Click Chemistry-Enabled Parallel Synthesis of N-Acyl Sulfonamides and Their Evaluation as Carbonic Anhydrase Inhibitors
Abstract
1. Introduction
2. Results and Discussion
2.1. Parallel Synthesis
2.2. Differential Scanning Fluorimetry
2.3. Esterase Activity Inhibition Screening and IC50 Determination
2.4. In Silico Studies
3. Materials and Methods
General Procedure for the Preparations of Libraries 4 and 7 (CuAAC and Subsequent Acylation)
- Step 1: Copper(I)-catalyzed azide–alkyne cycloaddition.
- Step 2: Acylation of Sulfonamides.
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A Major Update to the DrugBank Database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef]
- Ammazzalorso, A.; De Filippis, B.; Giampietro, L.; Amoroso, R. N-Acylsulfonamides: Synthetic Routes and Biological Potential in Medicinal Chemistry. Chem. Biol. Drug Des. 2017, 90, 1094–1105. [Google Scholar] [CrossRef]
- Francisco, K.R.; Varricchio, C.; Paniak, T.J.; Kozlowski, M.C.; Brancale, A.; Ballatore, C. Structure Property Relationships of N-Acylsulfonamides and Related Bioisosteres. Eur. J. Med. Chem. 2021, 218, 113399. [Google Scholar] [CrossRef]
- Amador, R.; Tahrioui, A.; Barreau, M.; Lesouhaitier, O.; Smietana, M.; Clavé, G. N-Acylsulfonamide: A Valuable Moiety to Design New Sulfa Drug Analogues. RSC Med. Chem. 2023, 14, 1567–1571. [Google Scholar] [CrossRef]
- Meanwell, N.A. Synopsis of Some Recent Tactical Application of Bioisosteres in Drug Design. J. Med. Chem. 2011, 54, 2529–2591. [Google Scholar] [CrossRef] [PubMed]
- Volochnyuk, D.M.; Ryabukhin, S.V.; Moroz, Y.S.; Savych, O.; Chuprina, A.; Horvath, D.; Zabolotna, Y.; Varnek, A.; Judd, D.B. Evolution of Commercially Available Compounds for HTS. Drug Discov. Today 2019, 24, 390–402. [Google Scholar] [CrossRef]
- Manallack, D.T.; Prankerd, R.J.; Nassta, G.C.; Ursu, O.; Oprea, T.I.; Chalmers, D.K. A Chemogenomic Analysis of Ionization Constants—Implications for Drug Discovery. ChemMedChem 2013, 8, 242–255. [Google Scholar] [CrossRef]
- Gavrylenko, O.V.; Vashchenko, B.V.; Naumchyk, V.; Chuk, O.; Kuchuk, O.; Pogribna, A.; Konovets, A.I.; Brovarets, V.S.; Zozulya, S.A.; Radchenko, D.S.; et al. Expanding Chemical Space of N-Acyl Sulfonamides for Carbonic Anhydrase Inhibitor Discovery. Eur. J. Med. Chem. 2025, 302, 118296. [Google Scholar] [CrossRef]
- Rostovtsev, V.V.; Green, L.G.; Fokin, V.V.; Sharpless, K.B. A Stepwise Huisgen Cycloaddition Process: Copper(I)-Catalyzed Regioselective “Ligation” of Azides and Terminal Alkynes. Angew. Chemie Int. Ed. 2002, 41, 2596–2599. [Google Scholar] [CrossRef]
- Hein, J.E.; Fokin, V.V. Copper-Catalyzed Azide–Alkyne Cycloaddition (CuAAC) and beyond: New Reactivity of Copper(i) Acetylides. Chem. Soc. Rev. 2010, 39, 1302. [Google Scholar] [CrossRef] [PubMed]
- Haldón, E.; Nicasio, M.C.; Pérez, P.J. Copper-Catalysed Azide–Alkyne Cycloadditions (CuAAC): An Update. Org. Biomol. Chem. 2015, 13, 9528–9550. [Google Scholar] [CrossRef] [PubMed]
- Meldal, M.; Tornøe, C.W. Cu-Catalyzed Azide−Alkyne Cycloaddition. Chem. Rev. 2008, 108, 2952–3015. [Google Scholar] [CrossRef]
- Wang, X.; Huang, B.; Liu, X.; Zhan, P. Discovery of Bioactive Molecules from CuAAC Click-Chemistry-Based Combinatorial Libraries. Drug Discov. Today 2016, 21, 118–132. [Google Scholar] [CrossRef] [PubMed]
- Castro, V.; Rodríguez, H.; Albericio, F. CuAAC: An Efficient Click Chemistry Reaction on Solid Phase. ACS Comb. Sci. 2016, 18, 1–14. [Google Scholar] [CrossRef]
- Gao, P.; Song, S.; Pannecouque, C.; De Clercq, E.; Zhan, P.; Liu, X. Rapid Identification of Novel Indolylarylsulfone Derivatives as Potent HIV-1 NNRTIs via Miniaturized CuAAC Click-Chemistry-Based Combinatorial Libraries. RSC Med. Chem. 2025, 16, 157–167. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem in 2021: New Data Content and Improved Web Interfaces. Nucleic Acids Res. 2021, 49, D1388–D1395. [Google Scholar] [CrossRef]
- Gaulton, A.; Bellis, L.J.; Bento, A.P.; Chambers, J.; Davies, M.; Hersey, A.; Light, Y.; McGlinchey, S.; Michalovich, D.; Al-Lazikani, B.; et al. ChEMBL: A Large-Scale Bioactivity Database for Drug Discovery. Nucleic Acids Res. 2012, 40, D1100–D1107. [Google Scholar] [CrossRef]
- Naud, J.; Lemke, C.; Goudreau, N.; Beaulieu, E.; White, P.D.; Llinàs-Brunet, M.; Forgione, P. Potent Triazolyl-Proline-Based Inhibitors of HCV NS3 Protease. Bioorg. Med. Chem. Lett. 2008, 18, 3400–3404. [Google Scholar] [CrossRef]
- Moeker, J.; Peat, T.S.; Bornaghi, L.F.; Vullo, D.; Supuran, C.T.; Poulsen, S.-A. Cyclic Secondary Sulfonamides: Unusually Good Inhibitors of Cancer-Related Carbonic Anhydrase Enzymes. J. Med. Chem. 2014, 57, 3522–3531. [Google Scholar] [CrossRef]
- Sharp, P.P.; Garnier, J.M.; Hatfaludi, T.; Xu, Z.; Segal, D.; Jarman, K.E.; Jousset, H.; Garnham, A.; Feutrill, J.T.; Cuzzupe, A.; et al. Design, Synthesis, and Biological Activity of 1,2,3-Triazolobenzodiazepine BET Bromodomain Inhibitors. ACS Med. Chem. Lett. 2017, 8, 1298–1303. [Google Scholar] [CrossRef] [PubMed]
- Garrido-González, J.J.; Medrano-Uribe, K.; Rosso, C.; Humbrías-Martín, J.; Dell’Amico, L. Photocatalytic Synthesis and Functionalization of Sulfones, Sulfonamides and Sulfoximines. Chem.–A Eur. J. 2024, 30, e202401307. [Google Scholar] [CrossRef]
- Gokcen, T.; Gulcin, I.; Ozturk, T.; Goren, A.C. A Class of Sulfonamides as Carbonic Anhydrase I and II Inhibitors. J. Enzyme Inhib. Med. Chem. 2016, 31, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Cecchi, A.; Hulikova, A.; Pastorek, J.; Pastoreková, S.; Scozzafava, A.; Winum, J.-Y.; Montero, J.-L.; Supuran, C.T. Carbonic Anhydrase Inhibitors. Design of Fluorescent Sulfonamides as Probes of Tumor-Associated Carbonic Anhydrase IX That Inhibit Isozyme IX-Mediated Acidification of Hypoxic Tumors. J. Med. Chem. 2005, 48, 4834–4841. [Google Scholar] [CrossRef]
- Begines, P.; Bonardi, A.; Nocentini, A.; Gratteri, P.; Giovannuzzi, S.; Ronca, R.; Tavani, C.; Luisa Massardi, M.; López, Ó.; Supuran, C.T. Design and Synthesis of Sulfonamides Incorporating a Biotin Moiety: Carbonic Anhydrase Inhibitory Effects, Antiproliferative Activity and Molecular Modeling Studies. Bioorg. Med. Chem. 2023, 94, 117467. [Google Scholar] [CrossRef]
- Almatary, A.M.; Husseiny, W.M.E.; Selim, K.B.; Eisa, H.M.H. Nitroimidazole-Sulfonamides as Carbonic Anhydrase IX and XII Inhibitors Targeting Tumor Hypoxia: Design, Synthesis, Molecular Docking and Molecular Dynamics Simulation. J. Mol. Struct. 2022, 1264, 133260. [Google Scholar] [CrossRef]
- Khushal, A.; Mumtaz, A.; Shadoul, W.A.; Zaidi, S.H.M.; Rafique, H.; Munir, A.; Maalik, A.; Shah, S.J.A.; Baig, A.; Khawaja, W.; et al. Synthesis, Carbonic Anhydrase II/IX/XII Inhibition, DFT, and Molecular Docking Studies of Hydrazide-Sulfonamide Hybrids of 4-Methylsalicyl- and Acyl-Substituted Hydrazide. Biomed Res. Int. 2022, 2022, 5293349. [Google Scholar] [CrossRef] [PubMed]
- Cheng, T.; Wang, N.; Wen, R.; Wang, S.; Zhang, H.; Cheng, M. Discovery of Non-Sulfonamide Carbonic Anhydrase IX Inhibitors through Structure-Based Virtual Screening. Phys. Chem. Chem. Phys. 2024, 26, 8767–8774. [Google Scholar] [CrossRef]
- Güttler, A.; Eiselt, Y.; Funtan, A.; Thiel, A.; Petrenko, M.; Keßler, J.; Thondorf, I.; Paschke, R.; Vordermark, D.; Bache, M. Betulin Sulfonamides as Carbonic Anhydrase Inhibitors and Anticancer Agents in Breast Cancer Cells. Int. J. Mol. Sci. 2021, 22, 8808. [Google Scholar] [CrossRef]
- Massah, A.R.; Adibi, H.; Khodarahmi, R.; Abiri, R.; Majnooni, M.B.; Shahidi, S.; Asadi, B.; Mehrabi, M.; Zolfigol, M.A. Synthesis, in Vitro Antibacterial and Carbonic Anhydrase II Inhibitory Activities of N-Acylsulfonamides Using Silica Sulfuric Acid as an Efficient Catalyst under Both Solvent-Free and Heterogeneous Conditions. Bioorg. Med. Chem. 2008, 16, 5465–5472. [Google Scholar] [CrossRef]
- Guan, S.-S.; Cheng, C.-C.; Ho, A.-S.; Wang, C.-C.; Luo, T.-Y.; Liao, T.-Z.; Chang, J.; Wu, C.-T.; Liu, S.-H. Sulfonamide Derivative Targeting Carbonic Anhydrase IX as a Nuclear Imaging Probe for Colorectal Cancer Detection in Vivo. Oncotarget 2015, 6, 36139–36155. [Google Scholar] [CrossRef]
- Behnke, C.A.; Le Trong, I.; Godden, J.W.; Merritt, E.A.; Teller, D.C.; Bajorath, J.; Stenkamp, R.E. Atomic Resolution Studies of Carbonic Anhydrase II. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 616–627. [Google Scholar] [CrossRef]
- Kazokaitė, J.; Niemans, R.; Dudutienė, V.; Becker, H.M.; Leitāns, J.; Zubrienė, A.; Baranauskienė, L.; Gondi, G.; Zeidler, R.; Matulienė, J.; et al. Novel Fluorinated Carbonic Anhydrase IX Inhibitors Reduce Hypoxia-Induced Acidification and Clonogenic Survival of Cancer Cells. Oncotarget 2018, 9, 26800–26816. [Google Scholar] [CrossRef]
- Available online: https://www.molsoft.com (accessed on 10 February 2025).
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High Performance Molecular Simulations through Multi-Level Parallelism from Laptops to Supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Valdés-Tresanco, M.S.; Valdés-Tresanco, M.E.; Valiente, P.A.; Moreno, E. Gmx_MMPBSA: A New Tool to Perform End-State Free Energy Calculations with GROMACS. J. Chem. Theory Comput. 2021, 17, 6281–6291. [Google Scholar] [CrossRef]
- Armarego, W.L.F.; Chai, C. Purification of Laboratory Chemicals, 5th ed.; Elsevier: Oxford, UK, 2003. [Google Scholar]
- Mahon, B.P.; Bhatt, A.; Socorro, L.; Driscoll, J.M.; Okoh, C.; Lomelino, C.L.; Mboge, M.Y.; Kurian, J.J.; Tu, C.; Agbandje-McKenna, M.; et al. The Structure of Carbonic Anhydrase IX Is Adapted for Low-PH Catalysis. Biochemistry 2016, 55, 4642–4653. [Google Scholar] [CrossRef]
- Banerjee, A.L.; Swanson, M.; Mallik, S.; Srivastava, D.K. Purification of Recombinant Human Carbonic Anhydrase-II by Metal Affinity Chromatography without Incorporating Histidine Tags. Protein Expr. Purif. 2004, 37, 450–454. [Google Scholar] [CrossRef]
- Wu, G.; Yuan, Y.; Hodge, C.N. Determining Appropriate Substrate Conversion for Enzymatic Assays in High-Throughput Screening. SLAS Discov. 2003, 8, 694–700. [Google Scholar] [CrossRef]
- Abagyan, R.; Totrov, M.; Kuznetsov, D. ICM—A New Method for Protein Modeling and Design: Applications to Docking and Structure Prediction from the Distorted Native Conformation. J. Comput. Chem. 1994, 15, 488–506. [Google Scholar] [CrossRef]
- Abagyan, R.; Totrov, M. Biased Probability Monte Carlo Conformational Searches and Electrostatic Calculations for Peptides and Proteins. J. Mol. Biol. 1994, 235, 983–1002. [Google Scholar] [CrossRef] [PubMed]






| # | Compound | CA-II | hCA-IX | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ΔTm, Median | IC50, µM | Docking Result | MM/GB(PB)SA, kcal/mol | ΔTm, Median | IC50, µM | Docking Result | MM/GB(PB)SA, kcal/mol | ||||
| Parameter “9” a | Parameter “12” b | Parameter “9” a | Parameter “12” b | ||||||||
| 1 | 4{17,7,27} | N/A c | 6.07 |  | −43.59 | −37.69 | 1.44 | 23.4 |  | −68.09 | −47.24 |
| 2 | 7{2,21,60} | N/A c | 8.74 |  | −70.35 | −46.40 | 1.17 | 17.3 |  | −61.09 | −38.36 |
| 3 | 4{16,7,28} | −2.55 | 1.70 | 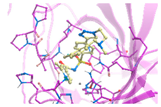 | −66.01 | −43.91 | 1.18 | 7.13 | 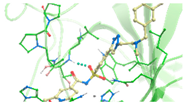 | −60.09 | −38.98 |
| 4 | 4{16,7,19} | −4.95 | 0.20 |  | −67.26 | −42.44 | 1.17 | 1.11 |  | −52.6 | −39.19 |
| 5 | 4{18,7,32} | N/A c | >100 |  | −36.74 | −33.74 | 0.99 | 84.2 d |  | −65.25 | −39.84 |
| # | Compound | hCA-II Residues | hCA-IX Residues |
|---|---|---|---|
| 1 | 4{17,7,27} | W5, F20, H64, A65, H96, V121, F131, L198, T200, P201, P202 | H64, S65, H94, V121, V131, D132, L135, L198, T200, P202 |
| 2 | 7{2,21,60} | N62, H64, H96, V121, F131, L141, V143, L198, T200 | N62, H64, S65, H96, V121, L198, T200, N244 |
| 3 | 4{16,7,28} | H64, A65, H96, V121, F131, G132, V135, Q136, L198, T200, P202, L204 | H64, S65, T69, L91, Q92, H96, V121, V131, L198, T200 |
| 4 | 4{16,7,19} | F20, H64, A65, H94, H96, V121, F131, L198, T200, P201, P202 | H64, S65, L91, Q92, H96, V121, V131, L135, L198, T200, P202 |
| 5 | 4{18,7,32} | W5, L60, H64, A65, E69, I91, Q92, H94, H96, V121, L198, T200 | H64, S65, T69, L91, Q92, H96, V121, L123, R130, V131, L198, T200 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Gavrylenko, O.V.; Vashchenko, B.V.; Naumchyk, V.; Sosunovych, B.S.; Chuk, O.; Hrabovskyi, O.; Kuchuk, O.; Pogribna, A.; Nikitin, S.O.; Konovets, A.I.; et al. Click Chemistry-Enabled Parallel Synthesis of N-Acyl Sulfonamides and Their Evaluation as Carbonic Anhydrase Inhibitors. Molecules 2026, 31, 318. https://doi.org/10.3390/molecules31020318
Gavrylenko OV, Vashchenko BV, Naumchyk V, Sosunovych BS, Chuk O, Hrabovskyi O, Kuchuk O, Pogribna A, Nikitin SO, Konovets AI, et al. Click Chemistry-Enabled Parallel Synthesis of N-Acyl Sulfonamides and Their Evaluation as Carbonic Anhydrase Inhibitors. Molecules. 2026; 31(2):318. https://doi.org/10.3390/molecules31020318
Chicago/Turabian StyleGavrylenko, Oleksii V., Bohdan V. Vashchenko, Vasyl Naumchyk, Bohdan S. Sosunovych, Oleksii Chuk, Oleksii Hrabovskyi, Olga Kuchuk, Alla Pogribna, Sergiy O. Nikitin, Anzhelika I. Konovets, and et al. 2026. "Click Chemistry-Enabled Parallel Synthesis of N-Acyl Sulfonamides and Their Evaluation as Carbonic Anhydrase Inhibitors" Molecules 31, no. 2: 318. https://doi.org/10.3390/molecules31020318
APA StyleGavrylenko, O. V., Vashchenko, B. V., Naumchyk, V., Sosunovych, B. S., Chuk, O., Hrabovskyi, O., Kuchuk, O., Pogribna, A., Nikitin, S. O., Konovets, A. I., Brovarets, V. S., Zozulya, S. A., Radchenko, D. S., Grygorenko, O. O., & Moroz, Y. S. (2026). Click Chemistry-Enabled Parallel Synthesis of N-Acyl Sulfonamides and Their Evaluation as Carbonic Anhydrase Inhibitors. Molecules, 31(2), 318. https://doi.org/10.3390/molecules31020318







