Abstract
A plethora of molecular and functional studies in tetrapods has led to the discovery of multiple taste 1 receptor (T1R) genes encoding G-protein coupled receptors (GPCRs) responsible for sweet (T1R2 + T1R3) and umami (T1R1 + T1R3) taste. In fish, the T1R gene family repertoires greatly expanded because of several T1R2 gene duplications, and recent studies have shown T1R2 functional divergence from canonical mammalian sweet taste perceptions, putatively as an adaptive mechanism to develop distinct feeding strategies in highly diverse aquatic habitats. We addressed this question in the carnivore fish gilthead seabream (Sparus aurata), a model species of aquaculture interest, and found that the saT1R gene repertoire consists of eight members including saT1R1, saT1R3 and six saT1R2a-f gene duplicates, adding further evidence to the evolutionary complexity of fishT1Rs families. To analyze saT1R taste functions, we first developed a stable gene reporter system based on Ca2+-dependent calcineurin/NFAT signaling to examine specifically in vitro the responses of a subset of saT1R heterodimers to L-amino acids (L-AAs) and sweet ligands. We show that although differentially tuned in sensitivity and magnitude of responses, saT1R1/R3, saT1R2a/R3 and saT1R2b/R3 may equally serve to transduce amino acid taste sensations. Furthermore, we present preliminary information on the potential involvement of the Gi protein alpha subunits saGαi1 and saGαi2 in taste signal transduction.
1. Introduction
Perception of sweet, umami and bitter taste is mediated by two distinct G protein-coupled receptor (GPCR) families, namely by taste 1 receptor (T1R) and T2R, mostly expressed in taste buds [1,2,3]. In most vertebrates, the T1R gene repertoire is relatively conserved and consists of three ancient duplicated genes that diverged before fish/tetrapod diversification, about 400 million years ago (MYA) [4,5]. The T1R family’s receptor subunits (T1R1, T1R2 and T1R3) dimerize to form functional receptors. The combination of T1R1/T1R3 forms the umami receptor and is activated by amino and nucleic acids, and the T1R2/T1R3 heterodimer responds to natural and artificial sugars including sweet proteins and D-amino acids [6]. Mammalian T1R transduction pathways of both sweet and umami sensations are subsequently initiated through multiple G-proteins [7,8]. Receptor activation leads to the release of Gα gustducin (Gαgust) from the beta-gamma subunits (Gβƴ) of the G-protein complex, and two parallel signaling cascades are then initiated which converge on common steps that mediate a rise in intracellular Ca2+ followed by neurotransmitter release. On one hand, Gβƴ dimer activates phospholipase Cβ2 (PLCβ2) to generate diacylglycerol and inositol triphosphate (IP3), which binds to its receptor IP3R in the endoplasmatic reticulum, triggering the release of Ca2+ stores and, finally, TRPM5 ion channel-mediated membrane depolarization [9,10]. On the other hand, Gαgust down-regulates adenylyl cyclases (ACs) leading to the inhibition of cAMP production. Decrease in cAMP levels promotes the downregulation of the cAMP-dependent protein kinase A (PKA), with consequent activation of IP3Rs and PLCβ2 signaling components, both inhibited by PKA in resting states. The upregulation of IP3Rs and PLCβ2 leads to intracellular Ca2+ release and downstream membrane depolarization [11,12,13]. Two seminal studies analyzing in silico genomes of fish models indicate that Gαgust is not present in teleost fish and other Gαi subunits have been proposed to play homologous functions [14,15].
While the existence of unique T1R1 and T1R3 genes seems to be a constant feature of vertebrate genomes, numerous T1R2 paralogs have been thus far identified in several fish species [4,16,17,18,19]. Studies on fish taste function using heterologous expression systems revealed that putative sweet T1R2 genes respond to L-amino acids (L-AAs) rather than sugars in omnivore species such as zebrafish (Danio rerio) and medaka (Oryzias latipes) [20], or preferentially to plant-specific fructose in the herbivorous grass carp (Ctenopharyngodon idellus) [19]. These observations indicate that T1R2 gene expansion may have served a key role in the evolution of species-specific taste adaptation to diverse habitats and diets. To further explore this hypothesis, the main purpose of this study was to describe and functionally characterize the first T1R gene repertoire in a carnivorous fish, in addition to a species with key relevance in Mediterranean aquaculture, the gilthead seabream Sparus aurata (sa). Furthermore, two novel Gαi subunits were tested as putative taste-associated proteins alongside the functional characterization of saT1Rs, by means of their pharmacological responses to different L-AAs and sweet tastants.
This information, besides its potential significant value for further clarifying evolutive aspects, is of great practical interest for aquaculture production. Linked to its important role in identifying nutrients and sources of metabolic energy, the T1R gene family is associated with the perception of attractive taste modalities. Therefore, understanding the molecular mechanisms that control feeding preferences and promote feed consumption by attractive taste sensations will contribute towards the production of more efficient species-tailored feeds, enabling a better utilization of the diets through the modulation of feeding behavior and food intake. Such knowledge may be particularly relevant during periods of depressed appetite associated to specific physiological or productive stressful events, or when using alternative protein ingredients with low palatability in fish feed formulations.
2. Results
2.1. Identification of saT1R Genes
Five saT1R genes were initially identified through blast searches of the preliminary seabream genome assembly (http://biocluster.her.hcmr.gr/myGenomeBrowser?portalname=Saurata_vI), subsequently cloned in their complete coding sequences [saT1R1 (846 AAs, XP_030277517), saT1R2a (826 AAs, XP_030278006), saT1R2b (824 AAs, XP_030278002), saT1R2c (817 AAs, MT892735), saT1R3 (856 AAs, XP_030274769)], and used in the pharmacological characterization of different receptor combinations (all except saT1R2c). During the final stages of this study, the genome assembly project was released on NCBI genebank [21] and a new blast search identified a total of seven saT1R-related complete coding sequences (CDS) derived from automated computational analysis (Gnomon). The seven saT1R genes included four out of the five initially cloned genes in the present study and three additional saT1R gene predictions, hereby referred to as saT1R2d (826 AAs, XM_030422143), saT1R2e (824 AAs, XM_030422144) and saT1R2f (821 AAs, XM_030422145), respectively. Therefore, information available up to date indicates that the saT1R gene repertoire comprises eight members: two one-to-one orthologs of T1R1 and T1R3 vertebrate genes (saT1R1 and saT1R3) and six saT1R2 duplicate members (saT1R2a-f) highly conserved to each other (nt identity from 83 to 92%), and moderately with respect to saT1R1 and saT1R3 (± 47% and 42% respectively). InterProScan scanned against InterPro’s signatures [22] saT1R protein alignments show the characteristic conserved domain architecture of family 3 (or C-type) GPCRs (Figure S1). This includes a large extracellular region containing the N-terminal, the Venus Flytrap (VF) and the 9-cystein-rich (CR) domains that structurally link to the heptahelical transmembrane (7TM) domain located upstream of the intracellular carboxyl-terminal domain [23,24].
2.2. Phylogenetic Analysis of saT1Rs and saGαi Genes
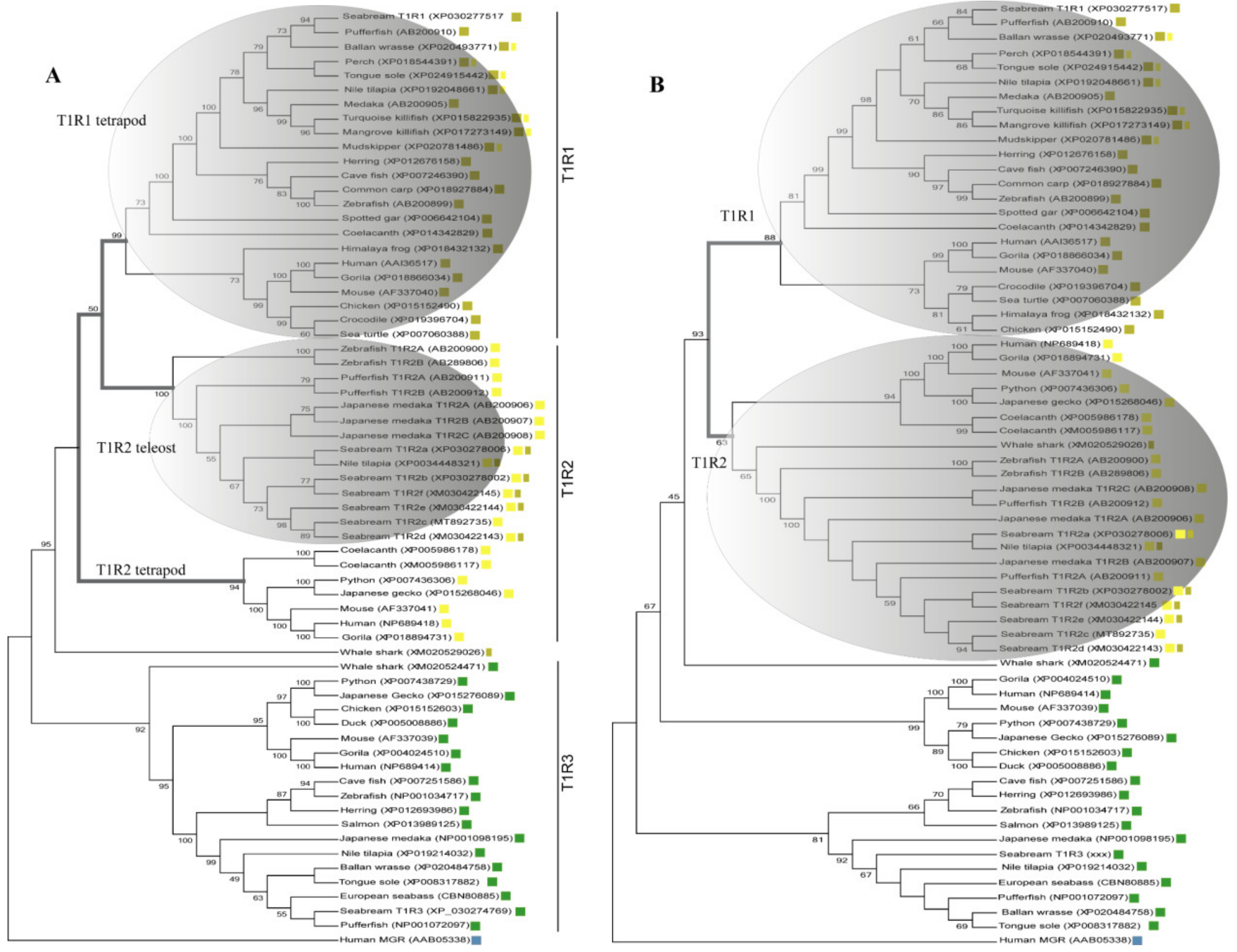
Inference of saT1R evolutionary relationships was performed using protein datasets including either the complete coding sequence (CDS-Tree) or the Venus Flytrap Domain (VFD-Tree) of annotated T1R ortholog sequences (64 sequences from 29 organisms). T1R phylogenetic trees from both CDS-Tree and VFD-Tree datasets, support the monophyly of T1R paralogs, with saT1R1, saT1R3 and saT1R2 genes clustering into separate clades, each comprising orthologs from birds, amphibians, reptiles and mammals (Figure 1A,B; Datasets S1, S2).
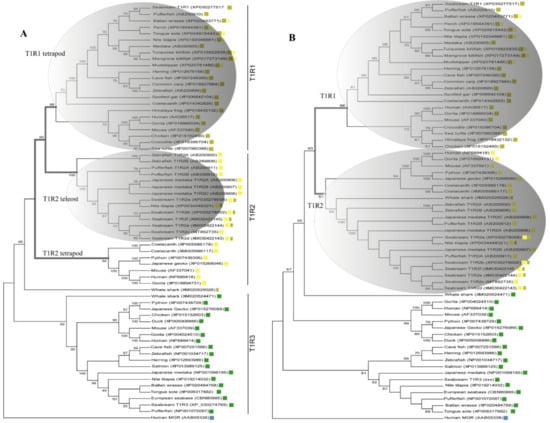
Figure 1.
Phylogenetic relationships inferred by Maximum Likelihood (ML) method of CDS-(A) and VFD-(B) trees using multiple alignments of deduced AA sequences of vertebrate T1Rs ortholog genes. Both evolutionary reconstructions cluster saT1R1, saT1R2a/b/c/d/e/f and saT1R3 genes in three distinct clades ( T1R1;
T1R1;  T1R2;
T1R2;  T1R3) each comprising their respective ortholog genes from mammals, amphibians, birds and reptiles. The (A) topology sets teleost T1R2 genes evolutionarily closer to T1R1 than T1R2 of tetrapod, while two compact T1R1 and T1R2 vertebrate clades are observed in the VFD (B) topology (grey line branches
T1R3) each comprising their respective ortholog genes from mammals, amphibians, birds and reptiles. The (A) topology sets teleost T1R2 genes evolutionarily closer to T1R1 than T1R2 of tetrapod, while two compact T1R1 and T1R2 vertebrate clades are observed in the VFD (B) topology (grey line branches  ).
).  T1R1 fish AA sequences as deduced from our phylogenetic reconstructions and NCBI-automate-annotated as vertebrate T1R2 orthologs.
T1R1 fish AA sequences as deduced from our phylogenetic reconstructions and NCBI-automate-annotated as vertebrate T1R2 orthologs.  T1R2 fish AA sequences as deduced from our phylogenetic reconstructions and NCBI-automate-annotated as vertebrate T1R1 orthologs. Robustness of the trees was estimated by 1000 random bootstrap replications. Only bootstrap values higher than 50% are shown. The human Metabotropic Glutamate Receptor 1 (MGR1;
T1R2 fish AA sequences as deduced from our phylogenetic reconstructions and NCBI-automate-annotated as vertebrate T1R1 orthologs. Robustness of the trees was estimated by 1000 random bootstrap replications. Only bootstrap values higher than 50% are shown. The human Metabotropic Glutamate Receptor 1 (MGR1;  ) was used for rooting the trees. GenBank T1Rs accession numbers are indicated next to species names.
) was used for rooting the trees. GenBank T1Rs accession numbers are indicated next to species names.
 T1R1;
T1R1;  T1R2;
T1R2;  T1R3) each comprising their respective ortholog genes from mammals, amphibians, birds and reptiles. The (A) topology sets teleost T1R2 genes evolutionarily closer to T1R1 than T1R2 of tetrapod, while two compact T1R1 and T1R2 vertebrate clades are observed in the VFD (B) topology (grey line branches
T1R3) each comprising their respective ortholog genes from mammals, amphibians, birds and reptiles. The (A) topology sets teleost T1R2 genes evolutionarily closer to T1R1 than T1R2 of tetrapod, while two compact T1R1 and T1R2 vertebrate clades are observed in the VFD (B) topology (grey line branches  ).
).  T1R1 fish AA sequences as deduced from our phylogenetic reconstructions and NCBI-automate-annotated as vertebrate T1R2 orthologs.
T1R1 fish AA sequences as deduced from our phylogenetic reconstructions and NCBI-automate-annotated as vertebrate T1R2 orthologs.  T1R2 fish AA sequences as deduced from our phylogenetic reconstructions and NCBI-automate-annotated as vertebrate T1R1 orthologs. Robustness of the trees was estimated by 1000 random bootstrap replications. Only bootstrap values higher than 50% are shown. The human Metabotropic Glutamate Receptor 1 (MGR1;
T1R2 fish AA sequences as deduced from our phylogenetic reconstructions and NCBI-automate-annotated as vertebrate T1R1 orthologs. Robustness of the trees was estimated by 1000 random bootstrap replications. Only bootstrap values higher than 50% are shown. The human Metabotropic Glutamate Receptor 1 (MGR1;  ) was used for rooting the trees. GenBank T1Rs accession numbers are indicated next to species names.
) was used for rooting the trees. GenBank T1Rs accession numbers are indicated next to species names.
Although the phylogenetic inferences deduced from CDS- and VFD-trees are generally in good agreement, the CDS-Tree sets tetrapod T1R1 and teleost T1R2 genes as sister groups (Figure 1A), while a compact T1R2 vertebrate clade is observed in the VFD-tree topology (Figure 1B).
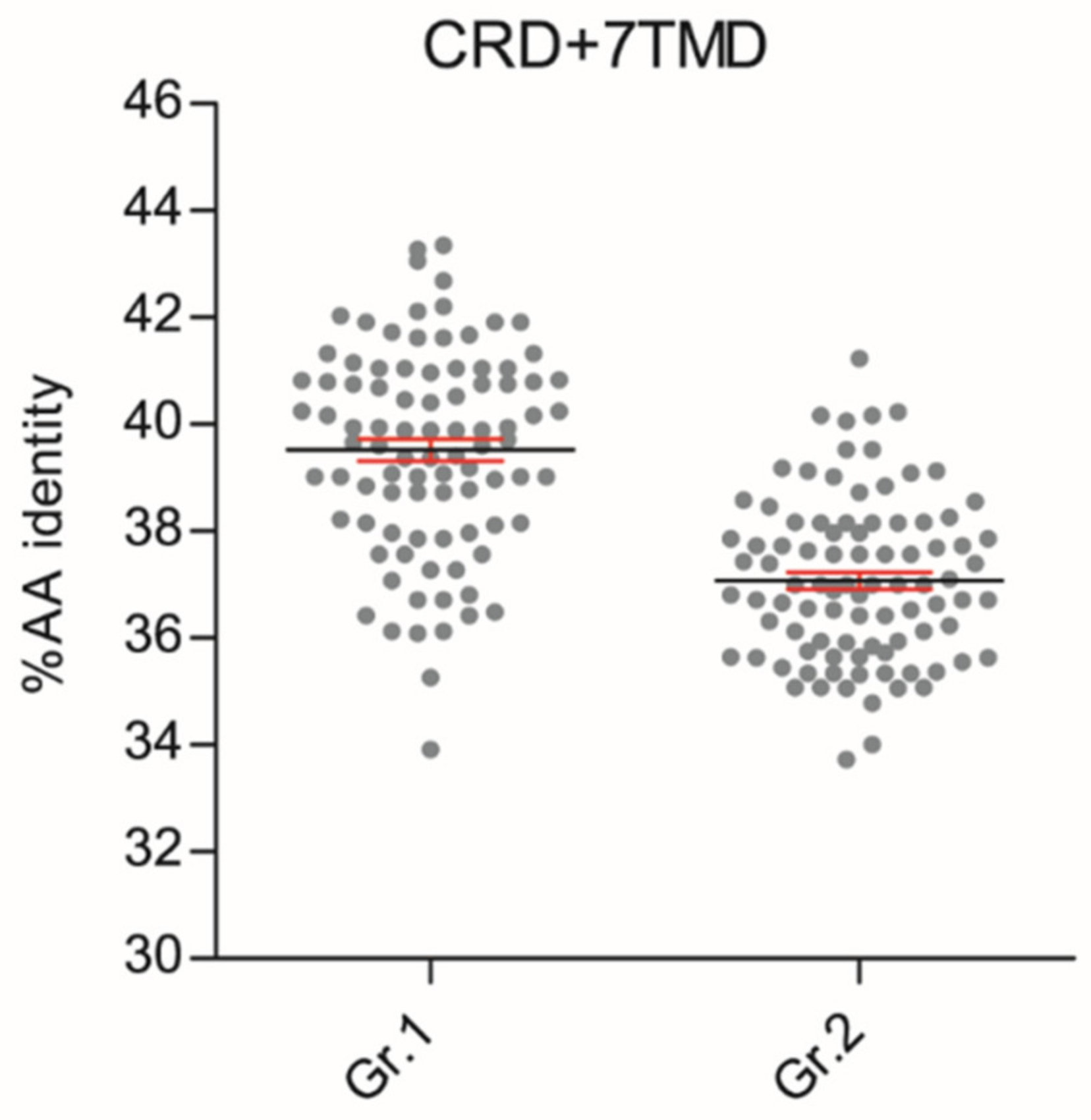
These phylogenetic reconstructions suggest that sequence conservation driving phylogenetic clustering of fish T1R2 with T1R1 tetrapod may be located outside the VFD. To substantiate this hypothesis, we carried out comparative sequence conservation analyses within the cysteine-rich and heptahelical transmembrane domains (CRD + 7TMD) of several T1R1 and T1R2 genes (Figure 2; Dataset S3). This analysis shows that fish T1R2 have significantly higher AA identity to T1R1 (39.52% ± 0.91) than to T1R2 (37.08% ± 0.58) of tetrapods (p < 0.0001) in these regions.
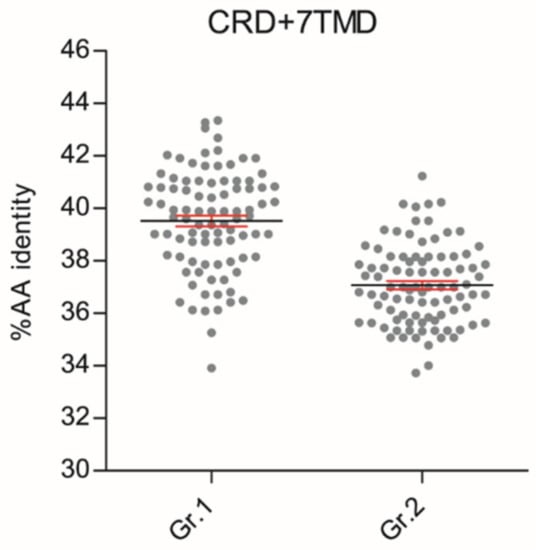
Figure 2.
Scatter plots of amino acid conservation within the 9-Cystein-Rich and heptahelical transmembrane domains (CRD + 7TMD, ± 348 AAs) of fish T1R2 versus tetrapod T1R1 (Gr.1) or tetrapod T1R2 (Gr.2). Each dot represents the percentage of AA sequence identities calculated in one-to-one combinatorial arrangements (n = 90) of fish T1R2s (n = 13) with T1R1 (n = 7) or T1R2 (n = 7) tetrapod sequences. T-test analysis of variance followed by Mann Whitney test was implemented to show significant differences between the two groups (p < 0.0001); percentage AA identity mean (black line) ± SEM (in red).
Moreover, our phylogenetic analyses refine the accuracy of fish T1R annotations, establishing new phylogenetic relationships for previously automatically annotated T1R1 and T1R2 fish sequences that unambiguously fall into opposite T1R orthology clades in both CDS- and VFD- datasets.
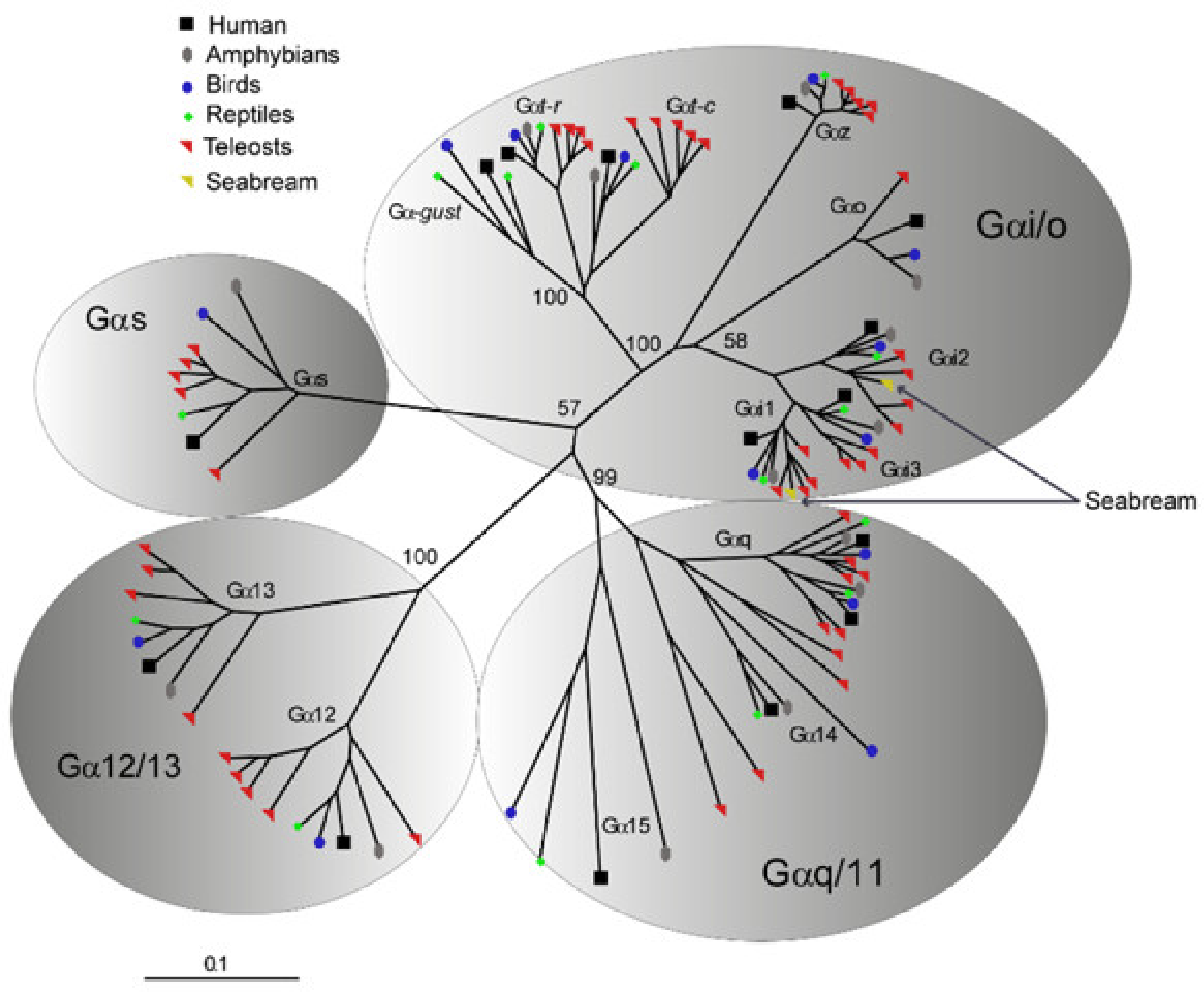
Phylogenetic analyses were additionally performed to validate the identity of saGαi1 and saGαi2 genes retrieved from NCBI (acc.ns. XP_030281956 and XP_030276216, respectively) and used for the in vitro transfections. Orthology inferences of saGαi CDSs were assessed using several vertebrate Gα-protein subunits (111 sequences from 18 organisms), representative of the four major classes Gs, Gi/Go, Gq/G11 and G12/G13 [25]. Results confirmed that saGαi1 and saGαi2 genes are members of the Gαi/o class, clustering closely with the respective vertebrate orthologs (Figure 3; Dataset S4).
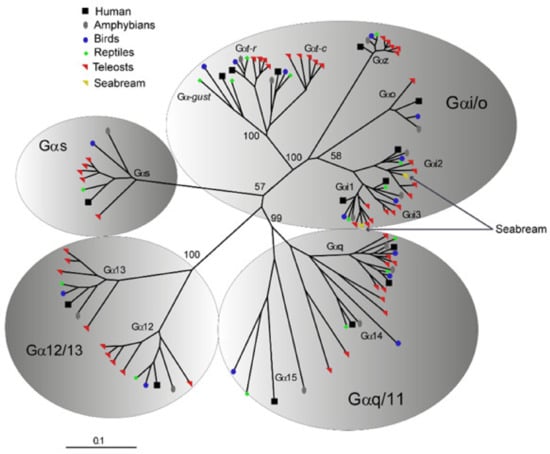
Figure 3.
Radial view representation of an unrooted Neighbor-Joining tree depicting the phylogenetic relationship among (Gα) protein subunits of Gs, Gi/Go, Gq/G11 and G12/G13 classes of representative vertebrate homologs. saGαi1 and saGαi2 AA sequences cluster in one-to-one ortholog relationships within the corresponding Gαi monophyletic clade, suggesting saGαi1-2 duplicated origin from ancient vertebrate whole genome duplications. Numbers above nodes indicate bootstrap values (based on 1000 replicates) that support the respective branch. The scale (lower left corner) indicates the mean number of AA substitutions per site.
2.3. Validation of pGL3-NFAT-luc Reporter Constructs for Intracellular Ca2+ Quantification in Transiently Transfected HEK293 Cells
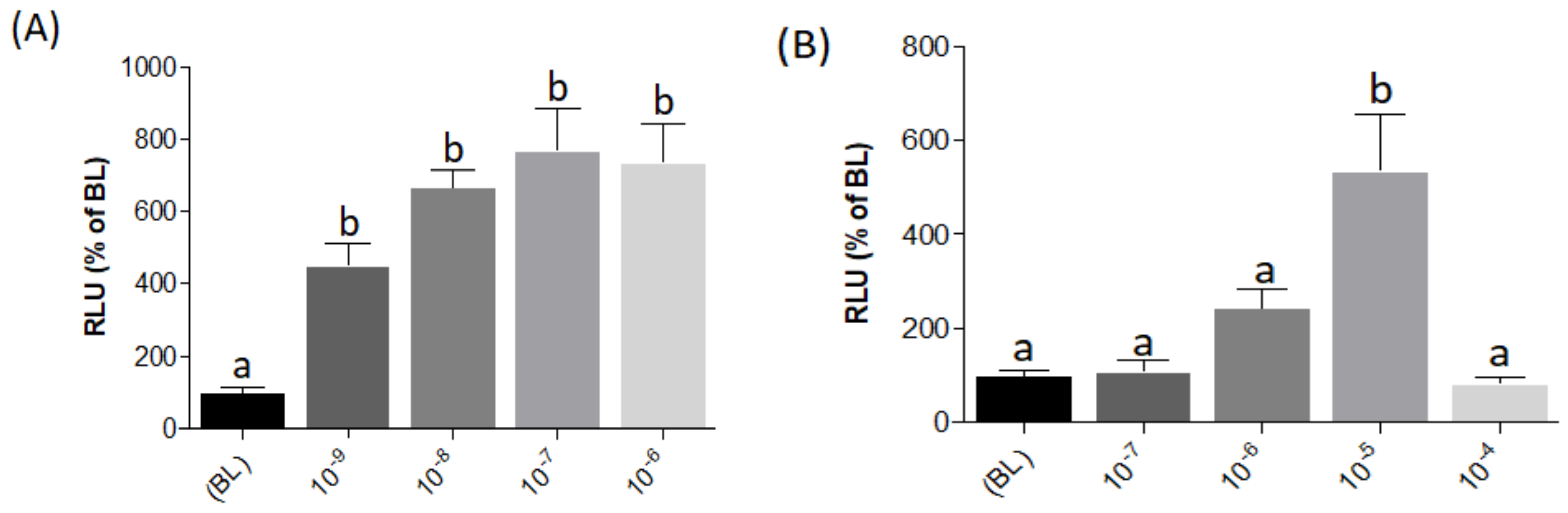
The luciferase reporter system used in the present study is based on a pGL3-plasmid including three (N)uclear (F)actor of (A)ctivated T cells (NFAT)-responsive elements located upstream of an interleukin-2 (IL2) minimal promoter driving expression of the firefly luciferase reporter gene (pGL3-NFAT-luc plasmid; Addgene (Watertown, MA, USA), cat. number. 17870). To evaluate pGL3-NFAT-luc construct responsiveness, transiently transfected HEK293 cells were stimulated with Phorbol 12-myristate 13-acetate (PMA) and ionomycin, two compounds known to modulate NFAT signaling by mobilization of intracellular Ca2+ storages [26,27,28,29]. Both compounds showed significant luminescence dose-response increases, indicating pGL3-NFAT-luc construct effectiveness for intracellular Ca2+ quantification (Figure 4A,B; Datasets S5 and S6) when expressed in a HEK293 heterologous cell system.
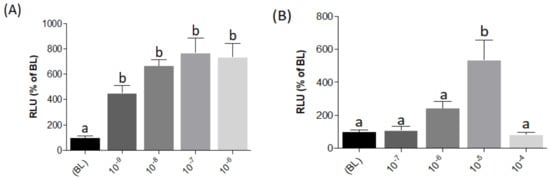
Figure 4.
Evaluation of pGL3-NFAT-luc construct responsiveness to 10-fold serial dilutions to PMA (A) and ionomycin (B) in transiently transfected HEK293 cells. RLU%: Relative Luminescence Unit percentage. Evaluation of L-AA responses was based on the RLU mean ± SEM of four independent determinations normalized to the mean response of the same transfections (n = 4) stimulated with assay medium (Dulbecco’s Modified Eagle Medium (DMEM) + 1% fetal bovine serum (FBS)). BL (Basal Levels) = 100% RLU. Different lowercase letters (a,b) on top of bars indicate significant differences (p < 0.05) between concentrations, assessed by one-way ANOVA followed by Tukey’s Multiple Comparison Test (GraphPad Prism version 5.00 Software, La Jolla, CA, USA).
2.4. Validation of Stable pGL3-NFAT-Luc-HEK293 Cell Lines
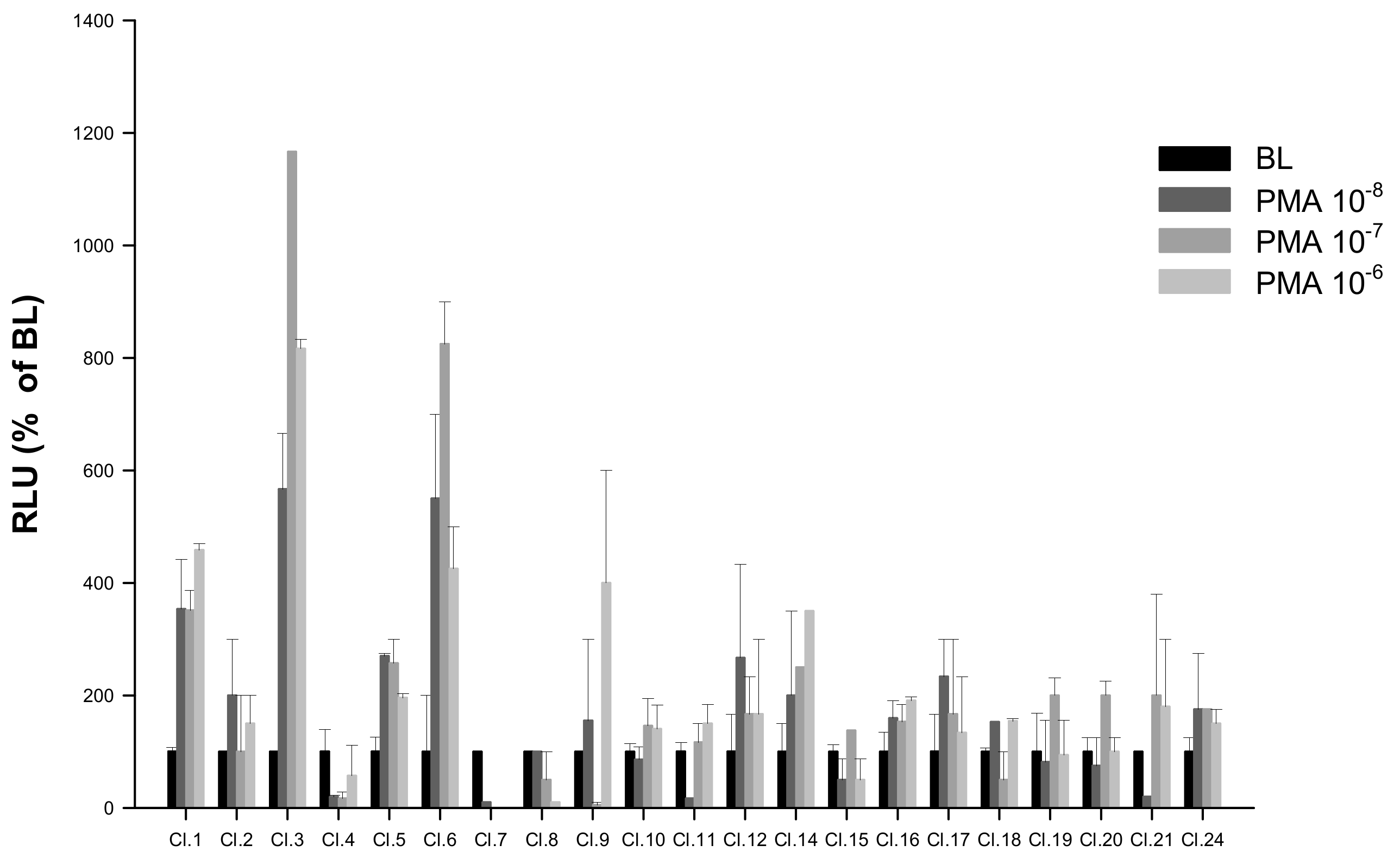
HEK293 cells lines stably expressing pGL3-NFAT-luc reporter construct were generated as described in Section 4.3. Among the 22 potentially stable pGL3-NFAT-luc-HEK293 clones screened by PMA, the clone showing highest luminescence responses (Cl.3) was selected for subsequent saT1Rs transient transfections (Figure 5; Dataset S5).
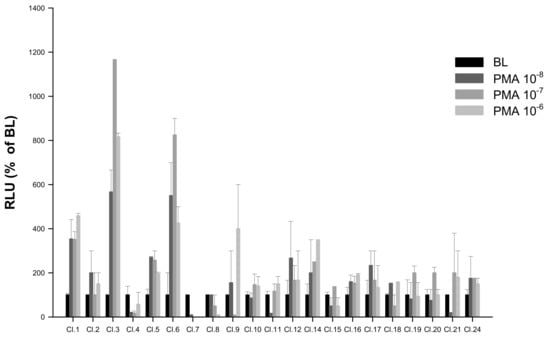
Figure 5.
Screening of putative stable pGL3-NFAT-luc-HEK293 clones by response to PMA (n = 4) at 10-fold serial dilutions tested over three concentrations (10−8, 10−7 and 10−6). Evaluation of PMA responses was based on the RLU mean ± SEM of four independent determinations normalized to the mean response of the same transfections stimulated with assay medium (DMEM +1% FBS). BL (Basal Levels) = 100% RLU. Clone 3 (Cl.3) was the most responsive to PMA-luminescence induction.
2.5. In Vitro Characterization of saT1: Responses to L-AAs
Consistent with the view that T1R proteins function as heterodimeric complexes, co-expression of T1R3 with either T1R1 or T1R2 has been largely reported in taste receptor cells of both mammals and fish [17,30,31]. Additionally, in vitro functional studies have been performed in several species, where it is generally assumed that the extent of T1R activation following L-AA and sugar stimuli is positively correlated to taste sensations [20,32,33]. However, as no information is available thus far on the activity of T1R receptors in a carnivorous fish species, we addressed this knowledge gap by characterizing the pharmacological responses of heterodimeric saT1R1/R3, saT1R2a/R3 and saT1R2b/R3 following transient transfections in stable (Cl.3) cell lines. Before cell culture experiments, expression of the initially identified saT1R genes was verified by qPCR and RT-PCR screening in several putative taste and non-taste tissues, and given that no evidence of saT1R2c transcriptional activity could be found (Figure S2), this saT1R2 duplicate was not considered for further functional investigation.
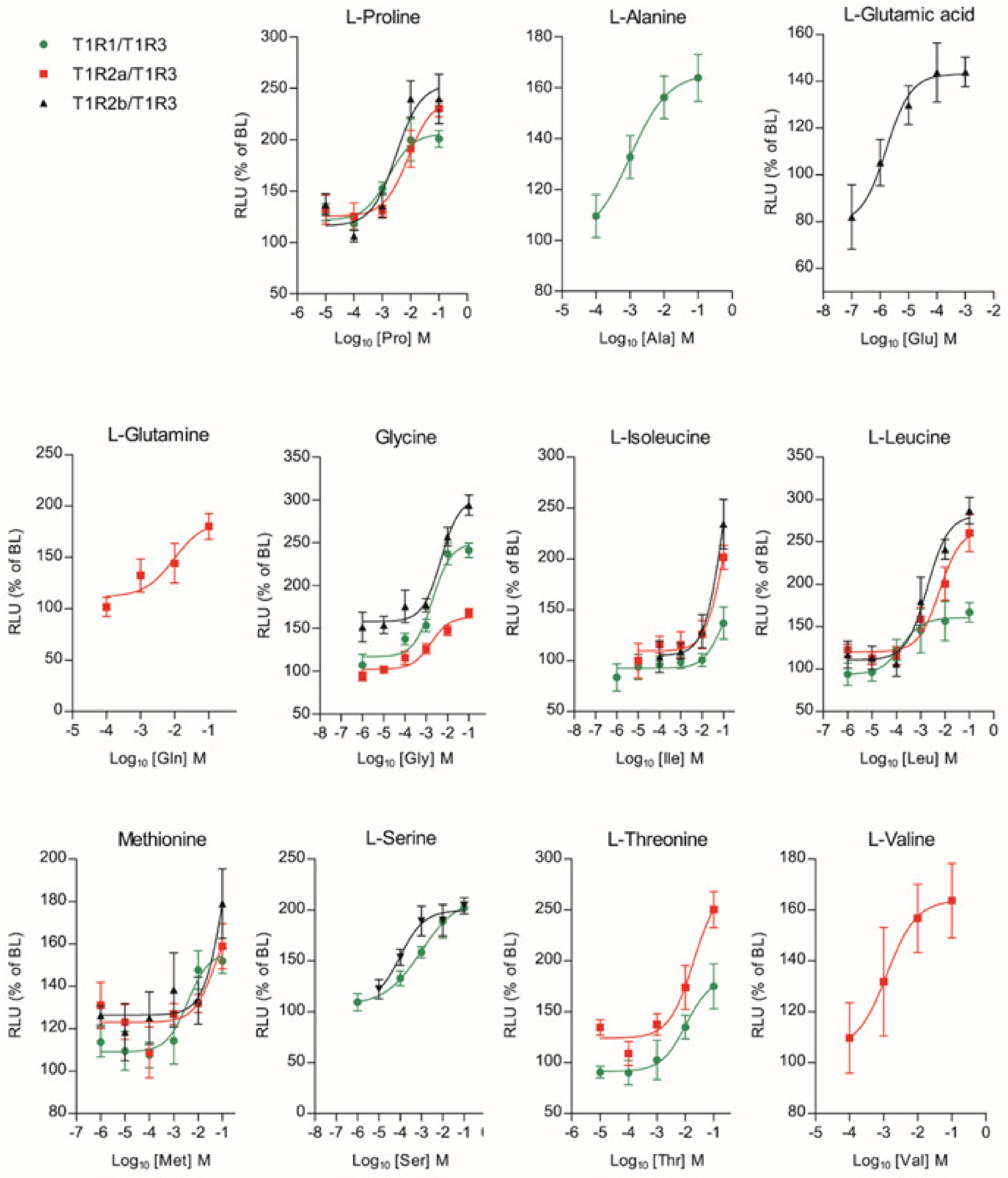
Among the full set of 20 L-AAs that were tested up to eight concentrations (10 nM to 100 mM), 11 of them generated a dose-response activation (Glutamic acid (Glu); Glutamine (Gln); Proline (Pro); Glycine (Gly); Alanine (Ala); Threonine (Thr); Serine (Ser); Valine (Val); Isoleucine (Ile); Leucine (Leu); Methionine (Met)), for at least one of the three saT1R heterodimers (Figure 6; Dataset S5).
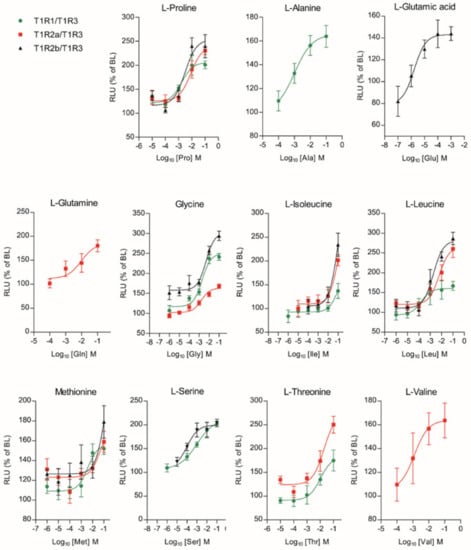
Figure 6.
Amino acid Log-dose response curves of activation in heterologous co-transfected (Cl.3) cells with saT1R1/3 (green dots), saT1R2a/3 (red squares) and saT1R2b/3 (black triangles) heterodimers. Evaluation of L-AA responses was based on the RLU mean ± SEM of four independent determinations normalized to the mean response of the same transfections stimulated with reduced assay medium (Basal Medium Eagle w/o L-glutamate (BME) + 1% FBS), and expressed as the percentage relative to basal levels (BL).
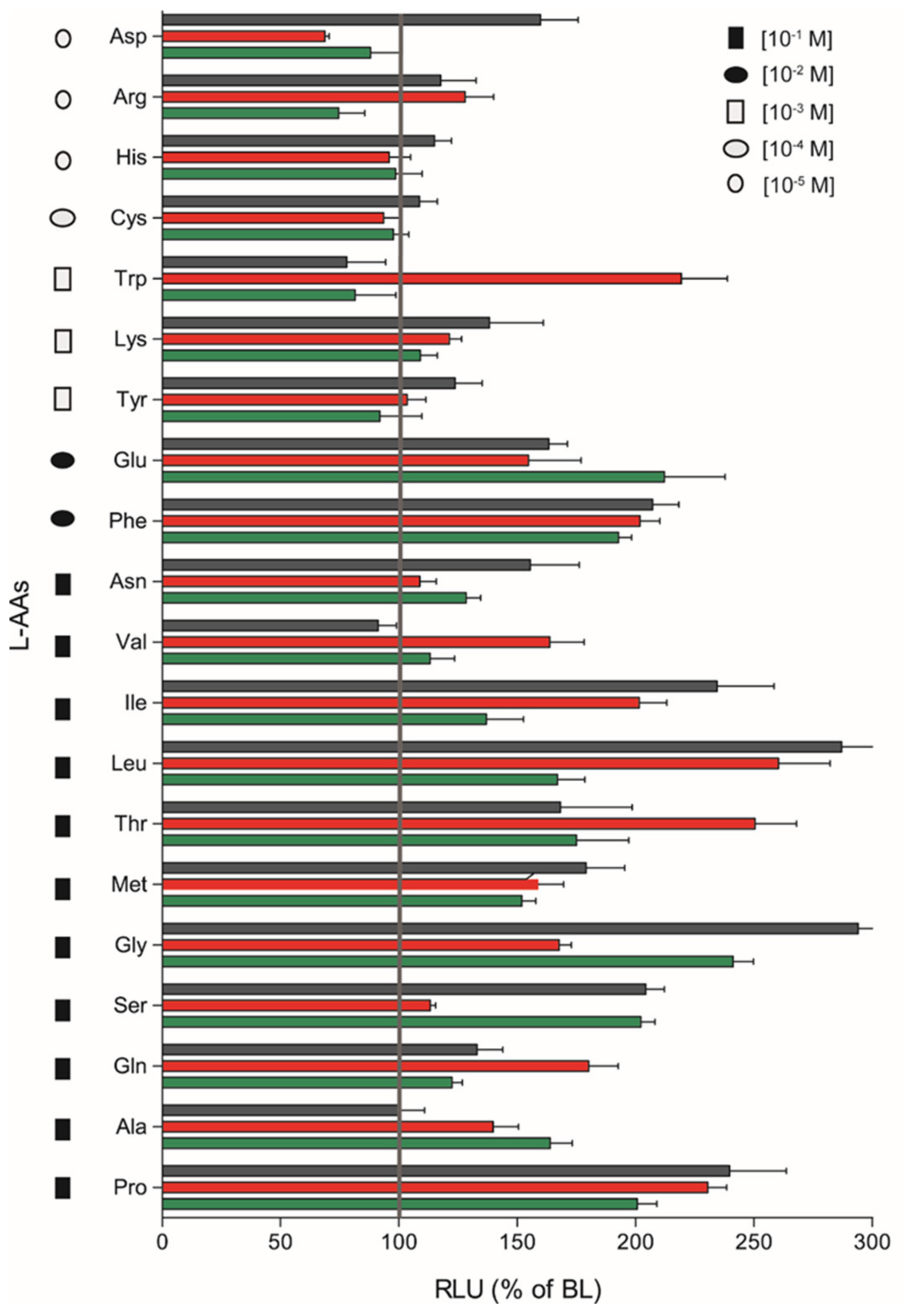
The remaining nine L-AAs (Arginine (Arg); Aspartic acid (Asp); Histidine (His); Asparagine (Asn); Cysteine (Cys); Lysine (Lys); Tryptophan (Trp); Tyrosine (Tyr); Phenylalanine (Phe)) did not respond in a dose-response manner, although significant stimulatory effects were observed for some heterodimers at a given AA concentration by plotting the magnitude of responses of saT1R1/R3, saT1R2a/R3 and saT1R2b/R3 at the maximal stimulation dosage (MSD) for each of the 20 L-AAs (Figure 7).
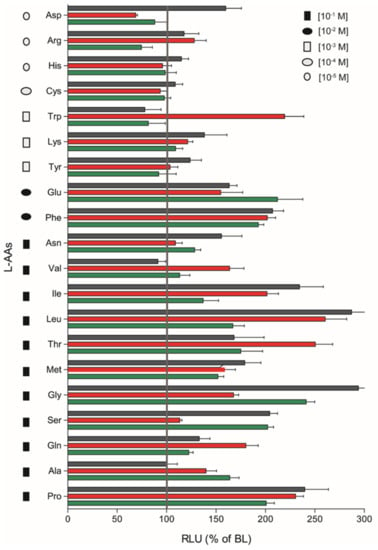
Figure 7.
Magnitudes of saT1R1/R3, saT1R2a/R3 and saT1R2b/R3 responses (green, red and black bars, respectively) to L-AAs based on RLU recorded at maximal stimulation dosages (MSD) within molar range concentrations of (10−1–10−5).
We evidenced fairly promiscuous profiles of activation, which suggests that the three taste-subunit combinations may equally serve to transduce L-AA taste sensations. saT1R2b/R3 was generally the most responsive heterodimer, showing the highest responses to Asp, His, Cys, Lys, Tyr, Phe, Asn, Ile, Leu, Met, Gly, Ser and Pro stimulations, followed by saT1R2a/R3 (Arg, Trp, Val, Thr, Gln) and saT1R1/R3 (Glu, Ala) (Figure 7; Table S1).
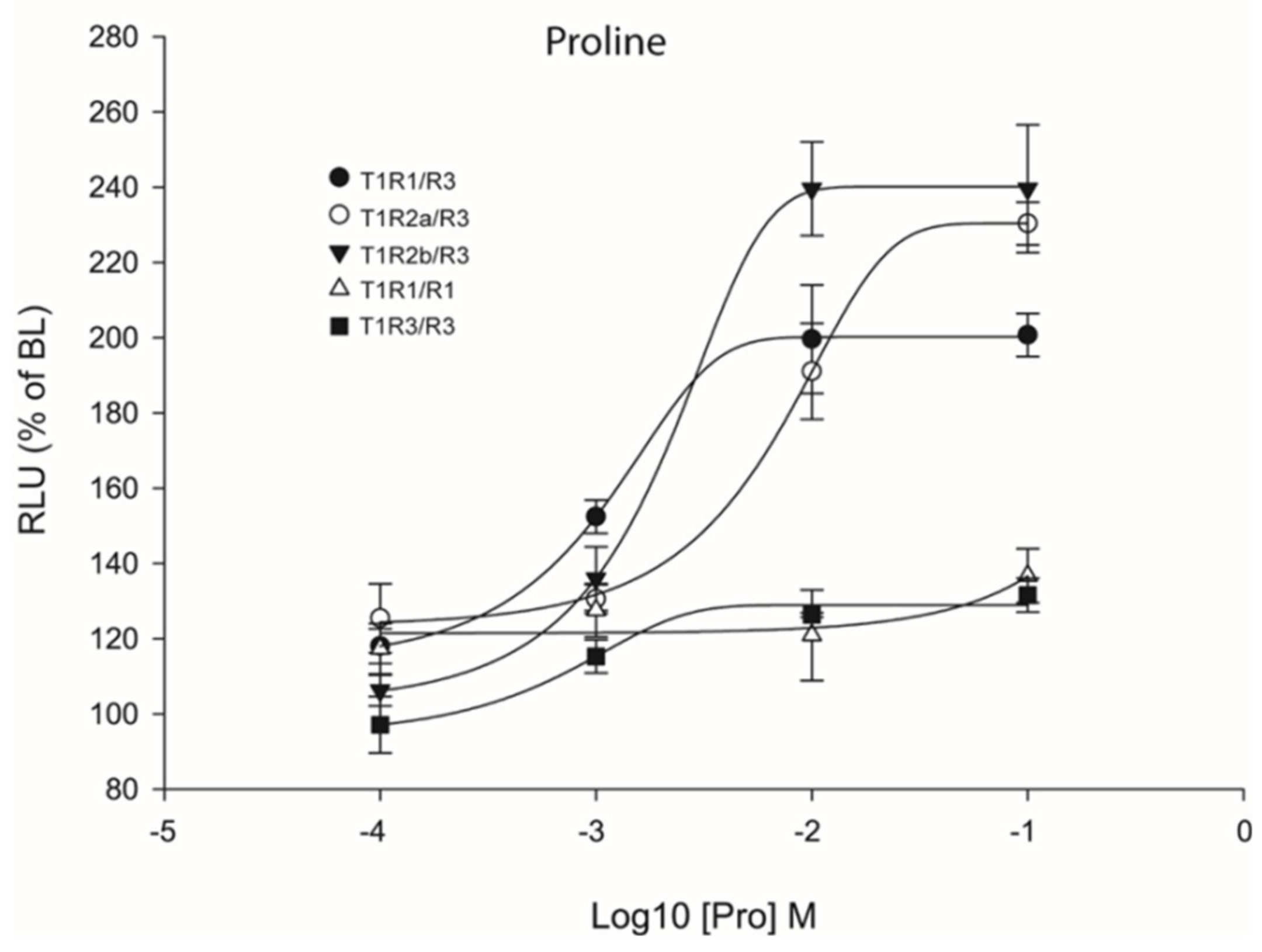
Previous studies have reported that heterodimeric coupling of saT1R complexes may not be the only mode of functional activation since these subunits, especially T1R3, can also couple as homodimers [30,34,35,36]. To analyze this possibility, transient transfections with double amounts of saT1R3 alone (or of saT1R1, as additional control) were also performed using L-Pro stimuli, as it presented highly reproducible dose responses for all three saT1R heterodimers (Figure 8; Dataset S5).
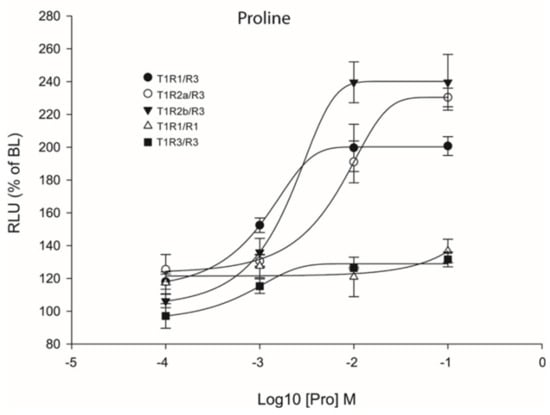
Figure 8.
Comparisons of proline Log-dose response curves of activation in heterologous transfections of hetero- and homo-dimeric saT1R combinations of saT1R1/R3 (black dots), saT1R2a/R3 (white dots), saT1R2b/3 (black triangles), saT1R1/R1 (white triangles) and saT1R3/R3 (black squares). Response is measured as RLU mean ± SEM of four independent determinations normalized to the mean response of the same transfections stimulated with reduced assay medium (BME + 1% FBS), expressed as percentage relative to basal levels (BL).
Our results showed that Cl.3 cells transiently transfected with only one saT1R subunit type did not respond to Pro stimulation. In addition, to reject the possibility that L-AA ligands could induce taste receptor-independent rises of Ca2+ [37], negative controls were also implemented by transfecting empty pcDNA™3 constructs, and potential non-specific luminescence signals were estimated at known MSDs for a subset of L-AAs (Pro, Ala, Gln, Ser, and Val) at 100 mM (Figure S3).
Furthermore, evaluation of half-maximal effective concentrations (EC50) of the L-AAs showing dose-response curves indicates important differences in the sensitivity of saT1R1/R3, saT1R2a/R3 and saT1R2b/R3. Although in the majority of cases direct comparison for a given L-AA was not possible for the three heterodimers, saT1R2b/R3 responded with the highest EC50 sensitivity recorded (Glu 1.68 × 10−6 and Ser 9.94 × 10−5 M, respectively; Table 1).

Table 1.
Evaluation of saT1R1/R3, saT1R2a/R3 and saT1R2b/R3 sensitivity, as deduced from ligand potency parameter EC50, recorded for 11 L-AAs producing a dose-response curve of activation.
On the other extreme, saT1R2a/R3 generally responded with the lowest sensitivity to L-AAs. Overall, and irrespectively of the specific heterodimer that was assessed, the following molar rank order of L-AA potencies as activators of saT1R-mediated taste responses was found: Glu > Ser > Leu > Ala > Pro > Val > Gly > Met > Gln > Thr > Ile (Table 1). It is noteworthy that the majority of EC50 sensitivity values recorded (in the range of 10−3-10−4 M) are in good agreement with those previously reported for T1R heterodimeric complexes of zebrafish and medaka using Ca2+-sensitive fluorescent dyes [20], further validating our methodological choice based on luciferase gene reporter systems.
2.6. In Vitro Characterization of saT1R: Responses to Natural Sugars
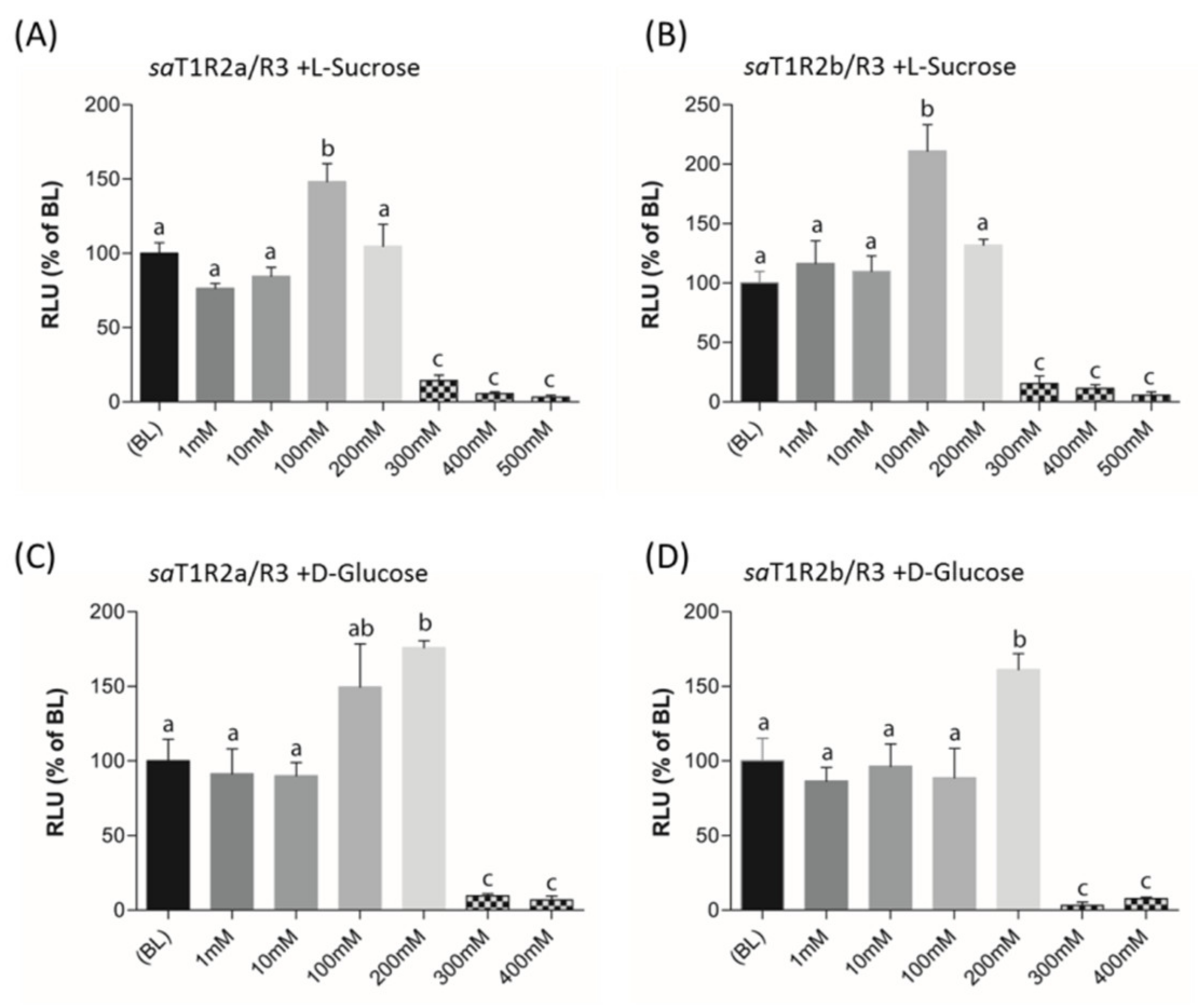
When L-Sucrose and D-Glucose were tested as potential ligands of putative sweet taste receptors saT1R2a/R3 and saT1R2b/R3, they elicited dose response patterns similar to those observed in mammalian T1R2/R3 [32,38]. Significant activations of both saT1R2a/R3 and saT1R2b/R3 were observed at 100 or 200 mM for L-Sucrose (Figure 8B, Figure 9A; Dataset S7) or D-Glucose (Figure 9C,D; Dataset S7), respectively, while at higher doses (300–500 mM), both sugars were toxic for the cells and/or inhibited their stimulation.
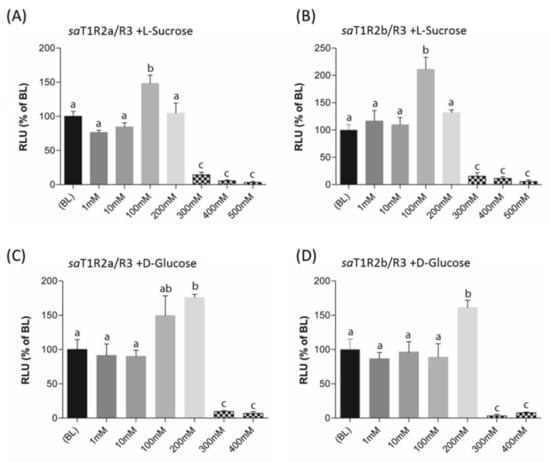
Figure 9.
saT1R2a/R3 and saT1R2b/R3 heterodimer responses to L-Sucrose (A,B) and D-glucose (C,D) at different concentrations (1–500 mM). Evaluation of sugar responses was based on the RLU mean ± SEM of four independent determinations (n = 4) normalized to the mean response of same transfections stimulated with reduced assay medium (BME + 1% FBS). BL (Basal Levels) = 100% RLU. Different lowercase letters (a,b,c) on top of bars indicate significant differences (p < 0.05) between concentrations, assessed by one-way ANOVA followed by Tukey’s Multiple Comparison Test (GraphPad Prism version 5.00).
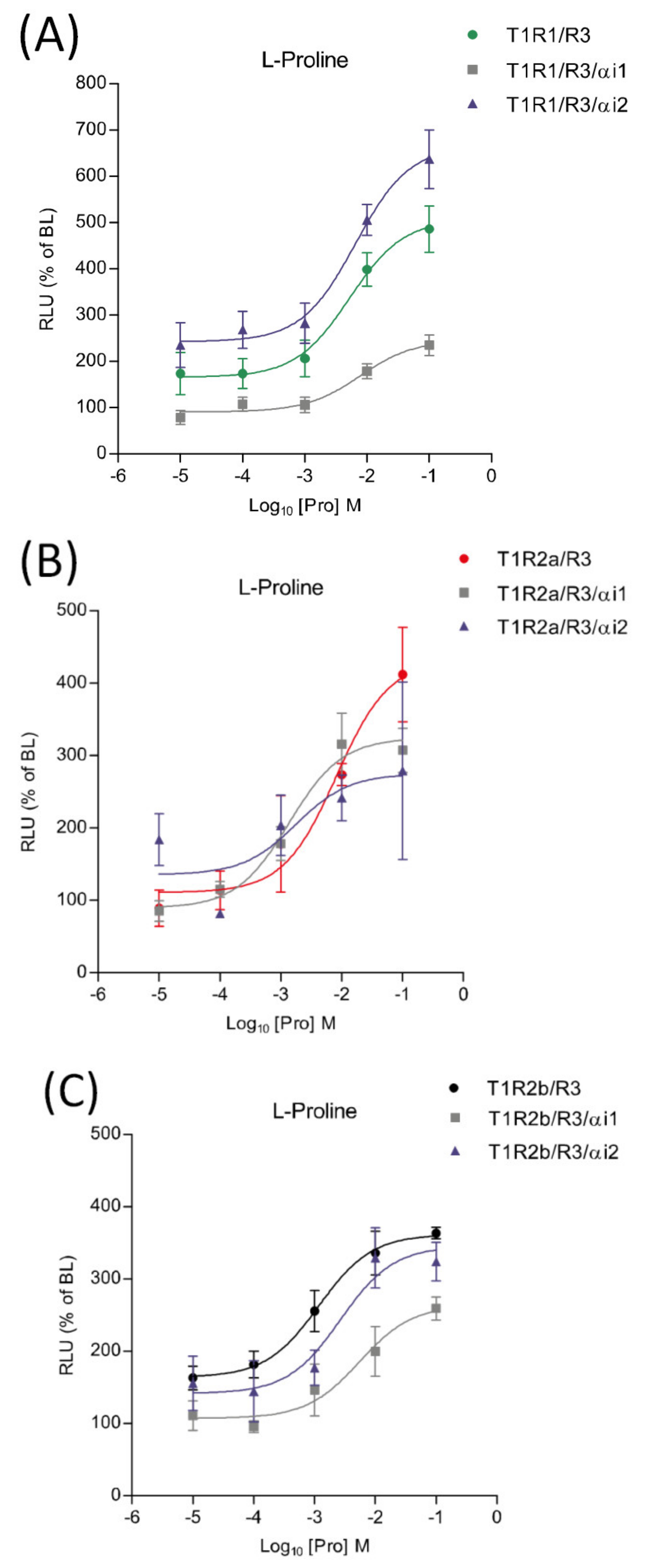
2.7. Pharmacological Responses to Proline in the Presence of saGαi1-2
The potential involvement of saGαi1-2 subunits in taste signaling pathways was investigated using transient heterologous expression of saT1R1/R3, saT1R2a/R3 and saT1R2b/R3 heterodimers in combination with saGαi1 or saGαi2 constructs, and using Pro as a standard ligand. When co-transfected with the saT1R1/R3 heterodimer, the two subunits triggered opposite effects: stimulatory in the case of saGαi2 and inhibitory for saGαi1 (Figure 10A; Dataset S5). On the other hand, both saGαi1-2 mediated inhibitory effects when co-transfected with the saT1R2b/R3 heterodimer (Figure 10C; Dataset S5). Due to the high variability recorded between independent determinations (n = 4), no clear profiles were observed for saT1R2a/R3/αi1 or /αi2 combinations, although both saGαi subunits show a general tendency towards inhibitory effects (Figure 10B; Dataset S5).
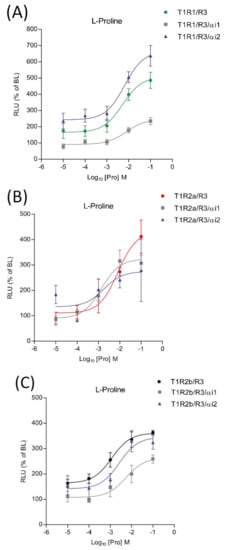
Figure 10.
Comparisons of proline Log-dose response curves of activation in heterologous transfections of saGαi1 and saGαi2 in combination with saT1R1/R3 (A), saT1R2a/R3 (B) and saT1R2b/3 (C). Evaluation of proline responses was based on the RLU mean ± SEM of four independent determinations normalized to the mean response of same transfections stimulated with reduced assay medium (BME + 1% FBS), expressed as the percentage relative to basal levels (BL).
3. Discussion
The phylogenetic tree reconstructions reported new putative T1R sequences identified in the present research. All ortholog proteins for each of the three T1R ancient paralogs showed similar conserved domains, as evidenced by their phylogenetic relationships. The earlier evolutionary emergence of T1R3 suggests that it might have been the first duplicated gene from a common T1R3/T1R1-2 ancestor.
The presence of multiple T1Rs in both fish and tetrapod genomes indicates that T1R duplications might have occurred before Actinopterygian/Sarcopterygian divergence, around 400 MYA [39]. Blast searches in ancient vertebrate genomes of Ciona intestinalis amphioxus, hagfish and lampreys failed to detect T1R distant homologs, while an elasmobranch genome (whale shark) exhibits two T1R genes. This suggests that the evolutionary expansion of T1R gene family might have taken place after jawless vertebrate radiation, predating the osteichthyes/chondrichthyes divergence. Furthermore, saT1R2 paralogs fall into a major clade including multiple T1R2 orthologs from other teleost species, suggesting that fish T1R2 expansion might have occurred in the common ancestor of extant teleost, in line with the teleost-specific (TS)–Whole Genome Duplication (WGD) hypothesis [40,41]. Yet, the presence of two T1R2 paralogs in the primitive land-vertebrate coelacanth, and the general consensus for singleton T1R1 and T1R3 loci in teleost genomes, also opens up alternative scenarios for an ancient T1R2 gene-specific duplication event predating Actinopterygii/Sarcopterygii divergence.
Both T1Rs phylogenies as deduced from CDS- and VFD-trees have been previously described [4,18], and possible functional implications can be ascertained based on these tree reconstructions.
The pivotal role of the class C-GPCR-VFD in ligand binding and transduction activation has been long recognized since the first solved crystal structure of the human MGR1. This protein is characterized by a dimeric bi-lobed protomer organization of the VFD domain that forms either an open “V” or a compact “U” dimer arrangement, reflecting resting or active states, respectively, upon glutamate binding [42]. Although attempts to purify and crystallize mammalian T1R-VFD have been unsuccessful until now, T1Rs are also likely to have a dynamic rearrangement of the dimeric VFD, based on sequence and protein structure similarities to MGR1. Recently, determination of the crystal structure of the heterodimeric T1R2a/3-VFDs in medaka indicated similar conformational changes as those underling human MGR1 activation [43], suggesting that the molecular structural basis for ligand recognition is presumably conserved within class C-GPCRs [44]. Moreover, protein modeling of medaka T1R2a/3-VFDs shows that ligand domains in this module might bind to different amino acids in a broad yet discriminating manner [45].
Although orthosteric sites of class C-GPCR have been thus far mainly identified in the VFD, the CRD and 7TM domains are also known to provide additional molecular basis for receptor activation via allosteric binding to endogenous and synthetic ligands, enabling to change the conformational state of the receptor, and thereby potentially modulating affinities and/or efficacies of orthosteric ligands [46]. For instance, mutational analyses and molecular modeling studies showed that several allosteric modulators bind to conserved pockets located in the CRD and 7TM domains of metabotropic, calcium sensing and sweet taste receptors [47,48,49,50,51].
Our T1R phylogenetic reconstructions based on CDS and VFD sequence datasets indicate that sequence conservation driving phylogenetic clustering of fish T1R2 with T1R1 tetrapod resides outside the VFD. Indeed, comparative sequence conservation analyses of (CRD + 7TMD) domains located downstream of the VFD show that fish T1R2 have significantly higher AA identity to T1R1 than to T1R2, suggesting that orthosteric and/or allosteric T1R2 responsive domains for L-AA recognition in fish might reside in these regions. To validate these predictions, dissection of conserved domains and generation of truncated versions of fish T1Rs should be performed to elucidate ligand binding specificity of individual protein modules. Such information would enable a better understanding of the structure-to-function link at the basis of umami and sweet T1R-mediated taste modalities in fish.
Evaluation of saT1Rs pharmacological responses to L-AAs and sweet ligands was conducted by systematic transient transfections of the heterodimers saT1R1/R3, saT1R2a/R3 or saT1R2b/R3 in HEK293 cells lines stably expressing pGL3-NFAT-luc reporter construct for intracellular Ca2+ /NFAT signaling detection. The basic principle in the use of NFAT luciferase constructs for quantification of saT1R responses lies in the role of Ca2+ as major secondary messenger in both taste transduction [52,53,54] and NFAT-mediated immune responses [55]. NFAT proteins reside ubiquitously in the cytosol in their inactive phosphorylated state [56]. Upon cell stimulation, increase of the Ca2+-dependent calcineurin dephosphorylates the serine-rich regions of NFAT regulatory domains, resulting in nuclear NFAT translocation and DNA binding to initiate and maintain different specific transcriptional programs [57,58,59,60]. Over the past decade, taste receptor functions have been mainly addressed by in vitro assay systems using Ca2+-sensitive fluorescent dyes, such as fluo-3, fluo-4 and fura-2 [20,38,61,62], or luminescence-based assays using jellyfish-derived apophotoproteins, such as aequorin, obelin and clytin as Ca2+ indicators [63], and more recently, the evaluation of taste receptor induced-calcium signaling in cell culture systems has been strengthened by the implementation of Förster resonance energy transfer (FRET) technologies [43,45]. To our knowledge, this is the first report employing a luciferese gene reporter system based on Ca2+-dependent calcineurin/NFAT signaling for taste receptor functional assays.
The in vitro characterization of saT1R responses to L-AAs and natural sugars indicate that T1R2-mediated sweet taste signaling has been conserved through the divergence between tetrapod and fish lineages. Nevertheless, T1R2 genes of some fish species have additionally acquired the ability for sensing AA compounds with considerably higher sensitivity than sugars, possibly as an adaptive mechanism to diversify feeding habits. In support of fish T1R2 adaptive functions, it was recently shown that a subset of four recently duplicated T1R2 paralogs in the herbivorous grass carp displayed enhanced T1R2s/T1R3 responses to plant-specific fructose. The authors suggest that T1R2 gene expansion in this species (possibly deriving from the extra lineage-specific genome duplication of cyprinids [64]) underlies taste adaptive strategies to dietary transition from carnivore to herbivore food habits [19]. Similarly, heterologous transfections of T1R2a/b/c/R3 dimeric complexes and testing with a broad range of L-AAs and natural and artificial sugars as potential ligands in the omnivorous zebrafish and medaka fish suggested that duplicated T1R2s in these species may have evolved for tuning a wide range of sensory modalities with a prominent sensitivity to amino acids [20]. Our data in a carnivorous species also support this idea.
Hence, current evidence suggests that expansion of T1R2 paralogs in fish genomes may have been particularly prone to positive selection, acting to improve fitness advantage in feeding adaptations to natural environments. This is particularly relevant in fishes, which are the largest and most diverse group of vertebrates, with nearly 30,000 species accounting for approximately half of all extant vertebrates [65,66]. Furthermore, fish inhabit almost every aquatic environment, many sharing the same ecological niche, often leading to the evolution of feeding specializations, for which variability in dietary preferences is key [67]. Other important fish gene duplications conferring adaptation to a wide variety of habitats have been reported for several gene families including, for instance, opsins [68,69], detoxification sulfotransferase (SULT) genes [70] or antifreeze glycoproteins (AFGP) [71], among others.
The development of a taste system with a broad spectrum and high sensitivity to detect amino acids in fish species is logically linked to the particularly high reliance on proteins rather than carbohydrates as a main source of metabolic energy, while glucose to satisfy the animal’s needs is produced mostly through gluconeogenesis from amino acids [72]. Such high protein requirements of fish are manifested by a striking 50–300% higher optimal dietary protein levels in aquaculture fish diets compared to terrestrial farm animals [73]. Within this high range, protein and AA requirements of different fish species can vary greatly; herbivorous and omnivorous species may require a diet with 25 to 35 percent crude protein, while carnivorous fish generally need higher amounts ranging from 35 to 50 percent of the total diet [74,75,76]. Furthermore, fish species can be very efficient in utilizing dietary amino acids for endogenous protein synthesis and deposition into body with high rates [77]. Nevertheless, precisely formulating diets in accordance to the species’ specific amino acid dietary requirements is a critical aspect given that AA deficiencies, or excesses, can impair key metabolic pathways, body homeostasis, immune responses, reproduction, welfare and growth [78]. In addition, importantly, high quality protein sources and supplemental AAs are expensive feed components that greatly influence production costs of global intensive aquaculture systems. An efficient utilization of dietary protein not only depends on its amino acid profile meeting a specific species’ life stage or physiological state requirements, but it is equally important to ensure a good acceptability of the feed and design appropriate feeding regimes that minimize feed waste. The need to substantially replace fish meal by alternative (non-capture fisheries based) protein sources in fish diets is a major focus of modern-day aquaculture [79,80]. Experimenting with mixtures of a wide range of alternative protein ingredients, combined with crystalline amino acid supplementation, has enabled great advances in this sustainability target. However, this is not always possible to accomplish in a cost-effective manner, particularly in carnivorous fish species. Difficulties are often associated to a lower acceptability (i.e., reduction of feed intake) of alternative proteins, particularly those of vegetable origin [81,82,83]. In this respect, an improved understanding of fish taste palatability and preferences will contribute towards achieving optimized feeding formulations tailored to species of aquaculture interest.
Two new saGαi genes have been additionally reported and phylogenetically characterized in this study. Taken together, our results suggest that despite their primary sequence conservation (83% identity), duplication of these saGαi genes might have provided genetic background for functional diversification. Furthermore, we provide preliminary data supporting the differential involvement of saGαi1 and saGαi2 subunits in taste transduction signal. While enhancement of the saGαi-mediated stimulatory effects on T1Rs are in good agreement with the known action of mammalian Gαgust in taste chemosensory transduction [34,84], it is challenging to speculate on the possible mechanisms involved in saGαi-mediated inhibitory effects. This certainly needs to be explored through further research, but existing literature enable us to formulate a hypothesis that could potentially explain these preliminary results. GPCRs have a functional versatility that enables them to activate more than one G protein type to change dynamically downstream signaling networks. For instance, the stimulatory α-subunit (Gαs) of the ubiquitously expressed Gs protein is known to have opposite effects to Gαi by mediating the activation of ACs, resulting in increases of intracellular cAMP and activation of PKA with consequent downregulation of PLCβ2 and IP3R components leading to declining Ca2+ levels [85,86]. In the context of our experiments, we can speculate that in vitro saGαi1 or saGαi2 overexpression might cause, directly or indirectly (and depending on the saT1R heterodimer type), functionally-impaired modulation of alternative Gα subunit proteins (such as Gαs), leading to intracellular Ca2+ decreases (as deduced from the reduced RLU levels). Indeed, recent studies based on computer modeling and bioluminescence FRET assays provided evidences for different Gαi and Gαs interacting interfaces in GPCR heteromeric complexes [87]. Furthermore, functional studies demonstrated the existence of cross-regulation leading to opposite downstream signals between Gi and Gs pathways [88].
4. Materials and Methods
4.1. In Silico Identification and Molecular Cloning of saT1Rs and saGαi Genes
In the initial phase of the study, pufferfish (Tetraodon negroviridis) T1R1 protein sequence (acc.no. AB200910) was used as a query against the preliminary seabream genome assembly (http://biocluster.her.hcmr.gr/myGenomeBrowser?portalname=Saurata_vI [21]), by translated tblastn search algorithm. Five unique saT1R gene fragments were found scattered throughout different contigs (Scaffold 199W17253) and subsequently analyzed using bioinformatics tools to predict coding sequences (https://blast.ncbi.nlm.nih.gov/Blast.cgi; http://web.expasy.org/translate), determine intron-exon boundaries (http://genes.mit.edu/GENSCAN.html), build alignments to identify multiple paralogs (https://www.ebi.ac.uk/Tools/msa/clustalo) and predict signal peptides as primary indication of functional genes (http://www.cbs.dtu.dk/services/SignalP). In silico saT1R gene fragments were identified as orthologs of medaka and zebrafish T1R1, T1R2a, T1R2b, T1R2c and T1R3 genes [20]. Four out of the five fragments (all except T1R2c) were further extended by 5′ and 3′ rapid amplification of cDNA ends (RACE) libraries prepared from tissue pools of lip, tongue, oral cavity epithelium and gill mRNAs (SMARTer ®® RACE 5′/3′ Amplification Kit, Clontech, Mountain View, CA). Amplification of saT1R CDS was performed using Long-Range PCR Kit (Qiagen, Toronto, CA); RT-PCR yields were subsequently gel purified (QIAquick, Qiagen, Toronto, ON, CA), cloned into pGEM-T Easy vectors (Promega, Madison, USA) and sequenced on both strands (University of Valencia, Valencia, Spain). saGαi1 and saGαi2 CDSs derived from NCBI automated predictions were also validated by conventional reverse transcription RT-PCR approaches and sequenced as described before. Primer sequences are available in electronic Supplementary Materials Table S2.
4.2. Phylogenetic Analyses
Multiple sequence alignments were generated using ClustalX V1.81 (Dublin, Ireland) [89] and Maximum Likelihood (ML) or Neighbor Joining (NJ) phylogenetic trees were constructed for each dataset based on the JTT matrix-based model, using MEGA5 (State College, PA, USA) [90,91] and NJplot (Villeurbanne, France) software’s [92]. Phylogenetic radial view of Gα proteins was performed by TreeView (Salisbury, UK) software [93] and cladograms robustness at each branching node was estimated by 1000 random bootstrap replications [94]. The human class (C) G protein-coupled metabotropic glutamate receptor 1 (MGR1, acc.no. AAB05338) was used as a distant out-group for rooting T1R phylogenetic trees.
4.3. Generation of Stable pGL3-NFAT-Luc HEK293 Cell Lines
HEK293 cells were cultured in standard medium DMEM (Gibco, Thermo Fisher, Saint Louis, MO, USA) containing 10% FBS, penicillin (100 U/mL) and streptomycin (100 g/mL) at 37 °C, with a humidified atmosphere at 5% CO2. pGL3-NFAT-luc-HEK293 stable clones were generated by co-transfections of pGL3-NFAT-luc and tgCMV/HyTK plasmids (50:1), the latter harboring the hygromycin phosphotransferase gene, using Lipofectamine LTX according to supplier’s protocols (Thermo Fisher, Saint Louis, MO, USA). Initially, HEK293 cells were selected using DMEM containing 400 μg/mL hygromycin B (Sigma, Darmstadt, Germany) in 24-well plates for two weeks; 24 colonies were further grown out in 96-well plates under reduced hygromycin selection (200 µg/mL) for three weeks. Potentially resistant NFAT-luc-HEK293 clones (n = 22) were validated after incubation in assay medium (DMEM + 1% FBS) containing PMA or Ionomycin (Sigma, Darmstadt, Germany) for 18 h (triplicates, 50,000 cells/well). Luciferase activity was quantified with ONE-Glo™ EX Reagent Kit (Promega, Madison, WI, USA) using a 96-microplate TECAN reader (Trading AG, Switzerland).
4.4. Transient Transfections and Stimulation Assays
saT1Rs (saT1R1, saT1R2a, saT1R2b saT1R3) and saGαi1-2 genes were RT-PCR amplified using 5’ and 3’ flanking primers carrying HindIII and XhoI restriction sites, respectively. Gel-purified fragments were cloned into pGEM-T Easy vectors, re-sequenced, digested and subcloned into pcDNA™3 (Invitrogen, Carlsbad, CA, USA). Effective saT1R cloning into pcDNA™3 was further verified by HindIII/XhoI digestion. Transient co-transfections were done in 6-well plates (confluence ~70–80%) using 100 μL of reduced serum medium (Opti-MEM, Gibco, Thermo Fisher, Saint Louis, MO, USA) containing 150 ng of each selected pcDNA™3 construct, 1:10 charge ratio of Enhanced Green Fluorescent expressing construct (EGFP; positive control), and pBluescript II SK (+) vector DNA (Addgene, Watertown, MA, USA) up to 1 µg of totally transfected DNA. Following 24 h incubation, cells were washed with fresh standard medium and plated in polylysine treated plates for additional 24 h. Cells were then stimulated with L-AAs and sugar compounds for 18 h in a reduced assay medium (BME w/o L-glutamate; Gibco, Thermo Fisher, Saint Louis, MO, USA) + 1% FBS), to prevent unspecific Ca2+/NFAT signaling activation and potential saT1R desensitization [95]. All L-AAs and sugars used for in vitro experiments were obtained from Merck KGaA (Darmstadt, Germany).
5. Conclusions
This study offers new important information for deciphering the molecular mechanisms underpinning the physiology of fish taste sensory modalities by reporting the first T1R gene repertoire in a carnivorous fish species. Additionally, it demonstrates that in vitro cell culture approaches using gene reporter systems based on Ca2+ dependent calcineurin/NFAT signaling can be readily used to test potency rankings and/or magnitude of saT1Rs responses to L-AA tastants and sugars. Through co-transfections of a subset of saT1Rs heterodimers, alone or in combination with the Gi alpha protein subunits saGαi1 and saGαi2, we found that L-AAs induce important taste stimulatory effects differentially mediated by saT1R1, saT1R2a and saT1R2b subunits. Furthermore, we show preliminary evidences that saGαi subunits can be involved in both stimulatory and inhibitory saT1R transduction signaling mechanisms. Our data strengthen information previously available in herbivorous and omnivorous fish showing that fish species possess multiple putative sweet dimeric receptors (TR2n/TR3), which are also activated by a broad spectrum of L-AAs with an even higher sensitivity than sugars. It is suggested that the expansion of TR2 in fish during the third-round genome duplication has provided novel genetic material to facilitate the adaptation to diverse environments and the development of feeding specializations. Finally, our study provides a platform to test saT1R-dependent diet selection in one of the most important marine species for Mediterranean aquaculture, and opens new opportunities to further optimize feeds by the use of targeted ingredients or additives susceptible to affect gustatory preferences.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/21/20/7732/s1.
Author Contributions
Conceptualization, S.M. and J.M.C.-R.; methodology, J.M.C.-R. and A.R.A.; investigation, A.R.A. and S.P.; data curation, A.R.A.; writing—original draft preparation, A.R.A.; writing—review and editing, A.R.A., J.M.C.-R. and S.M.; project administration, J.M.C.-R.; funding acquisition, S.M. and J.M.C.-R. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by LUCTA SA. Some collateral aspects were covered by the National Research Agency (AEI) [grant number: PID2019-103969RB-C33] to J.M.C.-R.
Acknowledgments
We thank Elisa Sánchez for technical assistance.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Roper, S. The cell biology of vertebrate taste receptors. Annu. Rev. Neurosci. 1989, 12, 329–353. [Google Scholar] [CrossRef] [PubMed]
- Hoon, M.A.; Adler, E.; Lindemeier, J.; Battey, F.; Ryba, N.J.P.; Zuker, C.S. Putative mammalian taste receptors: A class of taste-specific GPCRs with distinct topographic selectivity. Cell 1999, 96, 541–551. [Google Scholar] [CrossRef]
- Mombaerts, P. Genes and ligands for odorant, vomero nasal and taste receptors. Nat. Rev. Neurosci. 2004, 5, 263–278. [Google Scholar] [CrossRef] [PubMed]
- Shi, P.; Zhang, J. Contrasting modes of evolution between vertebrate sweet/umami receptor genes and bitter receptor genes. Mol. Biol. Evol. 2006, 23, 292–300. [Google Scholar] [CrossRef]
- Antinucci, M.; Risso, D.A. Matter of taste: Lineage-specific loss of function of taste receptor genes in vertebrates. Front. Mol. Biosci. 2017, 4, 81. [Google Scholar] [CrossRef] [PubMed]
- Yarmolinsky, D.A.; Zuker, C.S.; Ryba, N.J.P. Common sense about taste: From mammals to insects. Cell 2009, 139, 234–244. [Google Scholar] [CrossRef] [PubMed]
- Danilova, V.; Damak, S.; Margolskee, R.F.; Hellekant, G. Taste responses to sweet stimuli in alfa-gustducin knockout and wild-type mice. Chem. Senses 2006, 31, 573–580. [Google Scholar] [CrossRef]
- Ruiz, C.J.; Wray, K.; Delay, E.; Margolskee, R.F.; Kinnamon, S.C. Behavioral evidence for a role of a-gustducin in glutamate taste. Chem. Senses 2003, 28, 573–579. [Google Scholar] [CrossRef]
- Zhang, Y.; Hoon, M.A.; Chandrashekar, J.; Mueller, K. Coding of sweet, bitter, and umami tastes: Different receptor cells sharing similar signaling pathways. Cell 2003, 112, 293–301. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhao, Z.; Margolskee, R.; Liman, E. The transduction channel TRPM5 is gated by intracellular calcium in taste cells. J. Neurosci. 2007, 27, 5777–5786. [Google Scholar] [CrossRef]
- Clapp, T.R.; Trubey, K.R.; Vandenbeuch, A.; Stone, L.M.; Margolskee, R.F.; Chaudhari, N.; Kinnamon, S.C. Tonic activity of G alpha-gustducin regulates taste cell responsivity. FEBS Lett. 2008, 582, 3783–3787. [Google Scholar] [CrossRef] [PubMed]
- Finger, T.; Kinnamon, S. Taste isn’t just for taste buds anymore. F1000 Biol. Rep. 2011, 3, 20. [Google Scholar] [CrossRef]
- Taylor, C.W. Regulation of IP3 receptors by cyclic AMP. Cell Calcium 2017, 63, 48–52. [Google Scholar] [CrossRef] [PubMed]
- Oka, Y.; Korsching, S.I. Shared and unique G alpha proteins in the zebrafish versus mammalian senses of taste and smell. Chem. Senses 2011, 36, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Ohmoto, M.; Okada, S.; Nakamura, S.; Abe, K.; Matsumoto, I. Mutually exclusive expression of Gαia and Gα14 reveals diversification of taste receptor cells in zebrafish. J. Comp Neurol. 2011, 8, 1616–1629. [Google Scholar] [CrossRef]
- Hashiguchi, Y.; Furuta, Y.; Kaahara, R.; Nishida, M. Diversification and adaptive evolution of putative sweet taste receptors in three spine stickleback. Gene 2007, 396, 170–179. [Google Scholar] [CrossRef]
- Ishimaru, Y.; Okada, S.; Naito, H.; Nagai, T.; Yasuoka, A.; Matsumoto, I.; Abe, K. Two families of candidate taste receptors in fishes. Mech. Dev. 2005, 122, 1310–1321. [Google Scholar] [CrossRef]
- Picone, B.; Hesse, U.; Panji, S.; Van Heusden, P.; Jonas, M.; Christoffels, A. Taste and odorant receptors of the coelacanth—A gene repertoire in transition. J. Exp. Zool. B Mol. Dev. Evol. 2014, 322, 403–414. [Google Scholar] [CrossRef]
- Yuan, X.; Liang, X.F.; Cai, W.J.; He, S.; Guo, W.J.; Mai, K.S. Expansion of sweet taste receptor genes in grass carp (Ctenopharyngodon idellus) coincided with vegetarian adaptation. BMC Evol. Biol. 2020, 20, 25. [Google Scholar] [CrossRef]
- Oike, H.; Nagai, T.; Furuyama, A.; Okada, S.; Aihara, Y.; Ishimaru, Y.; Marui, T.; Matsumoto, I.; Misaka, T.; Abe, K. Characterization of ligands for fish taste receptors. J. Neurosci. 2007, 27, 5584–5592. [Google Scholar] [CrossRef]
- Pauletto, M.; Manousaki, T.; Ferraresso, S.; Babbucci, M.; Tsakogiannis, A.; Louro, B.; Vitulo, N.; Quoc, V.H.; Carraro, R.; Bertotto, D.; et al. Genomic analysis of Sparus aurata reveals the evolutionary dynamics of sex-biased genes in a sequential hermaphrodite fish. Commun. Biol. 2018, 1, 119. [Google Scholar] [CrossRef]
- Mulder, N.; Apweiler, R. InterPro and InterProScan: Tools for protein sequence classification and comparison. Methods Mol. Biol. 2007, 396, 59–70. [Google Scholar] [PubMed]
- Hammerland, L.G.; Krapcho, K.J.; Garrett, J.E.; Alasti, N.; Hung, B.C.; Simin, R.T.; Levinthal, C.; Nemeth, E.F.; Fuller, F.H. Domains determining ligand specificity for Ca2+ receptors. Mol. Pharmacol. 1999, 55, 642–648. [Google Scholar]
- Cao, J.; Huang, S.; Qian, J.; Huang, J.; Jin, L.; Su, Z.; Yang, J.; Liu, J. Evolution of the class C GPCR Venus flytrap modules involved positive selected functional divergence. BMC Evol. Biol. 2009, 9, 67. [Google Scholar] [CrossRef] [PubMed]
- Wettschureck, N.; Offermanns, S. Mammalian G proteins and their cell type specific functions. Physiol. Rev. 2005, 85, 1159–1204. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, C.M.; Putney, J.W. Differential effects of protein kinase C activation on calcium storage and capacitive calcium entry in NIH 3T3 Cells. J. Biol. Chem. 1996, 271, 21522–21528. [Google Scholar] [CrossRef]
- Garcia-Rodriguez, C.; Rao, A. Requirement for integration of phorbol 12-myristate 13-acetate and calcium pathways is preserved in the transactivation domain of NFAT1. Eur. J. Immunol. 2000, 30, 2432–2436. [Google Scholar] [CrossRef]
- San-Antonio, B.; Iniguez, M.A.; Fresno, M. Protein kinase C phosphorylates nuclear factor of activated T cells and regulates its transactivating activity. J. Biol. Chem. 2002, 277, 27073–27080. [Google Scholar] [CrossRef]
- Morgan, A.J.; Jacob, R. Lonomycin enhances Ca2+ influx by stimulating store-regulated cation entry and not by a direct action at the plasma membrane. Biochem. J. 1994, 300, 665–672. [Google Scholar] [CrossRef]
- Kim, M.R.; Kusakabe, Y.; Miura, H.; Shindo, Y.; Ninomiya, Y.; Hino, A. Regional expression patterns of taste receptors and gustducin in the mouse tongue. Biochem. Biophys. Res. Commun. 2003, 312, 500–506. [Google Scholar] [CrossRef]
- Okada, S. The taste system of small fish species. Biosci. Biotechnol. Biochem. 2015, 79, 1039–1043. [Google Scholar] [CrossRef]
- Nelson, G.; Hoon, M.A.; Chandrashekar, J.; Zhang, Y.; Ryba, N.J.; Zuker, C.S. Mammalian sweet taste receptors. Cell 2001, 106, 381–390. [Google Scholar] [CrossRef]
- Baldwin, M.W.; Toda, Y.; Nakagita, T.; O’Connell, M.J.; Klasing, K.C.; Misaka, T.; Edwards, S.V.; Liberles, S.D. Evolution of sweet taste perception in hummingbirds by transformation of the ancestral umami receptor. Science 2014, 345, 929–933. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.Q.; Zhang, Y.; Hoon, M.A.; Chandrashekar, J.; Erlenbach, I.; Ryba, N.; Zuker, S. The receptors for mammalian sweet and umami taste. Cell 2003, 115, 255–266. [Google Scholar] [CrossRef]
- Treesukosol, Y.; Smith, K.R.; Spector, A.C. The functional role of the T1R family of receptors in sweet taste and feeding. Physiol. Behav. 2011, 105, 14–26. [Google Scholar] [CrossRef] [PubMed]
- Bachmanov, A.A.; Beauchamp, G.K. Taste receptor genes. Annu. Rev. Nutr. 2007, 27, 389–414. [Google Scholar] [CrossRef]
- Wooding, S.; Kim, U.K.; Bamshad, M.J.; Larsen, J.; Jorde, L.B.; Drayna, D. Natural selection and molecular evolution in PTC, a bitter-taste receptor gene. Am. J. Hum. Genet. 2004, 74, 637–646. [Google Scholar] [CrossRef]
- Li, X.; Staszewski, L.; Xu, H.; Durick, K.; Zoller, M.; Adler, E. Human receptors for sweet and umami taste. Proc. Natl. Acad. Sci. USA 2002, 99, 4692–4696. [Google Scholar] [CrossRef]
- Broughton, R.E.; Betancur-R, R.; Li, C.; Arratia, G.; Ortí, G. Multi-locus phylogenetic analysis reveals the pattern and tempo of bony fish evolution. PLoS Curr. 2013. [Google Scholar] [CrossRef]
- Amores, A.; Force, A.; Yan, Y.-L.; Joly, L.; Amemiya, C.; Fritz, A.; Ho, R.K.; Langeland, J.; Prince, V.; Wang, Y.-L.; et al. Zebrafish Hox clusters and vertebrate genome evolution. Science 1998, 282, 1711–1714. [Google Scholar] [CrossRef]
- Glasauer, S.M.; Neuhauss, S.C. Whole-genome duplication in teleost fishes and its evolutionary consequences. Mol. Genet. Genom. 2014, 289, 1045–1060. [Google Scholar] [CrossRef]
- Kunishima, N.; Shimada, Y.; Tsuji, Y.; Sato, T.; Yamamoto, M.; Kumasaka, T.; Nakanishi, S.; Jingami, H.; Morikawa, K. Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor. Nature 2000, 407, 971–977. [Google Scholar] [CrossRef] [PubMed]
- Nango, E.; Akiyama, S.; Maki-Yonekura, S.; Ashikawa, Y.; Kusakabe, Y.; Krayukhina, E.; Maruno, T.; Uchiyama, S.; Nuemket, N.; Yonekura, K.; et al. Taste substance binding elicits conformational change of taste receptor T1r heterodimer extracellular domains. Sci. Rep. 2016, 6, 25745. [Google Scholar] [CrossRef] [PubMed]
- Behrens, M.; Briand, L.; de March, C.A.; Matsunami, H.; Yamashita, A.; Meyerhof, W.; Weyand, S. Structure-function relationships of olfactory and taste receptors. Chem. Senses 2018, 43, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Nuemket, N.; Yasui, N.; Kusakabe, Y.; Nomura, Y.; Atsumi, N.; Akiyama, S.; Nango, E.; Kato, Y.; Kaneko, M.K.; Takagi, J.; et al. Structural basis for perception of diverse chemical substances by T1r taste receptors. Nat. Commun. 2017, 8, 15530. [Google Scholar] [CrossRef] [PubMed]
- Chun, L.; Zhang, W.; Liu, J. Structure and ligand recognition of class C GPCRs. Acta Pharmacol. Sin. 2012, 33, 312–323. [Google Scholar] [CrossRef]
- Hu, J.; Reyes-Cruz, G.; Chen, W.; Jacobson, K.A.; Spiegel, A.M. Identification of acidic residues in the extracellular loops of the seven-transmembrane domain of the human Ca2+ receptor critical for response to Ca2+ and a positive allosteric modulator. J. Biol. Chem. 2002, 277, 46622–46631. [Google Scholar] [CrossRef]
- Hu, J.; Mc-Larnon, S.J.; Mora, S.; Jiang, J.; Thomas, C.; Jacobson, K.A.; Spiegel, A.M. A region in the seven-transmembrane domain of the human Ca2+ receptor critical for response to Ca2+. J. Biol. Chem. 2005, 280, 5113–5120. [Google Scholar] [CrossRef]
- Pagano, A.; Rüegg, D.; Litschig, S.; Stoehr, N.; Stierlin, C.; Heinrich, M.; Floersheim, P.; Prézeau, L.; Carroll, F.; Pin, J.-P.; et al. The non-competitive antagonists 2-methyl-6-(phenylethynyl) pyridine and 7-hydroxyiminocyclopropan[b]chromen-1a-carboxylic acid ethyl ester interact with overlapping binding pockets in the transmembrane region of group I metabotropic glutamate receptors. J. Biol. Chem. 2000, 275, 33750–33758. [Google Scholar] [CrossRef]
- Malherbe, P.; Kratochwil, N.; Zenner, M.T.; Piussi, J.; Diener, C.; Kratzeisen, C.; Fisher, C.; Porter, R.H.P. Mutational analysis and molecular modeling of the binding pocket of the metabotropic glutamate 5 receptor negative modulator 2-methyl-6-(phenylethynyl)-pyridine. Mol. Pharmacol. 2003, 64, 823–832. [Google Scholar] [CrossRef]
- Jiang, P.; Ji, Q.; Liu, Z.; Snyder, L.A.; Benard, L.M.; Margolskee, R.F.; Max, M. The cysteine-rich region of T1R3 determines responses to intensely sweet proteins. J. Biol. Chem. 2004, 279, 45068–45075. [Google Scholar] [CrossRef]
- Medler, K.F.; Kinnamon, S.C. Transduction mechanisms in taste cells. In Transduction Channels in Sensory Cells; Frings, S., Ed.; Wiley-VCH: Weinheim, Germany, 2004; pp. 53–177. [Google Scholar]
- Medler, K.F. Signaling mechanisms controlling taste cell function. Crit. Rev. Eukaryot. Gene Expr. 2008, 18, 125–137. [Google Scholar] [CrossRef]
- Medler, K.F. Calcium Signaling in Taste Cells. Biochim. Biophys Acta. 2015, 1853, 2025–2032. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Takahara, T.; Achiha, T.; Shibata, H.; Maki, M. Nanoluciferase reporter gene system directed by tandemly repeated pseudo-palindromic NFAT-response elements facilitates analysis of biological endpoint effects of cellular Ca2+ mobilization. Int. J. Mol. Sci. 2018, 19, 605. [Google Scholar] [CrossRef] [PubMed]
- Horsley, V.; Pavlath, G.K. NFAT: Ubiquitous regulator of cell differentiation and adaptation. J. Cell Biol. 2002, 156, 771–774. [Google Scholar] [CrossRef] [PubMed]
- Clipstone, N.A.; Crabtree, G.R. Identification of calcineurin as a key signalling enzyme in T-lymphocyte activation. Nature 1992, 357, 695–697. [Google Scholar] [CrossRef]
- Hogan, P.G.; Chen, L.; Nardone, J.; Rao, A. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev. 2003, 17, 2205–2232. [Google Scholar] [CrossRef]
- Macian, F. NFAT proteins: Key regulators of T-cell development and function. Nat. Rev. Immunol. 2005, 5, 472–484. [Google Scholar] [CrossRef]
- Muller, M.R.; Rao, A. NFAT, immunity and cancer: A transcription factor comes of age. Nat. Rev. Immunol. 2010, 10, 645–656. [Google Scholar] [CrossRef]
- Nakurawa, M.; Mori, T.; Hayashi, Y. Umami Changes Intracellular Ca2+ Levels Using Intracellular and Extracellular Sources in Mouse Taste Receptor Cells. Biosci. Biotechnol. Biochem. 2006, 70, 2613–2619. [Google Scholar]
- Medler, K.F. Calcium signaling in taste cells: Regulation required. Chem. Senses 2010, 35, 753–765. [Google Scholar] [CrossRef] [PubMed]
- Toda, Y.; Okada, S.; Misaka, T. Establishment of a new cell-based assay to measure the activity of sweeteners in fluorescent food extracts. J. Agric. Food Chem. 2011, 59, 12131–12138. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.T.; Li, J.T.; Zhang, X.F.; Sun, X.W. Transcriptome analysis reveals the time of the fourth round of genome duplication in common carp (Cyprinus carpio). BMC Genom. 2012, 13, 96. [Google Scholar] [CrossRef]
- Volff, J. Genome evolution and biodiversity in teleost fish. Heredity 2005, 94, 280–294. [Google Scholar] [CrossRef] [PubMed]
- Ravi, V.; Venkatesh, B. The divergent genomes of teleosts. Annu. Rev. Anim. Biosci. 2018, 6, 47–68. [Google Scholar] [CrossRef]
- Takahashi, R.; Watanabe, K.; Nishida, M.; Hori, M. Evolution of feeding specialization in Tanganyikan scale-eating cichlids: A molecular phylogenetic approach. BMC Evol. Biol. 2007, 7, 195. [Google Scholar] [CrossRef]
- Gojobori, J.; Innan, H. Potential of fish opsin gene duplications to evolve new adaptive functions. Trends Genet. 2009, 25, 198–202. [Google Scholar] [CrossRef]
- Rennison, D.J.; Owens, G.L.; Taylor, J.S. Opsin gene duplication and divergence in ray-finned fish. Mol. Phylogenet. Evol. 2012, 62, 986–1008. [Google Scholar] [CrossRef]
- Machado, H.E.; Jui, G.; Joyce, D.A.; Reilly, C.R., 3rd; Lunt, D.H.; Renn, S.C. Gene duplication in an African cichlid adaptive radiation. BMC Genom. 2014, 15, 161. [Google Scholar] [CrossRef]
- Chen, Z. Transcriptomic and genomic evolution under constant cold in Antarctic notothenioid fish. PNAS 2008, 105, 12944–12949. [Google Scholar] [CrossRef]
- Wilson, R. Utilization of dietary carbohydrate by fish. Aquaculture 1994, 124, 67–80. [Google Scholar] [CrossRef]
- Tacon, A.; Cowey, C. Protein and amino acid requirements. In Fish Energetics; Tytler, P., Calow, P., Eds.; Springer: Dordrecht, The Netherlands, 1985. [Google Scholar]
- Wilson, R.P. Protein and amino acid requirements of fishes. Annu. Rev. Nutr. 1986, 6, 225–244. [Google Scholar] [CrossRef] [PubMed]
- National Research Cuncil. Nutrient Requirements of Fish and Shrimp; The National Academies Press: Washington, DC, USA, 2011. [Google Scholar]
- Andersen, S.M.; Waagbø, R.; Espe, M. Functional amino acids in fish nutrition, health and welfare. Front Biosci. 2016, 8, 143–169. [Google Scholar]
- Kaushik, S.J.; Seiliez, I. Protein and amino acid nutrition and metabolism in fish: Current knowledge and future needs. Aquac. Res. 2010, 41, 322–332. [Google Scholar] [CrossRef]
- Wu, G. Functional amino acids in growth, reproduction, and health. Adv. Nutr. 2010, 1, 31–37. [Google Scholar] [CrossRef]
- Naylor, R.L.; Hardy, R.W.; Bureau, D.P.; Chiu, A.; Elliott, M.; Farrell, A.P.; Forster, I.; Gatlin, D.M.; Goldburg, R.J.; Hua, K.; et al. Feeding aquaculture in an era of finite resources. Proc. Natl. Acad. Sci. USA 2009, 106, 15103–15110. [Google Scholar] [CrossRef]
- Turchini, G.M.; Trushenski, J.T.; Glencross, B.D. Thoughts for the future of aquaculture nutrition: Realigning perspectives to reflect contemporary issues related to judicious use of marine resources in aquafeeds. N. Am. J. Aquac. 2019, 81, 13–39. [Google Scholar] [CrossRef]
- Gómez-Requeni, P.; Mingarro, M.; Calduch-Giner, J.A.; Medale, F.; Martin, S.A.; Houlihan, D.F.; Kaushik, S.; Pérez-Sánchez, J. Protein growth performance, amino acid utilisation and somatotropic axis responsiveness to fish meal replacement by plant protein sources in gilthead Sea bream (Sparus aurata). Aquaculture 2004, 232, 493–510. [Google Scholar] [CrossRef]
- Deng, J.; Mai, K.; Zhang, W.; Wang, X.; Xu, W.; Liufu, Z. Effects of replacing fish meal with soy protein concentrate on feed intake and growth of juvenile Japanese flounder, Paralichthys olivaceus. Aquaculture 2006, 258, 503–513. [Google Scholar] [CrossRef]
- Espe, M.; Lemme, A.; Petri, A.; El-Mowafi, A. Can Atlantic salmon (Salmo salar) grow on diets devoid of fish meal? Aquaculture 2006, 255, 255–262. [Google Scholar] [CrossRef]
- Roper, S.D. Signal transduction and information processing in mammalian taste buds. Pflugers Arch. 2007, 454, 759–776. [Google Scholar] [CrossRef]
- Gilman, A.G. G proteins: Transducers of receptor-generated signals. Ann. Rev. Biochem. 1987, 56, 615–649. [Google Scholar] [CrossRef]
- Rodbell, M. Nobel Lecture: Signal transduction: Evolution of an idea. Biosci. Rep. 1995, 15, 117–133. [Google Scholar] [CrossRef] [PubMed]
- Navarro, G.; Cordomí, A.; Zelman-Femiak, M.; Brugarolas, M.; Moreno, E.; Aguinaga, D.; Perez-Benito, L.; Cortés, A.; Casadó, V.; Mallol, J.; et al. Quaternary structure of a G-protein-coupled receptor heterotetramer in complex with Gi and Gs. BMC Biol. 2016, 14, 26. [Google Scholar] [CrossRef] [PubMed]
- Navarro, G.; Cordomí, A.; Brugarolas, M.; Moreno, E.; Aguinaga, D.; Pérez-Benito, L.; Ferre, S.; Cortés, A.; Casadó, V.; Mallol, J.; et al. Cross-communication between Gi and Gs in a G-protein-coupled receptor heterotetramer guided by a receptor C- terminal domain. BMC Biol. 2018, 16, 24. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jean Mougin, F.; Higgins, D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. CABIOS 1992, 8, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Perrière, G.; Gouy, M. WWW-Query: An on-line retrieval system for biological sequence banks. Biochimie 1996, 78, 364–369. [Google Scholar] [CrossRef]
- Page, R.D. Visualizing phylogenetic trees using TreeView. Curr. Protoc. Bioinform. 2002, 6. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Raghuwanshi, R.; Shen, W.; Montell, C. Food experience–induced taste desensitization modulated by the Drosophila TRPL channel. Nat. Neurosci. 2013, 16, 1468–1476. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).