Abstract
The in vitro cultures of Rindera graeca, a rare endemic plant, were developed as a sustainable source of phenolic acids. Various shoot and root cultures were established and scaled up in a sprinkle bioreactor. A multiplication rate of 7.2 shoots per explant was achieved. HPLC–PDA–ESI–HRMS analysis revealed the presence of rosmarinic acid (RA) and lithospermic acid B (LAB) as the main secondary metabolites in both the shoot and root cultures. The maximum RA (30.0 ± 3.2 mg/g DW) and LAB (49.3 ± 15.5 mg/g DW) yields were determined in root-regenerated shoots. The strongest free radical scavenging activity (87.4 ± 1.1%), according to 2,2-diphenyl-1-picrylhydrazyl-hydrate assay, was noted for roots cultivated in a DCR medium. The highest reducing power (2.3 µM ± 0.4 TE/g DW), determined by the ferric-reducing antioxidant power assay, was noted for shoots cultivated on an SH medium containing 0.5 mg/L 6-benzylaminopurine. A genetic analysis performed using random amplified polymorphic DNA and start codon targeted markers revealed genetic variation of 62.8% to 96.5% among the investigated shoots and roots. This variability reflects the capacity of cultivated shoots and roots to produce phenolic compounds.
Keywords:
antioxidants; phenolic compounds; micropropagation; hairy roots; anatomical roots; bioreactor; RAPD; SCoT 1. Introduction
The demand for plant-derived compounds for pharmaceutical and cosmetic industries is constantly increasing, accompanied by the growing threat of over-exploitation of natural resources. One solution to this great challenge may be found in plant biotechnology methods that allow high yields of bioactive compounds to be obtained in an environmentally independent manner while protecting ecosystem biodiversity [1,2,3].
Among the many and varied plant secondary metabolites, phenolic compounds are known to exert numerous beneficial effects on human health, including preventing cancer, diabetes, chronic degenerative diseases, and cardiovascular diseases, all of which are recognized to be associated with oxidative stress [4,5]. This efficacy of phenolic compounds is considered to be connected with their antioxidant potential, as they can delay, inhibit, or prevent biomolecule oxidation by, e.g., scavenging free radicals [6].
Previous studies have detailed a wide range of biotechnological approaches that can enhance the production of phenolic compounds via the cultivation of various plant parts, cells, tissues, and organs [7,8,9]. Although the biotechnological production of rosmarinic acid (RA; Figure 1a) has been intensively investigated, relatively few sources have examined the biotechnological efforts leading to enhanced production of lithospermic acid B (LAB; Figure 1b), also known as salvianolic acid B. Widely distributed in the plant kingdom, RA was found in all plants of the Boraginaceae family [10]. It is an ester of caffeic acid (CA) and 3,4-dihydroxyphenyllactic acid, while LA is a conjugate of RA and CA, and LAB is a dimer of RA [11,12].
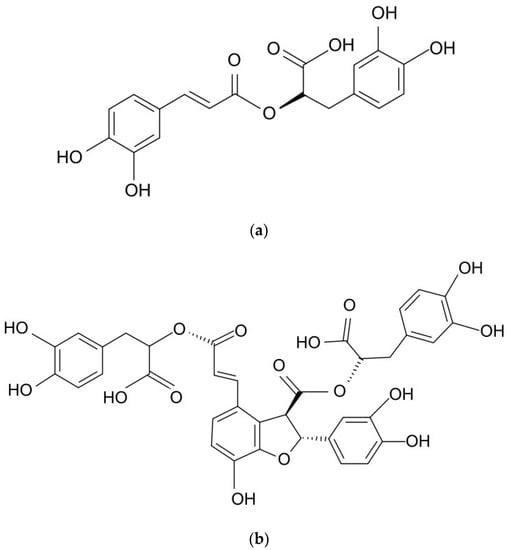
Figure 1.
Chemical structures of (a) rosmarinic acid (RA) and (b) lithospermic acid B (LAB).
Rindera graeca (A. DC.) Boiss. & Heldr. (synonyms: Cynoglossum graecum (A. DC.) Greuter & Burdet and Mattia graeca A. DC., Ref. [13]) belongs to the tribe Cynoglossae of Boraginaceae [14,15]. The genus Rindera comprises about 25 species that are predominantly distributed in Central and Western Asia [16]. R. graeca is a Greek endemic mountain species that grows from Peloponnesus (southern Greece) and Sterea Ellada toward North Pindos (northwest Greece) [17] on rocky slopes at altitudes of 1500–2300 m. In 1997, R. graeca was included in the IUCN Red List of Threatened Plants and marked as a rare species [18]. A few reports have examined the traditional usage of some Rindera species in Iranian and Turkish folk medicine, e.g., the seeds of R. lanata (Lam.) Bunge used to treat joint pain [19] and R. lanata and R. caespitosa (DC.) Gürke roots used as a poultice due to their anti-inflammatory properties [20]. Furthermore, a methanolic extract prepared from the aerial parts (leaves, stem, and flowers) of R. lanata was found to demonstrate antiviral activity against three different human rotavirus strains [21]. The authors noted the presence of twelve phenolic compounds, of which the major constituents were malic, quinic, and rosmarinic acids. In addition, essential oils distilled from aerial parts of R. lanata var. canescens demonstrated moderate antimicrobial potential [22]. In the chemical profile of the aerial parts of naturally grown R. graeca, nine phenolic compounds, including CA and its derivatives (chlorogenic acid, rabdosiin (a tetramer of CA), and its disodium salt), RA, salvianolic A acid, and LAB, were determined [23]. Later, these naturally occurring secondary metabolites were also identified in in vitro-cultivated R. graeca shoots and roots [24]. These were accompanied by rinderol, a furano-naphthoquinone, which was isolated for the first time from Onosma paniculata root extract by Dong et al. [25] and later from Cynoglossum columnae root in vitro cultures [26]. Next, the extract profiling of extracts derived from in vitro-cultivated R. graeca roots of different origins subjected to cold or drought stress was performed by our group using the HPLC–PDA–ESI–HRMS method, along with the quantitative determination of CA, RA, and LAB acids [27]. This study revealed that the major metabolite in all extracts was LAB.
The aim of the present study was to establish a system for the micropropagation of the rare, endemic boraginaceous species R. graeca to obtain plant material efficiently producing biologically active phenolic compounds, especially rosmarinic acid and lithospermic acid B. The genetic diversity of the cultivated shoot and root lines was assessed using RAPD (random amplified polymorphic DNA) and SCoT (start codon targeted) markers, and the antioxidant properties of the extracts obtained via biotechnological methods were evaluated. It also proposes a scale-up approach in the sprinkle bioreactor. This is the first detailed report on the establishment of R. graeca in vitro cultures for the production of phenolic compounds.
2. Results and Discussion
2.1. Micropropagation
Of the 30 R. graeca seeds used to initiate the in vitro culture, only three germinated. Two plantlets were used to micropropagate R. graeca, and the other served as the starting material for the initiation of callus tissue. The callus obtained on an MS medium supplemented with 0.5 mg/L NOA was transferred to a liquid hormone-free MS medium, and adventitious root development was observed within four weeks. To initiate the shoot cultures, the roots derived from the seed plantlets were cut off, and the shoots were placed on a DCR medium supplemented with 0.5 mg/L BAP. Next, micropropagation was carried out on various DCR medium modifications (Table S1) to induce auxiliary shoots. There are no data on the seed germination of this rare endemic species. However, for R. umbellate, it has been reported that its propagation in nature is limited by fungal infection and insufficient seed maturity [28]. The phenomenon of strong seed dormancy is not rare among Boraginaceous species. The strong dormancy at maturity of the seeds, probably maintained for a long time, has been described for Heliotropium europaeum, for example [29]. Another reason for limited seed germination may be the low viability of the seeds and the hard seed coat, as was reported for Lithospermum canescens [30].
The highest shoot induction rates were noted on the DCR medium supplemented with 0.5 mg/L BAP (DCR/BAP) and DCR/SV with the addition of (DCR/SV/KIN): 7.2 ± 3 and 7.1 ± 5, respectively (Figure 2).
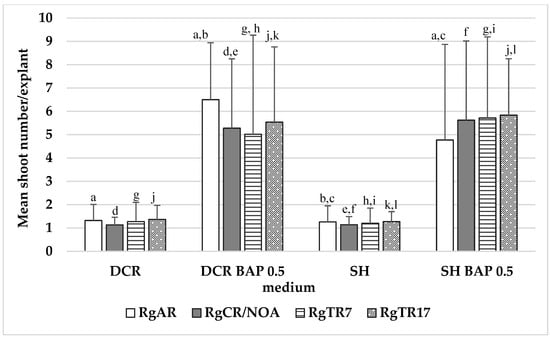
Figure 2.
The number of auxiliary shoots formed per explant in Rindera graeca shoot cultures. Shoots were obtained via regeneration from roots and were cultivated on DCR and SH without growth regulators or supplementation with 0.5 mg/L BAP. Values represent means ± SD, and those marked with the same letter are statistically significant (p < 0.05). RgAR—shoots regenerated from anatomical roots; RgCR/NOA—shoots regenerated from auxin-induced roots; RgTR7—shoots regenerated from RgTR7 hairy roots; RgTR17—shoots regenerated from RgTR17 hairy roots.
However, the quality of shoots developed on the DCR/BAP medium was superior to that formed on DCR/SV/KIN, in which the leaves in the rosettes were light green to yellow-green, which could be attributed to the reduced amount of salts and vitamins in the DCR/SV/KIN medium compared with the full-strength one (DCR). On the DCR/BAP medium, the shoots formed luxuriant rosettes composed of deep green leaves (Figure 3a). The approach of multiple shoot induction via direct organogenesis applied in the current study resulted in the growth of seven new shoots within four weeks, which seemed to be satisfactory [31]. When placed on the hormone-free DCR medium, the shoots spontaneously developed roots at their bases (Figure 3b).

Figure 3.
(a) Rosette of Rindera graeca shoots (RgS line) forming on DCR medium supplemented with 0.5 mg/L BAP; (b) roots spontaneously developing at the bases of RgCR/NOA line shoots cultivated on hormone-free DCR medium.
During the cultivation of both non-transformed and hairy roots, spontaneous shoot regeneration was observed; these were used to initiate cultures of regenerated shoots on solid DCR or SH media, which were plain or supplemented with 0.5 mg/L BAP. The presence of cytokinin was essential for shoot regeneration from the roots. The highest multiplication rate was noted for shoots regenerated from RgAR roots and cultivated on the DCR/BAP medium, which amounted to 6.5 ± 2.4 shoots per explant. The regenerants demonstrated a significantly lower (p < 0.05) multiplication rate than the shoots derived from seedlings (Figure 2). In addition, when placed on the hormone-free DCR medium, the shoots spontaneously developed roots at their bases (Figure 3b).
No studies so far have examined the micropropagation of R. graeca from existing meristems or its regeneration from roots; however, one paper examined the in vitro multiplication of R. umbellata (Waldst. & Kit.) Gürke [31]. The authors used immature embryos as the starting material for this; 72% germinated successfully and were efficiently propagated on an MS medium containing 1.0 mg/L BAP and 0.5 mg/L indole-3-acetic acid (IAA) supplemented with 0.1 M sucrose, followed by acclimatization to the greenhouse and field conditions. However, the authors found that the type of sugar and its concentration were the major factors influencing the number of explants forming buds, the most optimal values being 0.06 M sucrose or 0.3 M glucose.
In the reported up-to-date micropropagation protocols developed for Boraginaceous plants via direct organogenesis, the highest multiplication rates were associated with the use of thidiazuron (TDZ). However, TDZ does not appear to be the only treatment with beneficial effects on shoot regeneration and proliferation in Boraginaceous plants. In Table 1, the efforts that have been undertaken to obtain the highest micropropagation rates are summarized.

Table 1.
Summary of the most efficient culture conditions elaborated for micropropagation of various species of the Boraginaceae family.
2.2. Root Cultures
The R. graeca roots developed spontaneously on the RgS line shoots following transfer onto the hormone-free DCR medium. The roots were cut off and used to establish anatomical root RgAR cultures (Figure 4a). The second auxin-induced RgCR/NOA root line was initiated from roots developed on callus tissue induced by the addition of NOA 0.5 mg/L to the MS medium.
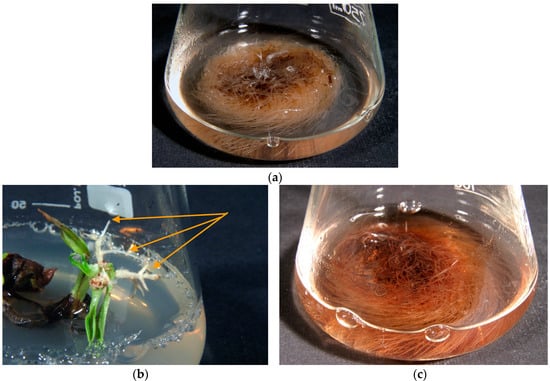
Figure 4.
(a) Rindera graeca anatomical roots growing in liquid hormone-free SH media; (b) hairy roots emerging from R. graeca leaves (indicated by arrows) inoculated with the Agrobacterium rhizogenes ATCC 15834 strain after 4 weeks of culture; (c) hairy roots of RgTR17 line cultivated in liquid hormone-free DCR medium for 4 weeks.
Following of transformation the using Agrobacterium rhizogenes ATCC 15834 strain, 60 roots emerged within two to four weeks after explant infection (Figure 4b). Finally, five hairy root line cultures were established on the basis of visual assessment. During the three subsequent passages, two root lines were chosen for further investigations on the basis of root growth capacity: RgTR7 and RgTR17 (Figure 4c). PCR analysis confirmed the incorporation of the rolB and rolC genes into RgTR7 and RgTR17 roots (Figure 5).
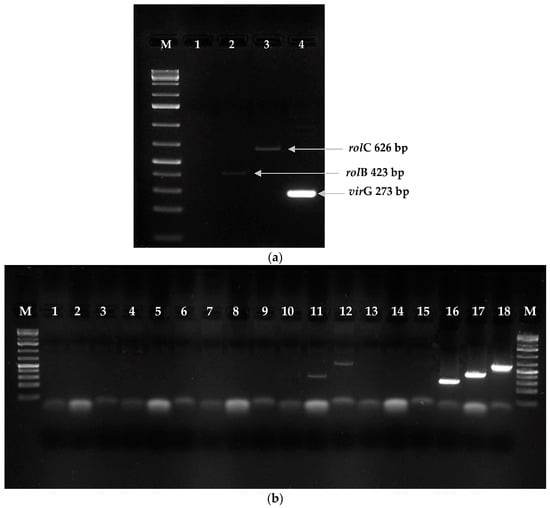
Figure 5.
Confirmation of genetic transformation by PCR analysis of genomic DNA isolated from Rindera graeca shoots regenerated from roots. (a) RgTR17 line: M—molecular weight marker (GeneRulerTM 1 kb Plus DNA ladder); lane 1—virG; lane 2—rolB (423 bp); lane 3—rolC (626 bp); lane 4—Agrobacterium rhizogenes 15834 plasmid—virG (273 bp). (b) M—molecular weight marker GeneRulerTM 100 bp DNA ladder; lanes 1–6: RgAR shoot line; lanes 7–9: RgCR/NOA shoot line; lanes 10–12: RgTR7 shoot line: 10—virG, 11—rolB (423 bp), 12—rolC (626 bp); lanes 13–15: RgCR/NOA shoot line; lanes 16–18: A. rhizogenes 15834 plasmid: lane 16—virG (273 bp), lane 17—rolB (423 bp), and lane 18—rolC (626 bp).
The non-transformed root and hairy root lines were cultivated in LS, SH, and the four modifications of the DCR medium (Figure 6). The highest root growth index was noted for the auxin-induced RgCR/NOA and RgTR7 root lines cultivated in DCR medium: 0.94 ± 0.035 and 0.94 ± 0.039, respectively. The DCR medium was chosen for further investigation. During cultivation roots, irrespectively the root line, in liquid hormone-free DCR or SH media in the dark, spontaneous regeneration of the shoots was observed (Figure 7a). On the basis of this observation, the cultures of shoots regenerating from roots on solid media were established at 16 h/8 h (light/dark) photoperiod (Figure 7b).
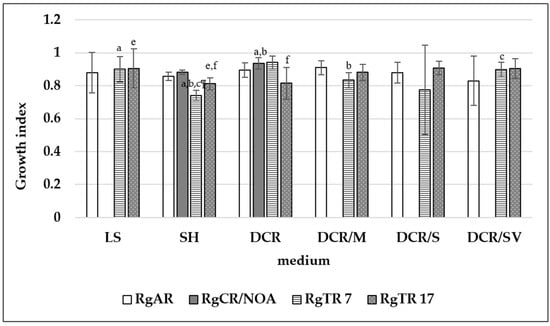
Figure 6.
Growth index of Rindera graeca non-transformed and hairy roots. Means denoted with the same letter are statistically significant (p < 0.05).

Figure 7.
Spontaneously regenerating shoots (indicated by arrows) form hairy roots of RgTR17 line during cultivation in the dark on liquid hormone-free DCR medium, 4th week of culture (a); shoots regenerating form explants of RgTR17 hairy root line when grown on solid SH supplemented with 0.5 mg/L BAP and cultivated under a 16/8h (light/dark) photoperiod, 4th week of culture (b).
2.3. Sprinkle Bioreactor Cultures
The R. graeca root cultures were scaled up in a nutrient sprinkle bioreactor. In the first batch, the root line RgTR17 was cultivated in the DCR medium; the fresh biomass rose from 15.23 g to 26.41 g, and the dry biomass increased from 1.68 g to 4.23 g. These values represented 1.7- and 2.5-fold increases in growth, respectively; this biomass growth was achieved as a result of 40 s/80 s intervals of medium sprinkle/pause. By contrast, the fresh biomass of roots increased from 2.35 g to 37.50 g (16-fold), and the dry biomass increased from 0.29 g to 4.83 g (16.7 times) with a 15 s/30 s interval of medium sprinkle/pause. The RgTR7 hairy root line was also cultivated in the DCR medium under 15 s/30 s sprinkle/pause intervals; however, these roots did not grow. As a result, the medium was changed from DCR to SH. Under these new conditions, i.e., the SH medium, the RgTR17 line roots demonstrated higher growth capacities than the RgTR7 roots (Figure 8a); their fresh and dry biomass increased 8.5- and 9.5-fold, while the RgTR7 roots increased 6.3- and 8.3-fold, respectively. The RgTR17 roots demonstrated 1.4-fold enhanced growth in the SH medium compared with the DCR medium. The RgAR roots were cultivated in the SH medium and demonstrated 9.8- and 7.6-fold increases in fresh and dry biomass, respectively. In addition, spontaneous shoot regeneration was observed during culture in the SH medium, irrespective of the root line (Figure 8b). The fresh and dry biomass of the regenerants were 17.6 g and 2.2 g for shoots derived from the roots of the RgTR17 line, 21.4 g and 2.5 g for the RgTR7 line, and 76.28 g and 10.65 g for the RgAR line. No shoot regeneration was observed when cultivating roots in the bioreactor using the DCR medium.

Figure 8.
(a) RgTR17 hairy roots cultivated in mist bioreactor in SH hormone-free medium for 3 weeks; (b) spontaneously regenerated shoots (indicated by arrows).
2.4. Genetic Analysis Using RAPD and SCoT Markers
Most studies on molecular markers applied to the identification of cultivars and varieties to date have focused on RAPD RFLP, SSRS, ISSR, or ALFP markers [40,41,42]. To assess genetic diversity, usually, two types of DNA primers are used, and we chose RAPD and SCoT ones. We expected that both types of primers would allow the generation of a sufficient number of bands to assess the genetic diversity among the shoot and root lines investigated in our study. In the RAPD analysis, 10 of the 20 primers (Table S2) showed well-resolved bands (Figure 9a,b). When SCoT primers were used to amplify all the genotypes, 11 of the 36 tested exhibited well-resolved bands (Figure 9c,d). Genetic analysis performed with RAPD and SCoT markers identified between 62.8% and 96.5% similarity depending on the regenerant shoot or root line (Table S3).
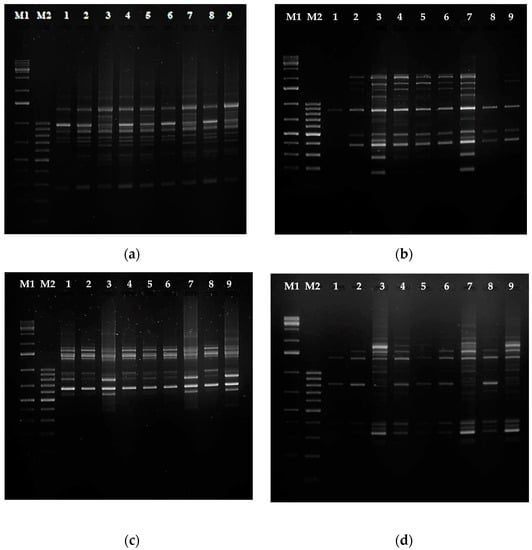
Figure 9.
PCR analysis of genomic DNA isolated from Rindera graeca shoot and root lines: (a) RAPD primer OPA7; (b) RAPD primer OPA11; (c) SCoT17 primer; (d) SCoT30 primer; M1—molecular weight marker GeneRulerTM 1 kb DNA ladder; M2—molecular weight marker GeneRulerTM 100 bp DNA ladder; lane 1—RgS shoots, lane 2—RgAR shoots, lane 3—RgAR roots, lane 4—RgCR/NOA shoots, lane 5—RgCR/NOA roots, lane 6—RgTR7 shoots, lane 7—RgTR7 roots, lane 8—RgTR17 shoots, lane 9—RgTR17 roots.
UPGMA analysis led to the identification of two major clusters. The first was divided into several sub-clusters; one of them comprised RgS, RgAR, RgCR/NOA, RgTR7, and RgTR17 shoots, while the other comprised RgCR/NOA shoots and roots. The second comprised roots of lines RgAR, RgTR7, and RgTR17 (Figure 10). Surprisingly, the hairy RgTR7 and anatomical RgAR root lines were found to be present in a common sub-cluster. The variation observed could have been caused by a combination of the genetic transformation and somaclonal variation imposed by the tissue culture conditions [43,44,45]. The RgCR/NOA root line was derived from calli obtained from a medium supplemented with NOA, and hence it is possible that the anatomical roots emerging from the base of the RgS shoots also appeared through some callus cells. It has been proposed that somaclonal variation could be connected to the presence of the callus stadium and changes in its ploidy levels; these have been reported to increase gradually during callus initiation but then return to low ploidy during prolonged cultivation [38,45]. Furthermore, it was described that in cultures of highly organized organs, such as stems, leaves, and roots, somaclonal variation is generated more often than in explants with existing meristems, i.e., shoot/root tips or lateral buds [46,47]. In hairy root lines, it is possible that the ROL genes of T-DNA, which could be randomly integrated with the plant genome, may be responsible for genetic variability [48]. This phenomenon could be also a result of chromosomal aberrations or gene silencing [49]. Many plant species have demonstrated spontaneous or induced shoot regeneration from hairy roots in vitro, as well as somatic embryogenesis [50]. An excised shoot can develop into a whole plant, which was also observed in the current study. A number of studies have reported regenerants from hairy roots developing hairy root syndrome, with the extent of the symptoms varying according to the number of incorporated ROL genes.
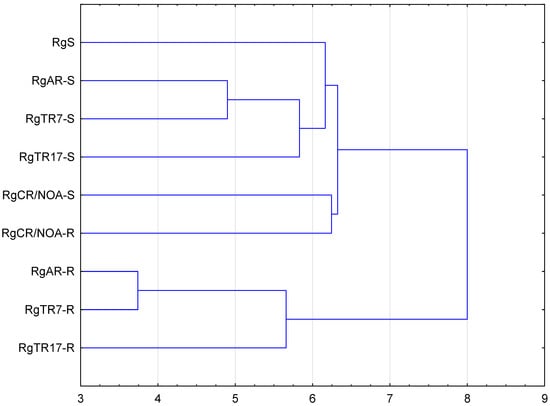
Figure 10.
UPGMA agglomeration analysis of Rindera graeca shoots and roots cultivated in vitro: RgS—shoots derived from seeds; RgCR/NOA-S—shoots derived from RgCR/NOA roots; RgCR/NOA-R—auxin-induced roots regenerated from callus; RgTR7-S—transgenic shoots derived from RgTR7 hairy roots; RgTR7-R—hairy roots of the RgTR7 line; RgTR17-S—transgenic shoots derived from RgTR17 hairy roots; RgTR17-R—hairy roots of the RgTR17 line.
In the current study, the morphology of shoots regenerated from R. graeca hairy roots did not differ from that of the non-transformed shoots, and this was in accordance with the results of the genetic analysis (Figure 7b and Figure 9). No morphological traits characteristic of shoots developed from hairy roots were observed (Figure S1), e.g., wrinkled leaves or dwarf and bushy phenotypes with short leaves attributed to the incorporation of ROL genes from A. rhizogenes T-DNA [51,52]. The absence of phenotypic changes between hairy roots and their regenerants could be explained by the fact that the TL- and TR-DNA regions of T-DNA are presented in the agropine-type bacterial strain and could be independently transferred to the nuclei of an infected plant cell and differently integrated into the plant genome. The TL-DNA ROL genes are crucial for the induction of hairy root syndrome [51,52], and the occurrence of full hairy root symptoms requires the expression of rolA, rolB, and rolC genes (see [51] and references therein). In the current study, rolB and rolC genes were confirmed in both the hairy roots and the derived regenerants. Both genes are believed to be necessary to induce significant improvements in root growth, with rolB playing a key role in root formation and rolC stimulating lateral branching [51]. Although the presence of rolA was not checked, the morphological features of the roots and shoot indicated its absence, as no bushy plantlets with wrinkled leaves were observed. Similarly, the successful incorporation of rolB and rolC genes into the regenerants was demonstrated by the increased formation of adventitious roots; various changes in leaf morphology for rolB; and reduced flower size, altered leaf morphology (small with wrinkled edges), and enhanced formation of auxiliary shoots among others for rolC. None of the mentioned symptoms were observed in the regenerants in the present study, as all three rol genes are needed for the full hairy root phenotype to develop [51,52]. In addition, the lack of any visible morphological traits of the hairy root phenotype could be attributed to the weak expression of rolB and rolC in the current experiment (Figure 5), probably due to the position effect and promotor activity modulation, as reported by Schmülling et al. [51] and Mauro et al. [52]. Similarly, the hairy root lines RgTR7 and RgTR17 did not differ morphologically from the non-transformed lines, demonstrating similar low ROL gene expression to the regenerants.
2.5. Phenolic Compounds Production
Although several publications have examined the in vitro production of RA and/or LAB as well as various other phenolic compounds, in Boraginaceous plant cultures, the majority have employed cultures of undifferentiated cells, i.e., callus and suspension cultures [53,54,55,56,57,58,59,60,61]. The number of studies that have described the accumulation of RA and/or LAB in root cultures of Boraginaceae plants is limited. There is one report on Eritrichum sericeum (Lehm.) DC. adventitious root cultures [62] and another on Lithospermum erythrorhizon callus suspension hairy root cultures [63]. Most of the literature concerning RA and LAB production in plant cultures has been dedicated to various members of the Lamiaceae family [7,8,9]. However, our group recently reported a comprehensive analysis of changes in the phytochemical profiles of the R. graeca RgAR, RgTR7, and RgTR17 root lines subjected to drought and cold stress [27].
In the present study, the presence of RA and LAB were confirmed in R. graeca shoots and roots cultivated in vitro in flasks and a sprinkle bioreactor by comparing their MS1 and MS2 fragmentation ion and retention times with the reference standards in negative ionization mode (Table 2, Figure 11, Table S4). LA was not detected.

Table 2.
Phenolic acid content (mg/g DW) in shoots and roots of Rindera graeca cultivated in vitro.
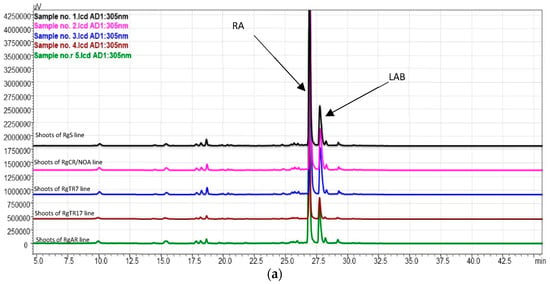
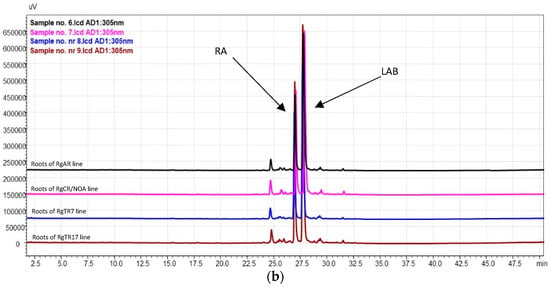
Figure 11.
HPLC–PDA chromatograms (wavelength 305 nm) of (a) shoot samples and (b) root samples. RA—rosmarinic acid; LAB—lithospermic acid B.
These findings are consistent with those of Naliwajski et al. [27] and Graikou et al. [24], who investigated in vitro cultures of R. graeca, as well as those of Ganos et al. [23], who examined the aerial parts of wild-grown R. graeca.
The highest yield of RA was detected in shoots regenerated from RgAR roots during cultivation in the SH medium in the sprinkle bioreactor: 30.0 ± 3.2 mg/g DW. This value was over 1.5-fold higher than that in flask cultures of the same shoot line in the same medium and almost 2.4-fold more than that in RgS shoots (Table 2). Among all the investigated root lines, the maximum RA content was determined in the RgAR roots cultivated in flasks in the DCR medium (14.4 ± 5.9 mg/g DW). In the bioreactor, the highest yield of RA was detected in RgAR roots during cultivation in the SH medium (5.6 ± 0.7 mg/g DW) (Table 2). In addition, in our previous investigation on R. graeca root cultures, we observed differences in the RA biosynthetic capacities among three tested root lines: RgAR, RgTR7, and RgTR17. The highest RA content was determined in the hairy roots of the RgTR7 line (33.69 mg/g DW) [27].
Weremczuk-Jeżyna et al. [64] reported RA production of up to 18.7 mg/g DW and LAB production of up to 5.9 mg/g DW in Dracocephalum forestii (Lamiaceae) shoots cultivated in flask cultures and RA and LAB production of 17.9 and 6.5 mg/g DW, respectively, in a sprinkle bioreactor [65]. However, transformed shoots of D. forestrii accumulated higher amounts of RA of up to 24.80 mg/g DW [66].
RA can be detected in all parts of the intact plant. It can also be produced by biotechnological methods, with levels as high as 36% DW reported in suspension cultures of Salvia officinalis L. and up to 14% DW in Ocimum basilicum L. hairy root cultures (see [10] and references therein).
Thus, the results of the present study are consistent with those of previous reports.
LAB was detected in both the shoots and the roots (Table 2). Its highest yield (49.3 ± 15.5 mg/g DW) was determined in RgAR regenerants cultivated in the DCR/BAP medium. This yield was significantly higher than that of the other regenerants, including the RgS shoot line. In the cultures carried out in the bioreactor, LAB was detected only in regenerated shoots of RgAR and RgTR17 lines, and only when SH medium was applied; in such cases, the yield in RgAR was over threefold higher than in the RgTR17 regenerates.
The R. graeca hairy roots in the present study demonstrated a fourfold higher LAB yield (11.7 mg/g DW) and twofold higher RA yield (19.97 mg/g DW) compared with previously studied D. forestii hairy cultures [67]. However, the highest yield of LAB determined under the conditions of the current study was over nine times lower (11.7 versus 106.07 mg/g DW) than that reported previously by our group [27]. This may be attributed to differences in environmental conditions, including the in vitro culture maintenance protocol, as this is known to have a strong influence on the profile of secondary metabolites.
Similar results have been observed in in Salvia miltiorrhiza Bunge hairy root cultures, which demonstrated an increase in LAB content accompanied by a decrease in RA content (Table 2) [68], as well in L. erythrorhizon cell and hairy root cultures [56]. Fujita et al. [69] report an appreciably higher amount of LAB than RA in cultivated cells, with the choice of medium having a marked effect; in this case, LAB production was almost eightfold higher in production medium M9 compared with LS. In hairy root cultures, the authors observed a higher accumulation of LAB than RA, which was only present in trace amounts, as well as LA production. While RA was distributed mainly in the aerial parts, LAB and LA were observed in the underground ones. LA was not detected in the shoots. In the present study, the content of the investigated phenolic compounds also depended on the culture conditions. Higher RA concentrations were observed in the shoots during cultivation in the SH medium; however, higher RA concentrations were only observed in the roots when they were cultivated in the DCR medium (Table 2). LA was detected in both the shoot and the root cultures, although it was observed in the roots only when they were grown in the DCR medium. It is not surprising that both investigated phenolic compounds were observed in hairy roots, as they possess all the enzymes that mimic the biosynthetic pathways of the parent plant [70]. Modern biotechnology methods allow considerably higher RA and caffeic acid derivatives to be obtained from plant material cultivated in vitro than from parent plants. In hairy root cultures of Dracocephalum kotschyi, depending on the root line, the RA content ranged from 10 to 1500 µg/g DW, which was 15 times higher than that from the roots of the intact plant [71]. In E. sericeum, RA accumulation amounted to 2.04% DW in callus cultures and 4.50% DW in adventitious roots. In comparison, RA was only present at 0.07% DW in the aerial and underground parts of the native plant and at 1.10% DW in L. erythrorhizon callus, compared with 0.28% and 0.66% in the aerial and underground parts of the native plant [62]. Similarly, Bryukhanov et al. [59] reported a 112-fold higher content of RA in cell culture than in native E. sericeum plant roots. In addition, in Martensia maritima, another borage species, the level of RA amounted to 0.74% DW in the callus tissue, compared with 0.11%, 0.20%, and 0.16% DW in the stems, leaves, and roots of the native plant [59].
2.6. Antioxidant Capacity of Plant Extracts
Significantly higher total phenolic compound (TPC) content was determined in extracts derived from shoots cultivated in untreated SH or SH supplemented with BAP (Table 3) compared with other plant extracts. The highest TPC yield (up to 264.7 ± 32.8 GAE mg/g DW) was noted in RgCR/NOA shoots maintained in the SH/BAP medium. In the root cultures, significantly higher TPC was noted in roots cultivated in the SH medium than in DCR, with the highest concentration amounting to 45.7 ± 8.1 GAE mg/g DW in the RgAR line. The detected TPC levels were remarkably higher in the shoots than in the roots (Table 3). The antioxidant level of the R. graeca plant material was determined using DPPH and FRAP assays. In the former, DPPH radical scavenging by an antioxidant resulted in a loss of absorbance directly proportional to the potential of the antioxidant, while in the latter, a Fe3+ complex with tripyridyltriazine was reduced to an intense blue Fe2+ complex by an antioxidant in an acidic medium, with the depth of color indicating the strength of the antioxidant [72].

Table 3.
Antioxidant capacity of Rindera graeca shoot and root extracts.
In the present study, the strongest DPPH free radical scavenging activity was noted for extracts prepared from roots cultivated in the DCR medium; among shoot cultures, the highest ability was observed for the extracts derived from shoots grown in the SH/BAP medium (Table 3). The antioxidant potential ranged from 32.8% to 87.4% depending on the root line and from 27.9% to 85.6% depending on the shoot line. The strongest reducing power determined in the FRAP assay was demonstrated for extracts prepared from shoots, which were also cultivated in the SH/BAP medium (Table 3). The estimated correlation between the DPPH radical scavenging activity and the TPC, RA, and LAB concentration amounted to r2 = 0.86, 0.42, and 0.41, respectively. By contrast, the results of the FRAP method demonstrated only a weak correlation with TPC (r2 = 0.16); however, higher correlations were observed with the RA and LAB content: r2 = 0.18 and −0.14, respectively. These findings indicate that the antioxidant potential determined by the DPPH and FRAP assays could be influenced by the presence of other groups of secondary metabolites reported for R. graeca, including flavonoids and shikonin-type naphthoquinones [24,27]. Although shikonin itself was not detected, four shikonin derivatives were previously reported [73]. Shikonin/alkannin and their derivatives, bearing both the quinone and hydroquinone moieties, are known to possess substantial antioxidant properties [74,75]. A structure–activity relationship study of selected shikonin/alkannin derivatives revealed that the side chain-OH group at C-1′ positively influenced the antioxidant capacity [76] and that a naphthoquinone moiety was essential for such activity [74]. Nevertheless, among the shoot cultures in the present study, significantly higher antioxidant activity was observed in extracts derived from cultures grown with BAP; these conditions offered a favorable multiplication rate (Figure 2) and shoot quality (Figure 3a). These findings are consistent with those reported in investigations of R. graeca aerial parts [23]. They are also similar to those found for in vitro Mentha haplocalyx cultures [23], in which both crude extracts and those derived from the shoots were subjected to phytochemical analysis. She et al. [23] isolated eight polyphenolic acids from the chlorophyll fraction of this species, among them LAB, RA, and LA, and demonstrated that LAB exerted higher DPPH scavenging activity than RA and LA.
This also indicates that a group of secondary metabolites other than investigated phenolic acids could a play role in the antioxidant power of the investigated extracts. This complex phenomenon was also described by Krolicka et al. [77] in in vitro Drosera aliciae Raym.-Hamet cultures.
3. Materials and Methods
3.1. Seed Germination
The seeds of R. graeca were collected and donated by Prof. Ioanna Chinou from the University of Athens in 2006, and the in vitro cultures were initiated the same year. Prior to sterilization, the seeds were placed at −20 °C for 72 h, and after sterilization, they were gently cut with a sterile scalpel. The seeds were surface-sterilized with 90% ethanol for 30 s and treated with 5% natrium hypochlorite for 30 min. Finally, they were rinsed three times with sterile water. A hormone-free, half-strength basal MS [78] medium (½ MS) containing 3% sucrose was used for the germination process. The pH of the medium was adjusted to 5.8, and it was autoclaved. Finally, 30 seeds of R. graeca were used to initiate the culture. This culture was carried out in 100 mL Erlenmeyer flasks containing 30 mL of the ½ MS solid medium at 25 ± 1 °C in the dark.
3.2. Shoot and Root Cultures
3.2.1. Shoot Micropropagation
The explants used as the source for the experiments were four-week-old seedlings of R. graeca. Micropropagation was performed using solid DCR [79] with four modifications: full strength (DCR), half the level of macroelements (DCR/M), half-strength of macro- and microelements (DCR/S), and half-strength of basal salts and vitamins (DCR/SV). The media were supplemented with 6-benzylaminopurine (BAP) or kinetin at doses of 0.5 or 1 mg/L before sterilization. Three shoots per flask were placed into a 300 mL Erlenmeyer flask containing 50 mL of one of the modified DCR media described above. Each DCR modification was supplemented with 3% sucrose, and the pH was adjusted to 5.7 prior to autoclaving. The experiments were carried out over three subsequent four-week passages, and each group was composed of five flasks with four to five explants per flask. The shoot cultures were performed for four weeks with a 16 h/8 h (light/dark) photoperiod with light provided by cool-white fluorescent lamps (40 µM/m2/s) at a temperature of 25 ± 1 °C. The multiplication rate was calculated after the four-week culture and expressed as the number of newly developed auxiliary shoots per explant ± SD.
3.2.2. Hairy Root Cultures
The four-week-old shoots were infected with Agrobacterium rhizogenes strain ATCC 15834, as described by Pietrosiuk et al. [80]. On the basis of the visual evaluation, out of 60 roots emerging from the inoculation sites, two hairy root lines were chosen for further investigation. After excision, to remove the bacteria from their tissues, the roots were placed into 100 mL Erlenmeyer flasks containing 30 mL of liquid hormone-free LS medium [81] supplemented with 3% sucrose and Cefotaxime (500 mg/L), and they were cultivated for two weeks. The pH of the medium was adjusted to 5.6 prior to autoclaving. This procedure was repeated three times. The putative hairy roots were cultivated in a liquid hormone-free LS medium to examine the growth parameters. For this purpose, the LS medium and the four DCR medium modifications were used. The roots were cultivated on a 105 rpm gyratory shaker (INFORS, Bottmingen, Switzerland) in 250 mL Erlenmeyer flasks containing 50 mL of the medium, with a subculture to fresh medium every four weeks; the incubation was performed at 23 ± 1 °C in the dark. To determine the growth of roots, they were gently pressed onto filter paper to remove the medium and weighted, and their fresh weight (FW) was recorded. The root growth index was calculated by subtracting the fresh weight at the time of inoculation (time zero) from that at the time of harvest (day 28th of culture) and dividing by the weight at the time of harvest. The growth index was calculated on the basis of the results of five subsequent passages (five flasks per medium variant). The material was later lyophilized under vacuum at 0.340 mbar and an initial product temperature of −20 °C (lyophilizer Christ ALPHA1-4 LSC, Osterode am Harz, Germany) until a constant weight was achieved, that is, 24–48 h, and weighed to determine the dry weight (DW). On the basis of the root biomass growth results, the DCR medium variant was chosen for further cultivation. However, after the sprinkle bioreactor experiments, the root growth was also investigated in an SH [82] liquid hormone-free medium (50 mL in 250 mL Erlenmeyer flasks) for three passages with five replicates. The SH medium was supplemented with 3% sucrose, and its pH was adjusted to 5.7 prior to autoclaving.
3.2.3. Non-Transformed Root Cultures
Two non-transformed root cultures were established. The first one was initiated from roots that spontaneously developed at the bases of shoots cultivated on a solid hormone-free DCR medium over four weeks of culture (RgAR). The second root line was induced by auxin and originated from callus tissue obtained from one-week-old hypocotyls of R. graeca (RgCR/NOA). The callus culture was initiated by an MS medium supplemented with 0.5 mg/L 2-naphthoxyacetic acid (NOA) and 3% sucrose; the pH was adjusted to 5.8 prior to autoclaving. The callus tissue developed on the hypocotyl over four weeks of culture, and next was transferred to a liquid hormone-free MS medium. Root development was observed for four weeks of culture. The roots were then transferred to 250 mL Erlenmeyer flasks containing 50 mL of the liquid hormone-free DCR or SH medium (each supplemented with 3% sucrose, pH adjusted to 5.7 prior to autoclaving) for further cultivation. The cultures were maintained at 23 ± 1 °C in the dark at 105 rpm on an INFORS gyratory shaker. The growth index of auxin-induced roots was determined as described in Section 3.2.2.
3.2.4. Regeneration of Shoots from Roots
During the cultivation of all root clones, it was noted that the roots were able to spontaneously regenerate shoots. This phenomenon prompted an investigation of shoot regenerant cultures. The regeneration of shoots from untransformed and hairy roots was performed on solid DCR or SH media. Five 2–3 cm long root fragments were placed on the media in five 300 mL Erlenmeyer flasks containing 50 mL of solid DCR or SH media. The cultures were cultivated for four weeks under a 16/8 h (light/dark) photoperiod with light provided by cool-white fluorescent lamps (40 µM/m2/s) and at a temperature of 25 ± 1 °C. The regenerated shoots were then excised from root tissue and placed on a solid DCR or SH medium, either plain or supplemented with 0.5 mg/L BAP. Each medium was supplemented with 3% sucrose, and their pH was adjusted to 5.7 prior to autoclaving. The experiments were carried out over three subsequent four-week passages. After this time, the number of regenerants per flask was calculated. Four shoots derived from each root line per flask were placed in each flask, and each passage consisted of five flasks.
3.2.5. Root Cultures in Sprinkle Bioreactor
To increase the culture scale, the RgAR line and two hairy root lines, RgTR7 and RgTR17, all after their third passage, were chosen for subculture in a bioreactor. For cultivation, hormone-free DCR and SH media were used. Each medium was supplemented with 3% sucrose, and their pH was adjusted to 5.7 prior to autoclaving. The batch cultures in the bioreactor lasted five weeks. The culture itself was performed in a 5.0 L (working volume 4.0 L) nutrient sprinkle bioreactor consisting of an immobilized glass cylinder (Büchi, Germany). In its interior, a stainless-steel net (the location of the culture) was situated 20 cm above the bottom of the growth bioreactor vessel, and the bottom of the growth chamber was used as a reservoir for the liquid nutrient medium. A Masterflex L/S peristaltic pump (Cole-Parmer, Vernon Hills, IL, USA) provided recirculation and the dosage of the liquid nutrient medium through a polypropylene dispersal nozzle situated 100 mm from the bottom of the growth cylinder. In a single cycle, the medium sprinkled cultivated plant material and was then returned to the medium reservoir. The operating time of the pump for medium delivery was set at 40 s, with 80 s breaks between each delivery; this was later changed to 15 s of medium delivery with 30 s breaks between each delivery. The nutrient was maintained at a constant temperature of 24 ± 1 °C by a Thermamix thermostat (Sartorius BBI Systems GmbH, Melsungen, Germany). The roots were grown in the dark. After five weeks of culture, the plant material was harvested, and the fresh and dry weight (g per bioreactor) was measured.
3.3. PCR Analysis
3.3.1. The Confirmation of Transformation
Successful transformation of the roots and root-derived shoots was confirmed at the molecular level by the determination of the T-DNA fragment of the Ri plasmid in the plant tissue by polymerase chain reaction (PCR). The Ri plasmid was isolated from 24 h cultures of the A. rhizogenes ATCC 15834 strain using the Plasmid Mini AX Kit (A&A Biotechnology, Gdańsk, Poland) according to the manufacturer’s instructions. The oligonucleotide primers for PCR detection of the homologous sequences to rolB (5′-GCTCTTGCAGTGCTAGATTT-3′ and 5′-GAAGGTGCAAGCTACCTCTC-3′) and rolC (5′-CTCCTGACATCAAACTCGTC-3′ and 5′-TGCTTCGAGTTATGGGTACA-3′) were designed by Królicka et al. [78]. In addition, whether the investigated organs were free of A. rhizogenes was investigated using primers homologous to virG (5′-ACTGAATATCAGGCAACGCC-3′ and 5′-GCGTCAAAGAAATAGCCAGC-3′) [83]; although virG is present in the Ri plasmid, it is not transferred with T-DNA to the plant cell [84]. The PCR reaction was performed with the Phire® Plant Direct PCR Kit (Thermo Scientific, Waltham, MA, USA) using the dilution protocol specified by the manufacturers. PCR analysis was carried out as follows: one cycle of initial denaturation at 98 °C for 5 min followed by 35 cycles at 98 °C for 5 s, 62 °C for 5 s, and 72 °C for 20 s and a final extension at 72 °C for 1 min. The Thermal Cycler Vario (Applied Biosystems, by Life Technologies, Austin, TX, USA) was used for amplification. The amplification products were resolved by electrophoresis on a 1% agarose gel (Prona, Burgos, Spain) with 1X TBE buffer, stained with SimplySafe (EURx, Gdańsk, Poland), and visualized under UV light (Gel-X Gel Analysis Systems, STI, Poznań, Poland). The size of amplification products was estimated for a molecular weight marker GeneRulerTM 1 kb Plus DNA ladder (Thermo Scientific, Waltham, MA, USA).
3.3.2. Genetic Analysis Using Random Amplified Polymorphic DNA (RAPD) and Start Codon Targeted (SCoT) Markers
The genetic diversity of the in vitro cultivated donor plantlets, and both the non-transformed and transformed shoot regenerants cultivated on the DCR medium with 0.5 mg/L BAP, was confirmed using random amplified polymorphic DNA (RAPD) primers and start codon targeted (SCoT) markers according to Collard and Mackill [85]. The genetic diversity of all investigated root lines was also estimated. The RAPD analysis used a set of 20 arbitrary primers (OPA, Operon Technologies Inc., Alameda, CA, USA) (Table S2), while the SCoT analysis employed 36 arbitrary primers designed by Collard and Mackill [85]. All primers were synthesized by Sigma-Aldrich (Merck KGaA, Darmstadt, Germany). The genomic DNA from the non-transformed and transformed shoot and root tissues was isolated using the CTAB/PVP method developed by Pirttilä et al. [86].
Each PCR reaction (final volume 10 µL) contained: 10X NH4 Complete Reaction Buffer, 2 mM dNTP Mix, 0.5 U SuperHotTaq DNA Polymerase (BIORON, Römerberg, Germany), 10 µM OPA/SCoT primers, 10% PVP, 25 mM MgCl2, and 25 ng of genomic DNA. All reagents were manufactured by ThermoFisher Scientific (Waltham, MA, USA). The PCR analysis for SCoT primers was performed under the following conditions: initial denaturation at 94 °C for 3 min followed by 35 cycles at 94 °C for 1 min, 51 °C for 1 min, and 72 °C for 2 min and a final extension at 72 °C for 5 min. For the OPA primers (Table S2), PCR analysis was carried out as follows: initial denaturation at 94 °C for 3 min followed by 35 cycles at 94 °C for 1 min, 44 °C for 1 min, and 72 °C for 2 min and a final extension at 72 °C for 7 min. The Thermal Cycler Vario (Applied Biosystems, by Life Technologies, Austin, TX, USA) was used for amplification.
The amplification products, stained with SimplySafe were resolved by electrophoresis on 1.5% agarose gel with 1X TBE buffer and visualized under UV light. The size of amplification products was estimated on the basis of 1 kb and 100 bp DNA ladder molecular weight markers (GeneRulerTM, Thermo Scientific, Waltham, MA, USA). The presence or absence of bands between samples was scored, and bands with the same mobility were counted as identical bands. DNA bands characterized by low visual intensity and those that could not be distinguished as present or not were considered ambiguous markers and were not scored. The resulting bands were scored as present (1) or absent (0) and entered in a computer file as a binary matrix, one for each molecular marker. Then, the matrices were analyzed using FreeTree software [87]. The Dice similarity coefficient was calculated and expressed as a percentage [87]. Hierarchical grouping was performed according to coefficients using the unweighted pair group method of arithmetic means (UPGMA); a UPGMA dendrogram was drawn using STATISTICA 13.1 PL software.
3.4. Phytochemical Investigation
All chemicals and reagents, if not stated otherwise, were purchased from Sigma-Aldrich (Merck KGaA, Darmstadt, Germany) or Avantor Performance Materials Poland S.A. (Gliwice, Poland). The standard compounds of lithospermic acid (LA) and lithospermic acid B (LAB) were manufactured by Phyto-Lab and purchased from LCG Standards (Poland); rosmarinic acid (RA) and caffeic acid were purchased from Sigma-Aldrich (Merck KGaA, Darmstadt, Germany).
3.4.1. Sample Preparations
Lyophilized and powdered plant material was used for the phytochemical investigations.
For the HPLC analysis, 100 mg of plant material dry mass was used. The plant material was extracted with 1 mL of 100% methanol for 15 min using an ultrasonic bath (Sonorex Bandelin, Berlin, Germany), then placed on an INFORS gyratory shaker (105 rpm) and incubated overnight at 23 ± 1 °C in the dark. The samples were then centrifuged, and the supernatant was collected. The extraction step was then repeated, and the supernatants were combined and evaporated until dry. The resulting mixtures were stored at −20 °C for analysis.
3.4.2. HPLC–UV–DAD Analysis of Phenolic Acids
The contents of caffeic acid, RA, LA, and LAB were analyzed by HPLC–UV–DAD. Before the HPLC analysis, the samples were re-dissolved in 100% HPLC-grade methanol (1 mL), and 20 μL was included in the HPLC analysis; the setup comprised a DIONEX (Sunnyvale, CA, USA) system with a UVD 340 S diode array detector and an automated sample injector (ASI-100). A NovaPak Phenyl column (150 × 3.9 mm; Waters, Milford, MA, USA) was used for the separation and determination of the phenolic acids in the plant extracts. Water acidified with 0.04 M H3PO4 (solvent A), acetonitrile (solvent B), and methanol (solvent C) were used as mobile phases. The gradient elution applied was as follows: 0–3 min—85% A, 7% B, 8% C; 3–13 min—70% A, 15% B, 15% C; 13–30 min—60% A, 20% B, 20% C. Eluting substances were visualized by UV absorption at 220 nm.
3.4.3. LCMS-IT-TOF Analysis
The sample analysis was performed according to the method developed by Naliwajski et al. [27]. A Shimadzu Prominence high-performance liquid chromatograph (HPLC) was used, coupled with an LCMS-IT-TOF mass spectrometer (Shimadzu, Kyoto, Japan), equipped with an ion trap (IT), a time-of-flight (TOF) detector, and an electrospray ionization (ESI) source. Mass spectra were recorded in positive ion mode using LCMSsolution software (Shimadzu, Kyoto, Japan). A Kinetex C18 column, 2.6 µm, 2.1 × 100 mm (Phenomenex, Torrance, CA, USA), was used with an injection volume of 3 µL, an oven column temperature of 40 °C, a flow rate of 0.2 mL/min, an analysis duration of 75 min, and PDA detection at wavelengths λ = 200–800 nm. The mobile phase consisted of (A) water with the addition of 0.2% CHCOOH and (B) methanol. The following gradient was applied: 0–10 min 5% B, 10–30 min 5→50% B, 30–35 min 50→50% B, 35–55 min 50→95% B, 55–60 min 95% B, 60–62 min 95→5% B, equilibrium time—13 min in 5% B.
In addition, the following conditions for the mass spectrometer were used: polarity positive and negative, mass range m/z 100–1000 Da in both modes, ion accumulation time: 10 ms in MS1 experiments and 25 ms in MS2 experiments, interface temperature: 220 °C, heat block temperature: 220 °C, nebulizing gas flow: 1.5 L/min, drying gas pressure: 100 kPa, IS: +4.5 kV, collision energy in MS2 experiments: 25–35%. The calibration mixture was used to calibrate the TOF detector of the LCMS-IT-TOF mass spectrometer.
3.4.4. Determination of Total Phenolic Compound Content (TPC)
TFC was estimated according to Pękal and Pyrzynska [88] with slight modifications. TFC was determined based on aluminum complex formation, while TPC was estimated by a Folin–Ciocalteu assay. Briefly, for the TPC assay, 20 µL of plant extract diluted in 80% methanol was added to a 96-well plate, followed by 40 µL of 0.2 N Folin–Ciocalteu reagent and 160 µL of Na2CO3. The absorbance was measured after 30 min of incubation at 765 nm. The results were expressed as mg of gallic acid equivalent (GAE)/g DW of plant biomass. Each batch sample was measured in four repetitions, and the mean value was taken. The results were calculated and expressed as mg of gallic acid equivalent (GAE) (mg GAE/g DW) using the regression equation determined from the standard curve: y = 0.0053x + 0.1413, r2 = 0.9918. For all assays, an Epoch BioTek (Agilent, Santa Clara, CA USA) microplate reader was used.
3.5. Antioxidant Activity Analysis
The DPPH assay was performed according to Fukumoto and Mazza [89] with some modifications. Briefly, 200 µL of 2,2-diphenyl-1-picrylhydrazyl-hydrate (DPPH) was added to each 20 µL plant sample diluted in 80% methanol. The mixture was incubated for 60 min at room temperature in the dark, and the absorbance was measured at 517 nm. The radical scavenging activity was expressed as a percentage of DPPH inhibition, calculated according to the formula:
where A0 is the absorbance of the control (DPPH solution + 80% methanol) and As is the absorbance of the sample.
% Inhibition = [(A0 − As)/A0] × 100%
The ferric-reducing antioxidant power (FRAP) assay was carried out according to Benzie and Strain [90], as modified by Bolanos de la Torre et al. [91]. Briefly, 300 µL of FRAP reagent was added to 10 µL plant samples diluted in 80% methanol. The absorbance of the reaction mixture was then detected at 593 nm after 30 min of incubation at room temperature in the dark. Each batch sample was measured in four repetitions, and the mean value was calculated. The results were calculated using the regression equation determined from the standard curve—y = 816.59x, r2 = 0.9997—and expressed as mg of Trolox equivalents (TE)/g DW.
3.6. Statistical Analysis
All the analyses were performed with appropriate experimental and analytical replicates. The statistical significance between means was assessed using the Kruskal–Wallis one-way analysis of variance performed with STATISTICA 13.1 PL software. The significance between groups was further estimated using the Mann–Whitney U test. A probability of p < 0.05 was considered significant. Pair-wise metabolite–antioxidant effects correlations were calculated by Pearson’s correlation coefficient test.
4. Conclusions
The biotechnological approaches developed in the current study establish a platform for the sustainable production of bioactive compounds with respect to the protection of plant kingdom biodiversity, which is one of the most important challenges of current times. Our findings indicate that Rindera graeca, a rare endemic plant, could be efficiently micropropagated in in vitro cultures. The highest shoot multiplication rates, 7.2 ± 3 and 7.1 ± 5, were obtained on two modifications of a DCR medium supplemented with 0.5 mg/L BAP or kinetin 0.5 mg/L, respectively. Moreover, the shoot and root cultures of this species, both transformed and non-transformed, could serve as sustainable sources of rosmarinic acid and lithospermic acid B. The production capacity depended on the shoot or root line and the culture conditions (type of medium, flask/bioreactor cultivation). Significantly higher levels of RA and LAB were detected in the shoot cultures than in the root ones. The maximum yield of RA was detected in shoots regenerated from RgAR roots during cultivation in the SH medium in the sprinkle bioreactor, i.e., 30.0 ± 3.2 mg/g DW. Among all the investigated root lines, the highest RA accumulation was determined in RgAR roots cultivated in flasks in the DCR medium (14.4 ± 5.9 mg/g DW). While in the bioreactor, the highest yield of RA was detected in RgAR roots during cultivation in the SH medium (5.6 ± 0.7 mg/g DW). The antioxidant potential ranged from 27.9% to 87.4% (DPPH radical scavenging) and from 0.3 to 2.3 µM TE/g DW (FRAP assay), depending on the shoot/root line and the medium used. The antioxidant capacity of the plant extracts did not depend on the content of the investigated phenolic acids but presumably would be affected by the presence of other secondary metabolites, e.g., other phenolic compounds. Genetic analysis carried out using RAPD and SCoT markers demonstrated genetic variability, which reflects the biosynthetic capacity of the cultivated shoot and root lines obtained via biotechnological methods. Our findings demonstrate that the conditions of the in vitro cultures significantly influenced the chemical profile of the investigated phenolic acids in cultivated organs. Furthermore, in vitro R. graeca cultures could be considered a source of both rosmarinic acid and lithospermic acid B. Nevertheless, further detailed investigations are needed, e.g., the identification of molecular targets influencing biosynthesis efficiency.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/molecules28124880/s1, Figure S1: Shoots regenerated from root explants of RgCR/NOA and RgTR17 root lines cultivated on various solid media; Table S1: Auxiliary shoot formation of Rindera graeca explants derived from seedlings after 4 weeks of culture on various modifications of DCR medium; Table S2: OPA primer sequence used to estimate genetic diversity of cultured in vitro shoot and root lines of Rindera graeca in RAPD reaction; Table S3: Dice similarity coefficient [%]; Table S4: Mass spectra of rosmarinic acid and lithospermic acid B detected in examined extracts of shoot and root Rindera graeca lines.
Author Contributions
Conceptualization, K.S.-B., Methodology, K.S.-B., M.G., Ł.K., B.W. and M.K.; validation, K.S.-B. and B.W.; Formal analysis, K.S.-B., I.C., K.G., M.J. and A.P.; Investigation, K.S.-B., M.G., Ł.K., M.K., B.W., A.P., M.J., E.S., P.S. and P.Z.; Resources, I.C. and A.P.; Data curation, K.S.-B. and B.W.; Writing—original draft preparation, K.S.-B.; Writing—review and editing, A.P., I.C., K.G., Ł.K. and M.J.; Visualization, M.G., B.W. and Ł.K.; Supervision, K.S.-B.; Project administration, K.S.-B.; Funding acquisition, A.P. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Statutory Budget (FW21/N/2018; 2019; 2020; 2021) of the Medical University of Warsaw and carried out with the use of CePT infrastructure financed by the European Union—the European Regional Development Fund within the Operational Program Innovative Economy for 2007–2013.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Acknowledgments
Conflicts of Interest
The authors declare no conflict of interest.
Sample Availability
Not applicable.
References
- Wilson, S.A.; Roberts, S.C. Recent advances towards development and commercialization of plant cell culture processes for the synthesis of biomolecules. Plant Biotechnol. J. 2012, 10, 249–268. [Google Scholar] [CrossRef] [PubMed]
- Ochoa-Villarreal, M.; Howat, S.; Hong, S.M.; Jang, M.O.; Jin, Y.W.; Lee, E.K.; Loake, G.J. Plant cell culture strategies for the production of natural products. BMB Rep. 2016, 49, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Yue, W.; Ming, Q.; Lin, B.; Rahman, K.; Zheng, C.; Han, T.; Qin, L. Medicinal plant cell suspension cultures: Pharmaceutical applications and high-yielding strategies for the desired secondary metabolites. Crit. Rev. Biotechnol. 2014, 36, 215–232. [Google Scholar] [CrossRef]
- Kumar, N.; Goel, N. Phenolic acids: Natural versatile molecules with promising therapeutic applications. Biotechnol. Rep. 2019, 24, e00370. [Google Scholar] [CrossRef]
- Anantharaju, P.G.; Gowda, P.C.; Vimalambike, M.G.; Madhunapantula, S.V. An overview on the role of dietary phenolics for the treatment of cancers. Nutr. J. 2016, 15, 99. [Google Scholar] [CrossRef]
- Dai, J.; Mumper, R.J. Plant phenolics: Extraction, analysis and their antioxidant and anticancer properties. Molecules 2010, 15, 7313–7352. [Google Scholar] [CrossRef] [PubMed]
- Dias, M.I.; Sousa, M.J.; Alves, R.C.; Ferreira, I.C.F.R. Exploring plant tissue culture to improve the production of phenolic compounds: A review. Ind. Crops Prod. 2016, 82, 9–22. [Google Scholar] [CrossRef]
- Bulgakov, V.P.; Vereshchagina, Y.V.; Veremeichik, G.N. Anticancer polyphenols from cultured plant cells: Production and new bioengineering strategies. Curr. Med. Chem. 2018, 25, 4671–4692. [Google Scholar] [CrossRef]
- Swamy, M.K.; Sinniah, U.R.; Ghasemzadeh, A. Anticancer potential of rosmarinic acid and its improved production through biotechnological interventions and functional genomics. Appl. Microbiol. Biotechnol. 2018, 102, 7775–7793. [Google Scholar] [CrossRef]
- Petersen, M. Rosmarinic acid: New aspects. Phytochem. Rev. 2013, 12, 207–227. [Google Scholar] [CrossRef]
- Petersen, M.; Simmonds, M.S.J. Rosmarinic acid. Phytochemistry 2003, 62, 121–125. [Google Scholar] [CrossRef]
- Bulgakov, V.P.; Inyushkina, Y.V.; Fedoreyev, S.A. Rosmarinic acid and its derivatives: Biotechnology and applications. Crit. Rev. Biotechnol. 2012, 32, 203–217. [Google Scholar] [CrossRef]
- Royal Botanic Gardens, Kew, Plants of the World Online. Available online: https://powo.science.kew.org/ (accessed on 20 October 2022).
- Weigend, M.; Luebert, F.; Selvi, F.; Brokamp, G.; Hilger, H.H. Multiple origins for Hound’s tongues (Cynoglossum L.) and Navel seeds (Omphalodes Mill.)—the phylogeny of the borage family (Boraginaceae s.str.). Mol. Phylogenet. Evol. 2013, 68, 604–618. [Google Scholar] [CrossRef]
- Pourghorban, Z.; Salmaki, Y.; Weigend, M. Phylogenetic relationships within the subtribe Cynoglossinae (Cynoglossoideae: Boraginaceae): New insights from nuclear and plastid DNA sequence data. Plant Syst. Evol. 2020, 306, 1–15. [Google Scholar] [CrossRef]
- Bigazzi, M.; Nardi, E.; Selvi, F. Palynological contribution to the systematics of Rindera and the allied genera Paracaryum and Solenanthus (Boraginaceae-Cynoglosseae). Willdenowia 2006, 36, 37–46. [Google Scholar] [CrossRef]
- Kamari, G.; Blanché, C.; Siljak- Yakovlev, S. Mediterranean chromosome number reports—24. Flora Mediterr. 2014, 24, 286–291. [Google Scholar] [CrossRef]
- Walter, K.S.; Gillett, H.J. 1997 IUCN Red List of Threatened Plants; Walter, K.S., Gillett, H.J., Eds.; Complied by the World Conservation Monitoring Centre; IUCN—The World Conservation Union: Gland, Switzerland; Cambridge, UK, 1998; ISBN 2-8317-0328-X. [Google Scholar]
- Mosaddegh, M.; Naghibi, F.; Moazzeni, H.; Pirani, A.; Esmaeili, S. Ethnobotanical survey of herbal remedies traditionally used in Kohgiluyeh va Boyer Ahmad province of Iran. J. Ethnopharmacol. 2012, 141, 80–95. [Google Scholar] [CrossRef]
- Altundag, E.; Ozturk, M. Ethnomedicinal studies on the plant resources of east Anatolia, Turkey. Procedia-Soc. Behav. Sci. 2011, 19, 756–777. [Google Scholar] [CrossRef]
- Civra, A.; Francese, R.; Sinato, D.; Donalisio, M.; Cagno, V.; Rubiolo, P.; Ceylan, R.; Uysal, A.; Zengin, G.; Lembo, D. In vitro screening for antiviral activity of Turkish plants revealing methanolic extract of Rindera lanata var. lanata active against human rotavirus. BMC Complement. Altern. Med. 2017, 17, 1–8. [Google Scholar] [CrossRef]
- Yücel, T.B.; Karaoğlu, Ş.A.; Yaylı, N. Antimicrobial activity and composition of Rindera lanata (LAM.) Bunge var. canescens (A.D.C.) Kosn. essential oil obtained by hydrodistillation and microwave assisted distillation. Rec. Nat. Prod. 2017, 11, 328–333. [Google Scholar]
- Ganos, C.; Aligiannis, N.; Chinou, I.; Naziris, N.; Chountoulesi, M.; Mroczek, T.; Graikou, K. Rindera graeca (Boraginaceae) phytochemical profile and biological activities. Molecules 2020, 25, 3625. [Google Scholar] [CrossRef]
- Graikou, K.; Damianakos, H.; Ganos, C.; Sykłowska-Baranek, K.; Jeziorek, M.; Pietrosiuk, A.; Roussakis, C.; Chinou, I. Chemical profile and screening of bioactive metabolites of Rindera graeca (A. DC.) Bois. & Heldr. (Boraginaceae) in vitro cultures. Plants 2021, 10, 834. [Google Scholar] [CrossRef]
- Dong, M.; Liu, D.; Li, Y.-H.H.; Chen, X.-Q.Q.; Luo, K.; Zhang, Y.-M.M.; Li, R.-T.T. Naphthoquinones from Onosma paniculatum with potential anti-inflammatory activity. Planta Med. 2017, 83, 631–635. [Google Scholar] [CrossRef] [PubMed]
- Jeziorek, M.; Damianakos, H.; Kawiak, A.; Laudy, A.E.; Zakrzewska, K.; Sykłowska-Baranek, K.; Chinou, I.; Pietrosiuk, A. Bioactive rinderol and cynoglosol isolated from Cynoglossum columnae Ten. in vitro root culture. Ind. Crop. Prod. 2019, 137, 446–452. [Google Scholar] [CrossRef]
- Naliwajski, M.R.; Wileńska, B.; Misicka, A.; Pietrosiuk, A.; Sykłowska-Baranek, K. HPLC-PDA-ESI-HRMS-based profiling of secondary metabolites of Rindera graeca anatomical and hairy roots treated with drought and cold stress. Cells 2022, 11, 931. [Google Scholar] [CrossRef]
- Perić, M.; Dmitrović, S.; Živković, S.; Filipović, B.; Skorić, M.; Simonović, A.; Todorović, S. In vitro growth, morphogenesis, and acclimatization of endangered Rindera umbellata (Waldst. & Kit.) Bunge. HortScience 2012, 47, 1123–1128. [Google Scholar] [CrossRef]
- Aliloo, A.A.; Darabinejad, S. Evaluation of different techniques for breaking seed dormancy of Heliotropium europaeum L. (Boraginaceae). J. Biol. Environ. Sci. 2013, 7, 87–91. [Google Scholar]
- Sollenberger, D.; Thomas, C.; Egerton-Warburton, L. Germination and propagation of Lithospermum canescens (Michx.) Lehm., a difficult-to-grow prairie species. Nativ. Plants J. 2022, 23, 309–323. [Google Scholar] [CrossRef]
- Rout, G.R.; Samantaray, S.; Das, P. In vitro manipulation and propagation of medicinal plants. Biotechnol. Adv. 2000, 18, 91–120. [Google Scholar] [CrossRef]
- Jiang, B.; Yang, Y.-G.G.; Guo, Y.-M.M.; Guo, Z.-C.C.; Chen, Y.-Z.Z.; Bo, J.; Yang, Y.-G.G.; Guo, Y.-M.M.; Guo, Z.-C.C.; Chen, Y.-Z.Z. Thidiazuron-induced in vitro shoot organogenesis of the medicinal plant Arnebia euchroma (Royle) Johnst. Vitr. Cell. Dev. Biol.-Plant 2005, 41, 677–681. [Google Scholar] [CrossRef]
- Malik, S.; Sharma, S.; Sharma, M.; Ahuja, P.S. Direct shoot regeneration from intact leaves of Arnebia euchroma (Royle) Johnston using thidiazuron. Cell Biol. Int. 2010, 34, 537–542. [Google Scholar] [CrossRef] [PubMed]
- Turker, A.U.; Yildirim, A.B.; Taş, İ.; Ucar Turker, A.; Yildirim, A.B.; Taş, İ. In vitro adventitious plant regeneration of Echium orientale L., an endemic plant: The evaluation of biological activities and phenolic content. Indian J. Biochem. Biophys. 2018, 55, 264–272. [Google Scholar]
- Parray, J.A.; Kamili, A.N.; Jan, S.; Mir, M.Y.; Shameem, N.; Ganai, B.A.; Abd-Allah, E.F.; Hashem, A.; Alqarawi, A.A. Manipulation of plant growth regulators on phytochemical constituents and DNA protection potential of the medicinal plant Arnebia benthamii. Biomed Res. Int. 2018, 2018, 6870139. [Google Scholar] [CrossRef]
- Shekhawat, M.S.; Shekhawat, N.S. Micropropagation of Arnebia hispidissima (Lehm). DC. and production of alkannin from callus and cell suspension culture. Acta Physiol. Plant. 2011, 33, 1445–1450. [Google Scholar] [CrossRef]
- Pal, M.; Chaudhury, A. High frequency direct plant regeneration, micropropagation and shikonin induction in Arnebia hispidissima. J. Crop. Sci. Biotechnol. 2010, 13, 13–20. [Google Scholar] [CrossRef]
- Zhou, R.; Zhang, Z. Organogenesis and pigmentation in cultures of Lithospermum erythrorhizon. J. Herbs. Spices Med. Plants 1993, 1, 57–63. [Google Scholar] [CrossRef]
- Sykłowska-Baranek, K.; Pietrosiuk, A.; Dłuska, H.; Furmanowa, M. Clonal multiplication of Lithospermum canescens (Michx.) Lehm. and Onosma paniculatum (Bur. and Franch). Herba Pol. 2004, 50, 55–64. [Google Scholar]
- Upadhyay, A.; Kadam, U.S.; Chacko, P.; Karibasappa, G.S. Microsatellite and RAPD analysis of grape (Vitis spp.) accessions and identification of duplicates/misnomers in germplasm collection. Indian J. Hortic. 2010, 67, 8–15. [Google Scholar]
- Chavhan, R.L.; Sable, S.; Narwade, A.V.; Hinge, V.R.; Kalbande, B.B.; Mukherjee, A.K.; Chakrabarty, P.K.; Kadam, U.S. Multiplex molecular marker-assisted analysis of significant pathogens of cotton (Gossypium sp.). Biocatal. Agric. Biotechnol. 2023, 47, 102557. [Google Scholar] [CrossRef]
- Hinge, V.R.; Shaikh, I.M.; Chavhan, R.L.; Deshmukh, A.S.; Shelake, R.M.; Ghuge, S.A.; Dethe, A.M.; Suprasanna, P.; Kadam, U.S. Assessment of genetic diversity and volatile content of commercially grown banana (Musa spp.) cultivars. Sci. Rep. 2022, 12, 1–16. [Google Scholar] [CrossRef]
- Miguel, C.; Marum, L. An epigenetic view of plant cells cultured in vitro: Somaclonal variation and beyond. J. Exp. Bot. 2011, 62, 3713–3725. [Google Scholar] [CrossRef]
- Wang, Q.W.L.; Wang, L. An evolutionary view of plant tissue culture: Somaclonal variation and selection. Plant Cell Rep. 2012, 31, 1535–1547. [Google Scholar] [CrossRef] [PubMed]
- dos Santos Ferreira, M.; de Jesus Rocha, A.; dos Santos Nascimento, F.; dos Santos Oliveira, W.D.; da Silva Soares, J.M.; Rebouças, T.A.; Morais Lino, L.S.; Haddad, F.; Ferreira, C.F.; Santos-Serejo, J.A.; et al. The role of somaclonal variation in plant genetic improvement: A systematic review. Agronomy 2023, 13, 730. [Google Scholar] [CrossRef]
- Krishna, H.; Alizadeh, M.; Singh, D. Somaclonal variations and their applications in horticultural crops improvement. 3 Biotech 2016, 6, 1–18. [Google Scholar] [CrossRef]
- Duta-Cornescu, G.; Constantin, N.; Pojoga, D.M.; Nicuta, D.; Simon-Gruita, A. Somaclonal variation—Advantage or disadvantage in micropropagation of the medicinal plants. Int. J. Mol. Sci. 2023, 24, 838. [Google Scholar] [CrossRef]
- Altamura, M.M. Agrobacterium rhizogenes rolB and rolD genes: Regulation and involvement in plant development. Plant Cell. Tissue Organ Cult. 2004, 77, 89–101. [Google Scholar] [CrossRef]
- Gantait, S.; Mukherjee, E. Hairy root culture technology: Applications, constraints and prospect. Appl. Microbiol. Biotechnol. 2021, 105, 35–53. [Google Scholar] [CrossRef]
- Roychowdhury, D.; Majumder, A.; Jha, S. Agrobacterium rhizogenes -mediated transformation in medicinal plants: Prospects and challenges. In Biotechnology for Medicinal Plants; Chandra, S., Lata, H.A.V., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 29–68. ISBN 9783642299742. [Google Scholar]
- Schmülling, T.; Schell, J.; Spena, A. Single genes from Agrobacterium rhizogenes influence plant development. EMBO J. 1988, 7, 2621–2629. [Google Scholar] [CrossRef]
- Mauro, M.L.; Costantino, P.; Bettini, P.P. The never ending story of rol genes: A century after. Plant Cell. Tissue Organ. Cult. 2017, 131, 201–212. [Google Scholar] [CrossRef]
- De-Eknamkul, W.; Ellis, B.E. Rosmarinic acid production and growth characteristics of Anchusa officinalis cell suspension cultures. Planta Med. 1984, 50, 346–350. [Google Scholar] [CrossRef]
- Mizukami, H.; Ogawa, T.; Ohashi, H.; Ellis, B.E. Induction of rosmarinic acid biosynthesis in Lithospermum erythorhizon cell suspension cultures by yeast extract. Plant Cell Rep. 1992, 11, 480–483. [Google Scholar] [CrossRef]
- Mizukami, H.; Tabira, Y.; Ellis, B.E. Methyl jasmonate-induced rosmarinic acid biosynthesis in Lithospermum erythrorhizon cell suspension cultures. Plant Cell Rep. 1993, 12, 706–799. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, H.; Zhao, P.; Yazaki, K.; Inoue, K. Regulation of lithospermic acid B and shikonin production in Lithospermum erythrorhizon cell suspension cultures. Chem. Pharm. Bull. 2002, 50, 1086–1090. [Google Scholar] [CrossRef]
- Mehrabani, M.; Shams-Ardakani, M.; Ghannadi, A.; Ghassemi, N.; Sajjadi-Jazi, S.E. Production of rosmarinic acid in Echium amoenum Fisch. and C.A. Mey cell cultures. Iran. J. Pharm. Res. 2005, 2, 111–115. [Google Scholar]
- Inyushkina, Y.V.; Bulgakov, V.P.; Veselova, M.V.; Bryukhanov, V.M.; Zverev, Y.F.; Lampatov, V.V.; Azarova, O.V.; Tchernoded, G.K.; Fedoreyev, S.A.; Zhuravlev, Y.N. High rabdosiin and rosmarinic acid production in Eritrichium sericeum callus cultures and the effect of the calli on masugi-nephritis in rats. Biosci. Biotechnol. Biochem. 2007, 71, 1286–1293. [Google Scholar] [CrossRef] [PubMed]
- Bryukhanov, V.M.; Bulgakov, V.P.; Zverev, Y.F.; Fedoreev, S.A.; Lampartov, V.; Veselova, M.V.; Azarova, O.V.; Zyablova, O.N.; Inyushkina, Y.V. An Eritrichium sericeum Lehm. (Boraginaceae) cell culture—A source of polyphenol compounds with pharmacological activity. Pharm. Chem. J. 2008, 42, 344–347. [Google Scholar] [CrossRef]
- Fedoreyev, S.A.; Inyushkina, Y.V.; Bulgakov, V.P.; Veselova, M.V.; Tchernoded, G.K.; Gerasimenko, A.V.; Zhuravlev, Y.N. Production of allantoin, rabdosiin and rosmarinic acid in callus cultures of the seacoastal plant Mertensia maritima (Boraginaceae). Plant Cell Tissue Organ. Cult. 2012, 110, 183–188. [Google Scholar] [CrossRef]
- Ogata, A.; Tsuruga, A.; Matsuno, M.; Mizukami, H. Elicitor-induced rosmarinic acid biosynthesis in Lithospermum erythrorhizon cell suspension cultures: Ctivities of rosmarinic acid synthase and the final two cytochrome P450-catalyzed hydroxylations. Plant Biotechnol. 2004, 21, 393–396. [Google Scholar] [CrossRef]
- Fedoreyev, S.A.; Veselova, M.V.; Krivoschekova, O.E.; Mischenko, N.P.; Denisenko, V.A.; Dmitrenok, P.S.; Glazunov, V.P.; Bulgakov, V.P.; Tchernoded, G.K.; Zhuravlev, Y.N. Caffeic acid metabolites from Eritrichium sericeum cell cultures. Planta Med. 2005, 71, 446–451. [Google Scholar] [CrossRef]
- Yamamoto, H.; Inoue, K.; Yazaki, K. Caffeic acid oligomers in Lithospermum erythrorhizon cell suspension cultures. Phytochemistry 2000, 53, 651–657. [Google Scholar] [CrossRef]
- Weremczuk-Jeżyna, I.; Kuźma, Ł.; Kiss, A.K.; Grzegorczyk-Karolak, I. Effect of cytokinins on shoots proliferation and rosmarinic and salvianolic acid B production in shoot culture of Dracocephalum forrestii W. W. Smith. Acta Physiol. Plant. 2018, 40, 189. [Google Scholar] [CrossRef]
- Weremczuk-Jeżyna, I.; Kochan, E.; Szymczyk, P.; Lisiecki, P.; Kuźma, Ł. The antioxidant and antimicrobial properties of phenol-rich extracts of Dracocephalum forrestii W. W. Smith shoot cultures grown in the nutrient sprinkle bioreactor. Phytochem. Lett. 2019, 30, 254–260. [Google Scholar] [CrossRef]
- Weremczuk-Jeżyna, I.; Skała, E.; Kuźma, Ł.; Kiss, A.K.; Grzegorczyk-Karolak, I. The effect of purine-type cytokinin on the proliferation and production of phenolic compounds in transformed shoots of Dracocephalum forrestii. J. Biotechnol. 2019, 306, 125–136. [Google Scholar] [CrossRef]
- Weremczuk-Jeżyna, I.; Skała, E.; Olszewska, M.A.; Kiss, A.K.; Balcerczak, E.; Wysokińska, H.; Kicel, A. The identification and quantitative determination of rosmarinic acid and salvianolic acid B in hairy root cultures of Dracocephalum forrestii W.W. Smith. Ind. Crops Prod. 2016, 91, 125–131. [Google Scholar] [CrossRef]
- Xiao, Y.; Gao, S.; Di, P.; Chen, J.; Chen, W.; Zhang, L. Lithospermic acid B is more responsive to silver ions (Ag+) than rosmarinic acid in Salvia miltiorrhiza hairy root cultures. Biosci. Rep. 2010, 30, 33–40. [Google Scholar] [CrossRef]
- Fujita, Y.; Hara, Y.; Ogino, T.; Suga, C. Production of shikonin derivatives by cell suspension cultures of Lithospermum erythrorhizon—I. Effects of nitrogen sources on the production of shikonin derivatives. Plant Cell Rep. 1981, 1, 59–60. [Google Scholar] [CrossRef]
- Talano, M.A.; Wevar Oller, A.L.; Gonzalez, P.S.; Agostini, E. Hairy roots, their multiple applications and recent patents. Recent Pat. Biotechnol. 2012, 6, 115–133. [Google Scholar] [CrossRef]
- Fattahi, M.; Nazeri, V.; Torras-Claveria, L.; Sefidkon, F.; Cusido, R.M.; Zamani, Z.; Palazon, J. A new biotechnological source of rosmarinic acid and surface flavonoids: Hairy root cultures of Dracocephalum kotschyi Boiss. Ind. Crops Prod. 2013, 50, 256–263. [Google Scholar] [CrossRef]
- Antolovich, M.; Prenzler, P.; Patsalides, E.; McDonald, S.; Robards, K. Methods for testing antioxidant activity. Analyst 2002, 127, 183–198. [Google Scholar] [CrossRef]
- Sykłowska-Baranek, K.; Pietrosiuk, A.; Kuźma, Ł.; Chinou, I.; Kongel, M.; Jeziorek, M. Establishment of Rindera graeca transgenic root culture as a source of shikonin derivatives. Planta Med. 2008, 74, 1161. [Google Scholar] [CrossRef]
- Assimopoulou, A.N.; Papageorgiou, V.P. Radical scavenging activity of Alkanna tinctoria root extracts and their main constituents, hydroxynaphthoquinones. Phyther. Res. 2005, 19, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Assimopoulou, A.N.; Boskou, D.; Papageorgiou, V.P. Antioxidant activities of alkannin, shikonin and Alkanna tinctoria root extracts in oil substrates. Food Chem. 2004, 87, 433–438. [Google Scholar] [CrossRef]
- Ordoudi, S.A.; Tsermentseli, S.K.; Nenadis, N.; Assimopoulou, A.N.; Tsimidou, M.Z.; Papageorgiou, V.P. Structure-radical scavenging activity relationship of alkannin/shikonin derivatives. Food Chem. 2011, 124, 171–176. [Google Scholar] [CrossRef]
- Krolicka, A.; Szpitter, A.; Maciag, M.; Biskup, E.; Gilgenast, E.; Romanik, G.; Kaminski, M.; Wegrzyn, G.; Lojkowska, E. Antibacterial and antioxidant activity of the secondary metabolites from in vitro cultures of the Alice sundew (Drosera aliciae). Biotechnol. Appl. Biochem. 2009, 53, 175–184. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Gupta, P.K.; Durzan, D.J. Shoot multiplication from mature trees of Douglas-fir (Pseudotsuga menziesii) and sugar pine (Pinus lambertiana). Plant Cell Rep. 1985, 4, 177–179. [Google Scholar] [CrossRef]
- Pietrosiuk, A.; Sykłowska-Baranek, K.; Wiedenfeld, H.; Wolinowska, R.; Furmanowa, M.; Jaroszyk, E. The shikonin derivatives and pyrrolizidine alkaloids in hairy root cultures of Lithospermum canescens (Michx.) Lehm. Plant Cell Rep. 2006, 25, 1052–1058. [Google Scholar] [CrossRef]
- Linsmaier, E.M.; Skoog, F. Organic growth factor requirements of tobacco tissue cultures. Physiol. Plant 1965, 18, 100–127. [Google Scholar] [CrossRef]
- Schenk, R.O.Y.U.; Hildebrandt, A.C. Medium and techniques for induction and growth of monocotyledonous and dicotyledonous plant cell cultures. Can. J. Bot. 1972, 50, 199–204. [Google Scholar] [CrossRef]
- Aoyama, T.; Hirayama, T.; Tamamoto, S.; Oka, A. Putative start codon TTG for the regulatory protein VirG of the hairy-root-inducing plasmid pRiA4. Gene 1989, 78, 173–178. [Google Scholar] [CrossRef]
- Sidwa-Gorycka, M.K.; Krolicka, A.; Orlita, A.; Malinski, E.; Golebiowski, M.; Kumirska, J.; Chromik, A.; Biskup, E.; Stepnowski, P.; Lojkowska, E. Genetic transformation of Ruta graveolens L. by Agrobacterium rhizogenes: Hairy root cultures a promising approach for production of coumarins and furanocoumarins. Plant Cell Tissue Organ 2009, 97, 59. [Google Scholar] [CrossRef]
- Collard, B.C.Y.; Mackill, D.J. Start Codon Targeted (SCoT) Polymorphism: A simple, novel DNA marker technique for generating gene-targeted markers in plants. Plant Mol. Biol. 2009, 27, 86–93. [Google Scholar] [CrossRef]
- Pirttilä, A.M.; Hirsikorpi, M.; Kämäräinen, T.; Jaakola, L.; Hohtola, A. m isolation methods for medicinal and aromatic plants. Plant Mol. Biol. Rep. 2001, 19, 273. [Google Scholar] [CrossRef]
- Pavlicek, A.; Hrda, S.; Flegr, J.; Pavlíček, A.; Hrdá, Š.; Flegr, J. FreeTree—Freeware program for construction of phylogenetic trees on the basis of distance data and bootstrap/jackknife analysis of the tree robustness. Aplication in the RAPD analysis of genus Frenkelia. Folia Biol. 1999, 45, 97–99. [Google Scholar]
- Pękal, A.; Pyrzynska, K. Evaluation of aluminium complexation reaction for flavonoid content assay. Food Anal. Methods 2014, 7, 1776–1782. [Google Scholar] [CrossRef]
- Fukumoto, L.R.; Mazza, G. Assessing antioxidant and prooxidant activities of phenolic compounds. J. Agric. Food Chem. 2000, 48, 3597–3604. [Google Scholar] [CrossRef]
- Benzie, I.F.F.; Strain, J.J. The ferric of plasma (FRAP) as measure of “antioxidant power”: The FRAP assay. Anal. Chem. 1996, 239, 70–76. [Google Scholar] [CrossRef]
- Bolanos de la Torre, A.A.; Henderson, T.; Nigam, P.S.; Owusu-Apenten, R.K. A universally calibrated microplate ferric reducing antioxidant power (FRAP) assay for foods and applications to Manuka honey. Food Chem. 2015, 174, 119–123. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).