Evaluating the Effectiveness of Reference Solvent Solubility Calculations for Binary Mixtures Based on Pure Solvent Solubility: The Case of Phenolic Acids
Abstract
1. Introduction
2. Results and Discussion
2.1. Solubility Measurements of Caffeic and Ferulic Acids
2.2. Identifying an Optimal Predictive Model via the DOO-IT Framework
2.3. Model of Phenolic Acids Solubility in Binary Mixtures
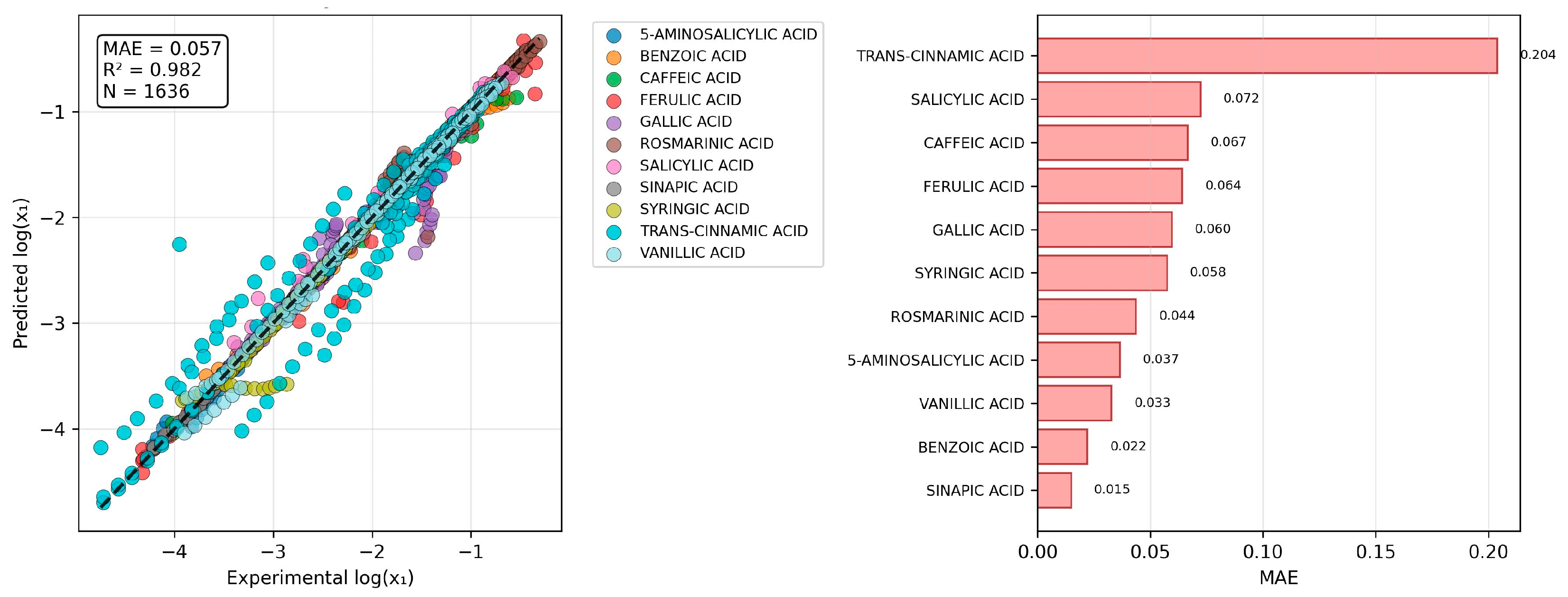
2.4. The Challenge of Predicting Binary Mixture Solubility
2.5. The Spectrum of Model Generalization
- (i)
- Defined application domains with clear boundaries for the chemical space, where predictions are valuable;
- (ii)
- Representative training data covering the operational chemical space;
- (iii)
- Incorporating regular model updates while new data are available to maintain chemical space coverage;
- (iv)
- Clear uncertainty quantification by implementing applicability domain checks to flag extrapolation attempts;
- (v)
- Ensuring that the users of model applications understand the models’ limitations and appropriate use cases.
3. Materials and Methods
3.1. Materials
3.2. Solubility Determination Procedure
3.3. COSMO-RS Computations
3.4. Reference Solvent Computations
3.5. Molecular Descriptors
3.6. Machine Learning Protocol
3.6.1. Core Algorithm and Data Preprocessing
3.6.2. Dual-Objective Optimization Protocol
3.6.3. Iterative Model Refinement and Candidate Selection
3.6.4. A Data-Driven, Multi-Criteria Framework for Model Selection Based on Stability and Performance
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mei, X.; Tang, Q.; Huang, G.; Long, R.; Huang, H. Preparation, structural analysis and antioxidant activities of phosphorylated (1 → 3)-β-D-glucan. Food Chem. 2020, 309, 125791. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Mai, D.H.A.; Sarwar, A.; Lee, E.Y. Reconstructing ethanol oxidation pathway in Pseudomonas putida KT2440 for bio-upgrading of ethanol to biodegradable polyhydroxybutanoates. Int. J. Biol. Macromol. 2022, 222, 902–914. [Google Scholar] [CrossRef]
- Páscoa, R.N.M.J.; Magalhães, L.M.; Lopes, J.A. FT-NIR spectroscopy as a tool for valorization of spent coffee grounds: Application to assessment of antioxidant properties. Food Res. Int. 2013, 51, 579–586. [Google Scholar] [CrossRef]
- Muszynska, B.; Sulkowska-Ziaja, K.; Ekiert, H. Phenolic acids in selected edible Basidiomycota species: Armillaria mellea, Boletus badius, and Pleurotus ostreatus. Acta Sci. Pol. 2013, 12, 107–116. [Google Scholar]
- Lekmine, S.; Benslama, O.; Kadi, K.; Brik, A.; Djeffali, O.; Ounissi, M.; Slimani, M.; Ola, M.S.; Eldahshan, O.A.; Martín-García, A.I.; et al. Preliminary Investigation of Astragalus arpilobus subsp. hauarensis: LC-MS/MS Chemical Profiling, In Vitro Evaluation of Antioxidant, Anti-Inflammatory Properties, Cytotoxicity, and In Silico Analysis against COX-2. Antioxidants 2024, 13, 654. [Google Scholar] [CrossRef]
- Zambonin, L.; Caliceti, C.; Vieceli Dalla Sega, F.; Fiorentini, D.; Hrelia, S.; Landi, L.; Prata, C. Dietary phenolic acids act as effective antioxidants in membrane models and in cultured cells, exhibiting proapoptotic effects in leukaemia cells. Oxid. Med. Cell. Longev. 2012, 2012, 839298. [Google Scholar] [CrossRef]
- Choi, S.; Han, S.; Lee, S.; Kim, J.; Kim, J.; Kang, D.-K. Synergistic Antioxidant and Anti-Inflammatory Effects of Phenolic Acid-Conjugated Glutamine–Histidine–Glycine–Valine (QHGV) Peptides Derived from Oysters (Crassostrea talienwhanensis). Antioxidants 2024, 13, 447. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, D.; Proença, C.; Varela, C.; Janela, J.; Tavares da Silva, E.J.; Fernandes, E.; Roleira, F.M.F. New phenolic cinnamic acid derivatives as selective COX-2 inhibitors. Design, synthesis, biological activity and structure-activity relationships. Bioorg. Chem. 2019, 91, 103179. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Fu, X.; Liu, T.; Zheng, Y.; Chen, J.; Luo, F. Phenolic composition, antioxidant, antibacterial and anti-inflammatory activities of leaf and stem extracts from Cryptotaenia japonica Hassk. Ind. Crops Prod. 2018, 122, 522–532. [Google Scholar] [CrossRef]
- Rampelotto, C.R.; Pereira, V.G.; da Silva Silveira, L.; Rossato, A.; Machado, A.K.; Sagrillo, M.R.; Gündel, A.; Burger, M.E.; Schaffazick, S.R.; de Bona da Silva, C. Ferulic acid-loaded nanocapsules: Evaluation of mucosal interaction, safety and antioxidant activity in human mononucleated cells. Toxicol. Vitr. 2022, 78, 105259. [Google Scholar] [CrossRef]
- Vilas-Boas, S.M.; Alves, R.S.; Brandão, P.; Campos, L.M.A.; Coutinho, J.A.P.; Pinho, S.P.; Ferreira, O. Solid-liquid phase equilibrium of trans-cinnamic acid, p-coumaric acid and ferulic acid in water and organic solvents: Experimental and modelling studies. Fluid Phase Equilib. 2020, 521, 112747. [Google Scholar] [CrossRef]
- Nordström, F.L.; Rasmuson, Å.C. Solubility and melting properties of salicylic acid. J. Chem. Eng. Data 2006, 51, 1668–1671. [Google Scholar] [CrossRef]
- Razali, N.; Mat-Junit, S.; Abdul-Muthalib, A.F.; Subramaniam, S.; Abdul-Aziz, A. Effects of various solvents on the extraction of antioxidant phenolics from the leaves, seeds, veins and skins of Tamarindus indica L. Food Chem. 2012, 131, 441–448. [Google Scholar] [CrossRef]
- Jakopič, J.; Veberič, R.; Štampar, F. Ekstrakcija fenolnih snovi iz zelenih plodov oreha z različnimi topili. Acta Agric. Slov. 2009, 93, 11–15. [Google Scholar]
- Jouki, M.; Khazaei, N. Compare of extraction of phenolic compounds from Pistacia atlantica in different solvents. In Advances in Biomedical Research; Proceedings; Anninos, P., Rossi, M., Pham, T.D., Falugi, C., Bussing, A., Koukkou, M., Eds.; World Scientific and Engineering Academy and Society: Athens, Greece, 2010; pp. 361–365. [Google Scholar]
- De Fina, K.M.; Sharp, T.L.; Roy, L.E.; Acree, W.E. Solubility of 2-hydroxybenzoic acid in select organic solvents at 298.15 K. J. Chem. Eng. Data 1999, 44, 1262–1264. [Google Scholar] [CrossRef]
- Montenegro, I.; González, B.; Domínguez, Á.; Gómez, E. Solubility study of several polyphenolic compounds in pure and binary solvents. J. Chem. Thermodyn. 2025, 203, 107434. [Google Scholar] [CrossRef]
- Waszkowiak, K.; Gliszczyńska-Świgło, A. Binary ethanol–water solvents affect phenolic profile and antioxidant capacity of flaxseed extracts. Eur. Food Res. Technol. 2016, 242, 777–786. [Google Scholar] [CrossRef]
- Mitrović, J.; Nikolić, N.; Karabegović, I.; Savić, S.; Petrović, S.; Pešić, M.; Šimurina, O. Evaluation of the solvent effect on the extraction and antioxidant activity of phenolic compounds from the nettle (Urtica dioica L.) seeds: Application of PCA and regression analyses. J. Food Meas. Charact. 2024, 18, 6618–6626. [Google Scholar] [CrossRef]
- Shakeel, F.; Haq, N.; Siddiqui, N.A. Thermodynamic solubility and solvation behavior of ferulic acid in different (PEG-400 + water) binary solvent mixtures. Drug Dev. Ind. Pharm. 2019, 45, 1468–1476. [Google Scholar] [CrossRef]
- Mullins, E.; Liu, Y.A.; Ghaderi, A.; Fast, S.D. Sigma profile database for predicting solid solubility in pure and mixed solvent mixtures for organic pharmacological compounds with COSMO-based thermodynamic methods. Ind. Eng. Chem. Res. 2008, 47, 1707–1725. [Google Scholar] [CrossRef]
- Faraz, O.; Poustchi, M.; Nazari Denyani, E.; Movahedi, P.; Rajabi Kouchi, F.; Shahriari, R. Thermodynamic modeling of pharmaceuticals solubility in pure, mixed and supercritical solvents. J. Mol. Liq. 2022, 353, 118809. [Google Scholar] [CrossRef]
- Queimada, A.J.; Mota, F.L.; Pinho, S.P.; Macedo, E.A. Solubilities of Biologically Active Phenolic Compounds: Measurements and Modeling. J. Phys. Chem. B 2009, 113, 3469–3476. [Google Scholar] [CrossRef] [PubMed]
- Yousefi Seyf, J.; Shojaeian, A. Measurement of the solubility and density of the saturated solution of 3-aminophenol in different pure solvents from 283.1 to 323.1 K: Correlative to pure predictive thermodynamic modeling and molecular dynamic simulation. J. Mol. Liq. 2023, 392, 123478. [Google Scholar] [CrossRef]
- Alsenz, J.; Kuentz, M. From Quantum Chemistry to Prediction of Drug Solubility in Glycerides. Mol. Pharm. 2019, 16, 4661–4669. [Google Scholar] [CrossRef]
- Klamt, A. Prediction of the mutual solubilities of hydrocarbons and water with COSMO-RS. Fluid Phase Equilib. 2003, 206, 223–235. [Google Scholar] [CrossRef]
- Mohan, M.; Demerdash, O.N.; Simmons, B.A.; Singh, S.; Kidder, M.K.; Smith, J.C. Physics-Based Machine Learning Models Predict Carbon Dioxide Solubility in Chemically Reactive Deep Eutectic Solvents. ACS Omega 2024, 9, 19548–19559. [Google Scholar] [CrossRef]
- Mu, Y.; Dai, T.; Fan, J.; Cheng, Y. Prediction of acetylene solubility by a mechanism-data hybrid-driven machine learning model constructed based on COSMO-RS theory. J. Mol. Liq. 2024, 414, 126194. [Google Scholar] [CrossRef]
- Qin, H.; Wang, K.; Ma, X.; Li, F.; Liu, Y.; Ji, X. Predicting the solubility of CO2 and N2 in ionic liquids based on COSMO-RS and machine learning. Front. Chem. 2024, 12, 1480468. [Google Scholar] [CrossRef]
- Mac Fhionnlaoich, N.; Zeglinski, J.; Simon, M.; Wood, B.; Davin, S.; Glennon, B. A hybrid approach to aqueous solubility prediction using COSMO-RS and machine learning. Chem. Eng. Res. Des. 2024, 209, 67–71. [Google Scholar] [CrossRef]
- Li, M.; Chen, H.; Zhang, H.; Zeng, M.; Chen, B.; Guan, L. Prediction of the Aqueous Solubility of Compounds Based on Light Gradient Boosting Machines with Molecular Fingerprints and the Cuckoo Search Algorithm. ACS Omega 2022, 7, 42027–42035. [Google Scholar] [CrossRef] [PubMed]
- Gheta, S.K.O.; Bonin, A.; Gerlach, T.; Göller, A.H. Predicting absolute aqueous solubility by applying a machine learning model for an artificially liquid-state as proxy for the solid-state. J. Comput. Aided. Mol. Des. 2023, 37, 765–789. [Google Scholar] [CrossRef]
- Oliveira, G.; Wegner, P.H.; de Lima Carvalho, P.V.; Voll, F.A.P.; de Paula Scheer, A.; de Pelegrini Soares, R.; Farias, F.O. Machine learning-enhanced COSMO-SAC for accurate solubility predictions. Fluid Phase Equilib. 2026, 600, 114535. [Google Scholar] [CrossRef]
- Wang, J.; Song, Z.; Chen, L.; Xu, T.; Deng, L.; Qi, Z. Prediction of CO2 solubility in deep eutectic solvents using random forest model based on COSMO-RS-derived descriptors. Green Chem. Eng. 2021, 2, 431–440. [Google Scholar] [CrossRef]
- Ferreira, M.L.; Araújo, J.M.M.; Vega, L.F.; Pereiro, A.B. Understanding the Absorption of Fluorinated Gases in Fluorinated Ionic Liquids for Recovering Purposes Using Soft-SAFT. J. Chem. Eng. Data 2022, 67, 1951–1963. [Google Scholar] [CrossRef]
- Shakeel, F.; Salem-Bekhit, M.M.; Haq, N.; Siddiqui, N.A. Solubility and thermodynamics of ferulic acid in different neat solvents: Measurement, correlation and molecular interactions. J. Mol. Liq. 2017, 236, 144–150. [Google Scholar] [CrossRef]
- Ji, W.; Meng, Q.; Ding, L.; Wang, F.; Dong, J.; Zhou, G.; Wang, B. Measurement and correlation of the solubility of caffeic acid in eight mono and water+ethanol mixed solvents at temperatures from (293.15 to 333.15) K. J. Mol. Liq. 2016, 224, 1275–1281. [Google Scholar] [CrossRef]
- Cysewski, P.; Jeliński, T.; Przybyłek, M.; Gliniewicz, N.; Majkowski, M.; Wąs, M. Navigating the Deep Eutectic Solvent Landscape: Experimental and Machine Learning Solubility Explorations of Syringic, p-Coumaric, and Caffeic Acids. Int. J. Mol. Sci. 2025, 26, 10099. [Google Scholar] [CrossRef]
- Cysewski, P.; Jeliński, T.; Giniewicz, J.; Kaźmierska, A.; Przybyłek, M. Duality of Simplicity and Accuracy in QSPR: A Machine Learning Framework for Predicting Solubility of Selected Pharmaceutical Acids in Deep Eutectic Solvents. Molecules 2025, 30, 4361. [Google Scholar] [CrossRef]
- Tropsha, A. Best Practices for QSAR Model Development, Validation, and Exploitation. Mol. Inform. 2010, 29, 476–488. [Google Scholar] [CrossRef] [PubMed]
- Golbraikh, A.; Tropsha, A. Beware of q2! J. Mol. Graph. Model. 2002, 20, 269–276. [Google Scholar] [CrossRef]
- Netzeva, T.I.; Worth, A.P.; Aldenberg, T.; Benigni, R.; Cronin, M.T.D.; Gramatica, P.; Jaworska, J.S.; Kahn, S.; Klopman, G.; Marchant, C.A.; et al. Current Status of Methods for Defining the Applicability Domain of (Quantitative) Structure-Activity Relationships. Altern. Lab. Anim. 2005, 33, 155–173. [Google Scholar] [CrossRef] [PubMed]
- Rakhimbekova, A.; Akhmetshin, T.N.; Minibaeva, G.I.; Nugmanov, R.I.; Gimadiev, T.R.; Madzhidov, T.I.; Baskin, I.I.; Varnek, A. Cross-validation strategies in QSPR modelling of chemical reactions. SAR QSAR Environ. Res. 2021, 32, 207–219. [Google Scholar] [CrossRef] [PubMed]
- Sushko, I.; Novotarskyi, S.; Körner, R.; Pandey, A.K.; Cherkasov, A.; Li, J.; Gramatica, P.; Hansen, K.; Schroeter, T.; Müller, K.R.; et al. Applicability domains for classification problems: Benchmarking of distance to models for ames mutagenicity set. J. Chem. Inf. Model. 2010, 50, 2094–2111. [Google Scholar] [CrossRef] [PubMed]
- Sahigara, F.; Mansouri, K.; Ballabio, D.; Mauri, A.; Consonni, V.; Todeschini, R. Comparison of different approaches to define the applicability domain of QSAR models. Molecules 2012, 17, 4791–4810. [Google Scholar] [CrossRef]
- Meinshausen, N.; Bühlmann, P. Stability Selection. J. R. Stat. Soc. Ser. B Stat. Methodol. 2010, 72, 417–473. [Google Scholar] [CrossRef]
- Martin, T.M.; Harten, P.; Young, D.M.; Muratov, E.N.; Golbraikh, A.; Zhu, H.; Tropsha, A. Does Rational Selection of Training and Test Sets Improve the Outcome of QSAR Modeling? J. Chem. Inf. Model. 2012, 52, 2570–2578. [Google Scholar] [CrossRef]
- Klamt, A. Conductor-like screening model for real solvents: A new approach to the quantitative calculation of solvation phenomena. J. Phys. Chem. 1995, 99, 2224–2235. [Google Scholar] [CrossRef]
- Klamt, A. COSMO-RS: From Quantum Chemistry to Fluid Phase Thermodynamics and Drug Design, 1st ed.; Elsevier: Amsterdam, The Netherlands, 2005; ISBN 9780444519948. [Google Scholar]
- Klamt, A.; Eckert, F.; Hornig, M.; Beck, M.E.; Bürger, T. Prediction of aqueous solubility of drugs and pesticides with COSMO-RS. J. Comput. Chem. 2002, 23, 275–281. [Google Scholar] [CrossRef]
- Klamt, A.; Eckert, F.; Arlt, W. COSMO-RS: An alternative to simulation for calculating thermodynamic properties of liquid mixtures. Annu. Rev. Chem. Biomol. Eng. 2010, 1, 101–122. [Google Scholar] [CrossRef]
- Dassault Systèmes. COSMOtherm; Version 24.0.0; BIOVIA: San Diego, CA, USA, 2024. [Google Scholar]
- Jeliński, T.; Przybyłek, M.; Różalski, R.; Romanek, K.; Wielewski, D.; Cysewski, P. Tuning Ferulic Acid Solubility in Choline-Chloride- and Betaine-Based Deep Eutectic Solvents: Experimental Determination and Machine Learning Modeling. Molecules 2024, 29, 3841. [Google Scholar] [CrossRef]
- Cysewski, P.; Jeliński, T.; Przybyłek, M.; Mai, A.; Kułak, J. Experimental and Machine-Learning-Assisted Design of Pharmaceutically Acceptable Deep Eutectic Solvents for the Solubility Improvement of Non-Selective COX Inhibitors Ibuprofen and Ketoprofen. Molecules 2024, 29, 2296. [Google Scholar] [CrossRef]
- Cysewski, P.; Jeliński, T.; Przybyłek, M. Exploration of the Solubility Hyperspace of Selected Active Pharmaceutical Ingredients in Choline- and Betaine-Based Deep Eutectic Solvents: Machine Learning Modeling and Experimental Validation. Molecules 2024, 29, 4894. [Google Scholar] [CrossRef]
- Przybyłek, M.; Kowalska, A.; Tymorek, N.; Dziaman, T.; Cysewski, P. Thermodynamic Characteristics of Phenacetin in Solid State and Saturated Solutions in Several Neat and Binary Solvents. Molecules 2021, 26, 4078. [Google Scholar] [CrossRef]
- Cysewski, P.; Przybyłek, M.; Kowalska, A.; Tymorek, N. Thermodynamics and intermolecular interactions of nicotinamide in neat and binary solutions: Experimental measurements and COSMO-RS concentration dependent reactions investigations. Int. J. Mol. Sci. 2021, 22, 7365. [Google Scholar] [CrossRef]
- Buchowski, H.; Ksiazczak, A.; Pietrzyk, S. Solvent activity along a saturation line and solubility of hydrogen-bonding solids. J. Phys. Chem. 1980, 84, 975–979. [Google Scholar] [CrossRef]
- Buchowski, H.; Kosiński, J.J.; Ksia̧zczak, A. Activity of solvent and solubility. J. Phys. Chem. 1988, 92, 6104–6107. [Google Scholar] [CrossRef]
- Jouyban, A.; Acree, W.E. Mathematical derivation of the Jouyban-Acree model to represent solute solubility data in mixed solvents at various temperatures. J. Mol. Liq. 2018, 256, 541–547. [Google Scholar] [CrossRef]
- Jouyban, A.; Acree, W.E. In silico prediction of drug solubility in water-ethanol mixtures using Jouyban-Acree model. J. Pharm. Pharm. Sci. 2006, 9, 262–269. [Google Scholar] [PubMed]
- Jouyban, A. In silico prediction of drug solubility in water-dioxane mixtures using the Jouyban-Acree model. Pharmazie 2007, 62, 46–50. [Google Scholar] [PubMed]
- Jouyban, A. Review of the cosolvency models for predicting solubility of drugs in water-cosolvent mixtures. J. Pharm. Pharm. Sci. 2008, 11, 32–58. [Google Scholar] [CrossRef]
- Jouyban, A. Review of the cosolvency models for predicting drug solubility in solvent mixtures: An update. J. Pharm. Pharm. Sci. 2019, 22, 466–485. [Google Scholar] [CrossRef]
- Aydi, A.; Claumann, C.A.; Wüst Zibetti, A.; Abderrabba, M. Differential Scanning Calorimetry Data and Solubility of Rosmarinic Acid in Different Pure Solvents and in Binary Mixtures (Methyl Acetate + Water) and (Ethyl Acetate + Water) from 293.2 to 313.2 K. J. Chem. Eng. Data 2016, 61, 3718–3723. [Google Scholar] [CrossRef]
- Wüst Zibetti, A.; Aydi, A.; Claumann, C.A.; Eladeb, A.; Adberraba, M. Correlation of solubility and prediction of the mixing properties of rosmarinic acid in different pure solvents and in binary solvent mixtures of ethanol + water and methanol + water from (293.2 to 318.2) K. J. Mol. Liq. 2016, 216, 370–376. [Google Scholar] [CrossRef]
- Matsuda, H.; Kaburagi, K.; Matsumoto, S.; Kurihara, K.; Tochigi, K.; Tomono, K. Solubilities of salicylic acid in pure solvents and binary mixtures containing cosolvent. J. Chem. Eng. Data 2009, 54, 480–484. [Google Scholar] [CrossRef]
- Zhang, Y.; Guo, F.; Cui, Q.; Lu, M.; Song, X.; Tang, H.; Li, Q. Measurement and Correlation of the Solubility of Vanillic Acid in Eight Pure and Water + Ethanol Mixed Solvents at Temperatures from (293.15 to 323.15) K. J. Chem. Eng. Data 2016, 61, 420–429. [Google Scholar] [CrossRef]
- Shakeel, F.; Haq, N.; Alam, P.; Jouyban, A.; Ghoneim, M.M.; Alshehri, S.; Martinez, F. Solubility of sinapic acid in some (ethylene glycol + water) mixtures: Measurement, computational modeling, thermodynamics, and preferential solvation. J. Mol. Liq. 2022, 348, 118057. [Google Scholar] [CrossRef]
- Sandeepa, K.; Ravi Kumar, K.; Neeharika, T.S.V.R.; Satyavathi, B.; Thella, P.K. Solubility Measurement and Thermodynamic Modeling of Benzoic Acid in Monosolvents and Binary Mixtures. J. Chem. Eng. Data 2018, 63, 2028–2037. [Google Scholar] [CrossRef]
- Marden, J.W.; Dover, M.V. The solubilities of several substances in mixed nonaqueous solutions. J. Am. Chem. Soc. 1916, 38, 1235–1245. [Google Scholar] [CrossRef][Green Version]
- Moradi, M.; Mazaher Haji Agha, E.; Hemmati, S.; Martinez, F.; Kuentz, M.; Jouyban, A. Solubility of 5-aminosalicylic acid in {N-methyl-2-pyrrolidone + ethanol} mixtures at T = (293.2 to 313.2) K. J. Mol. Liq. 2020, 306, 112774. [Google Scholar] [CrossRef]
- Shakeel, F.; Haq, N.; Alanazi, F.K.; Alanazi, S.A.; Alsarra, I.A. Solubility of sinapic acid in various (Carbitol + water) systems: Computational modeling and solution thermodynamics. J. Therm. Anal. Calorim. 2020, 142, 1437–1446. [Google Scholar] [CrossRef]
- Noubigh, A.; Akermi, A. Solubility and Thermodynamic Behavior of Syringic Acid in Eight Pure and Water + Methanol Mixed Solvents. J. Chem. Eng. Data 2017, 62, 3274–3283. [Google Scholar] [CrossRef]
- Dali, I.; Aydi, A.; Alberto, C.C.; Wüst, Z.A.; Manef, A. Correlation and semi-empirical modeling of solubility of gallic acid in different pure solvents and in binary solvent mixtures of propan-1-ol + water, propan-2-ol + water and acetonitrile + water from (293.2 to 318.2) K. J. Mol. Liq. 2016, 222, 503–519. [Google Scholar] [CrossRef]
- Noubigh, A.; Jeribi, C.; Mgaidi, A.; Abderrabba, M. Solubility of gallic acid in liquid mixtures of (ethanol + water) from (293.15 to 318.15) K. J. Chem. Thermodyn. 2012, 55, 75–78. [Google Scholar] [CrossRef]
- Noubigh, A.; Aydi, A.; Mgaidi, A.; Abderrabba, M. Measurement and correlation of the solubility of gallic acid in methanol plus water systems from (293.15 to 318.15) K. J. Mol. Liq. 2013, 187, 226–229. [Google Scholar] [CrossRef]
- Noubigh, A.; Akrmi, A. Temperature dependent solubility of vanillic acid in aqueous methanol mixtures: Measurements and thermodynamic modeling. J. Mol. Liq. 2016, 220, 277–282. [Google Scholar] [CrossRef]
- Noubigh, A.; Akremi, A. Solution thermodynamics of trans-Cinnamic acid in (methanol + water) and (ethanol + water) mixtures at different temperatures. J. Mol. Liq. 2019, 274, 752–758. [Google Scholar] [CrossRef]
- Haq, N.; Siddiqui, N.A.; Shakeel, F. Solubility and molecular interactions of ferulic acid in various (isopropanol + water) mixtures. J. Pharm. Pharmacol. 2017, 69, 1485–1494. [Google Scholar] [CrossRef]
- Shakeel, F.; Haq, N.; Salem-Bekhit, M.M.; Raish, M. Solubility and dissolution thermodynamics of sinapic acid in (DMSO + water) binary solvent mixtures at different temperatures. J. Mol. Liq. 2017, 225, 833–839. [Google Scholar] [CrossRef]
- Schölkopf, B.; Smola, A.J.; Williamson, R.C.; Bartlett, P.L. New support vector algorithms. Neural Comput. 2000, 12, 1207–1245. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.J.; Panaye, A.; Doucet, J.P.; Zhang, R.S.; Chen, H.F.; Liu, M.C.; Hu, Z.D.; Fan, B.T. Comparative study of QSAR/QSPR correlations using support vector machines, radial basis function neural networks, and multiple linear regression. J. Chem. Inf. Comput. Sci. 2004, 44, 1257–1266. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y. Support vector regression-based QSAR models for prediction of antioxidant activity of phenolic compounds. Sci. Rep. 2021, 11, 8806. [Google Scholar] [CrossRef] [PubMed]
- Cherkasov, A.; Muratov, E.N.; Fourches, D.; Varnek, A.; Baskin, I.I.; Cronin, M.; Dearden, J.; Gramatica, P.; Martin, Y.C.; Todeschini, R.; et al. QSAR modeling: Where have you been? Where are you going to? J. Med. Chem. 2014, 57, 4977–5010. [Google Scholar] [CrossRef]
- Gretton, A.; Borgwardt, K.M.; Rasch, M.J.; Schölkopf, B.; Smola, A. A kernel two-sample test. J. Mach. Learn. Res. 2012, 13, 723–773. [Google Scholar]
- Muandet, K.; Fukumizu, K.; Sriperumbudur, B.; Schölkopf, B. Kernel mean embedding of distributions: A review and beyond. Found. Trends Mach. Learn. 2017, 10, 1–141. [Google Scholar] [CrossRef]
- Scikit-Learn Developers. StandardScaler—Scikit-Learn 1.7.2 Documentation. Available online: https://scikit-learn.org/stable/modules/generated/sklearn.preprocessing.StandardScaler.html (accessed on 18 September 2025).
- Scikit-Learn Developers. 7.3.1. Standardization, or Mean Removal and Variance Scaling—Scikit-Learn User Guide. Available online: https://scikit-learn.org/stable/modules/preprocessing.html (accessed on 18 September 2025).
- Optuna Developers. Multi-Objective Optimization with Optuna—Optuna Documentation (Stable). Available online: https://optuna.readthedocs.io/en/stable/tutorial/20_recipes/002_multi_objective.html (accessed on 18 September 2025).
- Yanase, T. Announcing Optuna 3.2. Optuna Blog (Medium). Available online: https://medium.com/optuna/announcing-optuna-3-2-cfbfbe104d5f (accessed on 18 September 2025).
- Akiba, T.; Sano, S.; Yanase, T.; Ohta, T.; Koyama, M. Optuna: A Next-generation Hyperparameter Optimization Framework. In Proceedings of the ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Anchorage, AK, USA, 4–8 August 2019; ACM: New York, NY, USA, 2019; pp. 2623–2631. [Google Scholar]
- Breiman, L.; Friedman, J.H.; Olshen, R.A.; Stone, C.J. Classification and Regression Trees; Routledge: London, UK, 2017; ISBN 9781351460491. [Google Scholar]
- Hastie, T.; Tibshirani, R.; Friedman, J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction, 2nd ed.; Springer: New York, NY, USA, 2009. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Sugiura, N. Further analysis of the data by Akaike’s information criterion and the finite corrections. Commun. Stat.-Theory Methods 1978, 7, 13–26. [Google Scholar] [CrossRef]
- Hurvich, C.M.; Tsai, C.-L. Regression and Time Series Model Selection in Small Samples. Biometrika 1989, 76, 297–307. [Google Scholar] [CrossRef]
- Python Software Foundation. Python 3.10 Documentation. Available online: https://docs.python.org/3.10/ (accessed on 18 September 2025).
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Pandas Development Team. Pandas-dev/Pandas: Pandas; Version 2.3.0; Zenodo: Geneva, Switzerland, 2025; Available online: https://zenodo.org/records/15597513 (accessed on 18 September 2025).



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cysewski, P.; Jeliński, T.; Rozalski, R.; Lesniewski, F.; Przybyłek, M. Evaluating the Effectiveness of Reference Solvent Solubility Calculations for Binary Mixtures Based on Pure Solvent Solubility: The Case of Phenolic Acids. Molecules 2025, 30, 4444. https://doi.org/10.3390/molecules30224444
Cysewski P, Jeliński T, Rozalski R, Lesniewski F, Przybyłek M. Evaluating the Effectiveness of Reference Solvent Solubility Calculations for Binary Mixtures Based on Pure Solvent Solubility: The Case of Phenolic Acids. Molecules. 2025; 30(22):4444. https://doi.org/10.3390/molecules30224444
Chicago/Turabian StyleCysewski, Piotr, Tomasz Jeliński, Rafal Rozalski, Fabian Lesniewski, and Maciej Przybyłek. 2025. "Evaluating the Effectiveness of Reference Solvent Solubility Calculations for Binary Mixtures Based on Pure Solvent Solubility: The Case of Phenolic Acids" Molecules 30, no. 22: 4444. https://doi.org/10.3390/molecules30224444
APA StyleCysewski, P., Jeliński, T., Rozalski, R., Lesniewski, F., & Przybyłek, M. (2025). Evaluating the Effectiveness of Reference Solvent Solubility Calculations for Binary Mixtures Based on Pure Solvent Solubility: The Case of Phenolic Acids. Molecules, 30(22), 4444. https://doi.org/10.3390/molecules30224444









