MicroRNAs Involved in Anti-Tumour Immunity
Abstract
:1. Introduction
2. miRNAs and Innate Immunity
2.1. Macrophages
2.2. NK Cells
3. miRNAs and Adaptive Immunity
3.1. B Cells
Related Signalling Pathways in Lymphoma
3.2. T Cells
3.3. Dendritic Cells
4. miRNAs Related to Anti-Tumour Immunity
4.1. CNS Involvement
4.2. Cancer in Blood Components
4.3. Gynaecological Cancers
4.4. Cancers of the GI Tract
4.5. Hepatocellular Cancers
4.6. Other Cancers
5. Discussion
6. Current Studies Targeting miRNAs for Therapeutic Purposes
7. Future Challenges
References
- Li, S. Q.; Chen, F. J.; Cao, X.F. Distinctive microRNAs in esophageal tumor: Early diagnosis, prognosis judgment, and tumor treatment. Dis. Esophagus 2012. [Google Scholar] [CrossRef]
- Cho, W.C. MicroRNAs in cancer—from research to therapy. Biochim. Biophys. Acta 2010, 1805, 209–217. [Google Scholar]
- Cho, W.C. MicroRNAs: Potential biomarkers for cancer diagnosis, prognosis and targets for therapy. Int. J. Biochem. Cell Biol 2010, 42, 1273–1281. [Google Scholar]
- Bavan, L.; Midwood, K.; Nanchahal, J. microRNA epigenetics. BioDrugs 2011, 25, 27–41. [Google Scholar]
- Lodish, H. F.; Zhou, B.; Liu, G. Micromanagement of the immune system by miRNAs. Immunology 2008, 8, 120–130. [Google Scholar]
- Sonkoly, E.; Stahle, M.; Pivarcsi, A. MicroRNAs and immunity: novel players in the regulation of normal immune function and inflammation. Semin. Cancer Biol 2008, 18, 131–140. [Google Scholar]
- Asirvatham, A. J.; Magner, W. J.; Tomasi, T.B. miRNA regulation of cytokine genes. Cytokine 2009, 45, 58–69. [Google Scholar]
- Holmstrom, K.; Pedersen, A. W.; Claesson, M.H. Identification of a microRNA signature in dendritic cell vaccines for cancer therapy. Hum. Immunol 2010, 71, 67–73. [Google Scholar]
- O’Neill, L.A. How Toll-Like receptors signal: What we know and what we don’t know. Curr. Opin. Immunol 2006, 18, 3–9. [Google Scholar]
- Zhou, R.; O’Hara, S. P.; Chen, X.M. MicroRNA regulation of innate immune responses in epithelial cells. Cell Mol. Immunol 2011, 8, 371–379. [Google Scholar]
- Takeuchi, O.; Akira, S. Toll-Like receptor signaling. In Dendritic Cell Interactions with Bacteria; Rescigno, M., Ed.; University Press: Cambridge, UK, 2007; pp. 25–50. [Google Scholar]
- Gantier, M. P.; Stunden, H. J.; McCoy, C.E. A miR-19 regulon that controls NF-κB signaling. Nucleic Acids Res 2012, 40, 8048–8058. [Google Scholar]
- Squadrito, M. L.; Pucci, F.; Maqri, L. miR-511–3p modulates genetic programs of tumor-associated macrophages. Cell Rep 2012, 1, 141–154. [Google Scholar]
- Taganov, K. D.; Boldin, M. P.; Chang, K.J. NF-kappaB-Dependent induction of microRNA miR-146, an inhibitor targeted to signalling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA 2006, 103, 12481–12486. [Google Scholar]
- Jennewein, C.; von Knethen, A.; Schmid, T. MicroRNA-27b contributes to lipopolysaccharidemediated peroxisome proliferator-activated receptor gamma (PPARgamma) mRNA destabilization. J. Biol. Chem 2010, 285, 11846–11853. [Google Scholar]
- Bazzoni, F.; Rossato, M.; Fabbri, M. Induction and regulatory function of miR-9 in human monocytes and neutrophils exposed to proinflammatory signals. Proc. Natl. Acad. Sci. USA 2009, 106, 5282–5287. [Google Scholar]
- Ueda, R.; Kohanbash, G.; Sasaki, K. Dicer-Regulated microRNAs 222 and 339 promote resistance of cancer cells to cytotoxic T-lymphocytes by down-regulation of ICAM-1. Proc. Natl. Acad. Sci. USA 2009, 106, 10746–10751. [Google Scholar]
- Tili, E.; Michaille, J. J.; Cimino, A. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J. Immunol 2007, 179, 5082–5089. [Google Scholar]
- Jing, Q.; Huang, S.; Guth, S. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell 2005, 120, 623–634. [Google Scholar]
- Wang, P.; Gu, Y.; Zhang, Q. Identification of resting and Type I IFN-activated human NK cell miRNomes reveals MicroRNA-378 and MicroRNA-30e as negative regulators of NK Cell cytotoxicity. J. Immunol 2012, 189, 211–221. [Google Scholar]
- Kim, T. D.; Lee, S. U.; Yun, S. Human microRNA-27a* targets Prf1 and GzmB expression to regulate NK-cell cytotoxicity. Blood 2011, 118, 5476–5486. [Google Scholar]
- Gong, J.; Liu, R.; Zhuang, R. miR-30c-1* promotes natural killer cell cytotoxicity against human hepatoma cells by targeting the transcription factor HMBOX1. Cancer Sci 2012, 103, 645–652. [Google Scholar]
- Mosser, D.M. The many faces of macrophage activation. J. Leukoc. Biol 2003, 73, 209–212. [Google Scholar]
- Lisnic, V. J.; Krmpotic, A.; Jonjic, S. Modulation of natural killer cell activity by virus. Curr. Opin. Microbiol 2010, 13, 530–539. [Google Scholar]
- Rao, D. S.; O’Connell, R. M.; Chaudhuri, A.A. MicroRNA-34a perturbs B lymphocyte development by repressing the forkhead box transcription factor Foxp1. Immunity 2010, 33, 48–59. [Google Scholar]
- Ventura, A.; Young, A. G.; Winslow, M.M. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell 2008, 132, 875–876. [Google Scholar]
- Koralov, S. B.; Muljo, S. A.; Galler, G.R. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell 2008, 132, 860–874. [Google Scholar]
- Davidson-Moncada, J.; Papavasiliou, F. N.; Tam, W. MicroRNAs of the immune system: Roles in inflammation and cancer. Ann. N. Y. Acad. Sci 2010, 1183, 183–194. [Google Scholar]
- Lin, Y.C. C-Myb is an evolutionary conserved miR-150 target and miR-150/c-Myb interaction is important for embryonic development. Mol. Biol. Evol 2008, 25, 2189–2198. [Google Scholar]
- Zhou, B.; Wang, S.; Mayr, C. miR-150, a microRNA expressed in mature B and T cells, blocks early B cell development when expressed prematurely. Proc. Natl. Acad. Sci. USA 2007, 104, 7080–7085. [Google Scholar]
- Baltimore, D.; Boldin, M. P.; O’Connell, R.M. MicroRNAs: New regulators of immune cells development and function. Nat. Immunol 2008, 9, 839–845. [Google Scholar]
- Costinean, S.; Zanesi, N.; Pekarsky, Y. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR-155 transgenic mice. Proc. Natl. Acad. Sci. USA 2006, 103, 7024–7029. [Google Scholar]
- Costinean, S.; Sandhu, S. K.; Pedersen, I.M. Src homology 2 domain-containing inositol-5-phosphatase and CCAAT enhancer-binding protein beta are targeted by miR-155 in B cells of Emicro-MiR-155 transgenic mice. Blood 2009, 114, 1374–1382. [Google Scholar]
- Chaudhuri, A. A.; So, A. Y.; Mehta, A. Oncomir miR-125b regulates hematopoiesis by targeting the gene Lin28A. Proc. Natl. Acad. Sci. USA 2012, 109, 4233–4238. [Google Scholar]
- Nana-Sinkam, S. P.; Croce, C.M. MicroRNA in chronic lymphocytic leukemia: Transitioning from laboratory-based investigation to clinical application. Cancer Genet. Cytogenet 2010, 203, 127–133. [Google Scholar]
- Pallasch, C. P.; Patz, M.; Park, Y.J. miRNA deregulation by epigenetic silencing disrupts suppression of the oncogene PLAG1 in chronic lymphocytic leukemia. Blood 2009, 114, 3255–3264. [Google Scholar]
- Patz, M.; Pallasch, C. P.; Wendtner, C.M. Critical role of microRNAs in chronic lymphocytic leukemia: Overexpression of the oncogene PLAG1 by deregulated miRNAs. Leuk. Lymphoma 2010, 51, 1379–1381. [Google Scholar]
- Tan, L. P.; Wang, M.; Robertus, J.L. miRNA profiling of B-cell subsets: Specific miRNA profile for germinal center B cells with variation between centroblasts and centrocytes. Lab. Invest 2009, 89, 708–716. [Google Scholar]
- Robertus, J. L.; Harms, G.; Blokzijl, T. Specific expression of miR-17–5p and miR-127 in testicular and central nervous system diffuse large B-cell lymphoma. Mod. Pathol 2009, 22, 547–555. [Google Scholar]
- Iqbal, J.; Shen, Y.; Liu, Y. Genome-Wide miRNA profiling of mantle cell lymphoma reveals a distinct subgroup with poor prognosis. Blood 2012, 119, 4939–4948. [Google Scholar]
- Vigorito, E.; Perks, K. L.; Abreu-Goodger, C. MicroRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity 2007, 27, 847. [Google Scholar]
- Matsuyama, H.; Suzuki, H. I.; Nishimori, H. miR-135b mediates NPM-ALK-driven oncogenicity and renders IL-17-producing immunophenotype to anaplastic large cell lymphoma. Blood 2011, 118, 6881–6892. [Google Scholar]
- Himmelreich, H.; Mathys, A.; Wodnar-Filipowicz, A. Post-transcriptional regulation of ULBP1 ligand for the activating immunoreceptor NKG2D involves 3' untranslated region. Hum. Immunol 2011, 72, 470–478. [Google Scholar]
- Cubillos-Ruiz, J. R.; Baird, J. R.; Tesone, A.J. Reprogramming tumor-associated dendritic cells in vivo using miRNA mimetics triggers protective immunity against ovarian cancer. Cancer Res 2012, 72, 1683–1693. [Google Scholar]
- Zheng, J.; Jiang, H. Y.; Li, J. MicroRNA-23b promotes tolerogenic properties of dendritic cells in vitro through inhibiting Notch1/NF-κB signalling pathways. Allergy 2012, 67, 362–370. [Google Scholar]
- Turner, M. L.; Schnorfeil, F. M.; Brocker, T. MicroRNAs regulate dendritic cell differentiation and function. J. Immunol 2011, 187, 3911–3917. [Google Scholar]
- Nahid, M. A.; Satoh, M.; Chan, E.K. MicroRNA in TLR signaling and endotoxin tolerance. Cell Mol. Immunol 2011, 8, 388–403. [Google Scholar]
- Tserel, L.; Runnel, T.; Kisand, K. MicroRNA expression profiles of human blood monocyte-derived dendritic cells and macrophages reveal miR-511 as putative positive regulator of Toll-like receptor 4. J. Biol. Chem 2011, 286, 26487–26495. [Google Scholar]
- Hoefig, K. P.; Heissmeyer, V. MicroRNAs grow up in the immune system. Curr. Opin. Immunol 2008, 20, 281–287. [Google Scholar]
- Basso, K.; Sumazin, P.; Morozov, P. Identification of the human mature B cell miRNome. Immunity 2009, 30, 744–752. [Google Scholar]
- Medina, P. P.; Nolde, M.; Slack, F.J. OncomiR addiction in an in vivo model of miR-21-induced pre-B-cell lymphoma. Nature 2010, 467, 86–90. [Google Scholar]
- Jiang, B. H.; Liu, L.Z. PI3K/PTEN signaling in tumorigenesis and angiogenesis. Biochim. Biophys. Acta 2008, 1784, 150–158. [Google Scholar]
- Hafsi, S.; Pezzino, F. M.; Candido, S. Gene alterations in the PI3K/PTEN/AKT pathway as a mechanism of drug-resistance (review). Int. J. Oncol 2012, 40, 639–644. [Google Scholar]
- Yu, K.; Shi, C.; Toral-Barza, L. Beyond rapalog therapy: Preclinical pharmacology and antitumor activity of WYE-1251332, an ATP-competitive and specific inhibitor of mTORC1 and mTORC2. Cancer Res 2010, 70, 621–631. [Google Scholar]
- Rao, E.; Jiang, C.; Ji, M. The miRNA-17-92 cluster mediates chemoresistance and enhances tumor growth in mantle cell lymphoma via PI3K/AKT pathway activation. Leukemia 2012, 26, 1064–1072. [Google Scholar]
- Hart, D.N. Dendritic cells: Unique leukocyte populations which control the primary immune response. Blood 1997, 90, 3245–3287. [Google Scholar]
- Palucka, K.; Ueno, H.; Fay, J. Dendritic cells and immunity against cancer. J. Intern. Med 2011, 269, 64–73. [Google Scholar]
- Visone, R.; Croce, C.M. Keynote lecture: MiRNAs and cancer. Am. J. Pathol 2009, 174, 1131–1138. [Google Scholar]
- Bonifer, C.; Bowen, D.T. Epigenetic mechanisms regulating normal and malignant haematopoiesis: New therapeutic targets for clinical medicine. Expert Rev. Mol. Med 2010, 15, 1–21. [Google Scholar]
- Fabbri, M.; Croce, C. M.; Calin, G.A. MicroRNAs in the ontogeny of leukemias and lymphomas. Leuk. Lymphoma 2009, 50, 160–170. [Google Scholar]
- Cho, W.C. OncomiRs: The discovery and progress of microRNAs in cancers. Mol. Cancer 2007, 6, 60. [Google Scholar]
- Goldhoff, P.; Rubin, J.B. Dicer and microRNAs regulate glioma immunoresistance. Immunotherapy 2010, 2, 91–92. [Google Scholar]
- Mi, S.; Lu, J.; Sun, M. MicroRNA expression signatures accurately discriminate acute lymphoblastic leukemia from acute myeloid leukemia. Proc. Natl. Acad. Sci. USA 2007, 104, 19971–19976. [Google Scholar]
- Mavrakis, K. J.; Wolfe, A. L.; Oricchio, E. Genome-Wide RNA-mediated interference screen identifies miR-19 targets in Notch-induced T-cell acute lymphoblastic leukaemia. Nat. Cell Biol 2010, 12, 372–379. [Google Scholar]
- Cocco, C.; Canale, S.; Frasson, C. Interleukin-23 acts as antitumor agent on childhood B-acute lymphoblastic leukemia cells. Blood 2010, 116, 3887–3898. [Google Scholar]
- Canale, S.; Cocco, C.; Frasson, C. Interleukin-27 inhibits pediatric B-acute lymphoblastic leukemia cell spreading in a preclinical model. Leukemia 2011, 25, 1815–1824. [Google Scholar]
- Zhang, H.; Luo, X. Q.; Feng, D.D. Upregulation of microRNA-125b contributes to leukemogenesis and increases drug resistance in pediatric acute promyelocytic leukemia. Mol. Cancer 2011, 10, 108. [Google Scholar]
- Zhou, M.; Liu, Z.; Zhao, Y. MicroRNA-125b confers the resistance of breast cancer cells to paclitaxel through suppression of pro-apoptotic Bcl-2 antagonist killer 1 (Bak1) expression. J. Biol. Chem 2010, 285, 21496–21507. [Google Scholar]
- Wang, H.; Tan, G.; Dong, L. Circulating MiR-125b as a marker predicting chemoresistance in breast cancer. PLoS One 2012, 7, e34210. [Google Scholar]
- Xu, J.; Li, Y.; Wang, F. Suppressed miR-424 expression via upregulation of target gene Chk1 contributes to the progression of cervical cancer. Oncogene 2012. [Google Scholar] [CrossRef]
- Wang, F.; Li, Y.; Zhou, J. miR-375 is down-regulated in squamous cervical cancer and inhibits cell migration and invasion via targeting transcription factor SP1. Am. J. Pathol 2011, 179, 2580–2588. [Google Scholar]
- Qiang, R.; Wang, F.; Shi, L.Y. Plexin-B1 is a target of miR-214 in cervical cancer and promotes the growth and invasion of HeLa cells. Int. J. Biochem. Cell Biol 2011, 43, 632–641. [Google Scholar]
- Zhang, T.; Liu, M.; Wang, C. Down-regulation of MiR-206 promotes proliferation and invasion of laryngeal cancer by regulating VEGF expression. Anticancer Res 2011, 31, 3859–3863. [Google Scholar]
- Chen, Z. L.; Zhao, X. H.; Wang, J.W. microRNA-92a promotes lymph node metastasis of human esophageal squamous cell carcinoma via E-cadherin. J. Biom. Chem 2011, 286, 10725–10734. [Google Scholar]
- Xu, X.; Chen, Z.; Zhao, X. MicroRNA-25 promotes cell migration and invasion in esophageal squamous cell carcinoma. Biochem. Biophys. Res. Commun 2012, 421, 640–645. [Google Scholar]
- Lin, R. J.; Xiao, D. W.; Liao, L.D. MiR-142–3p as a potential prognostic biomarker for esophageal squamous cell carcinoma. J. Surg. Oncol 2012, 105, 175–182. [Google Scholar]
- Kimura, S.; Naqanuma, S.; Susuki, D. Expression of microRNAs in squamous cell carcinoma of human head and neck and the esophagus: miR-205 and miR-21 are specific markers for HNSCC and ESCC. Oncol. Rep 2010, 23, 1625–1633. [Google Scholar]
- Kano, M.; Seki, N.; Kikkawa, N. miR-145, miR-133a and miR-133b: Tumor-suppressive miRNAs target FSCN1 in esophageal squamous cell carcinoma. Int. J. Cancer 2010, 127, 2804–2814. [Google Scholar]
- Zhang, B. G.; Li, J. F.; Yu, B.Q. MicroRNA-21 promotes tumor proliferation and invasion in gastric cancer by targeting PTEN. Oncol. Rep 2012, 27, 1019–1026. [Google Scholar]
- Li, J.; Guo, Y.; Liang, X. MicroRNA-223 functions as an oncogene in human gastric cancer by targeting FBXW7/hCdc4. J. Cancer Res. Clin. Oncol 2012, 138, 763–774. [Google Scholar]
- Xu, Y.; Zhao, F.; Wang, Z. MicroRNA-335 acts as a metastasis suppressor in gastric cancer by targeting Bcl-w and specificity protein 1. Oncogene 2012, 31, 1398–1407. [Google Scholar]
- Zheng, B.; Liang, L.; Huang, S. MicroRNA-409 suppresses tumour cell invasion and metastasis by directly targeting radixin in gastric cancers. Oncogene 2011. [Google Scholar] [CrossRef]
- Li, C.; Nie, H.; Wang, M. MicroRNA-409–3p regulates cell proliferation and apoptosis by targeting PHF10 in gastric cancer. Cancer Lett 2012, 320, 189–197. [Google Scholar]
- Li, C. L.; Nie, H.; Wang, M. MicroRNA-155 is downregulated in gastric cancer cells and involved in cell metastasis. Oncol. Rep 2012, 27, 1960–1966. [Google Scholar]
- Zheng, B.; Liang, L.; Wang, C. MicroRNA-148a suppresses tumor cell invasion and metastasis by downregulating ROCK1 in gastric cancer. Clin. Cancer Res 2011, 17, 7574–7583. [Google Scholar]
- Guo, S. L.; Peng, Z.; Yang, X. MiR-148a promoted cell proliferation by targeting p27 in gastric cancer cells. Int. J. Biol. Sci 2011, 7, 567–574. [Google Scholar]
- Kong, W. Q.; Bai, R.; Liu, T. MicroRNA-182 targets cAMP-responsive element-binding protein 1 and suppresses cell growth in human gastric adenocarcinoma. FEBS J 2012, 279, 1252–1260. [Google Scholar]
- Fornari, F.; Milazzo, M.; Chieco, P. In hepatocellular carcinoma miR-519d is up-regulated by p53 and DNA hypomethylation and targets CDKN1A/p21, PTEN, AKT3 and TIMP2. J. Pathol. 2012. [Google Scholar] [CrossRef]
- Wu, N.; Liu, X.; Xu, X. MicroRNA-373, a new regulator of protein phosphatase 6, functions as an oncogene in hepatocellular carcinoma. FEBS. J 2011, 278, 2044–2050. [Google Scholar]
- Jia, X. Q.; Cheng, H. Q.; Qian, X. Lentivirus-mediated overexpression of microRNA-199a inhibits cell proliferation of human hepatocellular carcinoma. Cell Biochem. Biophys 2012, 62, 237–244. [Google Scholar]
- Zheng, F.; Liao, Y. J.; Cai, M.Y. The putative tumour suppressor microRNA-124 modulates hepatocellular carcinoma cell aggressiveness by repressing ROCK2 and EZH2. Gut 2012, 61, 278–289. [Google Scholar]
- Hou, Y. Y.; Cao, W. W.; Li, L. MicroRNA-519d targets MKi67 and suppresses cell growth in the hepatocellular carcinoma cell line QGY-7703. Cancer Lett 2011, 307, 182–190. [Google Scholar]
- Hodzic, J.; Giovannetti, E.; Calvo, B.D. Regulation of deoxycytidine kinase expression and sensitivity to gemcitabine by microRNA-330 and promoter methylation in cancer cells. Nucleosides Nucleotides Nucleic Acid 2011, 30, 1214–1222. [Google Scholar]
- Fuse, M.; Kojima, S.; Enokida, H. Tumor suppressive microRNAs (miR-222 and miR-31) regulate molecular pathways based on miRNA expression signature in prostate cancer. J. Hum. Genet. 2012. [Google Scholar] [CrossRef]
- Yin, Q.; Wang, X.; Fewell, C. MicroRNA miR-155 inhibits bone morphogenetic protein (BMP) signaling and BMP-mediated Epstein-Barr virus reactivation. J. Virol 2010, 84, 6318–6327. [Google Scholar]
- Ghiringhelli, H.; Rebe, C.; Hichami, A. Immunomodulation and anti-inflammatory roles of polyphenols as anticancer agents. Anitcancer Agents Med. Chem 2012, 12, 852–873. [Google Scholar]
- Iorio, M. V.; Croce, C.M. MicroRNA dysregulation in cancer: Diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol. Med 2012, 4, 143–159. [Google Scholar]
- Lin, Z.; Flemington, E.K. miRNAs in pathogenesis of oncogenic human viruses. Cancer Lett 2010, 305, 186–199. [Google Scholar]
- Yanaihara, N.; Caplen, N.; Bowman, E. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 2006, 9, 189–98. [Google Scholar]
- Schotte, D.; De Menezes, R. X.; Akbari Moqadam, F. MicroRNAs characterize genetic diversity and drug resistance in pediatric acute lymphoblastic leukemia. Haematologica 2011, 96, 703–11. [Google Scholar]
- Zhang, H.; Luo, X. Q.; Zhang, P. MicroRNA patterns associated with clinical prognostic parameters and CNS relapse prediction in pediatric acute leukemia. PLoS One 2009, 4, e7826. [Google Scholar]
- Cocco, C.; Airoldi, I. Cytokines and microRNA in pediatric B-acute lymphoblastic leukemia. Cytokine Growth Factor Rev 2011, 22, 149–156. [Google Scholar]
- Gilabert-Estelles, J.; Braza-Boils, A.; Ramon, L.A. Role of microRNAs in gynecological pathology. Curr. Med. Chem 2012, 19, 2406–2413. [Google Scholar]
- Tasawa, H.; Kagawa, S.; Fujiwara, T. MicroRNAs as potential target gene in cancer gene therapy of gastrointestinal tumors. Expert Opin. Biol. Ther 2011, 11, 145–155. [Google Scholar]
- Gordonpour, A.; Nam, R. K.; Sugar, L. MicroRNAs in prostate cancer: From biomarkers to molecularly-based therapeutics. Prostate Cancer Prostatic Dis. 2012. [Google Scholar] [CrossRef]
- Cho, W.C. Circulating microRNAs as minimally invasive biomarkers for cancer theragnosis and prognosis. Front. Genet 2011, 2, 7. [Google Scholar]
- Corsini, L. R.; Bronte, G.; Terrasi, M. The role of microRNAs in cancer: Diagnostic and prognostic biomarkers and targets of therapies. Experts Opin. Ther. Targets 2012, 16, S103–S109. [Google Scholar]
- Rodriques, A. S.; Dinis, J.; Gromicho, M. Genomics and cancer drug resistance. Curr. Pharm. Biotechnol 2012, 13, 651–673. [Google Scholar]
- Nakanishi, T.; Ross, D.D. Breast cancer resistance protein (BCRP/ABCG2): Its role in multidrug resistance and regulation of its gene expression. Clin. J. Cancer 2012, 31, 73–99. [Google Scholar]
- Natarajan, K.; Xie, Y.; Baer, M.R. Role of breast cancer resistance protein (BCRP/ABCG2) in cancer drug resistance. Biochem. Pharmacol 2012, 83, 1084–1103. [Google Scholar]
- Davis, S.; Lollo, B.; Freier, S. Improved targeting of miRNA with antisense oligonucleotides. Nucleic Acid Res 2006, 34, 2294–2304. [Google Scholar]
- Stenvang, J.; Petri, A.; Lindow, M. Inhibition of microRNA function by antimiR oligonucleotides. Silence 2012, 3, 1. [Google Scholar]
- Thorsen, S. B.; Obad, S.; Jensen, N.F. The therapeutic potential of MicroRNAs in cancer. Cancer J 2012, 18, 275–284. [Google Scholar]
- Cheng, C. J.; Slack, F.J. The duality of OncomiR addiction in the maintenance and treatment of cancer. Cancer J 2012, 18, 232–237. [Google Scholar]
- Calin, G. A.; Cimmino, A.; Fabbri, M. MiR-15a and miR-16–1 cluster functions in human leukemia. Proc. Natl. Acad. Sci. USA 2008, 105, 5166–5171. [Google Scholar]
- Si, M. L.; Zhu, S.; Wu, H. miR-21-mediated tumor growth. Oncogene 2007, 26, 2799–2803. [Google Scholar]
- Krutzfeldt, J.; Rajewsky, N.; Braich, R. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 2005, 438, 685–689. [Google Scholar]
- Weiler, J.; Hunziker, J.; Hall, J. Anti-miRNA oligonucleotides (AMOs): Ammunition to target miRNAs implicated in human disease? Gene Ther 2006, 13, 496–502. [Google Scholar]
- Chen, F.; Zhu, H. H.; Zhou, L.F. Inhibition of c-FLIP expression by miR-512–3p contributes to taxol-induced apoptosis in hepatocellular carcinoma cells. Oncol. Rep 2010, 23, 1457–1462. [Google Scholar]
- Kosaka, N.; Lguchi, H.; Ochiya, T. Circulating microRNA in body fluid: A new potential biomarker for cancer diagnosis and prognosis. Cancer Sci 2010, 101, 2087–2092. [Google Scholar]

| miRNAs | Target gene | Reference | ||
|---|---|---|---|---|
| Innate | ||||
| Macrophages | miR-511-3p | MRC1 | [13] | |
| miR-146a | TRAF6, IRAK1 | [14] | ||
| miR-27b | [15] | |||
| miR-9 | NF-κB1 | [16] | ||
| miR-222, miR-339 | ICAM-1 | [17] | ||
| miR-125b | TNF-α | [18] | ||
| miR-16 | [19] | |||
| NK cells | miR-378 | GZMB | [20] | |
| miR-30e | PRF | [20] | ||
| miR-27a * | PRF1 & GZMB | [21] | ||
| miR-30c-1 * | HMBOX1 | [22] |
| miRNAs | Target gene | Reference | ||
|---|---|---|---|---|
| Adaptive | ||||
| B cells | Pro B cells | miR-34a | FOXP1 | [25] |
| miR-17~92 | BIM | [26,27] | ||
| miR-150 | c-MYB | [28–30] | ||
| Pre B cells | miR-155 | BIC | [31,32] | |
| miR-155 | SHIP & C/EBPβ | [33] | ||
| miR-125b | LIN28A | [34] | ||
| Immature B cells | miR-15a, miR-16-1 | [35] | ||
| miR-181a, miR-181b, miR-107, miR-424 | PLAG1 | [36,37] | ||
| Naïve B cells | miR-17-5p, miR-106a, miR-181b | [38] | ||
| miR-17-5p, miR-127 | [39] | |||
| miR-17~92, miR-106a-363, miR-106b-25 | [40] | |||
| Mature B cells | miR-155 | PU.1 | [41] | |
| T cells | miR-135b | FOXO1, STAT6 & GATA3 | [42] | |
| miR-140-5p, miR-409-3p, miR-433-3p, miR-650 | ULBP1 | [43] | ||
| DCs | miR-155, miR-146a, miR-125a-5p | [8] | ||
| miR-155 | AGO2, AGO4 | [44] | ||
| miR-23b | NOTCH1 | [45] | ||
| miR-146a, miR-155, miR-132 | [46,47] | |||
| miR-511 | [48] |
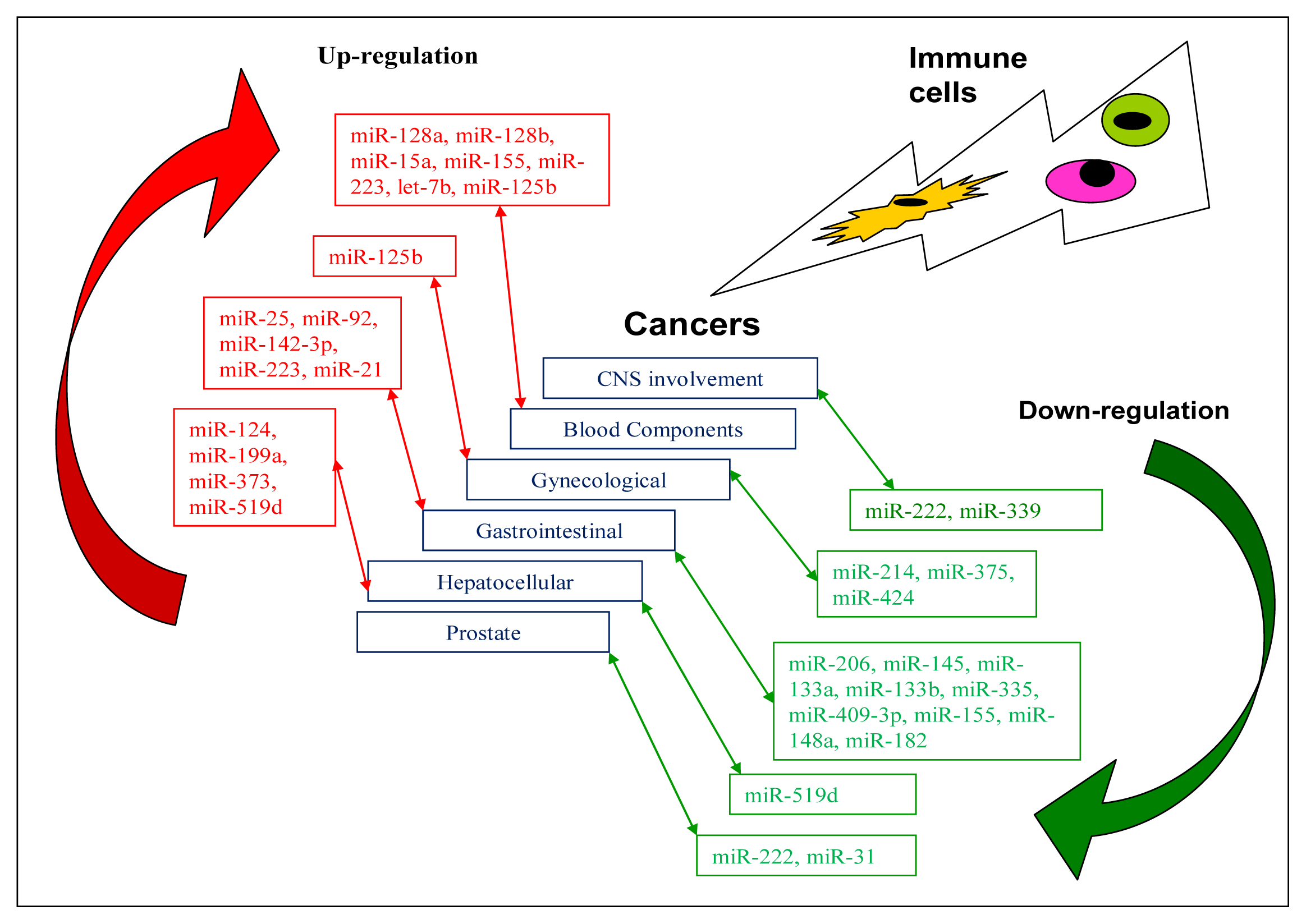
| Tumour type | Up-regulated | Down-regulated | Brief description of miRNAs | Target genes | References |
|---|---|---|---|---|---|
| CNS involvement | |||||
| CNS glioblastoma | miR-222miR-339 | Possible therapeutic targets | ICAM1 | [17,62] | |
| Blood component | |||||
| ALL | miR-128a miR-128b | Possible diagnostic markers | [63] | ||
| miR-19 | Oncomir | PTEN, BIM | [64] | ||
| miR-15a | BCL2 | [65] | |||
| miR-155 | [66] | ||||
| AML | miR-223, let-7b | Possible diagnostic markers | [63] | ||
| APL | miR-125b | Potential therapeutic target | BAK1 | [67] | |
| Gynaecological | |||||
| Breast cancer | miR-125b | [68,69] | |||
| Cervical | miR-424 | Tumour suppressor | CHK1, p-CHK1 | [70] | |
| miR-375 | SP1 | [71] | |||
| miR-214 | PLEXIN-B1 | [72] | |||
| Gastrointestinal | |||||
| Laryngeal squamous cell carcinoma | miR-206 | Tumour suppressor miRNA | VEGF | [73] | |
| Oesophagus squamous cell carcinoma | miR-92 | CDH1 | [74] | ||
| miR-25 | CDH1 | [75] | |||
| miR-142-3p | Potential prognostic marker | [76] | |||
| miR-21 | Oncomir | [77] | |||
| miR-145, miR133a, miR-133b | Tumour suppressor miRNAs | FSCN1 | [78] | ||
| Gastric cancer | miR-21 | PTEN | [79] | ||
| miR-223 | FBXW7/hCDC4 | [80] | |||
| miR-335 | Metastasis suppressor miRNA | SP1, BCL-W | [81] | ||
| miR-409-3p | RDX | [82] | |||
| miR-409-3p | PHF10 | [83] | |||
| miR-155 | Tumour suppressor miRNA | SMAD2 | [84] | ||
| miR-148a | ROCK1 | [85] | |||
| miR-148a | P27 or CDKN1B | [86] | |||
| miR-182 | Tumour suppressor miRNA | CREB1 | [87] | ||
| Hepatocellular cancer (HCC) | |||||
| HCC | miR-519d | Oncomir | PTEN, AKT3, TIMP2 | [88] | |
| miR-373 | PPP6C | [89] | |||
| miR-199a | HIF-1α | [90] | |||
| miR-124 | ROCK2, EZH2 | [91] | |||
| HCC cell line QGY-7703 | miR-519d | Tumour suppressor miRNA | MKI67 | [92] | |
| Other cancers | |||||
| Lung cancer | miR-330 | DCK | [93] | ||
| Prostate cancer | miR-222 miR-31 | Tumour suppressor miRNAs | [94] | ||
| Colon cancer | miR-155 | Oncomir | [95] |
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Yu, H.W.H.; Sze, D.M.Y.; Cho, W.C.S. MicroRNAs Involved in Anti-Tumour Immunity. Int. J. Mol. Sci. 2013, 14, 5587-5607. https://doi.org/10.3390/ijms14035587
Yu HWH, Sze DMY, Cho WCS. MicroRNAs Involved in Anti-Tumour Immunity. International Journal of Molecular Sciences. 2013; 14(3):5587-5607. https://doi.org/10.3390/ijms14035587
Chicago/Turabian StyleYu, Hong W. H., Daniel M. Y. Sze, and William C. S. Cho. 2013. "MicroRNAs Involved in Anti-Tumour Immunity" International Journal of Molecular Sciences 14, no. 3: 5587-5607. https://doi.org/10.3390/ijms14035587
APA StyleYu, H. W. H., Sze, D. M. Y., & Cho, W. C. S. (2013). MicroRNAs Involved in Anti-Tumour Immunity. International Journal of Molecular Sciences, 14(3), 5587-5607. https://doi.org/10.3390/ijms14035587






