Hidaka, H. A newly synthesized selective casein kinase i inhibitor, n-(2-aminoethyl)-5-chloroisoquinoline-8-sulfonamide, and affinity purification of casein kinase i from bovine testis. J. Biol. Chem. 1989, 264, 4924–4927. [Google Scholar]
- Klimczak, L.J.; Cashmore, A.R. Purification and characterization of casein kinase i from broccoli. Biochem. J. 1993, 293, 283–288. [Google Scholar] [CrossRef]
- Liu, W.; Xu, Z.H.; Luo, D.; Xue, H.W. Roles of oscki1, a rice casein kinase i, in root development and plant hormone sensitivity. Plant. J. 2003, 36, 189–202. [Google Scholar] [CrossRef] [PubMed]
- Badura, L.; Swanson, T.; Adamowicz, W.; Adams, J.; Cianfrogna, J.; Fisher, K.; Holland, J.; Kleiman, R.; Nelson, F.; Reynolds, L.; et al. An inhibitor of casein kinase i epsilon induces phase delays in circadian rhythms under free-running and entrained conditions. J. Pharmacol. Exp. Ther. 2007, 322, 730–738. [Google Scholar] [CrossRef]
- Saito, A.N.; Matsuo, H.; Kuwata, K.; Ono, A.; Kinoshita, T.; Yamaguchi, J.; Nakamichi, N. Structure-function study of a novel inhibitor of the casein kinase 1 family in arabidopsis thaliana. Plant. Direct 2019, 3, e00172. [Google Scholar] [CrossRef] [PubMed]
- Graves, P.R.; Roach, P.J. Role of cooh-terminal phosphorylation in the regulation of casein kinase i delta. J. Biol. Chem. 1995, 270, 21689–21694. [Google Scholar] [CrossRef] [PubMed]
- Dahan, J.; Wendehenne, D.; Ranjeva, R.; Pugin, A.; Bourque, S. Nuclear protein kinases: Still enigmatic components in plant cell signalling. New Phytol. 2010, 185, 355–368. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Ding, Y. Mlk1 and mlk2 integrate gibberellins and circadian clock signaling to modulate plant growth. Plant. Signal. Behav. 2018, 13, e1439654. [Google Scholar] [CrossRef]
- Su, Y.; Wang, S.; Zhang, F.; Zheng, H.; Liu, Y.; Huang, T.; Ding, Y. Phosphorylation of histone h2a at serine 95: A plant-specific mark involved in flowering time regulation and h2a.Z deposition. Plant. Cell 2017, 29, 2197–2213. [Google Scholar] [CrossRef]
- Heazlewood, J.L.; Durek, P.; Hummel, J.; Selbig, J.; Weckwerth, W.; Walther, D.; Schulze, W.X. Phosphat: A database of phosphorylation sites in arabidopsis thaliana and a plant-specific phosphorylation site predictor. Nucleic Acids Res. 2008, 36, D1015–D1021. [Google Scholar] [CrossRef]
- Gao, J.; Agrawal, G.K.; Thelen, J.J.; Xu, D. P3db: A plant protein phosphorylation database. Nucleic Acids Res. 2009, 37, D960–D962. [Google Scholar] [CrossRef]
- Arsova, B.; Schulze, W.X. Current status of the plant phosphorylation site database phosphat and its use as a resource for molecular plant physiology. Front. Plant Sci. 2012, 3, 132. [Google Scholar] [CrossRef]
- Gallien, S.; Duriez, E.; Crone, C.; Kellmann, M.; Moehring, T.; Domon, B. Targeted proteomic quantification on quadrupole-orbitrap mass spectrometer. Mol. Cell. Proteom. 2012, 11, 1709–1723. [Google Scholar] [CrossRef] [PubMed]
- Luger, K. Structure and dynamic behavior of nucleosomes. Curr. Opin. Genet. Dev. 2003, 13, 127–135. [Google Scholar] [CrossRef]
- Zhang, B.; Dong, Q.; Su, H.; Birchler, J.A.; Han, F. Histone phosphorylation: Its role during cell cycle and centromere identity in plants. Cytogenet. Genome Res. 2014, 143, 144–149. [Google Scholar] [CrossRef] [PubMed]
- Sawicka, A.; Seiser, C. Sensing core histone phosphorylation—A matter of perfect timing. Biochim. Et Biophys. Acta 2014, 1839, 711–718. [Google Scholar] [CrossRef]
- Rossetto, D.; Avvakumov, N.; Cote, J. Histone phosphorylation: A chromatin modification involved in diverse nuclear events. Epigenetics 2012, 7, 1098–1108. [Google Scholar] [CrossRef]
- Bigeard, J.; Rayapuram, N.; Pflieger, D.; Hirt, H. Phosphorylation-dependent regulation of plant chromatin and chromatin-associated proteins. Proteomics 2014, 14, 2127–2140. [Google Scholar] [CrossRef]
- Lopez, R.; Sarg, B.; Lindner, H.; Bartolome, S.; Ponte, I.; Suau, P.; Roque, A. Linker histone partial phosphorylation: Effects on secondary structure and chromatin condensation. Nucleic Acids Res. 2015, 43, 4463–4476. [Google Scholar] [CrossRef]
- Perez-Cadahia, B.; Drobic, B.; Khan, P.; Shivashankar, C.C.; Davie, J.R. Current understanding and importance of histone phosphorylation in regulating chromatin biology. Curr. Opin. Drug Discov. Dev. 2010, 13, 613–622. [Google Scholar]
- Moraes, I.; Casas-Mollano, J.A. Histone h3 phosphorylation in plants and other organisms. In Epigenetics in Plants of Agronomic Importance: Fundamentals and Applications; Alvarez-Venegas, R.l., Penã, C.D.l., Casas-Mollano, J.A., Eds.; Springer: Cham/Heidelberg, Germany; New York, NY, USA; Dordrecht, The Netherlands; London, UK, 2014; pp. 47–70. [Google Scholar]
- Mizoi, J.; Kanazawa, N.; Kidokoro, S.; Takahashi, F.; Qin, F.; Morimoto, K.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Heat-induced inhibition of phosphorylation of the stress-protective transcription factor dreb2a promotes thermotolerance of arabidopsis thaliana. J. Biol. Chem. 2019, 294, 902–917. [Google Scholar] [CrossRef]
- Park, Y.I.; Do, K.H.; Kim, I.S.; Park, H.H. Structural and functional studies of casein kinase i-like protein from rice. Plant. Cell Physiol. 2012, 53, 304–311. [Google Scholar] [CrossRef]
- Zhao, S.; Jiang, Y.; Zhao, Y.; Huang, S.; Yuan, M.; Zhao, Y.; Guo, Y. Casein kinase1-like protein2 regulates actin filament stability and stomatal closure via phosphorylation of actin depolymerizing factor. Plant. Cell 2016, 28, 1422–1439. [Google Scholar] [CrossRef] [PubMed]
- Min, L.; Hu, Q.; Li, Y.; Xu, J.; Ma, Y.; Zhu, L.; Yang, X.; Zhang, X. Leafy cotyledon1-casein kinase i-tcp15-phytochrome interacting factor4 network regulates somatic embryogenesis by regulating auxin homeostasis. Plant. Physiol. 2015, 169, 2805–2821. [Google Scholar] [CrossRef] [PubMed]
- Ni, W.; Xu, S.L.; Chalkley, R.J.; Pham, T.N.; Guan, S.; Maltby, D.A.; Burlingame, A.L.; Wang, Z.Y.; Quail, P.H. Multisite light-induced phosphorylation of the transcription factor pif3 is necessary for both its rapid degradation and concomitant negative feedback modulation of photoreceptor phyb levels in arabidopsis. Plant. Cell 2013, 25, 2679–2698. [Google Scholar] [CrossRef] [PubMed]
- Kwon, C.T.; Koo, B.H.; Kim, D.; Yoo, S.C.; Paek, N.C. Casein kinases i and 2alpha phosphorylate oryza sativa pseudo-response regulator 37 (osprr37) in photoperiodic flowering in rice. Mol. Cells 2015, 38, 81–88. [Google Scholar] [PubMed]
- Park, H.H. Casein kinase i-like protein linked to lipase in plant. Plant. Signal. Behav. 2012, 7, 719–721. [Google Scholar] [CrossRef]
- Pham, V.N.; Kathare, P.K.; Huq, E. Phytochromes and phytochrome interacting factors. Plant. Physiol. 2018, 176, 1025–1038. [Google Scholar] [CrossRef]
- Wang, Q.; Liu, Q.; Wang, X.; Zuo, Z.; Oka, Y.; Lin, C. New insights into the mechanisms of phytochrome-cryptochrome coaction. New Phytol. 2018, 217, 547–551. [Google Scholar] [CrossRef]
- Salome, P.A. Know your histone (zip) code: Flowering time and phosphorylation of histone h2a on serine 95. Plant. Cell 2017, 29, 2084–2085. [Google Scholar] [CrossRef]
- Huang, H.; Alvarez, S.; Bindbeutel, R.; Shen, Z.; Naldrett, M.J.; Evans, B.S.; Briggs, S.P.; Hicks, L.M.; Kay, S.A.; Nusinow, D.A. Identification of evening complex associated proteins in arabidopsis by affinity purification and mass spectrometry. Mol. Cell. Proteom. 2016, 15, 201–217. [Google Scholar] [CrossRef]
- Kim, J.I.; Park, J.E.; Zarate, X.; Song, P.S. Phytochrome phosphorylation in plant light signaling. Photochem. Photobiol. Sci. 2005, 4, 681–687. [Google Scholar] [CrossRef]
- Reischl, S.; Kramer, A. Kinases and phosphatases in the mammalian circadian clock. FEBS Lett. 2011, 585, 1393–1399. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Zhang, F.; Wang, S.; Su, Y.; Jiang, P.; Cheng, R.; Ji, X.; Hou, S.; Ding, Y. Mlk1 and mlk2 coordinate rga and cca1 activity to regulate hypocotyl elongation in arabidopsis thaliana. Plant. Cell 2017, 30, 67–82. [Google Scholar] [CrossRef] [PubMed]
- Ono, A.; Sato, A.; Fujimoto, K.J.; Matsuo, H.; Yanai, T.; Kinoshita, T.; Nakamichi, N. 3, 4-dibromo-7-azaindole modulates arabidopsis circadian clock by inhibiting casein kinase 1 activity. Plant. Cell Physiol. 2019, 60, 2360–2368. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; Wu, F.Q.; Wu, W.; Wang, H.J.; Zheng, X.M.; Zhang, Y.; Chen, X.; Zhou, K.; Jin, M.; Cheng, Z.; et al. Rice ltg1 is involved in adaptive growth and fitness under low ambient temperature. Plant. J. 2014, 78, 468–480. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.L.; Hsu, C.L.; Hu, C.W.; Ko, S.Y.; Hsieh, H.L.; Huang, H.C.; Juan, H.F. Integrating phosphoproteomics and bioinformatics to study brassinosteroid-regulated phosphorylation dynamics in arabidopsis. BMC Genom. 2015, 16, 533. [Google Scholar] [CrossRef]
- Probst, A.V.; Mittelsten Scheid, O. Stress-induced structural changes in plant chromatin. Curr. Opin. Plant. Biol. 2015, 27, 8–16. [Google Scholar] [CrossRef]
- Kim, J.M.; Sasaki, T.; Ueda, M.; Sako, K.; Seki, M. Chromatin changes in response to drought, salinity, heat, and cold stresses in plants. Front. Plant. Sci. 2015, 6, 114. [Google Scholar] [CrossRef]
- Sokol, A.; Kwiatkowska, A.; Jerzmanowski, A.; Prymakowska-Bosak, M. Up-regulation of stress-inducible genes in tobacco and arabidopsis cells in response to abiotic stresses and aba treatment correlates with dynamic changes in histone h3 and h4 modifications. Planta 2007, 227, 245–254. [Google Scholar] [CrossRef]
- Izabel, M.; Casas-Mollano, J.A. Histone h3 phosphorylation in plants and other organisms. In Epigenetics in Plants of Agronomic Importance: Fundamentals and Applications. Transcriptional Regulation and Chromatin Remodelling in Plants; Alvarez-Venegas, R.l., Penã, C.D.l., Casas-Mollano, J.A., Eds.; Springer: London, UK, 2014; pp. 47–65. [Google Scholar]
- Houben, A.; Demidov, D.; Caperta, A.D.; Karimi, R.; Agueci, F.; Vlasenko, L. Phosphorylation of histone h3 in plants—A dynamic affair. Biochim. Biophys. Acta 2007, 1769, 308–315. [Google Scholar] [CrossRef]
- Wirthmueller, L.; Asai, S.; Rallapalli, G.; Sklenar, J.; Fabro, G.; Kim, D.S.; Lintermann, R.; Jaspers, P.; Wrzaczek, M.; Kangasjarvi, J.; et al. Arabidopsis downy mildew effector harxl106 suppresses plant immunity by binding to radical-induced cell death1. New Phytol. 2018, 220, 232–248. [Google Scholar] [CrossRef]
- Li, Y.; Min, L.; Zhang, L.; Hu, Q.; Wu, Y.; Li, J.; Xie, S.; Ma, Y.; Zhang, X.; Zhu, L. Promoters of arabidopsis casein kinase i-like 2 and 7 confer specific high-temperature response in anther. Plant. Mol. Biol. 2018, 98, 33–49. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).