Abstract
Cadmium (Cd) is a toxic heavy metal element. It is relatively easily absorbed by plants and enters the food chain, resulting in human exposure to Cd. Italian ryegrass (Lolium multiflorum Lam.), an important forage cultivated widely in temperate regions worldwide, has the potential to be used in phytoremediation. However, genes regulating Cd translocation and accumulation in this species are not fully understood. Here, we optimized PacBio ISO-seq and integrated it with RNA-seq to construct a de novo full-length transcriptomic database for an un-sequenced autotetraploid species. With the database, we identified 2367 differentially expressed genes (DEGs) and profiled the molecular regulatory pathways of Italian ryegrass with Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis in response to Cd stress. Overexpression of a DEG LmAUX1 in Arabidopsis thaliana significantly enhanced plant Cd concentration. We also unveiled the complexity of alternative splicing (AS) with a genome-free strategy. We reconstructed full-length UniTransModels using the reference transcriptome, and 29.76% of full-length models had more than one isoform. Taken together, the results enhanced our understanding of the genetic diversity and complexity of Italian ryegrass under Cd stress and provided valuable genetic resources for its gene identification and molecular breeding.
1. Introduction
The genus Lolium L., belonging to the Poaceae family, is native to Europe, North Africa, and temperate Asia and has been introduced to almost all temperate regions in the world [1]. Italian ryegrass (L. multiflorum Lam.) and perennial ryegrass (L. perenne L.) are two important species, and both of them are valuable forage grasses. Because of their desirable characteristics, such as high yields, tolerance to grazing, rapid establishment, and high palatability and digestibility for ruminant animals, they have been cultivated through hybrid or molecular breeding in the last hundred years [2]. Moreover, molecular breeding has played an increasingly important role in breeding programs of late [3]. In many plants, it has shown promise in increasing yield as well as resistance to multiple biotic and abiotic stresses. However, molecular breeding depends on the availability of a reference genome. Until now, in species of the Lolium genus, only a draft genome of perennial ryegrass (L. perenne L.) was reported [4]. Additionally, there are still gaps in the genome, which limits the understanding of the molecular regulation mechanism, especially for closely related species, such as Italian ryegrass, so it provides little reference value. Italian ryegrass, also known as annual ryegrass, is used for turf, forage, and quick cover in the event of erosion [5], and it also has some new uses, such as lignocellulosic ethanol conversion programs [6] and phytoremediation [7].
Cadmium (Cd) is a widespread heavy metal element in nature, is highly toxic to almost all plants and animals, and is without any known biological functions [8]. Cd uptake by plants from Cd-contaminated soils happens relatively easily, resulting in human Cd exposure through the food chain [9]. Soil Cd pollution has drawn public attention due to its direct adverse impact on plant growth and the uncertainties regarding the safety of produce [10]. Many previous studies have shown that Cd exposure may cause a series of damages to plants [11], such as lipid peroxidation, enzyme inactivation, an overproduction of reactive oxygen species (ROSs), a disturbed assimilation of essential elements (for example, copper, iron, and zinc), activated programmed cell death (PCD), and membrane and DNA damage [12,13]. Additionally, these damages eventually lead to various toxic phenotypes, such as leaf yellowing, growth reduction, plant withering, or even death [14]. In recent years, common plant mechanisms, in response to Cd stress, have been revealed, such as Cd transporters, Cd cellular compartmentation, and chelation [15]. In Sedum alfredii, a hyperaccumulator, an apoplastic pathway mediated by abscisic acid (ABA), regulated the uptake of Cd [16]; in rice, several transporters participating in the absorption, transport, and distribution of Cd have been characterized, including OsHMA3 [8], OsNramp5 [17], and OsCAL1 [18]; in Arabidopsis thaliana, the ABC-type transporter AtABCC3 induced by Cd increased plant Cd tolerance mediated by phytochelatin [19].
RNA-seq, as a high-throughput next-generation sequencing (NGS) technique, has become an indispensable tool for transcriptome-wide analysis of differential gene expression and gene regulatory networks. It has recently been widely used for researching plants in response to various stresses, including Cd stress, in such plants as switchgrass [20], rice [21], and S. alfredii Hance [22]. However, the limitations of RNA-seq, i.e., short read lengths and amplification biases, restrict researchers to accurately obtain full-length transcripts and differential splicing of mRNA [23], particularly for polyploid species or species lacking a high-quality reference transcriptome [24]. As a third-generation sequencing technique, single-molecule real-time (SMRT) sequencing can overcome these limitations by generating long reads without further assembly. It is highly suitable for isoform discovery, in de novo transcriptome analysis [25]. Isoform sequencing (ISO-seq) from PacBio dependent on the SMRT sequencing platform has been used to analyze full-length transcriptomes in sorghum [23], maize [25], rice [26], Pteris vittata [27], and pepper [28].
Roots act as the first barrier to Cd exposure for plants grown in Cd-contaminated soils. Identifying Cd-inducible genes in Italian ryegrass roots can deepen our understanding of molecular regulatory pathways in response to Cd stress. In this study, we constructed a de novo full-length transcriptomic database for Italian ryegrass root with the ISO-seq technique. Integrated with RNA-seq, we identified the differentially expressed genes (DEGs) and analyzed the regulated networks related to Cd tolerance and translation. The function of a DEG LmAUX1 was validated in A. thaliana. The overexpression of this gene significantly decreases plant Cd tolerance and enhances plant Cd concentration. Thus, it is possible that this gene could be used for regulating Cd accumulation in Italian ryegrass.
2. Results
2.1. Assembly of the Full-Length Reference Transcriptomic Database of Italian Ryegrass
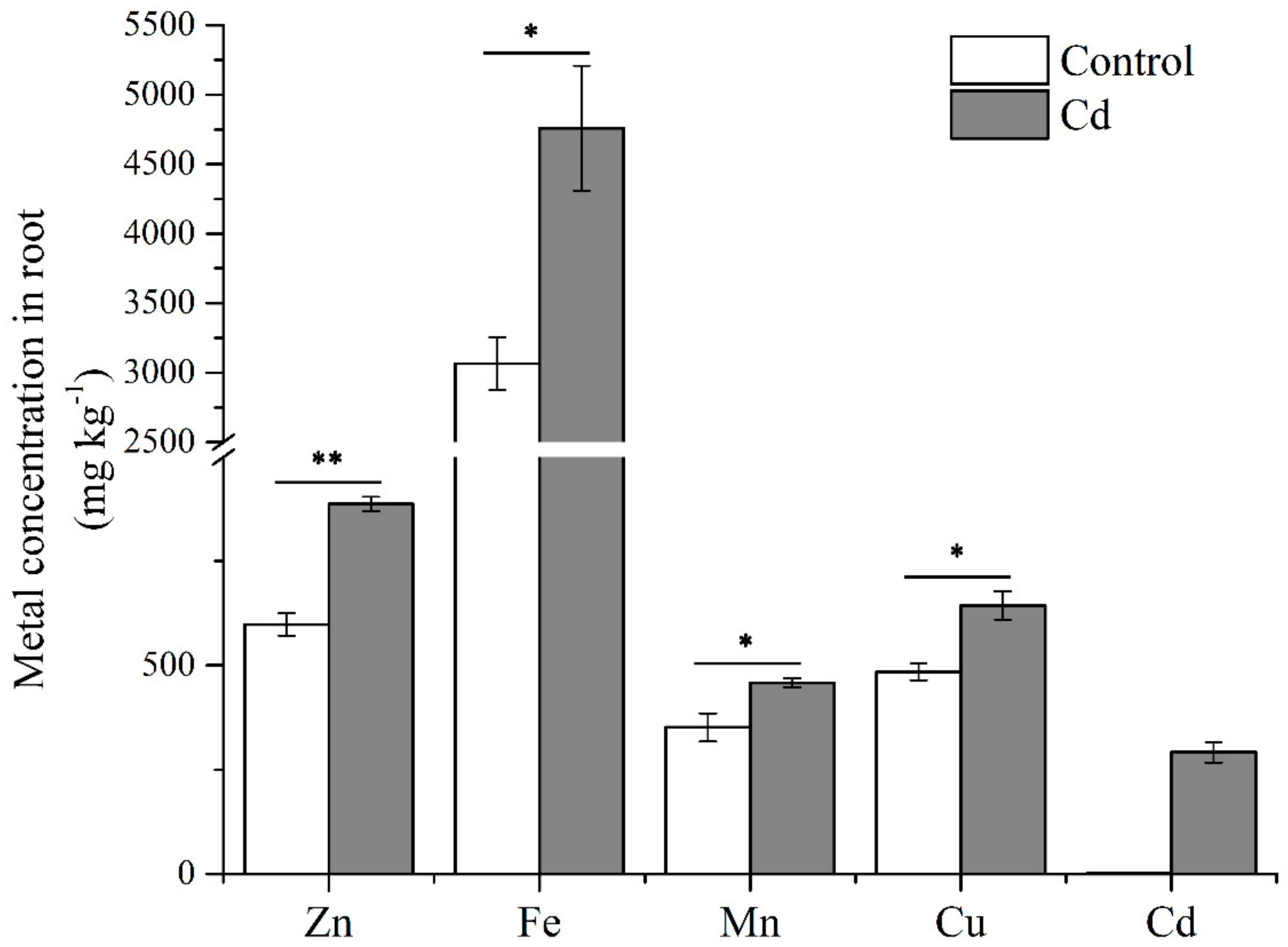
Roots are the first organ to contact Cd stress. To identify gene comportments that foremost regulated plant Cd uptake and tolerance, we treated Italian ryegrass with 50 µmol L−1 Cd for six hours. Metal concentrations of Italian ryegrass roots were significant affected by Cd (Figure 1). The concentrations of Zn, Fe, Mn, and Cu were significantly increased, and the Cd concentration reached 291.60 mg k−1. To profile the transcriptomic information, total RNA of Italian ryegrass roots was extracted after a six-hour treatment with Cd, and three control and three treated samples were then combined into one sample in equal amounts. The poly(A) RNA was enriched from the mixed RNA sample. Subsequently, according to the results of the pre-experiment, the length of Italian ryegrass mRNA was mostly enriched in a range less than 4 kb (Figure S1). We prepared two overlapping cDNA libraries with insert fragments of 1–4 kb and 3–10 kb, and 20 cycles LD-PCR cycles for a 3–10 kb library were amplified (1–4 kb library, 16 cycles). A library was sequenced by a SMRT cell with the PacBio Sequel platform (Pacific Biosciences, Menlo Park, CA, USA). In summary, we obtained 415,985 and 386,341 reads of inserts for the 1–4 kb library and 3–10 kb library, respectively. Furthermore, 69.06% and 40.47% of them were full-length reads, which had poly(A) tail signals, 5′ adaptor sequences, and 3′ adapter sequences. The average full-length non-chimeric (FLNC) reads were 1711 and 3540 base pairs (bps) for the 1–4 kb library and 3–10 kb library, respectively (Table S1). Usually, the FLNC reads of each cDNA library contain repetitive isoforms. According to the isoform-level clustering algorithm (ICE) analysis, the FLNC reads were aligned, and similarity sequences were assigned to a cluster. Each cluster was identified as a uniform isoform. The isoform sequences were then polished and integrated with non-full-length non-chimeric reads. Isoforms with a predicted accuracy of >99% were considered high-quality isoforms, and others were regarded as low-quality. The average lengths of consensus isoform reads were modified to 1756 and 3653 bps in the 1–4 kb and 3–10 kb libraries, respectively (Table S2).
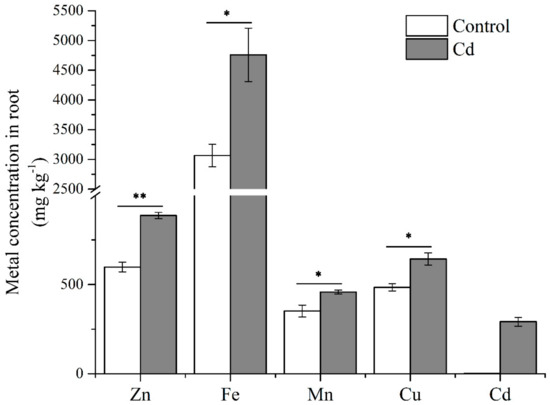
Figure 1.
Metal concentrations of Italian ryegrass roots. Data are means ±SE (n = 3). Asterisks indicate significant differences (* p < 0.05 and ** p < 0.01 by Duncan’s test).
To further improve the accuracy of the PacBio ISO-seq data, we sequenced six samples of the Italian ryegrass root with the same treatment as PacBio ISO-seq on the Illumina Hiseq X Ten platform (Illumina, San Diego, CA, USA) with 150 bp pair-ends. In total, each of RNA-seq samples generated more than 39,340,473 clean reads (Table S3). The low-quality isoforms generated by ICE proofread were corrected with RNA-seq short reads using LSC software (http://augroup.org/LSC/LSC/), and the high-quality isoforms and corrective low-quality isoforms were then combined as whole full-length isoforms. In standard ISO-seq analysis, a transcript may generate different isoforms, and these isoforms may be assigned to different libraries, so we removed redundant isoforms using CD-HIT-EST software (http://www.bioinformatics.org/cd-hit/). Finally, the non-redundant high-quality transcripts were obtained and considered the reference transcriptome with a size of 340 Mb.
2.2. Annotation of the Full Length Reference Transcriptome
All isoforms of the reference transcriptome were aligned to the protein and nucleotide databases including NCBI non-redundant proteins (NR), NCBI non-redundant nucleotides (NT), Swiss-Prot, Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), and KOG. As shown in Table 1, in total, 146,545 isoforms were annotated; there were 123,344 isoforms annotated in the GO database and 72,725 isoforms annotated in the KEGG database. The best harbored isoforms in the reference transcriptome were in the NR database, and 145,825 isoforms hit in the NR database.

Table 1.
Information of function annotation.
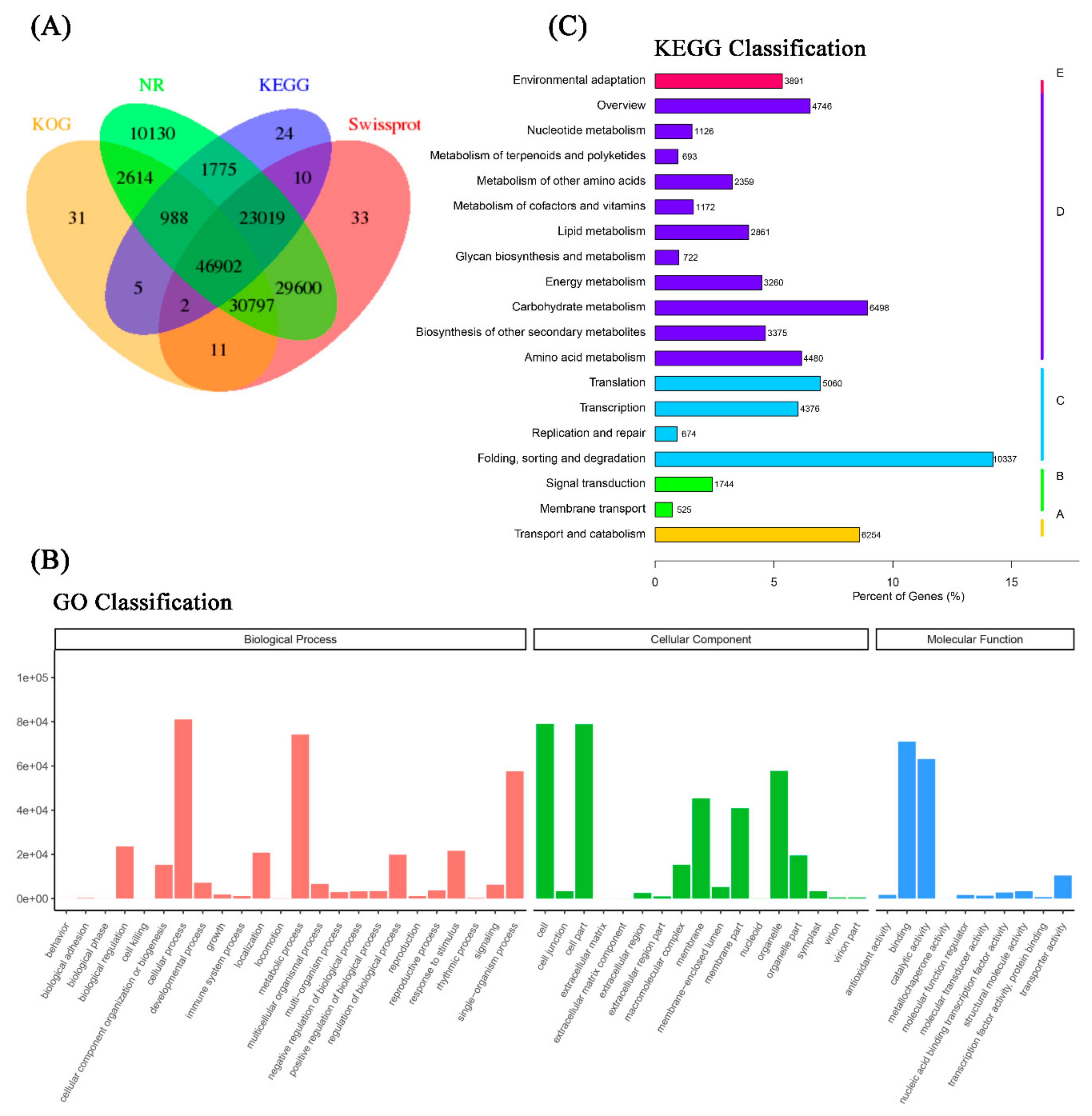
Overall, there were 30,797 isoforms mapped in at least three databases, and 46,902 isoforms hit four databases (KOG, NR, KEGG, and Swiss-Prot) (Figure 2A). For GO analysis, the isoforms were annotated in three categories—biological process (BP, 352,144 isoforms), cellular component (CC, 353,049 isoforms), and molecular function (MF, 156,073 isoforms). Within the functional classifications, cellular process (GO:0009987, 81,045 isoforms) and metabolic process (GO:0008152, 74,167 isoforms) were the two most functional terms in BP; cell (GO:0005623, 79,012 isoforms) and cell part (GO:0044464, 78,846 isoforms) were the two most functional terms in CC. Catalytic activity (GO:0003824, 63,137 isoforms) and binding (GO:0005488, 70,972 isoforms) were the most abundant functional terms in MF (Figure 2B). To further profile the pathways that all isoforms take part in, the KEGG Orthology identifiers (KOs) that annotated isoforms enriched in the KEGG database were classified in five metabolic pathways (Hierarchy 1). In cellular process KOs, the isoforms were mainly associated with transport and catabolism (6254 isoforms, 8.6%) pathways. There were 2269 isoforms (3.1%) that took part in environmental information processing KOs. In genetic information processing KOs, there were 20,447 isoforms (28.1%) that participated, and they were mainly focused on folding, sorting, and degradation pathways. More than 43.0% (31,292) isoforms hit for metabolism KOs, which were involved in many pathways, such as carbohydrate metabolism, amino acid metabolism, and global and overview maps. Further, less than 5.4% (3891) of isoforms were categorized into organismal system KOs and were involved in environmental adaptation pathways (Figure 2C).
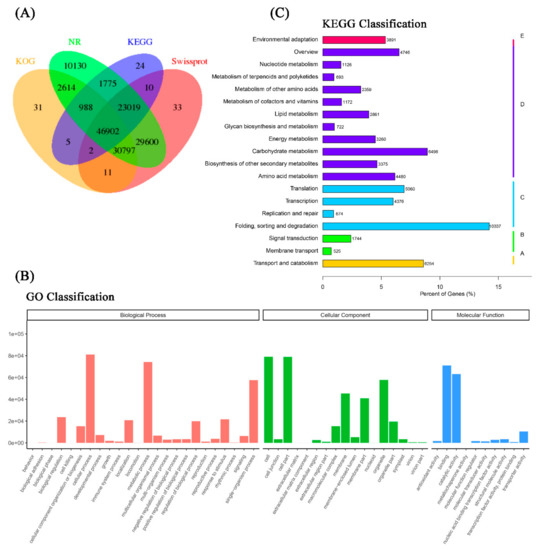
Figure 2.
Functional annotation of the full-length reference transcriptome. (A) Venn diagram of all isoforms from the reference transcriptome hits of KOG, non-redundant proteins (NR), Kyoto Encyclopedia of Genes and Genomes (KEGG), and SwissProt databases. (B) Gene ontology classification of all identified isoforms from the reference transcriptome. (C) KEGG classification of all identified isoforms from the reference transcriptome.
2.3. Identification and Functional Profiles of Differentially Expressed Genes
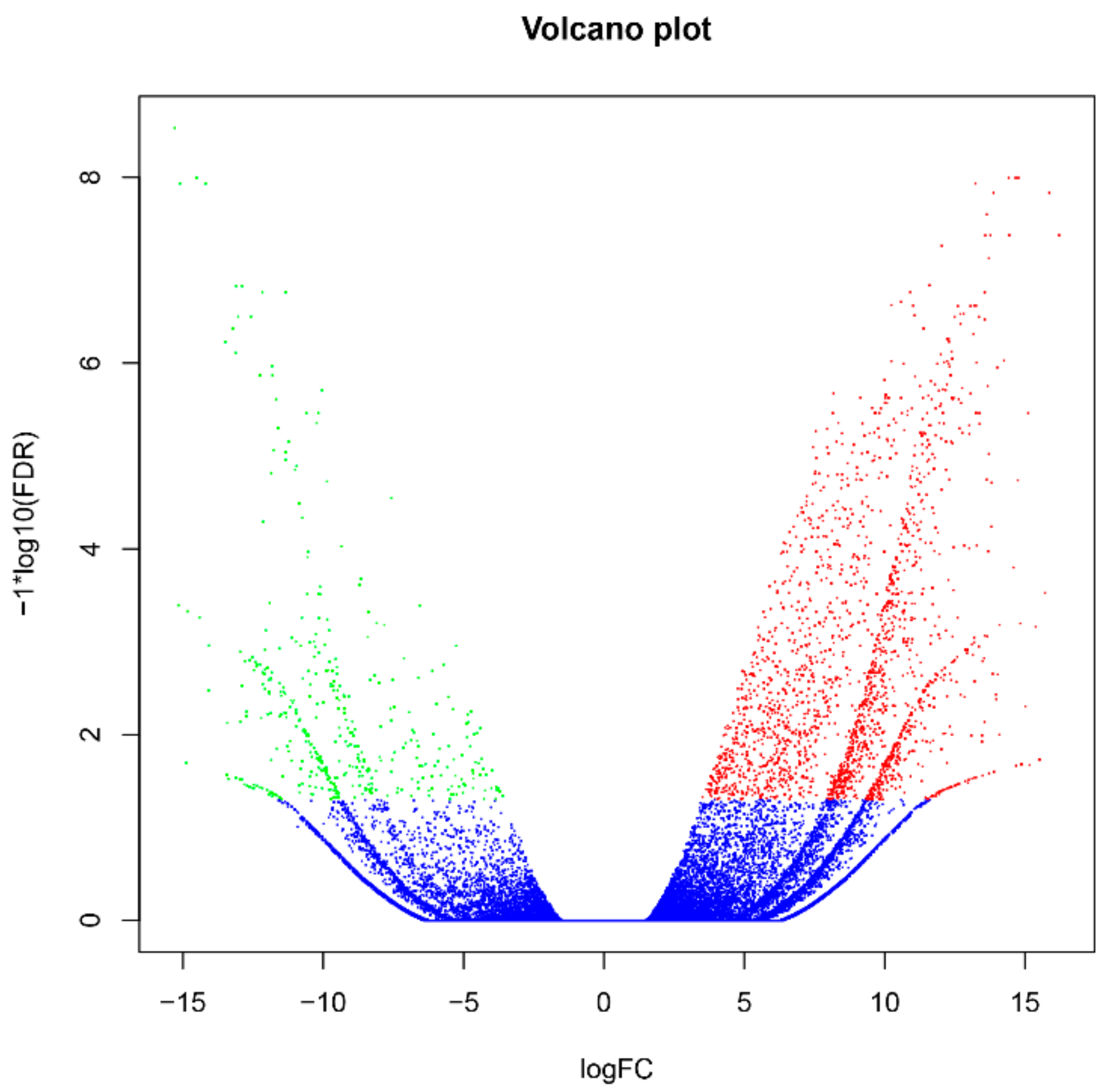
The filtered clean reads from Illumina RNA-seq were mapped to the reference transcriptome generated by PacBio ISO-seq. Of all the reads, 68.73–71.43% were mapped to the reference transcriptome, 9.22–10.29% were uniquely mapped reads, and 98.71–90.78% were multiple aligned reads (Table 2). The expression levels of all isoforms were calculated by RESM software and shown as FPKM values. With the edgeR package, the different expression levels of the isoforms were assessed according to the threshold (FDR values < 0.001). A total of 2367 differentially expressed genes (DEGs) were obtained in response to Cd stress, 1944 DEGs were significantly upregulated, 423 DEGs were significantly downregulated (Figure 3), and relative expression levels of 20 random selected DEGs were further verified using qRT-PCR (Figure S2). Specific primers were designed with Primer Premier 5.0 (PREMIER Biosoft International, Palo Alto, CA, USA) (Table S6).

Table 2.
Information of RNA-seq clean reads mapped with the reference transcriptome.

Figure 3.
Distribution and expression levels of differentially expressed genes (DEGs) in Italian ryegrass roots exposed or not to cadmium (Cd). The x-axis represents the log2 [Fold Change] values under the mean normalized expression of all isoforms (y-axis). Red dots indicate the upregulated DEGs, and green dots represent the down regulated DEGs.
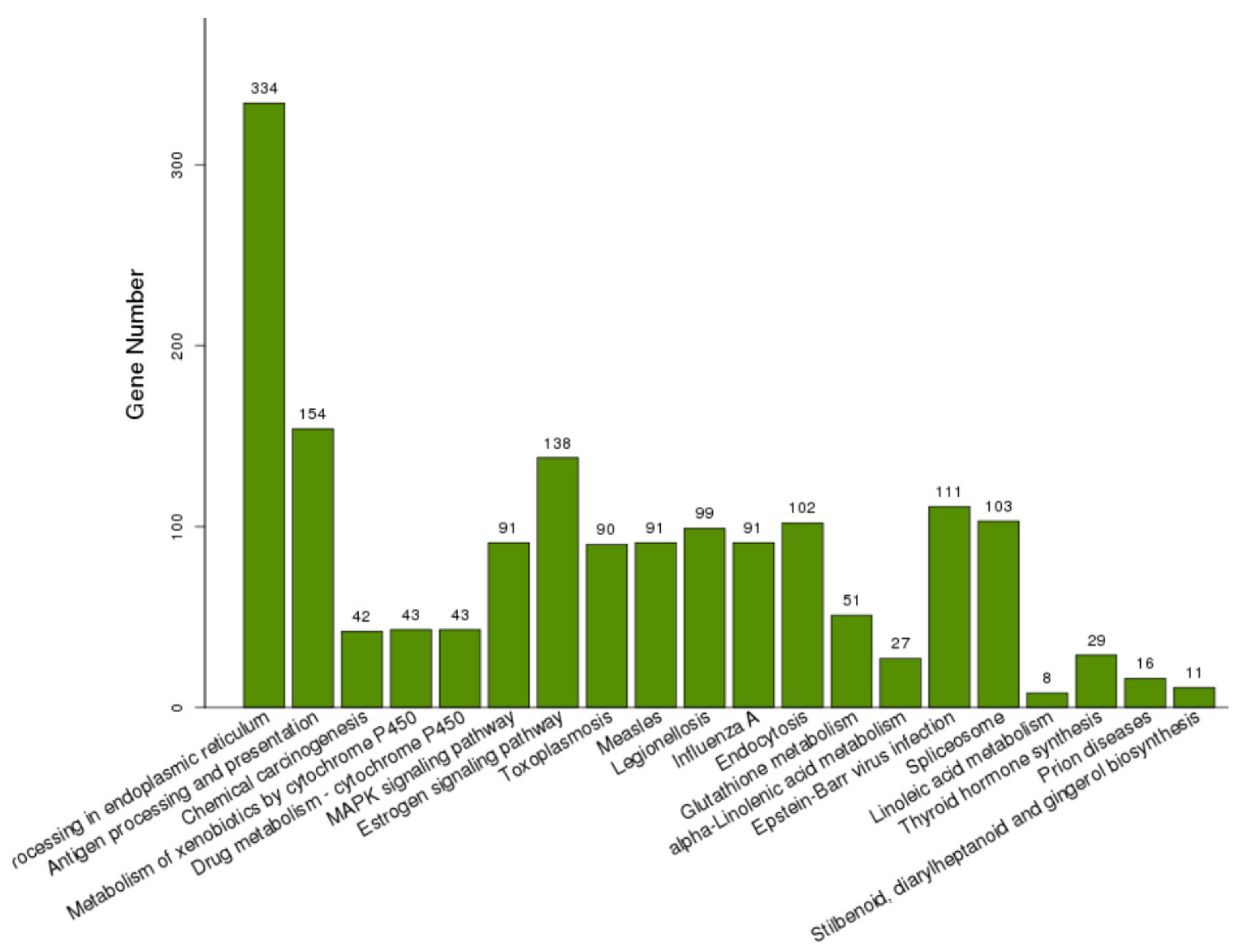
To characterize the functions of the DEGs, GO enrichment analysis was performed to categorize potential functions (Supplementary Materials Table S1). The DEGs significantly annotated in the top 20 terms in the BP, MF, and CC categories are shown in Table 3, Table 4 and Table 5. For the BP category, the four most enriched GO terms were associated with oxidation reduction processes (GO:0055114), response to heat (GO:0009408), protein folding (GO:0006457), and response to oxidative stress (GO:0006979). The two terms involved in unfolded protein binding (GO:0051082) and heme binding (GO:0020037) were assigned to the MF category. The extracellular regions (GO:0005576) and vacuoles (GO:0005773) were the two most important terms in the CC category. The DEGs were also subject to KEGG analysis, and 1094 DEGs were assigned to 205 pathways (Supplementary Materials Table S2). The DEGs significantly enriched in the top 20 significant KOs are shown in Figure 4. The highly enriched pathways of DEGs were protein processing in the endoplasmic reticulum (ko04141) and antigen processing and presentation (ko04612). The endocytosis (ko04144) and glutathione metabolism (ko00480) also appeared in the main KEGG pathways of the DEGs.

Table 3.
Gene Ontology (GO) analysis of DEGs significantly annotated in the biological process (BP).

Table 4.
GO analysis of DEGs significantly annotated in the molecular function (MF).

Table 5.
GO analysis of DEGs significantly annotated in the cellular component (CC).
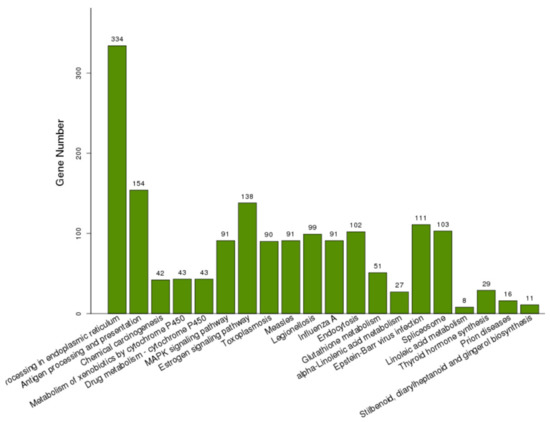
Figure 4.
KEGG pathway enrichment analysis based on the DEGs significantly enriched in KOs. The number of above each bar was the number of DEGs enriched in each KO.
2.4. Functional Validation of LmAUX1 in Response to Cd
To confirm whether these putative genes were associated in plant Cd tolerance, we cloned a significantly downregulated gene, according to the blastp results, and named it LmAUX1. We hypothesized that it may affect plant Cd uptake and distribution. The predicted CDS of this gene (Multiflorum_1-4k_c11285_f6p6_2149) was isolated and over-expressed in A. thaliana (Col-0 and aux1-7 mutant).
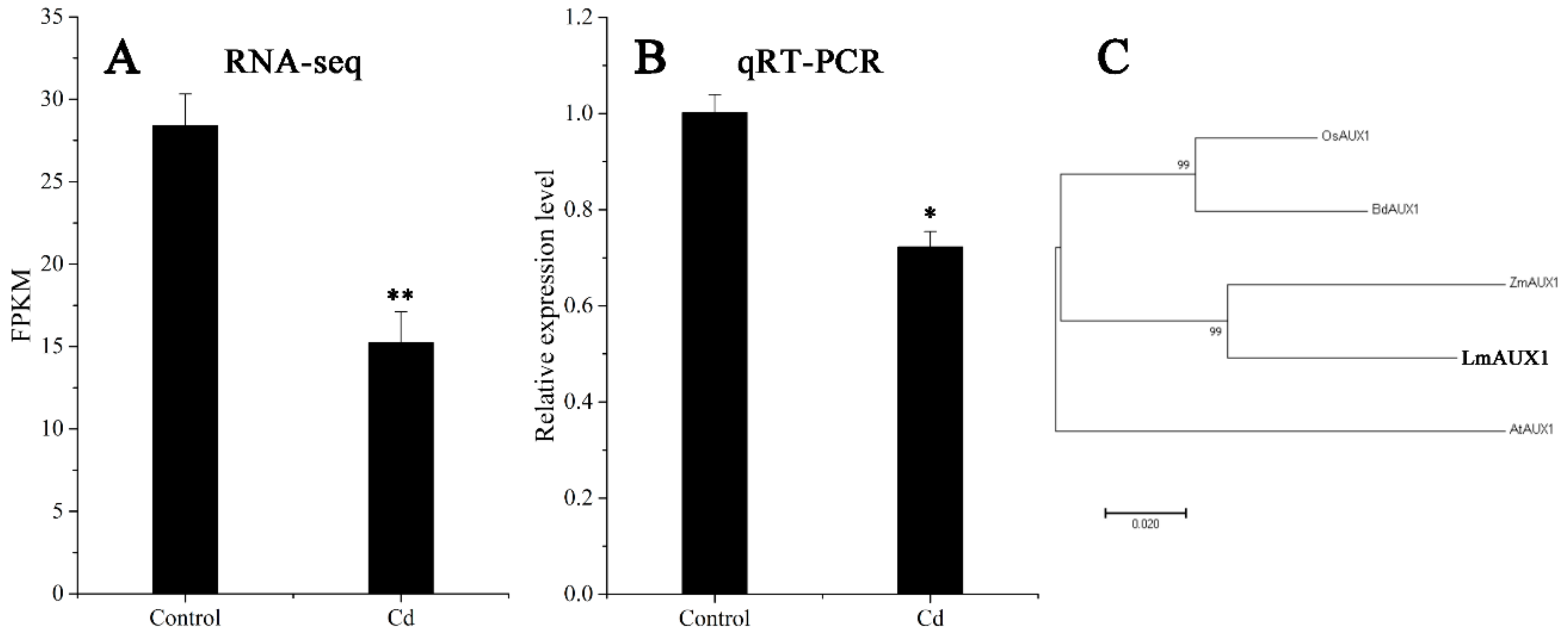
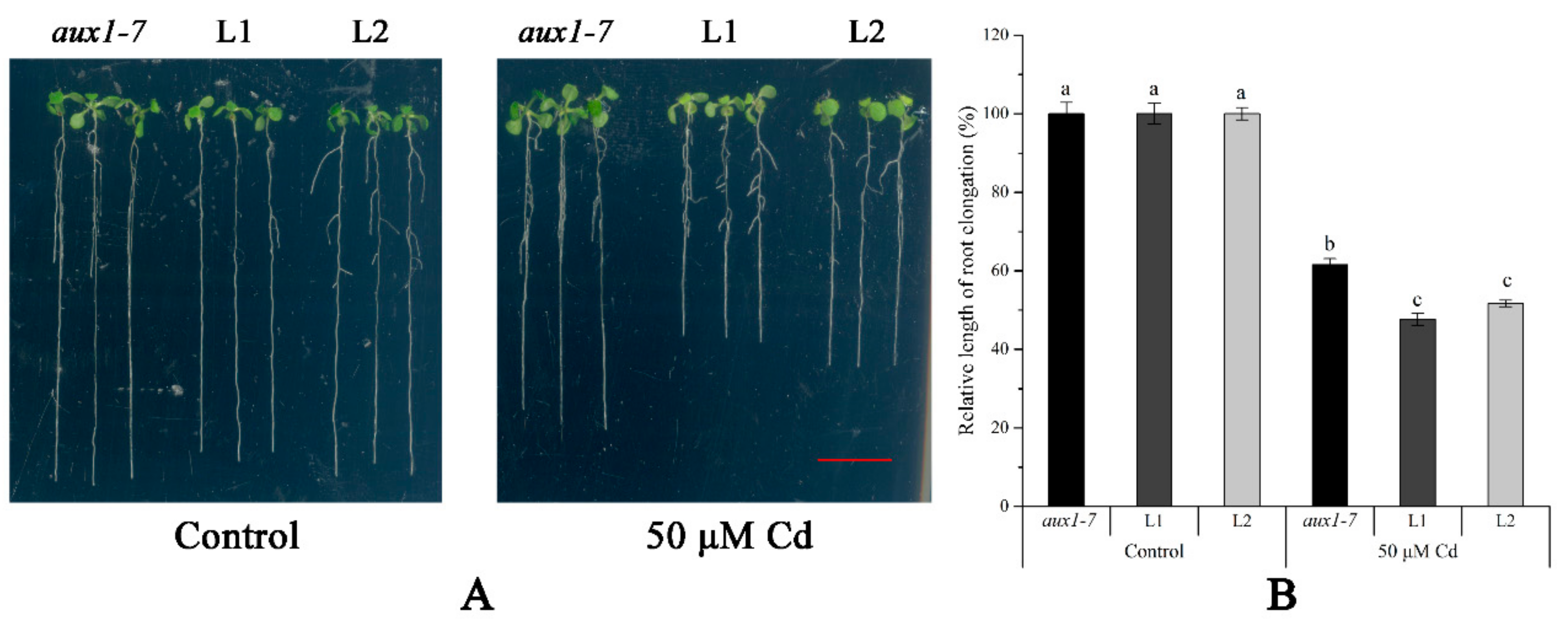
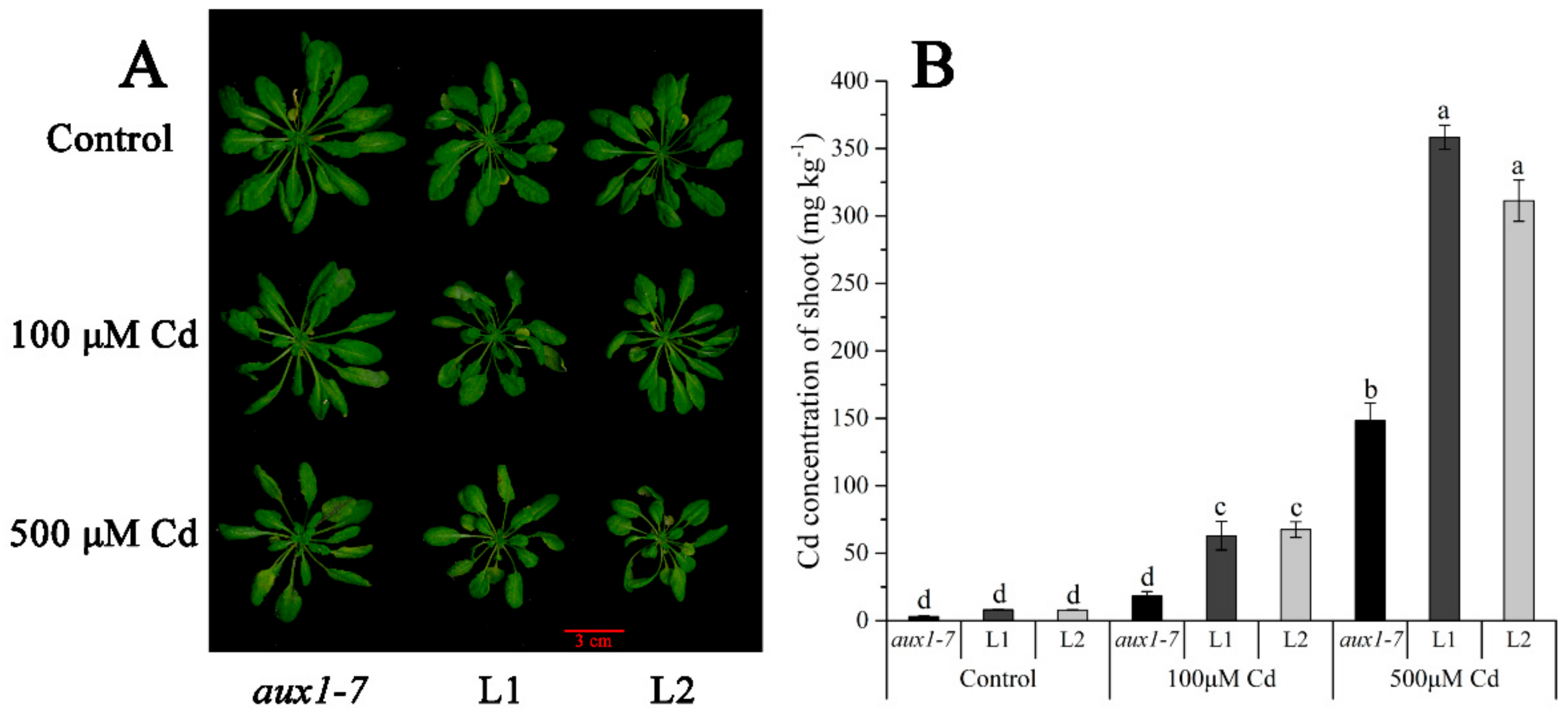
The FPKM values obtained from RNA-seq and relative expression levels verified by qRT-PCR of the DEG (Multiflorum_1-4k_c11285_f6p6_2149) are shown in Figure 5A,B, respectively. According to the analysis of the phylogenetic tree, it was a homologue gene of AUX1 in A. thaliana, Brachypodium distachyon, Zea mays, and rice (Figure 5C). Multiple transgenic lines of A. thaliana (Col-0 ecotype and aux1-7 mutant) were generated by the T-DNA-inserted fragment harboring 35S:lmAUX1. Two independent transgenic lines appeared as Cd-tolerance trails, respectively. The T3 generations of transgenic lines, the aux1-7 mutant, and Col-0 were treated with and without 50 µM CdCl2 in 1/2 MS medium (Figure 6A and Figure S3A), and the results clearly showed that the relative length of root elongation in the transgenic lines was significantly lower than those of the aux1-7 mutant and Col-0 (Figure 6B and Figure S3B). Col-0, the aux1-7 mutant, and the overexpression lines were grown in soils irrigated with water or with a 100 or 500 µM CdCl2 solution, respectively (Figure 7A), and the shoot Cd concentration of transgenic lines was significant increased compared to the aux1-7 mutant (Figure 7B) and Col-0 (Figure S4). The shoot Zn, Mn, and Cu concentrations of transgenic lines was not significant affected compared to the aux1-7 mutant and Col-0, but the Fe concentration of the transgenic lines was significant changed compared to the aux1-7 mutant (Figure S5) and Col-0 (Figure S6), except aux1-7 and its overexpression lines (L1 and L2) treated with 500 µM Cd.
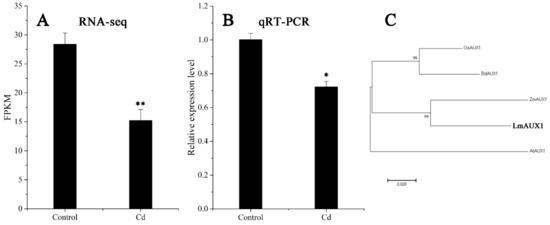
Figure 5.
Expression level and phylogenetic tree analysis of AUX1. (A) Expression level of LmAUX1 calculated by RNA-seq. Data are means ± SE (n = 3). Asterisks indicate significant differences (** p < 0.01 by Duncan’s test). (B) Relative expression level of LmAUX1 verified by qRT-PCR. Data are means ± SE (n = 4). Asterisks indicate significant differences (* p < 0.05 by Duncan’s test). (C) Phylogenetic tree of AUX1 proteins: the phylogenetic tree was constructed from an alignment of AUX1 proteins from five different plant species by neighbor-joining methods with bootstrapping analysis (1000 replicates), AtAUX1 (Arabidopsis thaliana, AT2G38120), BdAXU1 (Brachypodium distachyon, Bradi2g55340), LmAUX1 (Lolium multiflorum, Multiflorum_1-4k_c11285_f6p6_2149), OsAUX1 (Oryza sativa, Os01g63770), and ZmAXU1 (Zea mays, PWZ17958).
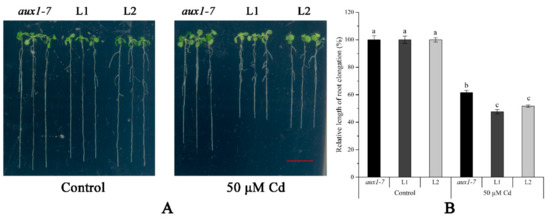
Figure 6.
The Cd tolerance experiment on plates of LmAUX1. LmAUX1 was transformed into the Arabidopsis thaliana aux1-7 mutant with the CaMV35S promoter, and two lines (L1 and L2) were used for the experiment. (A) Phenotype of plants grown in 1/2 MS medium with and without 50 μM Cd for 4–5 d. Scale bar = 1 cm. (B) Relative length of root elongation. Data are means ± SE (n ≥ 15); different letters above the bars were significantly different at p < 0.05 (Duncan’s test).
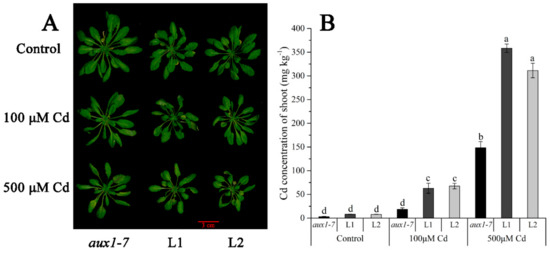
Figure 7.
Cd translocated experiment in soil of LmAUX1. LmAUX1 was transformed into the Arabidopsis thaliana aux1-7 mutant with the CaMV35S promoter, and two lines (L1 and L2) were used for the experiment. (A) Phenotype of plants grown in soils irrigated with 0, 100, or 500 μM Cd solution for 3–4 weeks. (B) The Cd concentration of shoot. Data are means ± SE (n = 3); different letters above the bars are significantly different at p < 0.05 (Duncan’s test).
2.5. Alternative Splicing Identification
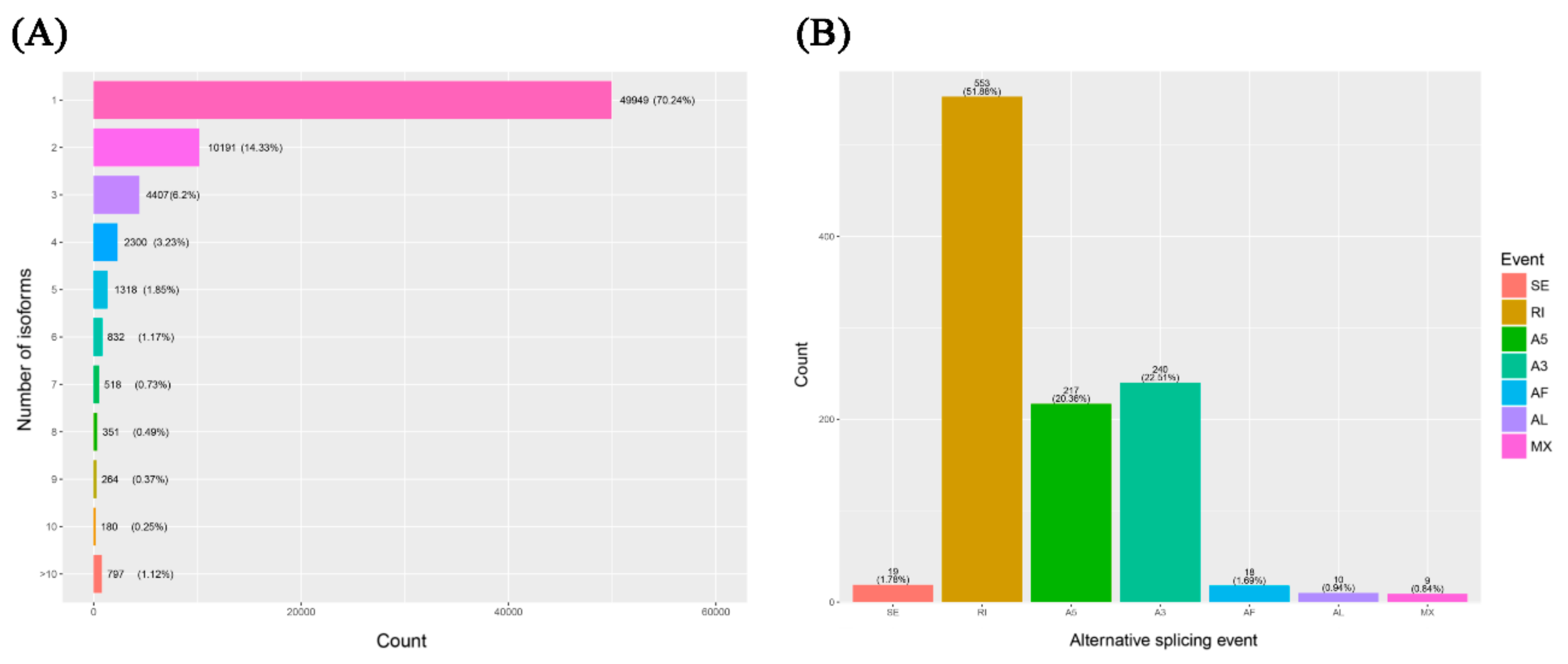
The inherent advantage of PacBio ISO-seq makes it possible to understand the complexity of alternative splicing (AS) at the whole transcriptome scale even without a reference genome. In this study, using the full-length reference transcriptomic database, we reconstructed the unique models of all transcripts by cogent software, named full-length UniTransModels. Transcript isoforms of Italian ryegrass were clustered by mapping individual ISO-seq consensus isoforms back to the reconstructed full-length UniTransModels. In summary, a total of 29.76% of the UniTransModels had more than one isoform, and almost half of the UniTransModels (14.33%) had two isoforms; there were still some UniTransModels with more than 10 isoforms (797, 1.12%) (Figure 8A). The different types of AS events were assessed based on the UniTransModels as the references instead of canonical genome-based AS events. Retained introns (RIs) were the majority of AS events, and together with alternative 5′ end or 3′ end AS events, these three types of AS events constituted more than 90% of detected events (Figure 8B).

Figure 8.
Alternative splicing (AS) analysis of the Lolium multiflorum full-length transcriptome using ISO-seq. (A) Distribution of isoform numbers for UniTransModels. (B) Numbers of different AS events detected in full-length transcriptome. SE: skipping exon; RI: retained intron; A5: alternative 5′ splice-site; A3: alternative 3′ splice-site; AF: alternative first exon; AL: alternative last exon; MX: mutually exclusive exons.
By aligning Illumina short reads to transcript models (UniTransModels), we could further validate the reliability of isoforms detected by the pipeline without a reference genome, and different AS events of some isoforms in UniTransModels in control and Cd stress treatments are illustrated in Figure S7.
3. Discussion
In the past decade, next-generation sequencing (NGS) technology has been widely used for genetic discovery and research [29], and the well-known short-read RNA-seq has become an integral part in evaluating whole RNA expression patterns [30]. However, the sequenced fragments of NGS are usually between 50 and 300 bp, and this was insufficient in the accurate reconstruction and reliable expression estimation of transcript variants, especially for species with low-quality genomes or even without reference genomes [31]. The studies of the species that lack reliable genetic information were seriously restricted in the precise understanding of the genetic diversity and whole expression patterns. Single-molecule long-read sequencing, which was a third sequencing technique, can identify the full structure of individual transcripts by full-length sequencing, which can uniquely reveal the complexity of the transcriptome [23,25].
Italian ryegrass is one of the most widely used forage species in the world [32]. For now, forage grasses are not only used for feedstuff and soil stability enhancer, but also potentially used for biofuel and bioremediation activities [5]. Herbaceous crops may play an important role in sustainable bioenergy feedstock in the future [33]. Italian ryegrass can provide substantial biomass feedstocks due to its high yields and rapid growth. A report indicated that Italian ryegrass was a high-potential bioethanol resource with a higher yield of bioethanol (84.6%) compared to switchgrass (77.7%) and silvergrass (64.3%) [6]. Molecular breeding in crops has shown promise in increasing yield and multiple stress in a changing climate and environment, and it may improve the desirable characteristics in the breeding of Italian ryegrass with molecular biology methods. Transcriptomic analyses are important for understanding gene function. The current knowledge about ryegrass is mainly dependent on gene expression sequenced NGS technologies [34,35]. As a result, the fragmentation of RNA information cannot fully characterize the complexity and diversity of the transcriptome.
In this study, we constructed the transcriptome of Italian ryegrass root using PacBio ISO-seq. We aimed to obtain full and accurate transcriptome information about ryegrass so as to unveil the various responsive networks under Cd stress. Given our pre-experiment, we constructed two overlapped libraries (inserted sizes of 1–4 kb and 3–10 kb), rather than the normal libraries, i.e., 1–4 kb and 4–10 kb libraries. In summary, we obtained 8.12 and 5.52 GB of raw data, respectively (Table S4). According to the SMRT standard protocol procedures, the raw data were then further filtered (Table S5 and Figure S8). The results of quality control indicated that, instead of using the multiple size-fractionated libraries method [25], shortages in the sequencing data between 3 and 4 kb were supplemented, and the overlapped strategy avoided the biases that shorter fragments of a unique library sequence were regnant (Figure S9). With further classification, clustering, polishing, and comprehensive filter procedures as well as integration with Illumina RNA-seq data, we obtained a 340 Mb full length reference transcriptome, and 146,545 isoforms were annotated with different protein and nucleotide databases. The various functions of Italian ryegrass transcripts were thus unveiled.
RNA-seq is one of the most well-known transcriptomic research pipelines. It is useful for understanding the expressed patterns and for identifying genes, be they of model or non-model species [36]. Cadmium is a toxic heavy metal element, causing serious harm to almost all plants, animals, and humans [8]. Understanding the regulatory mechanisms of genes controlled under Cd may contribute to precise breeding with safer forage or new uses of Italian ryegrass in phytoremediation. In this work, nine-day Italian ryegrass seedlings were treated with and without Cd treatment. After six hours, the concentrations of heavy metals were significant affected by Cd. The mRNAs of six Italian ryegrass roots with and without Cd treatment were sequenced with RNA-seq. Over 60 GB of raw data were obtained. Heat map analysis indicated a correlation of all samples, which were clustered as the control group and the Cd group (Figure S10). The data were aligned with the reference transcriptome generated by PacBio ISO-seq, and a large number of DEGs were generated, which provided valuable information on the transcriptional regulation of genes in responses to Cd stress in Italian ryegrass.
Gene Ontology (GO) enrichment analysis enhanced our understanding of DEGs. The DEGs of biological processes involved oxidation–reduction processes, protein folding, and responses to oxidative stress, which was present in the highest proportion. Cadmium can lead to a rise in ROSs and disturb redox homeostasis [37]. Overproduction of ROS can generate oxidative stress, causing protein carbonylation, which can lead to misfolded or inactivated proteins [38]. In addition, some DEGs are involved in responses to heat terms. Heat Shock Proteins (HSPs) play a crucial role in plant tolerance to multiple stress [39]. Some HSP/chaperone genes have been reported to contribute to the positive enhancement of plant Cd tolerance [20,40,41]. HSPs can also act as molecular chaperones to process the folding, assembly, translocation, and degradation of matched proteins [42]. For example, the HSP chaperones are responsible for the folding of some nascent proteins to alleviate endoplasmic reticulum (ER) stress [43], including kinases [44], transcription factors [45], and transporters [46]. Previous transcriptomic research on Cd-treated plants has also emphasized that these biological processes all serve as important functions in adaptation to biotic and abiotic stresses, but these are not limited to Cd stress [21,47]. In addition, some DEGs have been classified as contributing to cellular components, such as extracellular regions, vacuoles, and cell walls in common with other RNA-seq analyses [48]. Additionally, few Cd-responsive genes in Italian ryegrass have been cloned and characterized. Therefore, a list of DEGs as candidate genes could be helpful for further studies (Supplementary Materials Table S3).
In previous studies, the homeostasis of auxin was found to be disturbed by Cd in A. thaliana [49,50]. Additionally, our previous study also showed that Cd interfered with the auxin homeostasis (Figures S11 and S12), and the loss of function of the auxin transporter enhanced plant Cd tolerance (Figure S13). We transformed the lmAUX1 into the aux1-7 mutant, and the tropic movement of roots was regained (Figure S14). In the Cd tolerance on plates, the relative length of root elongation was significant inhibited. Additionally, soil experiments showed that the Cd concentration of shoots was significantly increased. These results indicate that LmAUX1 can regulate plant Cd tolerance and accumulation. In Italian ryegrass, the relative expression level of lmAUX1 was downregulated under Cd stress, so it is suggested that Italian ryegrass enhanced self-tolerance by decreasing the expression level of LmAUX1.
Alternative splicing (AS) is an important post-transcriptional regulatory mechanism for the diversity of transcriptomes and proteomes [51]. Recently, the versatile roles of AS in plants have been further clarified, such as the salt stress tolerance in A. thaliana and the mineral nutrient homeostasis in rice. In A. thaliana, the U1 spliceosomal protein, AtU1A, is responsible for recognizing the 5′ splice site in the initial step of pre-mRNA splicing. The 5′ splice site in the Arabidopsis aconitase (ACO1) pre-mRNA can be sequentially recognized by the U1 snRNA that is associated with the AtU1A protein. Splicing of ACO1 pre-mRNA is important for maintaining proper ROS levels in the cell and for contributing to salt tolerance [52]. Ser/Arg (SR)-rich proteins interact with pre-mRNA sequences and splicing factors during spliceosome assembly in order to perform essential functions in constitutive and AS. In rice, three SR protein-encoding genes (SR40, SCL57, and SCL25) regulate P uptake and remobilization between leaves and shoots of rice [53]. However, these studies have mainly focused on model plants using NGS techniques. For polyploid species or un-sequenced species, it is difficult to accurately identify transcript isoforms [24]. PacBio ISO-seq is a more reliable and superior strategy to understand the whole transcriptomes. It can directly generate accurate AS isoforms, rather than short read sequencing, to annotate a novel whole-genome assembly [54]. Here, we characterized AS in autotetraploid Italian ryegrass using PacBio ISO-seq. In common with results with respect to cotton [24], rice [26], strawberry [29], and switchgrass [55], the retained intron events contributed the most AS events. In addition, the percentage of unique isoforms is greater than those of other species; it appeared that polyploidy also played an important role in increasing the transcript diversity of Italian ryegrass [56]. Our results only scratch the surface of the AS in Italian ryegrass under Cd stress without the reference genome, and we expect to reveal more AS transcript-harboring genes under multiple stresses in the future.
4. Materials and Methods
4.1. Plant Materials, Growth Conditions, and Treatment
A Cd-tolerant ryegrass cultivar (“IdyII”) was chosen for this study [57]. The Italian ryegrass seeds were treated with 50% H2SO4 for 5 min, washed with deionized water thoroughly, subsequently disinfected with 30% NaClO for 15 min, and then rinsed with deionized water again. After that, the seeds were preliminarily germinated on a net floating on deionized water at 25 °C in the dark. Three days later, the germinated seeds were cultivated in a greenhouse with a day/night temperature of 25/20 °C, 65 ± 5% relative humidity, and a 12 h light/dark cycle with 150 µmol m−2 s−1 light intensity for two days. The seedlings with a uniform height of about 4 cm were transferred to a 1 L black plastic beaker filled with a 1/4 strength modified Hoagland solution with the following composition: 1.5 mmol L−1 KNO3, 1.0 mmol L−1 Ca(NO3)·4H2O, 0.5 mmol L−1 NH4H2PO4, 0.25 mmol L−1 MgSO4·H2O, 50 µmol L−1 KCl, 25 µmol L−1 H3BO3, 2.0 µmol L−1 MnSO4·H2O, 2.0 µmol L−1 ZnSO4·7H2O, 0.5 µmol L−1 CuSO4·5H2O, 0.5 µmol L−1 (NH4)6MO7O24·4H2O, and 20 µmol L−1 Fe(II)-EDTA-Na2. The pH value was adjusted to 6.5, and the nutrient solution was renewed every 3 d. Seedlings were continuously cultivated in the greenhouse. After pre-culture for 9 d, plants were treated with and without 50 µmol L−1 Cd supplied as CdCl2 for 6 h. There were three biological replicates for the assays. A black beaker containing seven seedlings represented a biological replicate. From each beaker, two seedlings used for PacBio ISO-seq and Illumina RNA-seq were chosen, respectively. Other seedlings were used for the determination of metal concentrations.
4.2. RNA Isolation, RNA-seq, and PacBio Full-Length ISO-SeqLibrary Preparation and Sequencing
Total RNAs of root samples were extracted using TRIzol (Invitrogen™, Life Technologies, Carlsbad CA, USA), and residual DNA was removed with RNase-free DNase I (New England BioLabs, Ipswich, MA, USA) according to the manufacturer’s instructions. The total RNA was quantified and assessed with an Agilent Bioanalyzer 2100 (Agilent Technologies, Palo Alto, CA, USA), and the RNA integrity numbers (RIN) of all samples were greater than 9.0. Total RNA samples were sent to Berry Genomics (http://www.berrygenomics.com) for sequencing.
The RNA-seq libraries were constructed using the TruSeq RNA Sample Prep Kit (Illumina, San Diego, CA, USA) as per the manufacturer’s instructions. The libraries were sequenced using Illumina Hiseq X Ten (Illumina, San Diego, CA, USA) as 150 bp paired-end reads.
For the PacBio full-length ISO-seq libraries, two equal mixed RNAs (2 μg, mixed with equal three controls and three-treatment total RNAs) were used for 1–4 and 3–10 kb cDNA library construction, respectively. Poly(A) RNA was isolated using the Poly(A) PuristTM Kit (Ambion, Inc., Austin, TX, USA)). RNA was reverse-transcribed into cDNA using the SMARTer™ PCR cDNA Synthesis Kit (Clontech Laboratories Inc., Palo Alto, California, USA). In PCR amplification, for 1–4 kb library construction, the LD-PCR included 16 cycles, and for 3–10 kb library, the LD-PCR included 20 cycles. The 1–4 and 3–10 kb cDNA fragments were generated by BluePippin size selection (Sage Science, Beverly, MA, USA). The size selected cDNA products were applied to constructed SMRTbell Template libraries (SMRTBell Template Prep Kit, Pacific Biosciences, Menlo Park, CA, USA). Each library used one SMRT cell sequenced on a PacBio sequel platform (Pacific Biosciences, Menlo Park, CA, USA).
4.3. Assembly of Reference Transcriptomic Database
Raw short reads of Illumina RNA-seq were filtered by removing reads containing adaptors, reads containing poly-N (>10%), and low-quality reads. Q20, Q30, GC-contents, and sequence duplication levels of the clean data were calculated. Clean reads were used for next analysis.
The standard protocol of ISO-seq (SMRT Analysis 2.3) was used to process the raw PacBio full-length ISO-seq data. Raw long reads were filtered, and the reads for which length was no more than 50 bp and of which accuracy was less than 0.75 were removed. The reads of inserts (ROIs) were obtained from the circular consensus sequences (CCS). After examining for 5′ and 3′ adaptors and poly(A) signals, full-length and non-full-length cDNA reads were defined, and chimeric reads were removed, which included sequencing primers. According to the isoform-level clustering algorithm (ICE), the full-length non-chimeric reads were aligned, and similarity sequences were assigned to a cluster. Each cluster was identified as a uniform isoform. Non-full-length cDNA reads were then applied to polish each cluster. The isoform sequences (with a predicted accuracy of >99%) were considered high-quality isoforms, and others were regarded as low-quality isoforms. The Illumina short reads were used to improve the low-quality isoform accuracies by LSC (LSC 2.0, http://augroup.org/LSC/LSC)). The modified low-quality isoforms and high-quality isoforms were combined as high-quality full-length transcripts. Finally, the redundancies were moved using CD-HIT-EST (CD-HIT-EST 4.6, http://www.bioinformatics.org/cd-hit, parameterization: c = 0.99, T = 6, G = 1, U = 10, s = 0.999, d = 40, and p = 1) to obtain non-redundant high-quality transcripts. The transcripts were considered the reference transcriptome and used for further analysis.
4.4. Annotation of Gene Function
To understand the functions of the reference transcriptome, the protein-coding sequences (CDS) of all transcripts were predicted by TransDecoder (Transdecoder 3.0.0, https://github.com/TransDecoder) at the default setting. The coding likelihood scores of six open reading frames (ORFs) with a length of coding protein sequences of more than 100 amino acids (aa) in a transcript were counted, and the highest score of ORFs was retained as the CDS. If a candidate ORF was found fully encapsulated by the coordinates of another candidate ORF, the longer one was reported [58]. All predicted CDSs were aligned to protein and nucleotide databases (NCBI non-redundant protein (NR), NCBI non-redundant nucleotide (NT), Swiss-Prot, Gene Ontology (GO), KEGG, and KOG) using blast.
4.5. Quantification of Gene Expression Levels
All clean reads of Illumina RNA-seq corresponding to each pooled sample were mapped against the assembled reference transcriptome using bowtie 2 (V2.1.0). The FPMK (fragments per Kilobase Million) values of each isoform were computed for each pooled sample and normalized using the RPKM (RSEM 1.2.15) [59].
4.6. Identification and Function Assessment of DEGs
EdgeR package (edgeR 3.14.0) was used to identify differentially expressed genes (DEGs) between the control and Cd treatment. Significant DEGs were identified with the threshold of FDR < 0.001 and log2 (FoldChange) > 1.
The GO enrichment analysis was implemented to assess the functions of DEGs using the topGO R package (topGO 2.24.0). The KEGG pathways were applied to enrich the DEGs to different biochemical metabolic pathways and signal transduction pathways with KOBAS software (KOBAS 2.0).
4.7. Validation of DEGs with qRT-PCR
The relative expression levels of 20 randomly selected DEGs were validated with qRT-PCR. New total RNAs were isolated from the plants re-cultivated as were previous samples used for sequencing. The cDNA was synthesized with the HiScript® Q RT SuperMix for qPCR (Vazyme, #R223-1, Nanjing, China). The qRT-PCR was carried out using the ChamQTM SYBR® Master Mix (Vazyme, #Q311-02/03, Nanjing, China) in the QuantStudio 5 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). All samples were normalized with the reference gene LmeEF1A(s), calculated using the 2−ΔΔCt method, and shown as mean ±SE. Four biological replicates were performed.
4.8. Vector Construction and Ectopic Overexpression of LmAUX1 in Arabidopsis Thaliana
To reveal the functions controlled by the genes of Italian ryegrass response to Cd stress, a DEG (Multiflorum_1-4k_c11285_f6p6_2149) was selected for functional characterization. A blastp search in the National Center for Biotechnology Information (NCBI) with predicted protein coding sequences of this gene as the default setting indicated that it coded an auxin transporter. Thus, it was named LmAUX1. The RNA-seq result showed that the relative expression level of this gene was significantly decreased in response to Cd stress, and this was validated with qRT-PCR. The phylogenetic tree, based on amino acid sequences of predicted protein coding sequences and AUX1 amino acid sequences of several other plant species, was constructed using MEGA 7 software (Molecular Evolutionary Genetics Analysis 7.0 software). Bootstrapping analysis was processed as the neighbor-joining method (1000 replicates).
The predicted CDSs of LmAUX1 were cloned into the pCAMBIA1305-eGFP vector with the CaMV35s promoter. The constructed vector was chemically transformed into the Agrobacterium tumefaciens strain “EHA105,” and the target gene was then transformed into A. thaliana (LmAUX1 was transformed into both the Col-0 ecotype and the aux1-7 mutant, respectively) by the floral dip method. Positive transgenic lines were screened with 25 mg L−1 hygromycin on the half-strength Murashige and Skoog solid medium supplied with 1% sucrose and 0.8% agar (pH 5.8) (1/2 MS medium). More than 15 transgenic lines were obtained, and two T3 lines of them were used for functional characterization. The LmAUX1 overexpression lines transformed into aux1-7 mutants named L1 and L2, and LmAUX1 overexpression lines were transformed into Col-0 ecotypes named OE1 and OE2.
For Cd tolerance experiments on plates, seeds were surface-sterilized with 10% NaClO for 15 min and plated on 1/2 MS medium. The seeds were vernalized for two days in darkness at 4 °C and then cultivated in a growth chamber with a 16/8 h light/dark cycle and a temperature of 23/20 °C for the day/night cycle. After 4–5 days, seedlings were transferred to 1/2 MS medium with and without 50 µM CdCl2. After five days, the seedlings were photographed, and the length of primary root elongation was measured using the ImageJ software. The experiments were repeated twice.
For Cd transferred experiments in soil, Col-0, aux1-7, and LmAUX1 overexpression lines (L1, L2, OE1, and OE2) were grown in soil under a long-day photoperiod (16/8 h light/dark) for two weeks. Subsequently, the plants were grown on soils irrigated with a 0, 100, or 500 μM Cd solution every two or three day for 3–4 weeks.
4.9. Determination of Metal Concentrations
The samples were dried at 70 °C for 3 d and then subjected to with mixed acid (HNO3 + HClO4 (85:15, v/v)) using a DigiBlock ED54-iTouch Digester (LabTech, Beijing, China). The samples were completely digested at 170 °C until they turned white; subsequently, the residual acid was volatilized, and all samples were re-dissolved in 10 mL 2.5% HNO3. The concentrations of metals were determined using am inductively coupled plasma optical emission spectrometer (ICP-OES, Perkin Elmer Optima 8000).
4.10. Microscopic Imaging of GUS Staining and Fluorescence of DII-VENUS
GUS staining was carried out according to the method reported in Hu et al. [50]. GUS activity in primary root apices in 5-day-old DR5::GUS seedlings treated with 0, 25, 50, 100 μM Cd for 3–4 days on 1/2 MS plates. GUS-stained images were observed under a stereo microscope (ZEISS Stemi 2000-C) and photographed by a CCD camera (Canon PowerShot A620).
Confocal microscopy was performed using a confocal laser scanning microscope (PerkinElmer, Waltham, MA, USA, UltraVIEW® VoX) according to the manufacturer’s instructions, excitation and emission wavelengths were 488 to 520 nm for DII-VENUS. Auxin signaling level in primary root apices in five-day-old Arabidopsis thaliana DII-VENUS seedlings treated with 0, 25, 50, 100 μM Cd for 3–4 days on 1/2 MS plates.
4.11. Alternative Splicing Identification
The reference transcriptome was re-constructed using Cogent (coding genome reconstruction tool, Cogent v1.4), and a set of transcripts models was then generated and named UniTransModels [60]. All isoforms of the reference transcriptome (transcripts before Cogent reconstruction) were then aligned to UniTransModels with GMAP software (GMAP v2017-06-20). At the same time, a gff file was generated, including the coordinate information of isoforms with respect to the UniTransModels. The transcripts were mapped to the UniTransModels and used for the splicing junctions assessed. The same splicing junctions of the transcripts were collapsed together, and these transcripts with different splicing junctions were identified as transcriptional isoforms of UniTransModels. Additionally, alternative splicing (AS) events were detected with the SUPPA pipeline (https://bitbucket.org/regulatorygenomicsupf/suppa/src/master/) using default settings [61].
The AS events were shown by sashimi plots generated using the IGV browser (IGV 2.5) with bam files and a gff file [62]. The bam files were obtained by mapping the Illumina short reads against UniTransModels using TopHat (TopHat 2.0.4). The FASTA file of the UniTransModels was then imported into IGV as a reference, and the corresponding mapped bam files and isoform gff files were loaded into IGV as tracks. After that, the sashimi plots were obtained from the bam file track. In the opened sashimi plot, the red peaks and junctions are from the Illumina bam file and the blue isoform bars are from the gff file.
4.12. Accession Numbers
The raw sequence data sequenced in this study have been deposited in the Genome Sequence Archive in the BIG Data Centre, Beijing Institute of Genomics (BIG), the Chinese Academy of Sciences (GSA, http://bigd.big.ac.cn/gsa), under accession number CRA001799.
5. Conclusions
In summary, according to the analyses of PacBio long read ISO-seq and Illumina short read RNA-seq data, we constructed a high-quality Italian ryegrass reference transcriptome and unveiled a comprehensive picture of the regulatory networks in Italian ryegrass under Cd stress. We then characterized a DEG (LmAUX1) expressed in A. thaliana, which significantly enhanced plant Cd accumulation. It is possible that this gene can be used to breed a high-Cd-accumulation Italian ryegrass cultivar. This work deepens our understanding of the mechanism of Italian ryegrass in response to Cd stress and contributes to its gene identification and molecular breeding.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/21/3/1067/s1.
Author Contributions
Conceptualization: Z.H., Q.C., and L.L.; Methodology: Z.H.; Formal Analysis: Z.H.; Investigation: Z.H., Y.Z., Y.H., Q.C., and T.Z.; Data Curation: Z.H.; Writing—Review and Editing: Z.H.; Supervision: Q.C.; Funding Acquisition: Q.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Jiangsu Science and Technology Support Program for Social Development, grant number BE2014709.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Charmet, G.; Balfourier, F.; Chatard, V. Taxonomic relationships and interspecific hybridization in the genus Lolium (grasses). Genet. Resour. Crop Evolut. 1996, 43, 319–327. [Google Scholar] [CrossRef]
- Aguirre, A.A.; Studer, B.; Frei, U.L.; übberstedt, T. Prospects for hybrid breeding in bioenergy grasses. BioEnergy Res. 2012, 5, 10–19. [Google Scholar] [CrossRef]
- Fè, D.; Ashraf, B.; Byrne, S.; Czaban, A.; Roulund, N.; Lenk, I.; Asp, T. Prospects for introducing genomic selection info forage grass breeding. In EGF at 50: The Future of European Grasslands. In Proceedings of the 25th General Meeting of the European Grassland Federation, Aberystwyth, Wales, 7–11 September 2014; Centre for Agriculture and Biosciences International: Wallingford, UK, 2014; pp. 830–832. Available online: https://www.cabdirect.org/cabdirect/abstract/20143369178 (accessed on 7 September 2014).
- Byrne, S.L.; Nagy, I.; Pfeifer, M.; Armstead, I.; Swain, S.; Studer, B.; Mayer, K.; Campbell, J.D.; Czaban, A.; Hentrup, S.; et al. A synteny-based draft genome sequence of the forage grass Lolium perenne. Plant J. 2015, 84, 816–826. [Google Scholar] [CrossRef] [PubMed]
- Inoue, M.; Stewart, A.; Cai, H.W. Italian ryegrass. In Genetics, Genomics and Breeding of Forage Crops; Kole, C., Yamada, T., Cai, H.W., Eds.; CRC Press: Boca Raton, FL, USA, 2013; pp. 36–57. [Google Scholar] [CrossRef]
- Yasuda, M.; Takenouchi, Y.; Nitta, Y.; Ishii, Y.; Ohta, K. Italian ryegrass (Lolium multiflorum Lam) as a high-potential bio-ethanol resource. BioEnergy Res. 2015, 8, 1303–1309. [Google Scholar] [CrossRef]
- Zhu, T.; Fu, D.; Yang, F. Effect of saponin on the phytoextraction of Pb, Cd and Zn from soil using Italian ryegrass. Bull. Environ. Contam. Toxicol. 2015, 94, 129–133. [Google Scholar] [CrossRef] [PubMed]
- Daisei, U.; Naoki, Y.; Izumi, K.; Feng, H.C.; Tsuyu, A.; Masahiro, Y.; Ma, J.F. Gene limiting cadmium accumulation in rice. Proc. Natl. Acad. Sci. USA 2010, 107, 16500–16505. [Google Scholar] [CrossRef]
- Clemens, S.; Ma, J.F. Toxic heavy metal and metalloid accumulation in crop plants and foods. Annu. Rev. Plant Biol. 2016, 67, 489–512. [Google Scholar] [CrossRef]
- Wang, P.; Chen, H.; Kopittke, P.M.; Zhao, F.J. Cadmium contamination in agricultural soils of China and the impact on food safety. Environ. Pollut. 2019, 249, 1038–1048. [Google Scholar] [CrossRef]
- Rani, A.; Kumar, A.; Lal, A.; Pant, M. Cellular mechanisms of cadmium-induced toxicity: A review. Int. J. Environ. Health Res. 2014, 24, 378–399. [Google Scholar] [CrossRef]
- Zhang, L.; Pei, Y.; Wang, H.; Jin, Z.; Liu, Z.; Qiao, Z.; Fang, H.; Zhang, Y. Hydrogen sulfide alleviates cadmium-induced cell death through restraining ROS accumulation in roots of Brassica rapa L. Ssp. Pekinensis. Oxid. Med. Cell. Longev. 2015, 2015, 804603. [Google Scholar] [CrossRef]
- Gupta, D.K.; Pena, L.B.; Romero-Puertas, M.C.; Hernandez, A.; Inouhe, M.; Sandalio, L.M. NADPH oxidases differentially regulate ROS metabolism and nutrient uptake under cadmium toxicity. Plant Cell Environ. 2016, 40, 509–526. [Google Scholar] [CrossRef] [PubMed]
- Rizwan, M.; Ali, S.; Abbas, T.; Zia-Ur-Rehman, M.; Hannan, F.; Keller, C.; Al-Wabel, M.I.; Ok, Y.S. Cadmium minimization in wheat: A critical review. Ecotoxicol. Environ. Saf. 2016, 130, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Aarts, M.G.M. The molecular mechanism of zinc and cadmium stress response in plants. Cell. Mol. Life Sci. 2012, 69, 3187–3206. [Google Scholar] [CrossRef]
- Tao, Q.; Jupa, R.; Liu, Y.; Luo, J.; Li, J.; Kováč, J.; Ki, B.; Li, Q.; Wu, K.; Liang, Y.; et al. Abscisic acid-mediated modifications of radial apoplastic transport pathway play a key role in cadmium uptake in hyperaccumulator Sedum alfredii. Plant Cell Environ. 2019, 42, 1425–1440. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, S.; Ishimaru, Y.; Igura, M.; Kuramata, M.; Abe, T.; Senoura, T.; Hase, Y.; Arao, T.; Nishizawa, N.K.; Nakanishi, H. Ion-beam irradiation, gene identification, and marker-assisted breeding in the development of low-cadmium rice. Proc. Natl. Acad. Sci. USA 2012, 109, 19166–19171. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.S.; Huang, J.; Zeng, D.L.; Peng, J.S.; Zhang, G.B.; Ma, H.L.; Guan, Y.; Yi, H.Y.; Fu, Y.L.; Han, B.; et al. A defensin-like protein drives cadmium efflux and allocation in rice. Nat. Commun. 2018, 9, 645. [Google Scholar] [CrossRef]
- Brunetti, P.; Zanella, L.; De Paolis, A.; Di Litta, D.; Cecchetti, V.; Falasca, G.; Barbieri, M.; Altamura, M.M.; Costantino, P.; Cardarelli, M. Cadmium-inducible expression of the ABC-type transporter AtABCC3 increases phytochelatin-mediated cadmium tolerance in Arabidopsis. J. Exp. Bot. 2015, 66, 3815–3829. [Google Scholar] [CrossRef]
- Song, G.; Yuan, S.; Wen, X.; Xie, Z.; Lou, L.; Hu, B.; Cai, Q.; Xu, B. Transcriptome analysis of Cd-treated switchgrass root revealed novel transcripts and the importance of HSF/HSP network in switchgrass Cd tolerance. Plant Cell Rep. 2018, 37, 1485–1497. [Google Scholar] [CrossRef]
- He, F.; Liu, Q.; Zheng, L.; Cui, Y.; Shen, Z.; Zheng, L. RNA-Seq analysis of rice roots reveals the involvement of post-transcriptional regulation in response to cadmium stress. Front. Plant Sci. 2015, 6, 1136. [Google Scholar] [CrossRef]
- Gao, J.; Sun, L.; Yang, X.; Liu, J.X. Transcriptomic analysis of cadmium stress response in the heavy metal hyperaccumulator Sedum alfredii Hance. PLoS ONE 2014, 8, e64643. [Google Scholar] [CrossRef]
- Abdel-Ghany, S.E.; Hamilton, M.; Jacobi, J.L.; Ngam, P.; Devitt, N.; Schilkey, F.; Ben-Hur, A.; Reddy, A.S. A survey of the sorghum transcriptome using single-molecule long reads. Nat. Commun. 2016, 7, 11706. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Wang, P.; Liang, F.; Ye, Z.; Li, J.; Shen, C.; Pei, L.; Wang, F.; Hu, J.; Tu, L.; et al. A global survey of alternative splicing in allopolyploid cotton: Landscape, complexity and regulation. New Phytol. 2018, 217, 163–178. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Tseng, E.; Regulski, M.; Clark, T.A.; Hon, T.; Jiao, Y.; Lu, Z.; Olson, A.; Stein, J.C.; Ware, D. Unveiling the complexity of the maize transcriptome by single-molecule long-read sequencing. Nat. Commun. 2016, 7, 11708. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Sun, M.; Wang, J.; Lei, M.; Li, C.; Zhao, D.; Huang, J.; Li, W.; Li, S.; Li, J.; et al. PacBio full-length cDNA sequencing integrated with RNA-seq reads drastically improves the discovery of splicing transcripts in rice. Plant J. 2018, 97, 296–305. [Google Scholar] [CrossRef]
- Yan, H.; Gao, Y.; Wu, L.; Wang, L.; Zhang, T.; Dai, C.; Xu, W.; Feng, L.; Ma, M.; Zhu, Y.G.; et al. Potential use of the Pteris vittata arsenic hyperaccumulation-regulation network for phytoremediation. J. Hazard. Mater. 2019, 368, 386–396. [Google Scholar] [CrossRef]
- Wang, J.; Deng, Y.; Zhou, Y.; Liu, D.; Yu, H.; Zhou, Y.; Lv, J.; Ou, L.; Li, X.; Ma, Y.; et al. Full-length mRNA sequencing and gene expression profiling reveal broad involvement of natural antisense transcript gene pairs in pepper development and response to stresses. Plant J. 2019. [Google Scholar] [CrossRef]
- Yuan, H.; Yu, H.; Huang, T.; Shen, X.; Xia, J.; Pang, F.; Wang, J.; Zhao, M. The complexity of the Fragaria x ananassa (octoploid) transcriptome by single-molecule long-read sequencing. Hortic. Res. 2019, 6, 46. [Google Scholar] [CrossRef]
- Anvar, S.Y.; Allard, G.; Tseng, E.; Sheynkman, G.M.; de Klerk, E.; Vermaat, M.; Yin, R.H.; Johansson, H.E.; Ariyurek, Y.; den Dunnen, J.T.; et al. Full-length mRNA sequencing uncovers a widespread coupling between transcription initiation and mRNA processing. Genome Biol. 2018, 19, 46. [Google Scholar] [CrossRef]
- Steijger, T.; Abril, J.F.; Engstrom, P.G.; Kokocinski, F.; Hubbard, T.; Guigo, R.; Harrow, J.; Bertone, P. Assessment of transcript reconstruction methods for RNA-seq. Nat. Methods 2013, 10, 1177–1184. [Google Scholar] [CrossRef]
- Hirata, M.; Cai, H.; Inoue, M.; Yuyama, N.; Miura, Y.; Komatsu, T.; Takamizo, T.; Fujimori, M. Development of simple sequence repeat (SSR) markers and construction of an SSR-based linkage map in Italian ryegrass (Lolium multiflorum Lam.). Theor. Appl. Genet. 2006, 113, 270–279. [Google Scholar] [CrossRef]
- Lee, D.K.; Aberle, E.; Anderson, E.K.; Anderson, W.; Baldwin, B.S.; Baltensperger, D.; Barrett, M.; Blumenthal, J.; Bonos, S.; Bouton, J. Biomass production of herbaceous energy crops in the United States: Field trial results and yield potential maps from the multiyear regional feedstock partnership. GCB Bioenergy 2018, 10, 698–716. [Google Scholar] [CrossRef]
- Duhoux, A.; Carrere, S.; Gouzy, J.; Bonin, L.; Delye, C. RNA-Seq analysis of rye-grass transcriptomic response to an herbicide inhibiting acetolactate-synthase identifies transcripts linked to non-target-site-based resistance. Plant Mol. Biol. 2015, 87, 473–487. [Google Scholar] [CrossRef] [PubMed]
- Knorst, V.; Byrne, S.; Yates, S.; Asp, T.; Widmer, F.; Studer, B.; Kolliker, R. Pooled DNA sequencing to identify SNPs associated with a major QTL for bacterial wilt resistance in Italian ryegrass (Lolium multiflorum Lam.). Theor. Appl. Genet. 2019, 132, 947–958. [Google Scholar] [CrossRef] [PubMed]
- Jain, M. A next-generation approach to the characterization of a non-model plant transcriptome. Curr. Sci. 2011, 101, 1435–1439. [Google Scholar]
- Chmielowska-Bąk, J.; Gzyl, J.A.; Rucińska-Sobkowiak, R.; Arasimowicz-Jelonek, M.; Deckert, J. The new insights into cadmium sensing. Front. Plant Sci. 2014, 5, 245. [Google Scholar] [CrossRef]
- Braconi, D.; Bernardini, G.; Santucci, A. Linking protein oxidation to environmental pollutants: Redox proteomic approaches. J. Proteom. 2011, 74, 2324–2337. [Google Scholar] [CrossRef]
- Wang, W.; Vinocur, B.; Shoseyov, O.; Altman, A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 2004, 9, 244–252. [Google Scholar] [CrossRef]
- Shim, D.; Hwang, J.U.; Lee, J.; Lee, S.; Choi, Y.; An, G.; Martinoia, E.; Lee, Y. Orthologs of the class A4 heat shock transcription factor HsfA4a confer cadmium tolerance in wheat and rice. Plant Cell 2009, 21, 4031–4043. [Google Scholar] [CrossRef]
- Cai, S.Y.; Zhang, Y.; Xu, Y.P.; Qi, Z.Y.; Li, M.Q.; Ahammed, G.J.; Xia, X.J.; Shi, K.; Zhou, Y.H.; Reiter, R.J. HsfA1a upregulates melatonin biosynthesis to confer cadmium tolerance in tomato plants. J. Pineal Res. 2017, 62, e12387. [Google Scholar] [CrossRef]
- Tamás, M.; Sharma, S.; Ibstedt, S.; Jacobson, T.; Christen, P. Heavy metals and metalloids as a cause for protein misfolding and aggregation. Biomolecules 2014, 4, 252–267. [Google Scholar] [CrossRef]
- Liu, L.; Cui, F.; Li, Q.; Yin, B.; Zhang, H.; Lin, B.; Wu, Y.; Xia, R.; Tang, S.; Xie, Q. The endoplasmic reticulum-associated degradation is necessary for plant salt tolerance. Cell Res. 2011, 21, 957. [Google Scholar] [CrossRef]
- Piper, P.; Truman, A.; Millson, S.; Nuttall, J. Hsp90 chaperone control over transcriptional regulation by the yeast Slt2(Mpk1)p and human ERK5 mitogen-activated protein kinases (MAPKs). Biochem. Soc. Trans. 2006, 34, 783–785. [Google Scholar] [CrossRef]
- Rodriguez, F.; Arsène-Ploetze, F.; Rist, W.; Rüdiger, S.; Schneider-Mergener, J.; Mayer, M.P.; Bukau, B. Molecular basis for regulation of the heat shock transcription factor σ32 by the DnaK and DnaJ chaperones. Mol. Cell 2008, 32, 347–358. [Google Scholar] [CrossRef] [PubMed]
- Kimura, T.; Kambe, T. The functions of metallothionein and ZIP and ZnT transporters: An overview and perspective. Int. J. Mol. Sci. 2016, 17, 336. [Google Scholar] [CrossRef]
- Chen, C.; Cao, Q.; Jiang, Q.; Li, J.; Yu, R.; Shi, G. Comparative transcriptome analysis reveals gene network regulating cadmium uptake and translocation in peanut roots under iron deficiency. BMC Plant Biol. 2019, 19, 35. [Google Scholar] [CrossRef]
- Lin, C.Y.; Trinh, N.N.; Fu, S.F.; Hsiung, Y.C.; Chia, L.C.; Lin, C.W.; Huang, H.J. Comparison of early transcriptome responses to copper and cadmium in rice roots. Plant Mol. Biol. 2013, 81, 507–522. [Google Scholar] [CrossRef]
- Yuan, H.M.; Huang, X. Inhibition of root meristem growth by cadmium involves nitric oxide-mediated repression of auxin accumulation and signalling in Arabidopsis. Plant Cell Environ. 2016, 39, 120–135. [Google Scholar] [CrossRef]
- Hu, Y.F.; Zhou, G.; Na, X.F.; Yang, L.; Nan, W.B.; Liu, X.; Zhang, Y.Q.; Li, J.L.; Bi, Y.R. Cadmium interferes with maintenance of auxin homeostasis in Arabidopsis seedlings. J. Plant Physiol. 2013, 170, 965–975. [Google Scholar] [CrossRef]
- Marquez, Y.; Brown, J.W.; Simpson, C.; Barta, A.; Kalyna, M. Transcriptome survey reveals increased complexity of the alternative splicing landscape. Arabidopsis Genome Res. 2012, 22, 1184–1195. [Google Scholar] [CrossRef]
- Gu, J.; Xia, Z.; Luo, Y.; Jiang, X.; Qian, B.; Xie, H.; Zhu, J.K.; Xiong, L.; Zhu, J.; Wang, Z.Y. Spliceosomal protein U1A is involved in alternative splicing and salt stress tolerance in Arabidopsis thaliana. Nucleic Acids Res. 2018, 46, 1777–1792. [Google Scholar] [CrossRef]
- Dong, C.; He, F.; Berkowitz, O.; Liu, J.; Cao, P.; Tang, M.; Shi, H.; Wang, W.; Li, Q.; Shen, Z.; et al. Alternative splicing plays a critical role in maintaining mineral nutrient homeostasis in rice (Oryza sativa). Plant Cell 2018, 30, 2267–2285. [Google Scholar] [CrossRef]
- Li, J.; Harata-Lee, Y.; Denton, M.D.; Feng, Q.; Rathjen, J.R.; Qu, Z.; Adelson, D.L. Long read reference genome-free reconstruction of a full-length transcriptome from Astragalus membranaceus reveals transcript variants involved in bioactive compound biosynthesis. Cell Discov. 2017, 3, 17031. [Google Scholar] [CrossRef]
- Zuo, C.; Blow, M.; Sreedasyam, A.; Kuo, R.C.; Ramamoorthy, G.K.; Torres-Jerez, I.; Li, G.; Wang, M.; Dilworth, D.; Barry, K.; et al. Revealing the transcriptomic complexity of switchgrass by PacBio long-read sequencing. Biotechnol. Biofuels 2018, 11, 170. [Google Scholar] [CrossRef]
- Yoo, M.J.; Liu, X.; Pires, J.C.; Soltis, P.S.; Soltis, D.E. Nonadditive gene expression in polyploids. Annu. Rev. Genet. 2014, 48, 485–517. [Google Scholar] [CrossRef]
- Fang, Z.; Lou, L.; Tai, Z.; Wang, Y.; Yang, L.; Hu, Z.; Cai, Q. Comparative study of Cd uptake and tolerance of two Italian ryegrass (Lolium multiflorum) cultivars. Peer J. 2017, 5, e3621. [Google Scholar] [CrossRef]
- Tian, J.; Feng, S.; Liu, Y.; Zhao, L.; Tian, L.; Hu, Y.; Yang, T.; Wei, A. Single-molecule long-read sequencing of Zanthoxylum bungeanum maxim. Transcriptome: Identification of aroma-related genes. Forests 2018, 9, 765. [Google Scholar] [CrossRef]
- Zhou, Y.; Tang, Q.; Wu, M.; Mou, D.; Liu, H.; Wang, S.; Zhang, C.; Ding, L.; Luo, J. Comparative transcriptomics provides novel insights into the mechanisms of selenium tolerance in the hyperaccumulator plant Cardamine hupingshanensis. Sci. Rep. 2018, 8, 2789. [Google Scholar] [CrossRef]
- Wu, T.D.; Reeder, J.; Lawrence, M.; Becker, G.; Brauer, M.J. GMAP and GSNAP for genomic sequence alignment: Enhancements to speed, accuracy, and functionality. In Statistical Genomics; Humana Press: New York, NY, USA, 2016; pp. 283–334. [Google Scholar] [CrossRef]
- Alamancos, G.P.; Pagès, A.; Trincado, J.L.; Bellora, N.; Eyras, E. Leveraging transcript quantification for fast computation of alternative splicing profiles. RNA 2015, 21, 1521–1531. [Google Scholar] [CrossRef]
- Thorvaldsdóttir, H.; Robinson, J.T.; Mesirov, J.P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 2013, 14, 178–192. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).