Abstract
Thiazolidinedione is a five-membered heterocycle that is widely used in drug discovery endeavors. In this study, we report the design, synthesis, and biological evaluation of a series of thiazolidinedione-based HDAC6 inhibitors. In particular, compound 6b exerts an excellent inhibitory activity against HDAC6 with an IC50 value of 21 nM, displaying a good HDAC6 selectivity over HDAC1. Compound 6b dose-dependently induces the acetylation level of α-tubulin via inhibition of HDAC6 in human neuroblastoma SH-SY5Y cell line. Moreover, compound 6b efficiently reverses methamphetamine-induced morphology changes of SH-SY5Y cells via regulating acetylation landscape of α-tubulin. Collectively, compound 6b represents a novel HDAC6-isoform selective inhibitor and demonstrates promising therapeutic potential for the treatment of methamphetamine addiction.
1. Introduction
Methamphetamine (METH) is a widely abused illicit psychostimulant that causes euphoria, hyperactivity, and wakefulness. METH abuse has dramatically risen over the last decade owing to its readily accessibility and powerful euphoric effect [1]. Acute overdose of METH induces cardiac arrhythmia, strokes, seizures, and hyperthermia, and repeated exposure to this substance leads to severe addiction, altered cognitive function, and neurological damage [2,3,4,5,6,7]. This chronic METH abuse is accompanied by significant changes in gene and protein expression within specific brain subregions such as dorsal striatum in the reward circuitry, suggesting that there is a clear connection between epigenetic mechanisms and METH addiction [8]. Therefore, a number of studies have been conducted to elucidate the epigenetic mechanisms underlying METH addiction [9,10]. Interestingly, it is reported that acute and chronic exposure to METH increases histone acetylation, implying that epigenetic enzymes including histone deacetylases (HDACs) participate in the mechanisms of METH addiction [11,12,13]. Moreover, it is known that administration of METH regulates cytoplasmic HDAC6 enzyme, altering epigenetic landscape of α-tubulin in endothelial cells [14]. Given the fact that HDAC enzymes play an important role in METH addition, we decided to investigate the therapeutic potential of HDAC inhibitors for the treatment of METH addiction.
HDAC is an enzyme that catalyzes the removal of acetyl groups from lysine residues at the N-terminal tails of histones [15]. These enzymes also regulate the acetylation status of non-histone proteins such as α-tubulin, Hsp90, and p53 [16]. There are eighteen isoforms of HDAC enzymes, further classified into four classes. These four identified classes are characterized by their distinct functions, substrate specificities, and subcellular locations. Accordingly, there has been great effort to develop isoform-selective HDAC inhibitors to elucidate the detailed functions of each isoform and to avoid unwanted side effects in chemotherapy [17,18].
2. Results and Discussion
We previously performed a structure-based virtual screening and discovered a novel class of anthraquinone-based HDAC6 inhibitors with a good HDAC6 selectivity over other HDAC isozymes (Figure 1) [19]. We found out that two carbonyl oxygens of anthraquinone played a pivotal role in exhibiting selective inhibition of HDAC6. These carbonyl groups recognized the unique rim and surface area of HDAC6, which is different from other HDAC isozymes. Despite the fact that this anthraquinone-based HDAC6 inhibitor displayed an excellent in vitro biological activity, its intrinsic poor solubility hampered further study for in vivo application. Thiazolidinedione (TZD) is a five-membered ring molecule containing two heteroatoms (nitrogen and sulfur) and two carbonyl groups. Thiazolidinedione and its derivatives have diverse biological activities including anti-microbial, anti-oxidant, anti-inflammatory, anti-diabetic, anti-cancer, and anti-tubercular activities, and a number of thiazolidinedione-based drugs, such as troglitazone and rosiglitazone, have been clinically approved for the treatment of type 2 diabetes [20,21]. The fact that thiazolidinedione is a privileged scaffold from clinically approved drugs with two carbonyl groups prompted us to explore the thiazolidinedione-based HDAC6 inhibitors.
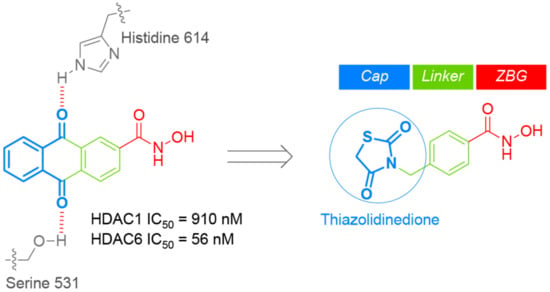
Figure 1.
Drug design of thiazolidinedione-based histone deacetylase 6 (HDAC6) inhibitors.
The synthesis of thiazolidinedione-based HDAC6 inhibitors 6a-b is illustrated in Scheme 1. The key intermediate 3a-b were synthesized following the previously reported procedure [22]. Briefly, the base-promoted N-alkylation reaction of thiazolidine-2,4-one (1) with methyl 3-(bromomethyl)benzoate (2a) or methyl 4-(bromomethyl)benzoate (2b) successfully provided compounds 3a-b in 53–55% yields. The initial attempt to directly convert ester 3a-b to hydroxamate 6a-b using hydroxylamine in the presence of sodium hydroxide in methanol was not fruitful due to the instability of thiazolidine-2,4-one ring in this reaction condition. That led us to follow an alternative synthetic route. Therefore, compound 3a-b were hydrolyzed into carboxylic acid 4a-b in the aqueous acidic condition. Having prepared carboxylic acid 4a-b, we then performed amide coupling reaction of 4a-b with NH2OTHP using EDC, HOBt, and TEA in DCM, which provided compound 5a-b in 43–57% yields. Finally, the removal of THP-protecting group from compound 5a-b was accomplished by the acidic hydrolysis and successfully afforded compound 6a-b in 36–42% yields.
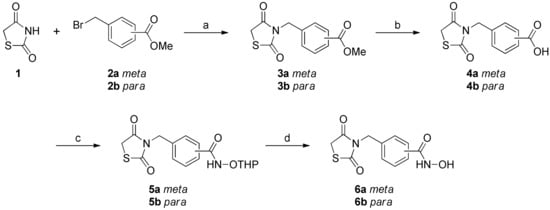
Scheme 1.
Synthesis of compounds 6a-ba. a Reagents and conditions: (a) K2CO3, acetone, reflux, 12 h, 53% for 3a, 55% for 3b; (b) 6N HCl reflux, 12 h, 76% for 4a, 89% for 4b; (c) EDC·HCl, HOBt, NH2OTHP, TEA, rt, 18 h, 43% for 5a, 57% for 5b; (d) HCl, Et2O, DCM, rt, 2 h, 36% for 6a, 42% for 6b.
With compound 6a-b in hand, we first measured in vitro inhibitory activity of meta-analogue 6a and para-analogue 6b against HDAC1 and HDAC6 enzymes. As shown in Table 1, para-analogue 6b led to higher binding affinity to HDAC1 and HDAC6 than meta-analogue 6a, in that para-analogue 6b furnished IC50 values of 388 nM and 21 nM against HDAC1 and HDAC6, respectively. This result directed us to synthesize more para-analogues derived from compound 6b.

Table 1.
In vitro inhibitory activity against HDAC1 and HDAC6 isotypes.
The synthetic route to compounds 8a-c as shown in Scheme 2 started from the intermediate 5a. Base-catalyzed alkylation reaction of 5a with allyl bromide, propyl iodide, and benzyl bromide in DMF led to compounds 7a-c in 14–27% yields. Subsequently, the cleavage of THP-protecting group from compounds 7a-c under acidic condition resulted in compounds 8a-c in 38–44% yields.
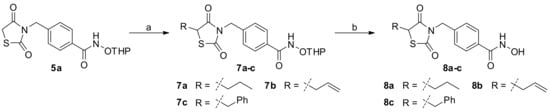
Scheme 2.
Synthesis of compounds 8a-ca. a Reagents and conditions: (a) R-Br, K2CO3, DMF, rt, 18 h, 14% for 7a, 27% for 7b, 23% for 7c (d) HCl in Et2O, DCM, rt, 2h, 38% for 8a, 40% for 8b, 44% for 8c.
Upon completion of synthesis, we examined inhibitory activities of 5-substituted thiazolidinedione-based HDAC inhibitors 8a-c against HDAC1 and HDAC6 isotypes. As shown in Table 1, 5-substituted thiazolidinedione-based HDAC inhibitors 8a-c led to a decrease in their binding affinity to both HDAC1 and HDAC6 isotypes, compared with their non-substituted parent compound 6b. The result indicated that the substitution of 2,4-thiazolidinedione at 5-position caused a negative effect on their proper binding to HDAC1 and HDAC6 enzymes. Interestingly, compound 6b (HDAC6 IC50 = 21 nM) displayed an excellent inhibitory activity against HDAC6, even better than FDA-approved drug SAHA (HDAC6 IC50 = 226 nM). Overall, compound 6b exhibited the most excellent inhibitory activity against HDAC1 and HDAC6 among newly synthesized analogues with a good HDAC6 selectivity over HDAC1. Accordingly, we chose compound 6b for further biological evaluation.
We next evaluated the effect of compound 6b on the cell viability of SH-SY5Y, which is a human neuroblastoma cell line (Figure 2). Although the exposure of SH-SY5Y cells with high concentrations (50, 70, and 100 µM) of 6b decreased the cell viability of SH-SY5Y in a dose-dependent manner, compound 6b exhibited no cytotoxic effect on cell viability up to 30 µM concentration. It has been well-reported that selective inhibition of HDAC6 isoform did not produce a lethal effect on cell viability, in contrast to inhibition of class I HDAC enzymes.
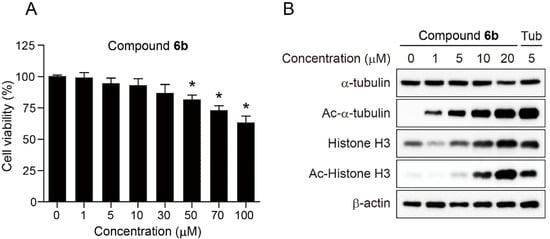
Figure 2.
Effect of compound 6b on the cell viability of SH-SY5Y and the acetylation status of α-tubulin and Histone H3. (A) SH-SY5Y cells were treated with the indicated concentrations of compound 6b for 24 h and the cell viability was measured using the colorimetric MTS assay. Data are presented as mean ± SD (n = 4). (B) SH-SY5Y cells were incubated with the indicated concentrations of compound 6b or Tubastatin A (Tub) for 24 h and the expression levels of α-tubulin, Ac-α-tubulin, Histone H3, and Ac-Histone H3 were analysed by Western blot. Tubastatin A was employed as a positive control and β-actin was used as a loading control. The p value was obtained by Student t-test. * p < 0.01 as compared to vehicle.
We further explored the precise cellular mechanism of compound 6b. Histone H3 and α-tubulin are well-known substrates of HDAC1 and HDAC6 [23,24]. Therefore, inhibition of HDAC1 and HDAC6 enzymes promotes the accumulation of acetylated Histone H3 and α-tubulin, respectively. Therefore, SH-SY5Y cells were incubated with compound 6b at various concentrations (0, 1, 5, 10, and 20 µM) for 24 h and the expression levels of α-tubulin, Ac-α-tubulin, Histone H3, and Ac-Histone H3 were analysed by Western blot. Highly selective HDAC6 inhibitor, Tubastatin A (Tub, 5 µM) was employed as a reference drug. As expected, compound 6b dose-dependently induced the acetylation of α-tubulin and Histone H3. The administration of 6b at relatively low concentration of 1 µM caused the acetylation of α-tubulin via inhibition of HDAC6. In contrast, 10 µM concentration of 6b was able to promote the acetylation of Histone H3 via inhibition of HDAC1, suggesting that compound 6b more efficiently inhibited HDAC6 than HDAC1 in this cellular setting. It is also worth noting that the protein level of Histone H3 increases in proportion to the concentration of compound 6b, implying that compound 6b might have an impact on the transcription factors. The expression level of internal standard, β-actin remained unchanged as expected. Taken together, the results indicated that compound 6b more potently suppressed HDAC6 enzyme activity than HDAC1 in SH-SY5Y cells, which is positively correlated with in vitro HDAC assay shown in Table 1.
There has been a report that methamphetamine promotes the acetylation of α-tubulin via the modification of HDAC6 activity, impacting cytoskeleton stability, cell motility, and polarity [14]. This cytoskeleton disarrangement is strongly associated with the disruption of blood brain barrier (BBB) integrity, allowing harmful substances in the bloodstream to leak into the central nervous system (CNS) [25]. This study prompted us to investigate the effect of methamphetamine on cell morphology. We first examined whether methamphetamine exerted any cytotoxic effect on cell viability prior to analysing the effect of methamphetamine (METH) on the cell morphology of SH-SY5Y cells (Figure 3). SH-SY5Y cells were incubated with methamphetamine (0, 0.1, and 1 mM) for 24 h and then analysed the cell viability using MTS assay. The assay revealed that methamphetamine exerted no toxicity up to 1 mM concentration for 24 h. We next investigated the effect of METH on the cell morphology and the reverse effect of compound 6b in preventing METH-induced cell morphology changes. Accordingly, SH-SY5Y cells were pretreated with 1 mM of METH for 4 h and then incubated with compound 6b (5, 10, and 20 µM) for 24 h. As expected, untreated SH-SY5Y cells (DMSO) had a normal elongated spindle morphology. Upon the treatment of cells with 1 mM of METH, a majority of SH-SY5Y cells showed a rounded shape with a shrunken morphology. Subsequently, the administration of compound 6b (5, 10, and 20 µM) led to a reversed effect on the cell morphology change from a rounded shape back to a spindle shape in a dose-dependent manner.
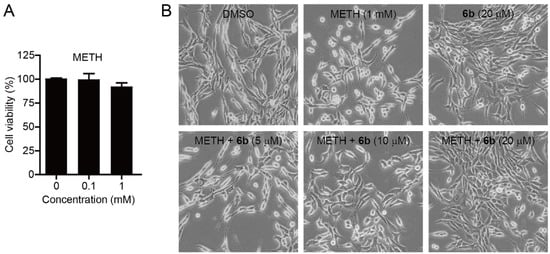
Figure 3.
Effect of methamphetamine (METH) and compound 6b on the cell morphology and viability of SH-SY5Y cells. (A) Effect of methamphetamine on the cell viability of SH-SY5Y. Cells were incubated with the indicated concentrations of methamphetamine (METH) for 24 h and cell viability was measured using the colorimetric MTS assay. Data are presented as mean ± SD (n = 4). (B) Bright-field images of SH-SY5Y cells. Cells were pretreated with methamphetamine (METH, 1 mM) for 4 h and then incubated with the indicated concentrations of compound 6b for 24 h.
We next explored the underlying molecular and biochemical mechanisms behind the morphological changes. α-Tubulin is the basic protein that constitutes the cytoskeleton and the dynamic acetylation and deacetylation of α-tubulin play an important role in the morphology of cells. SH-SY5Y cells were treated with methamphetamine at various concentrations (0, 0.5, 1, 2, and 3 mM) for 24 h and the expression levels of α-tubulin and Ac-α-tubulin were measured by Western blot (Figure 4). The result indicated that methamphetamine dose-dependently decreased the acetylation of α-tubulin, while the expression level of α-tubulin remained unchanged. We next investigated if compound 6b reversed the loss of the acetylation caused by methamphetamine. Therefore, SH-SY5Y cells were treated with methamphetamine (METH, 3 mM) for 4 h, followed by incubation with compound 6b for 24 h. As seen in Figure 4, treatment of cells with METH reduced the acetylation status of α-tubulin. Interestingly, the subsequent treatment of cells with compound 6b led to a significant increase in the acetylation of α-tubulin by reversing the METH-promoted loss of acetylation. Collectively, the results indicated that METH promoted the morphological changes of cells by disrupting the acetylation status of α-tubulin, which could be reversed by compound 6b probably via the inhibition of HDAC6 enzymes.
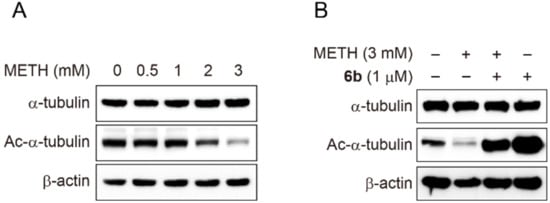
Figure 4.
Western blot analysis of α-tubulin acetylation status after the treatment with methamphetamine (METH) and compound 6b. (A) SH-SY5Y cells were incubated with the indicated concentrations of methamphetamine (METH) for 24 h and the expression levels of Ac-α-tubulin and α-tubulin were analysed by Western blot. β-actin was used as a loading control. (B) SH-SY5Y cells were pretreated with methamphetamine (METH, 3 mM) for 4 h before exposure to compound 6b (1 µM) for 24 h. The expression levels of Ac-α-tubulin and α-tubulin were analysed by Western blot. β-actin was used as a loading control.
To investigate the molecular basis of interactions between compound 6b and HDAC6 (PDB code: 5EF7), we performed in silico docking study (Figure 5 and Supplementary Figure S1). The docking simulation suggested that compound 6b well bound to the substrate binding pocket, validating its inhibitory potential to HDAC6. The hydroxamate group of compound 6b chelated the Zn2+ ion in the bottom of the pocket with its carbonyl oxygen (C=O) and hydroxyl oxygen (OH) in a bidentate fashion.
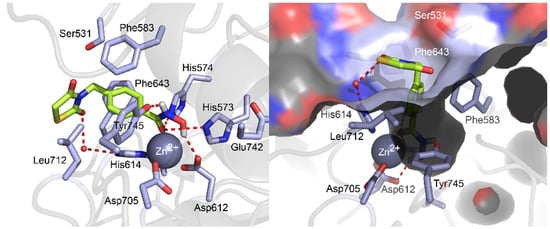
Figure 5.
Molecular docking pose of compound 6b in the binding pocket of HDAC6 (PDB code: 5EF7). The carbon, oxygen, nitrogen, sulfur, and hydrogen atoms of compound 6b are shown in lime, red, blue, yellow, and white, respectively. The side chains of the binding pocket are colored by atom type (carbon, light blue; oxygen, red; nitrogen, red) and labeled with their residue name. Water molecule is shown as a red sphere. The hydrogen bonds are shown in dashed lines. Molecular docking simulations were performed by Autodock 4.2 and docking poses were visualized using PyMOL1.3.
Additionally, the carbonyl oxygen (C=O) of the hydroxamate formed two hydrogen bonds with the side chain of His573 and His574, while the hydroxyl (OH) group and amine (NH) group of the hydroxamate formed hydrogen bonds with Asp612 and Tyr745, respectively. The middle phenyl ring of compound 6b were nicely positioned in the hydrophobic channel, forming π−π interactions with the lipophilic side chains of Phe583 and Phe643 residues. Thiazolidinedione ring was located in the rim of the substrate binding pocket, in that one carbonyl oxygen (C=O) of thiazolidinedione ring formed a hydrogen bonding interaction with a conserved water molecule and the hydrophobic sulfur and carbon atoms at the 1 and 5 position of the thiazolidinedione ring formed proximal Van der Waals interactions with the lipophilic side chain of Leu712. Collectively, the docking study illustrated that compound 6b well fitted into the active site of HDAC6 with diverse intermolecular interactions such as zinc chelation, hydrogen bonds, π−π interactions, and Van der Waals interactions and the estimated binding energy of compound 6b resulted in −7.02 kcal/mol.
3. Materials and Methods
3.1. Chemistry
3.1.1. General Methods and Materials
All reagents and solvents were purchased from commercial suppliers and used without further purification. All experiments dealing with moisture-sensitive compounds were carried out under argon atmosphere. Concentration or solvent removal under reduced pressure was carried out using rotary evaporator. Analytical thin layer chroatography was performed on precoated silica gel F254 TLC plates (E, Merck) with visualization under UV light or by staining using iodine. Column chromatography and medium pressure liquid chromatography (MPLC) was conducted on silica (Merck Silica Gel 40–63 µm) or performed by using a Biotage SP1 flash purification system with prepacked silica gel cartridges (Biotage). NMR analyses were carried out using a JNM-ECZ500R (500 MHz) manufactured by Jeol resonance. Chemical shifts are reported in parts per million (δ). The deuterium lock signal of the sample solvent was used as a reference, and coupling constants (J) are given in hertz (Hz). The splitting pattern abbreviations are as follows: s, singlet; d, doublet; t, triplet; q, quartet; dd, doublet of doublet; td, triplet of doublet; m, multiplet. The LC-QTOF-MS analysis was performed using an Agilent 6530 Accurate-Mass Q-TOF LC/MS System with Agilent 1290 Infinity LC (Agilent Technologies, Palo Alto, CA, USA). The guard column and the analytical column were Zorbax SB-C8 (3.5 μm, 2.1 × 30 mm, Agilent Technologies) and Zorbax SB-Aq (1.8 μm, 2.1 × 100 mm, Agilent Technologies), respectively, and were maintained at 40 °C. The mobile phase consisted of 0.1% formic acid in water (A) and 0.1% formic acid in acetonitrile (B). The gradient conditions were as follows: 0–30 min, 1–20% B; 30–40 min, 20–90% B; 40–45 min, 90% B; 45–47 min, 90–1% B; 47–52 min, 1% B at a flow rate of 400 μL/min. The MS system was operated using ESI in the positive and the negative ionization mode. The optimized conditions of the QTOF-MS system for both ionization modes were as follows: drying gas temperature, 300 °C; drying gas flow, 10 L/min; nebulization pressure, 45 psi; sheath gas temperature, 350 °C; sheath gas flow, 10 L/min; capillary voltage, 3500 V; nozzle voltage, 0 V; fragmentor voltage, 175 V; skimmer voltage, 65 V. The mass range was 50–1700 m/z and the scan rate was 2.00 spectra/sec for both MS and MS/MS analyses.
3.1.2. General Procedure for the Synthesis of Compounds 3a-b
The mixture of thiazolidine-2,4-one (10 mmol), methyl 4-(bromomethyl)benzoate or methyl 3-(bromomethyl)benzoate (11 mmol), and anhydrous K2CO3 (15 mmol) was refluxed in acetone (50 mL) overnight. The solid K2CO3 was filtered and the filtrate was evaporated to obtain the crude product, which was purified by MPLC to afford compound 3a-b in 53–55%.
Methyl 3-((2,4-dioxothiazolidin-3-yl)methyl)benzoate (3a)
53% Yield. 1H-NMR (500 MHz, CDCl3) δ 8.04 (s, 1H), 7.99 (d, J = 7.4 Hz, 1H), 7.59 (d, J = 7.4 Hz, 1H), 7.41 (t, J = 7.7 Hz, 1H), 4.81 (s, 2H), 3.97 (s, 2H), 3.91 (s, 3H).
Methyl 4-((2,4-dioxothiazolidin-3-yl)methyl)benzoate (3b)
55% Yield. 1H-NMR (500 MHz, CDCl3) δ 7.99 (d, J = 8.6 Hz, 2H), 7.44 (d, J = 8.0 Hz, 2H), 4.81 (s, 2H), 3.97 (s, 2H), 3.91 (s, 3H).
3.1.3. General Procedure for the Synthesis of Compounds 4a-b
A suspension of compound 3a or 3b (2 mmol) in 6N HCl (25 mL) was stirred at reflux for 12 h. The mixture was then cooled and kept at 4 °C for 2 h. The desired product precipitated which was filtered, washed with water (2 × 20 mL) and dried in vacuo to afford compound 4a-b in 76–89%.
3-((2,4-Dioxothiazolidin-3-yl)methyl)benzoic acid (4a)
76% Yield. 1H-NMR (500 MHz, CD3OD) δ 7.99 (s, 1H), 7.95 (d, J = 7.4 Hz, 1H), 7.58 (d, J = 7.4 Hz, 1H), 7.44 (t, J = 7.4 Hz, 1H), 4.81 (s, 2H), 4.13 (s, 2H).
4-((2,4-Dioxothiazolidin-3-yl)methyl)benzoic acid (4b)
89% Yield. 1H-NMR (500 MHz, CD3OD) δ 7.98 (d, J = 8.0 Hz, 2H), 7.42 (d, J = 8.0 Hz, 2H), 4.81 (s, 2H), 4.15 (s, 2H)
3.1.4. General Procedure for the Synthesis of Compounds 5a-b
To a solution of compound 4a or 4b (1.1 mmol), EDC·HCl (4.4 mmol), HOBt (2.2 mmol) in dry DCM was added triethylamine (7.7 mmol) and O-tetrahydropyran-2-ylhydroxylamine (1.4 mmol). The reaction mixture was stirred at room temperature for 18 h. Then, DCM was washed with brine solution. The organic layer was dried over Na2SO4, concentrated in vacuo. The product was purified by MPLC to afford 5a-b in 43–57% yield.
3-((2,4-Dioxothiazolidin-3-yl)methyl)-N-((tetrahydro-2H-pyran-2-yl)oxy)benzamide (5a)
43% Yield. 1H-NMR (500 MHz, CDCl3) δ 9.20 (s, 1H), 7.71 (s, 1H), 7.69 (d, J = 8.0 Hz, 1H), 7.50 (d, J = 7.4 Hz, 1H), 7.37 (t, J = 7.7 Hz, 1H), 5.06 (s, 1H), 4.76 (s, 2H), 3.98 (d, J = 11.5 Hz, 1H), 3.96 (s, 2H), 3.62 (t, J = 5.7 Hz, 1H), 1.81–1.87 (m, 3H), 1.56–1.65 (m, 3H).
4-((2,4-Dioxothiazolidin-3-yl)methyl)-N-((tetrahydro-2H-pyran-2-yl)oxy)benzamide (5b)
57% Yield. 1H-NMR (500 MHz, CDCl3) δ 8.86 (s, 1H), 7.71 (d, J = 8.0 Hz, 2H), 7.44 (d, J = 8.0 Hz, 2H), 5.06 (s, 1H), 4.79 (s, 2H), 3.99 (d, J = 8.6 Hz, 1H), 3.96 (s, 2H), 3.64 (dd, J = 6.3, 5.2 Hz, 1H), 1.83–1.92 (m, 3H), 1.59–1.66 (m, 3H).
3.1.5. General Procedure for the Synthesis of Compounds 7a-c
Compound 6a (0.22 mmol), alkyl halide (0.22 mmol) and anhydrous K2CO3 (0.22 mmol) were added into dry DMF (5 mL) and the mixture was stirred at room temperature for 18 h. Then DMF was evaporated in vacuo. The solid crude product was purified by MPLC to afford 7a-c in 14–27% yield.
4-((2,4-Dioxo-5-propylthiazolidin-3-yl)methyl)-N-((tetrahydro-2H-pyran-2-yl)oxy)benzamide (7a)
14% yield. 1H-NMR (500 MHz, CDCl3) δ 8.74 (s, 1H), 7.71 (d, J = 8.0 Hz, 2H), 7.43 (d, J = 8.0 Hz, 2H), 5.06 (s, 1H), 4.77 (dd, J = 20.6, 14.3 Hz, 2H), 4.21 (q, J = 4.4 Hz, 1H), 3.97–4.01 (m, 1H), 3.65 (t, J = 5.4 Hz, 1H), 2.15 (td, J = 9.5, 5.3 Hz, 1H), 1.79–1.91 (m, 4H), 1.59–1.68 (m, 3H), 1.38–1.50 (m, 2H), 0.95 (t, J = 7.2 Hz, 3H).
4-((5-Allyl-2,4-dioxothiazolidin-3-yl)methyl)-N-((tetrahydro-2H-pyran-2-yl)oxy)benzamide (7b)
27% yield. 1H-NMR (500 MHz, CDCl3) δ 9.09 (s, 1H), 7.70 (d, J = 8.0 Hz, 2H), 7.35–7.39 (m, 2H), 5.66–5.75 (m, 1H), 5.13–5.18 (m, 2H), 5.05 (s, 1H), 4.75 (dd, J = 24.1, 14.3 Hz, 2H), 4.26–4.30 (m, 1H), 3.99 (q, J = 10.1 Hz, 1H), 3.62 (t, J = 5.4 Hz, 1H), 2.87–2.92 (m, 1H), 2.56–2.63 (m, 1H), 1.81–1.87 (m, 3H), 1.53–1.64 (m, 3H).
4-((5-Benzyl-2,4-dioxothiazolidin-3-yl)methyl)-N-((tetrahydro-2H-pyran-2-yl)oxy)benzamide (7c)
23% yield. 1H-NMR (500 MHz, CDCl3) δ 8.81 (s, 1H), 7.69 (d, J = 8.0 Hz, 2H), 7.31 (d, J = 8.0 Hz, 2H), 7.27 (s, 3H), 7.17 (q, J = 2.9 Hz, 2H), 5.08 (s, 1H), 4.74 (dd, J = 22.6, 14.6 Hz, 2H), 4.50 (q, J = 4.4 Hz, 1H), 3.98–4.02 (m, 1H), 3.67 (t, J = 5.7 Hz, 1H), 3.49 (dd, J = 14.3, 4.0 Hz, 1H), 3.14 (dd, J = 14.3, 9.2 Hz, 1H), 1.85–1.91 (m, 3H), 1.62–1.68 (m, 3H).
3.1.6. General Procedure for the Synthesis of Compounds 6a-b and 8a-c
Compounds 5a-b or 7a-c (0.1 mmol) were dissolved in CH2Cl2 (4 mL). Then 2M HCl in diethyl ether (4 mL) was added dropwise. The reaction mixture was stirred for 2h at room temperature. The solvent was evaporated in vacuo. The crude was purified by MPLC to afford 6a-b and 8a-c in 36–44% yield.
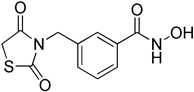
3-((2,4-Dioxothiazolidin-3-yl)methyl)-N-hydroxybenzamide (6a)
36% yield. 1H-NMR (500 MHz, CD3OD) δ 7.71 (s, 1H), 7.66 (d, J = 8.0 Hz, 1H), 7.52 (d, J = 8.0 Hz, 1H), 7.43 (t, J = 7.7 Hz, 1H), 4.80 (s, 2H), 4.13 (s, 2H) 13C-NMR (125 MHz, CD3OD) δ 173.96, 173.50, 167.80, 137.75, 132.73, 130.01, 127.98, 127.64, 45.55, 34.75. ESI MS (m/z) 267.04 [M+H]+.
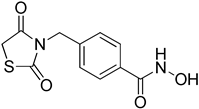
4-((2,4-Dioxothiazolidin-3-yl)methyl)-N-hydroxybenzamide (6b)
42% yield. 1H-NMR (500 MHz, CD3OD) δ 7.71 (d, J = 8.0 Hz, 2H), 7.42 (d, J = 8.0 Hz, 2H), 4.79 (s, 2H), 4.13 (s, 2H). 13C-NMR (125 MHz, CD3OD) δ 173.88, 173.43, 167.72, 140.76, 133.13, 129.44, 128.44, 45.48, 34.72. ESI MS (m/z) 267.04 [M+H]+.
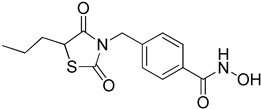
4-((2,4-Dioxo-5-propylthiazolidin-3-yl)methyl)-N-hydroxybenzamide (8a)
38% yield. 1H-NMR (500 MHz, CD3OD) δ 7.98 (d, J = 8.0 Hz, 2H), 7.41 (d, J = 8.6 Hz, 2H), 4.80 (s, 2H), 4.48–4.51 (m, 1H), 2.08–2.15 (m, 1H), 1.83–1.90 (m, 1H), 1.34–1.53 (m, 2H), 0.94–0.97 (m, 3H). 13C-NMR (125 MHz, CD3OD) δ 176.20, 172.88, 169.34, 142.09, 131.57, 131.09, 129.17, 50.89, 45.51, 35.87, 20.97, 13.76 ESI MS (m/z) 309.09 [M+H]+.
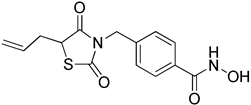
4-((5-Allyl-2,4-dioxothiazolidin-3-yl)methyl)-N-hydroxybenzamide (8b)
40% yield. 1H-NMR (500 MHz, CD3OD) δ 7.98 (d, J = 8.6 Hz, 2H), 7.41 (t, J = 7.7 Hz, 2H), 5.73–5.81 (m, 1H), 5.11–5.19 (m, 2H), 4.77–4.83 (m, 2H), 4.59 (q, J = 4.0 Hz, 1H), 2.87–2.91 (m, 1H), 2.62–2.68 (m, 1H) 13C-NMR (125 MHz, CD3OD) δ 175.56, 172.79, 169.34, 142.04, 133.49, 131.55, 131.05, 129.22, 120.10, 50.33, 45.53, 37.38. ESI MS (m/z) 307.08 [M+H]+.
4-((5-Benzyl-2,4-dioxothiazolidin-3-yl)methyl)-N-hydroxybenzamide (8c)
1H-NMR (500 MHz, CD3OD) 44% yield. δ 7.92 (d, J = 8.0 Hz, 2H), 7.18–7.24 (m, 7H), 4.83 (q, J = 4.0 Hz, 1H), 4.73 (dd, J = 22.3, 14.9 Hz, 2H), 3.41 (dd, J = 14.0, 4.3 Hz, 1H), 3.26 (q, J = 7.3 Hz, 1H). 13C-NMR (125 MHz, CD3OD) δ 175.37, 172.60, 169.39, 141.84, 136.97, 131.38, 131.03, 130.84, 129.54, 128.96, 128.44, 52.28, 45.47, 38.47. ESI MS (m/z) 357.09 [M+H]+.
3.2. Biology
3.2.1. Materials
Dulbecco’s Modified Eagle’s medium (DMEM) with L-glutamine was purchased from GenDEPOT (Barker, TX, USA) and fatal bovine serum (FBS) and penicillin and streptomycin were purchased Gibco BRL (Gaithersburg, MD, USA). Antibodies for α-tubulin, Ac-α-tubulin (Lys40), Histone H3, Ac-Histone H3 (Lys9), and β-actin were purchased from Cell Signaling Technology (Boston, MA, USA). Goat anti-rabbit IgG horseradish peroxidase conjugate was purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Cell Titer 96 Aqueous One Solution cell proliferation assay kit was purchased from Promega (Madison, WI, USA). Amersham ECL select Western blotting detection reagent was purchased from GE Healthcare. HDAC fluorogenic assay kits (HDAC1, #50061; HDAC6, #50076) were purchased from BPS Bioscience (San Diego, CA, USA). Tubastain A (Portland, OR, USA) was purchased from TCI chemicals. For in vitro studies, Methamphetamine (METH) was purchased from the Ministry of Food and Drug Safety (Cheongju, Korea).
3.2.2. Cell Culture
SH-SY5Y were grown in DMEM supplemented with streptomycin (500 mg/mL), penicillin (100 units/m), and 10% fetal bovine serum (FBS). The cells were incubated at 37 °C in a humidified atmosphere containing 5% CO2.
3.2.3. Assessment of Cell Morphology
SH-SY5Y cells (1 × 104 cells/well) were seeded in 6-well plate, and the cells were allowed to attach for 72 h. Culture medium was then changed to fresh medium containing METH (1 mM) for pretreat 4 h. After being preincubated for 4 h, add to compound 6b (5, 10, and 20 µM) for 24 h. Cell morphology was observed with inverted phase contrast microscope (Olympus, Tokyo, Japan) at 20× objective
3.2.4. HDAC Assay
Enzymatic HDAC assay was performed following manufacturer’s protocol (BPS Bioscience). Briefly, HDAC assay buffer (35 µL) was mixed with 5 µL of BSA (1 mg/mL) and 5 µL of HDAC substrate (200 µM) in 96-well black plate. Five microliters of HDAC1 enzyme (0.4 ng/μL) and HDAC6 enzyme (7 ng/µL) were added to the each well, followed by various concentrations of compound 6a-b, 8a-c and SAHA (5 µL), and then resulting mixture was incubated at 37 °C for 30 min. After incubation, 50 µL of undiluted 2 × HDAC developer was added to each well. After the mixture was incubated at RT for 15 min, fluorescence intensity was measured using a microplate reader at 360 nm excitation and 460 nm emission wavelengths.
3.2.5. Cell Proliferation Assay
SH-SY5Y (1 × 103 cells/well) were seeded in 96-well plate, the medium volume was brought to 100 µL, and the cells were allowed to attach for 14 h. Various concentrations of compound 6b (1, 5, 10, 30, 50, 70, and 100 µM) and METH (0.1 and 1 mM) were then added to the wells. Cells were then incubated at 37 °C for 24 h. Cell viability was determined using the Promega Cell Titer 96 Aqueous One Solution cell proliferation assay. Absorbance at 490 nm was read on Tecan Infinite F200 Pro plate reader, and values were expressed as percent of absorbance from cells incubated in DMSO alone.
3.2.6. Western Blot
The cells were washed and lysed in ice-cold lysis buffer (23 mM Tris-HCl pH 7.6, 130 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS) and 30 µg of lysate per lane was separated by SDS-PAGE, followed by transferring to a PVDF membrane (Bio rad, Hercules, USA). The membrane was blocked with 5% skim milk in TBST, and then incubated with primary antibody. After being treated with goat-anti rabbit secondary antibody (Santa Cruz, CA, USA) coupled horseradish peroxidase, proteins were visualized by ECL chemiluminescence according to the instructions of the manufacturer’s instruction (GE Healthcare, Chicago, IL, USA).
3.2.7. Statistical Analysis
Experiments were performed at least three times, with consistent results. The results are given as mean ± standard derivation (SD). The p-value was assessed using Student t-tests. Results were considered statistically significant at p < 0.01.
3.3. Docking Studies
In silico docking of compound 6b with the 3D coordinates of the X-ray crystal structure of HDAC6 (PDB code: 5EF7) was accomplished using the AutoDock 4.2 program downloaded from the Molecular Graphics Laboratory of the Scripps Research Institute. The AutoDock program was chosen because it uses a genetic algorithm to generate the poses of the ligand inside a known or predicted binding site utilizing the Lamarckian version of the genetic algorithm where the changes in conformations adopted by molecules after in situ optimization are used as subsequent poses for the offspring. In the docking experiments carried out, Gasteiger charges were placed on the X-ray structure of HDAC6 along with 6b using tools from the AutoDock suite. A grid box centered on the substrate binding pocket of HDACs enzyme with definitions of 50 × 50 × 50 points and 0.375 Å spacing was chosen for ligand docking experiments. The docking parameters consisted of setting the population size to 150, the number of generations to 27,000, and the number of evaluations to 2,500,000, while the number of docking runs was set to 100 with a cutoff of 1 Å for the root-mean-square tolerance for the grouping of each docking run. The docking pose of HDAC6 with compound 6b was depicted in Figure 5 and rendering of the picture was generated using PyMol (DeLanoScientific). Furthermore, we performed the self-docking experiment of the native ligand HPOB into HDAC6 and assessed the effectiveness of the docking simulations. The estimated binding energy of HPOB resulted in −7.85 kcal/mol and RMSD between the docked binding pose and the native binding pose led to 1.8 Å.
4. Conclusions
In the current study, we designed and synthesized a new series of thiazolidinedione-based HDAC6 inhibitors. Biological evaluation of these analogues illustrated that compound 6b furnished the most potent HDAC6 inhibition activity with an IC50 value of 21 nM, which is approximately 10-fold greater than FDA-approved drug, SAHA (HDAC6 IC50 = 226 nM). Exposure of SH-SY5Y cells with compound 6b more efficiently promoted the acetylation of α-tubulin than Histone H3, indicating that compound 6b suppressed the activity of HDAC6 enzyme more selectively than HDAC1. Furthermore, compound 6b reversed methamphetamine-induced morphology changes of SH-SY5Y cells in a dose-dependent manner. Western blot analysis revealed that biochemical mechanisms underlying methamphetamine-induced morphology changes are associated with disturbance of α-tubulin acetylation. Taken together, compound 6b represents a novel HDAC6-selective inhibitor, demonstrating promising therapeutic potential in methamphetamine addiction.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/20/24/6213/s1.
Author Contributions
Conceptualization, C.S., Y.J.O., B.P., S.L., C.-H.J., S.L., J.H.S., and Y.H.S.; methodology, C.S. and Y.J.O.; writing—original draft preparation, C.S., Y.J.O., and Y.H.S.; writing—review and editing, C.S., Y.J.O., B.P., S.L., C.-H.J., S.L., J.H.S., and Y.H.S.; project administration, Y.H.S.; funding acquisition, B.P., S.L., C.-H.J., S.L., J.H.S., and Y.H.S.
Acknowledgments
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2016R1A6A1A03011325 and 2016R1D1A1B01009559).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Galbraith, N. The methamphetamine problem: Commentary on psychiatric morbidity and socio-occupational dysfunction in residents of a drug rehabilitation centre. BJPsych Bull. 2015, 39, 218–220. [Google Scholar] [CrossRef] [PubMed]
- Courtney, K.E.; Ray, L.A. Methamphetamine: An update on epidemiology, pharmacology, clinical phenomenology, and treatment literature. Drug Alcohol Depend. 2014, 143, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, H.; Killinger, B.A.; Miller, C.V.; Moszczynska, A. Single and binge methamphetamine administrations have different effects on the levels of dopamine D2 autoreceptor and dopamine transporter in rat striatum. Int. J. Mol. Sci. 2014, 15, 5884–5906. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Chen, Y.Y.; Shen, Y.; Cao, X.; Li, A.; Liu, Q.; Li, Z.; Zhang, L.B.; Dai, W.; Tan, T.; et al. Methamphetamine abuse impairs motor cortical plasticity and function. Mol. Psychiatry 2017, 22, 1274–1281. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.L.; Bian, J.W.; Zheng, Z.J.; Zhao, L.; Han, S.; Sun, X.H.; Li, J.F.; Ni, G.X. Effects of methamphetamine abuse on spatial cognitive function. Sci. Rep. 2018, 8, 5502. [Google Scholar] [CrossRef] [PubMed]
- Groman, S.M.; Rich, K.M.; Smith, N.J.; Lee, D.; Taylor, J.R. Chronic Exposure to Methamphetamine Disrupts Reinforcement-Based Decision Making in Rats. Neuropsychopharmacology 2018, 43, 770–780. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.; Lee, J.H.; Seo, Y.H.; Jang, J.H.; Jeong, C.H.; Lee, S.; Jeong, G.S.; Park, B. Epicatechin Prevents Methamphetamine-Induced Neuronal Cell Death via Inhibition of ER Stress. Biomol. Ther. (Seoul) 2019, 27, 145–151. [Google Scholar] [CrossRef]
- Krasnova, I.N.; Cadet, J.L. Methamphetamine toxicity and messengers of death. Brain Res. Rev. 2009, 60, 379–407. [Google Scholar] [CrossRef]
- Anderson, E.M.; Penrod, R.D.; Barry, S.M.; Hughes, B.W.; Taniguchi, M.; Cowan, C.W. It is a complex issue: Emerging connections between epigenetic regulators in drug addiction. Eur. J. Neuro. Sci. 2019, 50, 2477–2491. [Google Scholar] [CrossRef]
- Godino, A.; Jayanthi, S.; Cadet, J.L. Epigenetic landscape of amphetamine and methamphetamine addiction in rodents. Epigenetics 2015, 10, 574–580. [Google Scholar] [CrossRef]
- Torres, O.V.; Ladenheim, B.; Jayanthi, S.; McCoy, M.T.; Krasnova, I.N.; Vautier, F.A.; Cadet, J.L. An Acute Methamphetamine Injection Downregulates the Expression of Several Histone Deacetylases (HDACs) in the Mouse Nucleus Accumbens: Potential Regulatory Role of HDAC2 Expression. Neurotox Res. 2016, 30, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Cadet, J.L. Epigenetics of Stress, Addiction, and Resilience: Therapeutic Implications. Mol. Neurobiol. 2016, 53, 545–560. [Google Scholar] [CrossRef] [PubMed]
- Kalda, A.; Heidmets, L.T.; Shen, H.Y.; Zharkovsky, A.; Chen, J.F. Histone deacetylase inhibitors modulates the induction and expression of amphetamine-induced behavioral sensitization partially through an associated learning of the environment in mice. Behav. Brain Res. 2007, 181, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, S.; Salta, S.; Summavielle, T. Methamphetamine promotes alpha-tubulin deacetylation in endothelial cells: The protective role of acetyl-l-carnitine. Toxicol. Lett. 2015, 234, 131–138. [Google Scholar] [CrossRef]
- Drazic, A.; Myklebust, L.M.; Ree, R.; Arnesen, T. The world of protein acetylation. Biochim. Biophys Acta. 2016, 1864, 1372–1401. [Google Scholar] [CrossRef]
- Glozak, M.A.; Sengupta, N.; Zhang, X.; Seto, E. Acetylation and deacetylation of non-histone proteins. Gene 2005, 363, 15–23. [Google Scholar] [CrossRef]
- Choi, M.A.; Park, S.Y.; Chae, H.Y.; Song, Y.; Sharma, C.; Seo, Y.H. Design, synthesis and biological evaluation of a series of CNS penetrant HDAC inhibitors structurally derived from amyloid-β probes. Sci. Rep. 2019, 9, 13187. [Google Scholar] [CrossRef]
- Lim, J.; Song, Y.; Jang, J.-H.; Jeong, C.-H.; Lee, S.; Park, B.; Seo, Y.H. Aspirin-inspired acetyl-donating HDACs inhibitors. Arch. Pharm. Res. 2018, 41, 967–976. [Google Scholar] [CrossRef]
- Song, Y.; Lim, J.; Seo, Y.H. A novel class of anthraquinone-based HDAC6 inhibitors. Eur. J. Med. Chem. 2019, 164, 263–272. [Google Scholar] [CrossRef]
- Sucheta; Tahlan, S.; Verma, P.K. Biological potential of thiazolidinedione derivatives of synthetic origin. Chem. Cent. J. 2017, 11, 130. [Google Scholar] [CrossRef]
- Saltiel, A.R.; Olefsky, J.M. Thiazolidinediones in the treatment of insulin resistance and type II diabetes. Diabetes 1996, 45, 1661–1669. [Google Scholar] [CrossRef]
- Ma, L.; Pei, H.; Lei, L.; He, L.; Chen, J.; Liang, X.; Peng, A.; Ye, H.; Xiang, M.; Chen, L. Structural exploration, synthesis and pharmacological evaluation of novel 5-benzylidenethiazolidine-2,4-dione derivatives as iNOS inhibitors against inflammatory diseases. Eur. J. Med. Chem. 2015, 92, 178–190. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, N.; Caron, C.; Matthias, G.; Hess, D.; Khochbin, S.; Matthias, P. HDAC-6 interacts with and deacetylates tubulin and microtubules in vivo. EMBO J. 2003, 22, 1168–1179. [Google Scholar] [CrossRef]
- Li, F.; Wu, R.; Cui, X.; Zha, L.; Yu, L.; Shi, H.; Xue, B. Histone Deacetylase 1 (HDAC1) Negatively Regulates Thermogenic Program in Brown Adipocytes via Coordinated Regulation of Histone H3 Lysine 27 (H3K27) Deacetylation and Methylation. J. Biol. Chem. 2016, 291, 4523–4536. [Google Scholar] [CrossRef]
- Strebl, M.G.; Campbell, A.J.; Zhao, W.N.; Schroeder, F.A.; Riley, M.M.; Chindavong, P.S.; Morin, T.M.; Haggarty, S.J.; Wagner, F.F.; Ritter, T.; et al. HDAC6 Brain Mapping with [(18)F]Bavarostat Enabled by a Ru-Mediated Deoxyfluorination. ACS Cent. Sci. 2017, 3, 1006–1014. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).