Characterization of a Functional Role of the Bradyrhizobium japonicum Isocitrate Lyase in Desiccation Tolerance
Abstract
:1. Introduction
2. Results
2.1. Purified AceA Has ICL Activity
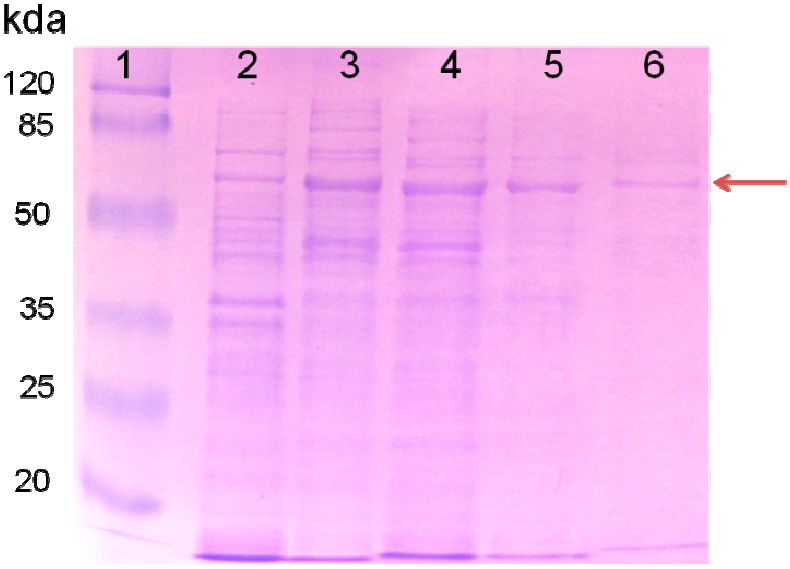
| Purification Steps | Soluble Protein (mg) | Total Activity (U) | Specific Activity (U/mg) |
|---|---|---|---|
| Cell-free extract | 45.2 | 36.2 | 0.8 |
| Filtered on Ni-NTA column | 2.3 | 3.0 | 1.3 |
| Purified ICL protein | 0.04 | 0.1 | 2.1 |
2.2. The aceA Mutant WC2455 Is Sensitive to Desiccation and Salt Stress

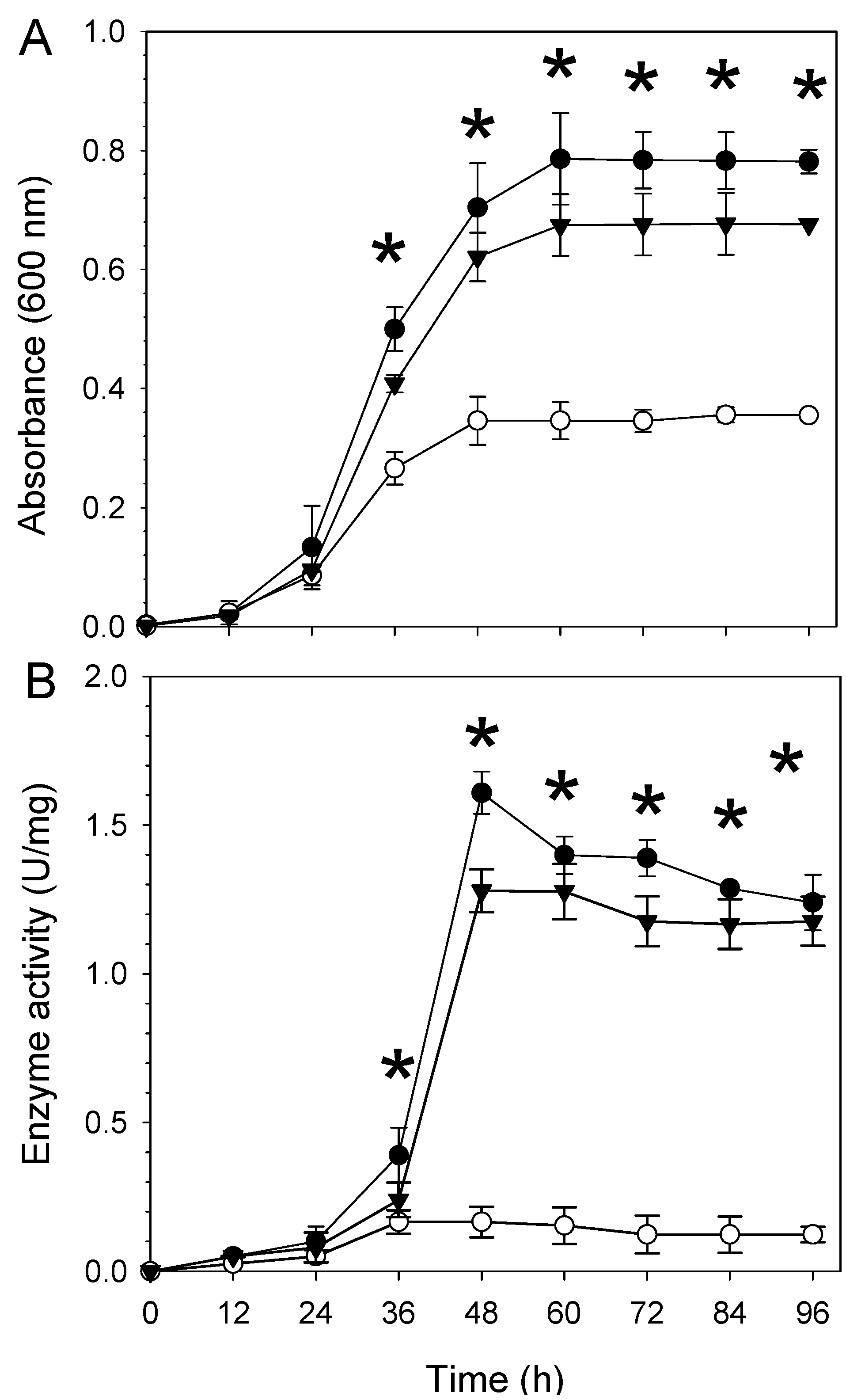
2.3. Involvement of AceA in Response to Desiccation Stress Is Independent of the Glyoxylate Pathway or the TCA Cycle
| Locus (Gene) | Gene Description a | Desiccation | |
|---|---|---|---|
| Wild Type | WC2455 | ||
| bll0452 (sucA) | alpha-ketoglutarate dehydrogenase | 1.0 | 1.0 |
| bll0455 (sucC) | succinyl-CoA synthetase beta chain | −1.3 | −1.7 |
| bll1474 (glcB) | malate synthase | −1.6 | 1.1 |
| blr0512 (sdhC) | succinate dehydrogenase cytochrome | 1.3 | 1.0 |
| blr2455 (aceA) | isocitrate lyase | 148.0 | ND b |
| blr5747 (icdA) | isocitrate dehydrogenase | −1.3 | 1.1 |
| blr6519 (fumC) | fumarase C | 1.6 | 1.2 |
2.4. Comparison of the Global Transcription Profiles in Wild Type and Mutant Strains
| Physiological Process | Locus (Gene ID) a | Description b | Fold Induction |
|---|---|---|---|
| Chaperones | bsl3986 (cspA) | cold shock protein | 1.7 |
| blr4637 | probable HspC2 heat shock protein | 1.6 | |
| blr4635 (groEL) | chaperonin GroEL | 1.5 | |
| blr4653 (dnaJ) | molecular chaperone DnaJ family | 1.7 | |
| bll5219 (hspD) | small heat shock protein | 2.1 | |
| blr5625 (groES) | 10 KD chaperonin | 2.5 | |
| blr5626 (groEL) | 60 KDA chaperonin | 2.1 | |
| Energy metabolism | blr1656 | putative glycosyl hydrolase | 2.0 |
| bll3998 | probable succinate-semialdehyde dehydrogenase | 2.3 | |
| blr4657 | beta-glucosidase | 1.6 | |
| bll4784 | aldehyde dehydrogenase | 1.8 | |
| blr6128 (cycB) | cytochrome c552 | 1.5 | |
| blr7040 (napC) | cytochrome C-type protein | 2.1 | |
| Heat shock response systems | blr0678 | heat shock protein 70 | 2.0 |
| Translation | bll5377 (rpsK) | 30S ribosomal protein S11 | 1.8 |
| bll5381 (rplO) | 50S ribosomal protein L15 | 1.6 | |
| bsl5382 (rpmD) | 50S ribosomal protein L30 | 1.6 | |
| bsl5391 (rpsQ) | 30S ribosomal protein S17 | 1.5 | |
| bsl5392 (rpmC) | 50S ribosomal protein L29 | 1.8 | |
| bll5397 (rplB) | 50S ribosomal protein L2 | 1.7 | |
| bll5415 (rplK) | 50S ribosomal Protein L11 | 2.4 |
2.5. The Inactivation of AceA Results in Delayed Soybean Nodulation
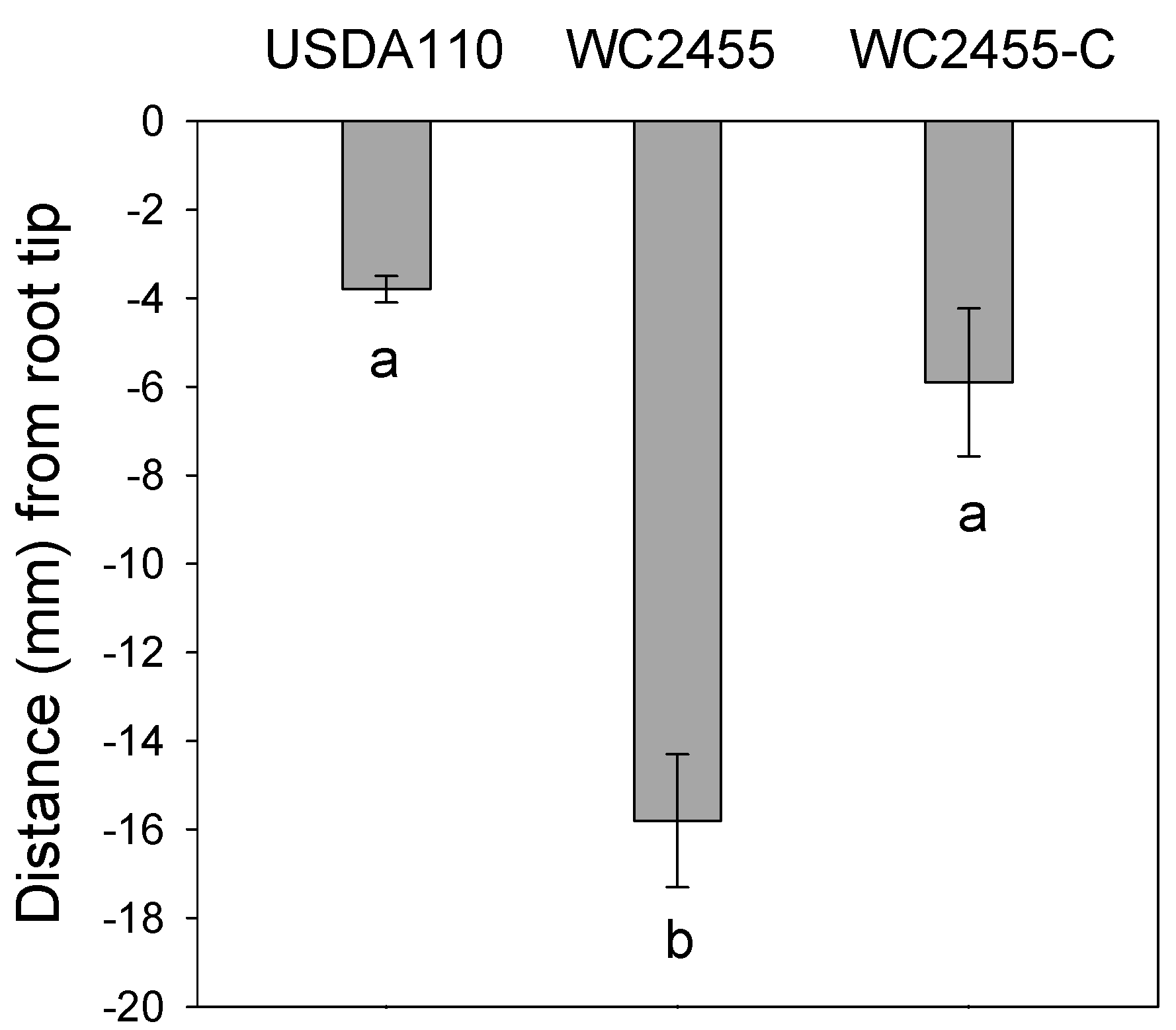
3. Discussion
4. Experimental Section
4.1. Bacterial Strains and Culture Conditions
4.2. Desiccation Stress Assay
| Strain or Plasmid | Genotypes, Relevant Characteristics | Source |
|---|---|---|
| B. japonicum strains | ||
| USDA110 | wild type | USDA-ARS (Beltsville, MD, USA) |
| WC2455 | aceA::Km | [25] |
| WC2455-C | WC2455 complemented strain | [6] |
| E. coli strains | ||
| DH5α | supE44 ∆lacU169 (ø 80lacZ∆M15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | [27] |
| RIL(DE3) | argU (AGA, AGG), ileY (AUA), leuW (CUA) | Agilent (La Jolla, CA, USA) |
| Plasmids | ||
| pTE3 | complementing plasmid, Tcr | [28] |
| pRK2073 | RK2, Tra+,Smr | [29] |
| pGEM-T easy | cloning vector | Promega (Madison, WI, USA) |
| pQE2 | expression vector, 6X His tag, T7 promoter | Qiagen (Valencia, CA, USA) |
| pGEM-T easy::aceA | pGEM T easy containing 1.5 kb fragment including entire aceA gene | This study |
| pTE-aceA | pTE3 containing 1.5 kb fragment of aceA | This study |
| pHis-aceA | pQE2 containing 1.8 kb fragment of aceA | This study |
4.3. Salt Stress Assay
4.4. RNA Isolation and Microarray Analysis
4.5. qRT-PCR Analysis
| Gene | Forward Sequence 5′–3′ | Reverse Sequence 5′−3′ |
|---|---|---|
| bll0452 | CGGCATCGACGACATCTACCTGAT | TCCAGATAGGGCTCGATGAAGTGC |
| bll0455 | GAGACAGAGGAAGACGCCAAGGAA | GCCATGCCGTAGAGCTTGATGATG |
| bll1474 | GCCTCCAAGCGCATCATGTTCATC | CATGTCGACGTTCCAGTCCTCGTA |
| blr0512 | TTCAAGGCCAATGAGCGCGAAG | ACGATCCAGATCAGCACCGTCA |
| blr2455 | GGCGACCAGTACAACAGCTT | GTCTCGATCCAGAGCAGGTC |
| blr5747 | TGTCGACCAAGAACACCATCCTCA | TAGTTCTTGCAGGCCCAGACATAGC |
| blr6519 | GGCCATTTCGAGCTCAACGTCTAC | CTGACGCAATGTTCGGTGAAGGAG |
| bll0631 | TCAACCTTCTGACGGTGAACGC | TGCAGCAATTGCGACAGACCTT |
4.6. ICL Enzyme Assay
4.7. Construction of pQE2::AceA Strain
4.8. Purification of AceA Protein
4.9. Nodulation Assay
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bashan, Y. Inoculants of plant growth-promoting bacteria for use in agriculture. Biotechnol. Adv. 1998, 16, 729–770. [Google Scholar] [CrossRef]
- Mary, P.; Ochin, D.; Tailliez, R. Rates of drying and survival of Rhizobium meliloti strains during storage at different relative humidities. Appl. Environ. Microbiol. 1985, 50, 207–211. [Google Scholar] [PubMed]
- Streeter, J.G. Factors affecting the survival of Bradyrhizobium applied in liquid cultures to soya bean [Glycine max (L.) Merr.] seeds. J. Appl. Microbiol. 2007, 103, 1282–1290. [Google Scholar] [CrossRef] [PubMed]
- Cytryn, E.J.; Sangurdekar, D.P.; Streeter, J.G.; Franck, W.L.; Chang, W.-S.; Stacey, G.; Emerich, D.W.; Joshi, T.; Xu, D.; Sadowsky, M.J. Transcriptional and physiological responses of Bradyrhizobium japonicum to desiccation-induced stress. J. Bacteriol. 2007, 189, 6751–6762. [Google Scholar] [CrossRef] [PubMed]
- Donati, A.J.; Jeon, J.M.; Sangurdekar, D.; So, J.-S.; Chang, W.-S. Genome-wide transcriptional and physiological responses of Bradyrhizobium japonicum to paraquat-mediated oxidative stress. Appl. Environ. Microbiol. 2011, 77, 3633–3643. [Google Scholar] [CrossRef] [PubMed]
- Jeon, J.M.; Lee, H.-I.; Donati, A.J.; So, J.-S.; Emerich, D.W.; Woo-Suk Chang, W.-S.C. Whole-genome expression profiling of Bradyrhizobium japonicum in response to hydrogen peroxide. Mol. Plant Microbe Interact. 2011, 24, 1472–1481. [Google Scholar] [CrossRef] [PubMed]
- Fang, F.C.; Libby, S.J.; Castor, M.E.; Fung, A.M. Isocitrate lyase (AceA) is required for Salmonella persistence but not for acute lethal infection in mice. Infect. Immun. 2005, 73, 2547–2549. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, M.C.; Fink, G.R. Life and death in a macrophage: Role of the glyoxylate cycle in virulence. Eukaryot. Cell 2002, 1, 657–662. [Google Scholar] [CrossRef] [PubMed]
- Dunn, M. Tricarboxylic acid cycle and anaplerotic enzymes in rhizobia. FEMS Microbiol. Rev. 1998, 22, 105–123. [Google Scholar] [CrossRef] [PubMed]
- Geddes, B.A.; Oresnik, I.J. Physiology, genetics, and biochemistry of carbon metabolism in the alphaproteobacterium Sinorhizobium meliloti. Can. J. Microbiol. 2014, 60, 491–507. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Xiao, X.; Li, J.; Luo, J.; Wang, F. Identification of genes regulated by changing salinity in the deep-sea bacterium Shewanella sp. WP3 using RNA arbitrarily primed PCR. Extremophiles 2006, 10, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, S.; Yamaoka, N.; Takada, Y.; Fukunaga, N. The cold-inducible icl gene encoding thermolabile isocitrate lyase of a psychrophilic bacterium, Colwellia maris. Microbiology 2002, 148, 2579–2589. [Google Scholar] [PubMed]
- Dunn, M.F.; Ramirez-Trujillo, J.A.; Hernandez-Lucas, I. Major roles of isocitrate lyase and malate synthase in bacterial and fungal pathogenesis. Microbiology 2009, 155, 3166–3175. [Google Scholar] [CrossRef] [PubMed]
- Munoz-Elias, E.J.; McKinney, J.D. Mycobacterium tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nat. Med. 2005, 11, 638–644. [Google Scholar] [CrossRef] [PubMed]
- Sharma, V.; Sharma, S.; Hoener, Z.U.; Bentrup, K.; McKinney, J.D.; Russell, D.G.; Sacchettini, J.C. Structure of isocitrate lyase, a persistence factor of Mycobacterium tuberculosis. Nat. Struct. Biol. 2000, 7, 663–668. [Google Scholar] [CrossRef] [PubMed]
- Idnurm, A.; Howlett, B.J. Isocitrate lyase is essential for pathogenicity of the fungus Leptosphaeria maculans to canola (Brassica napus). Eukaryot. Cell 2002, 1, 719–724. [Google Scholar] [CrossRef] [PubMed]
- Pradheep, S.A.; Narayanan, S.; Vasan, S.K.; Narayanan, P.R. Cloning and expression of aceA gene encoding isocitrate lyase from Mycobacterium tuberculosis. Curr. Sci. 2000, 79, 1585–1588. [Google Scholar]
- Nollen, E.A.; Morimoto, R.I. Chaperoning signaling pathways: Molecular chaperones as stress-sensing “heat shock” proteins. J. Cell Sci. 2002, 115, 2809–2816. [Google Scholar] [PubMed]
- Chang, W.-S.; Franck, W.L.; Cytryn, E.; Jeong, S.; Joshi, T.; Emerich, D.W.; Sadowsky, M.J.; Xu, D.; Stacey, G. An oligonucleotide microarray resource for transcriptional profiling of Bradyrhizobium japonicum. Mol. Plant Microbe Interact. 2007, 20, 1298–1307. [Google Scholar] [CrossRef] [PubMed]
- Tao, H.; Bausch, C.; Richmond, C.; Blattner, F.R.; Conway, T. Functional genomics: Expression analysis of Escherichia coli growing on minimal and rich media. J. Bacteriol. 1999, 181, 6425–6440. [Google Scholar] [PubMed]
- Fisher, M.A.; Plikaytis, B.B.; Shinnick, T.M. Microarray analysis of the Mycobacterium tuberculosis transcriptional response to the acidic conditions found in phagosomes. J. Bacteriol. 2002, 184, 4025–4032. [Google Scholar] [CrossRef] [PubMed]
- Hamel, R.; Appanna, V.D.; Viswanatha, T.; Puiseux-Dao, S. Overexpression of isocitrate lyase is an important strategy in the survival of Pseudomonas fluorescens exposed to aluminum. Biochem. Biophys. Res. Commun. 2004, 317, 1189–1194. [Google Scholar] [CrossRef] [PubMed]
- Karlekar, K.; Parekh, T.V.; Chhatpar, H.S. Salt mediated changes in some enzymes of carbohydrate-metabolism in halotolerant Cladosporium sphaerospermum. J. Biosci. 1985, 9, 197–201. [Google Scholar] [CrossRef]
- Sadowsky, M.J.; Tully, R.E.; Cregan, P.B.; Keyser, H.H. Genetic diversity in Bradyrhizobium japonicum serogroup 123 and its relation to genotype-specific nodulation of soybean. Appl. Environ. Microbiol. 1987, 53, 2624–2630. [Google Scholar] [PubMed]
- Franck, W.L.; Chang, W.-S.; Qiu, J.; Sugawara, M.; Sadowsky, M.J.; Stephanie, A.; Smith, S.A.S.; Gary Stacey, G.S. Whole-genome transcriptional profiling of Bradyrhizobium japonicum during chemoautotrophic growth. J. Bacteriol. 2008, 190, 6697–6705. [Google Scholar] [CrossRef] [PubMed]
- Bergersen, F.J. The growth of rhizobia in synthetic media. Aust. J. Biol. Sci. 1961, 14, 349–360. [Google Scholar]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 1989; Volume 3, p. A.10. [Google Scholar]
- Egelhoff, T.T.; Fisher, R.F.; Jacobs, T.W.; Mulligan, J.T.; Long, S.R. Nucleotide sequence of Rhizobium meliloti 1021 nodulation genes: nodD is read divergently from nodABC. DNA 1985, 4, 241–248. [Google Scholar] [CrossRef] [PubMed]
- Leong, S.A.; Ditta, G.S.; Helinski, D.R. Heme biosynthesis in Rhizobium. Identification of a cloned gene coding for delta-aminolevulinic acid synthetase from Rhizobium meliloti. J. Biol. Chem. 1982, 257, 8724–8730. [Google Scholar] [PubMed]
- Bittner, M.; Butow, R.; DeRisi, J.; Diehn, M.; Eberwine, J.; Epstein, C.B.; Glynne, R.; Grimmond, S.; Ideker, T.; Kacharmina, J.E.; et al. Expression analysis of RNA. In DNA Microarrays; Bowtell, D., Sambrook, J., Eds.; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 2003; pp. 101–288. [Google Scholar]
- Green, L.S.; Karr, D.B.; Emerich, D.W. Isocitrate dehydrogenase and glyoxylate cycle enzyme activities in Bradyrhizobium japonicum under various growth conditions. Arch. Microbiol. 1998, 169, 445–451. [Google Scholar] [CrossRef] [PubMed]
- Chell, R.M.; Sundaram, T.K.; Wilkinson, A.E. Isolation and characterization of isocitrate lyase from a thermophilic Bacillus sp. Biochem. J. 1978, 173, 165–177. [Google Scholar] [PubMed]
- Lee, H.-I.; Lee, J.-H.; Park, K.-H.; Sangurdekar, D.; Chang, W.-S. Effect of soybean coumestrol on Bradyrhizobium japonicum nodulation ability, biofilm formation, and transcriptional profile. Appl. Environ. Microbiol. 2012, 78, 2896–2903. [Google Scholar] [CrossRef] [PubMed]
- Donati, A.J.; Lee, H.-I.; Leveau, J.H.; Chang, W.-S. Effects of indole-3-acetic acid on the transcriptional activities and stress tolerance of Bradyrhizobium japonicum. PLoS ONE 2013, 8, e76559. [Google Scholar] [CrossRef] [PubMed]
- Broughton, W.J.; Dilworth, M.J. Control of leghaemoglobin synthesis in snake beans. Biochem. J. 1971, 125, 1075–1080. [Google Scholar]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeon, J.-M.; Lee, H.-I.; Sadowsky, M.J.; Sugawara, M.; Chang, W.-S. Characterization of a Functional Role of the Bradyrhizobium japonicum Isocitrate Lyase in Desiccation Tolerance. Int. J. Mol. Sci. 2015, 16, 16695-16709. https://doi.org/10.3390/ijms160716695
Jeon J-M, Lee H-I, Sadowsky MJ, Sugawara M, Chang W-S. Characterization of a Functional Role of the Bradyrhizobium japonicum Isocitrate Lyase in Desiccation Tolerance. International Journal of Molecular Sciences. 2015; 16(7):16695-16709. https://doi.org/10.3390/ijms160716695
Chicago/Turabian StyleJeon, Jeong-Min, Hae-In Lee, Michael J. Sadowsky, Masayuki Sugawara, and Woo-Suk Chang. 2015. "Characterization of a Functional Role of the Bradyrhizobium japonicum Isocitrate Lyase in Desiccation Tolerance" International Journal of Molecular Sciences 16, no. 7: 16695-16709. https://doi.org/10.3390/ijms160716695
APA StyleJeon, J.-M., Lee, H.-I., Sadowsky, M. J., Sugawara, M., & Chang, W.-S. (2015). Characterization of a Functional Role of the Bradyrhizobium japonicum Isocitrate Lyase in Desiccation Tolerance. International Journal of Molecular Sciences, 16(7), 16695-16709. https://doi.org/10.3390/ijms160716695





