Abstract
Rutaceae plants are known for being a rich source of coumarins. Preliminary molecular docking showed that there was no significant difference for coumarins in Clausena and Murraya, both of which had high scoring values and showed good potential inhibitory activity to the MAO-B enzyme. Overall, 32 coumarins were isolated from Murraya exotica L., including a new coumarin 5-demethoxy-10′-ethoxyexotimarin F (1). Their structures were elucidated on the basis of a comprehensive analysis of 1D and 2D NMR and HRMS spectroscopic data, and the absolute configurations were assigned via a comparison of the specific rotations and the ECD exciton coupling method. The potential of new coumarin (1) as a selective inhibitor of MAO-B was initially evaluated through molecular docking and pharmacophore studies. Compound (1) showed selectivity for the MAO-B isoenzyme and inhibitory activity in the sub-micromolar range with an IC50 value of 153.25 ± 1.58 nM (MAO-B selectivity index > 172).
1. Introduction
Depression, Alzheimer’s disease, and Parkinson’s disease, as three major neuropsychiatric diseases, have seriously affected human health and quality of life. Monoamine oxidase (MAO, EC 1.4.3.4) is a flavoenzyme bound to the mitochondrial outer membranes of the cells, which is responsible for the oxidative deamination of neurotransmitters and dietary amines. It exists in two isoforms, MAO-A and MAO-B. Although sharing 70% sequence identity, MAO-A and B displayed different substrate and inhibitor specificities; serotonin and norepinephrine are preferentially metabolized by MAO-A and phenylethylamine, benzylamine, and dopamine by MAO-B, whereas clorgyline and L-deprenyl selectively inhibit MAO-A and B, respectively. They are the well-known target for antidepressants, Parkinson’s disease, and neuroprotective drugs [1,2,3,4]. Monoamine oxidase inhibitors (MAOIs) were used for the treatment of various neurodegenerative disorders. However, they are associated with serious side effects and lack efficacy and selectivity for a single MAO isoform. It is commonly accepted that the discovery of MAOIs from herbal sources is an important strategy for drug design and development to treat various neurodegenerative diseases such as depression, anxiety, Parkinson’s disease, and Alzheimer’s disease [5].
Plant-derived coumarins are an important class of compounds due to their significant biological activities. Coumarins are abundantly found in species of the Rutaceae family, such as the genus Clausena and Murraya [6,7,8,9,10,11,12]. Our previous studies obtained a novel coumarin, anisucoumaramide, from Clausena anisum-olens, which was found to exhibit high selectivity and inhibitory activity in the nanomolar range against MAO-B with an IC50 value of 143.65 ± 0.90 nm [6]. Therefore, we hope to continue to obtain novel coumarins with similar activity from Rutaceae plants. Murraya exotica (Rutaceae) as a dwarf tree or an evergreen shrub was commonly cultivated in gardens as an ornamental plant in many tropical and subtropical regions. It is an important medicine used for treating fever, cough, and infectious wounds and eliminating pain from injury and trauma [13]. Previous studies have shown that several types of secondary coumarins in M. exotica are widely used in the medical, spice, and seasoning industries [8]. In this study, the MAO-B inhibitory potential of coumarins reported from Clausena and Murraya (Rutaceae) was predicted by molecular docking. The results showed that there was no significant difference between the two. Most of them had certain inhibitory potential, and about one-third of them had better scores. As a continuation of a search for more bioactive coumarins from Rutaceae species, 32 coumarins were isolated from Murraya exotica L., including a new coumarin 5-demethoxy-10′-ethoxyexotimarin F (1). It was preliminarily found that the new coumarin 1 may have high MAO-B selective inhibitory activity through molecular docking and pharmacophore model study. The determination of hMAO isoform activity displayed (1) showed selectivity for the MAO-B isoenzyme and inhibitory activity in the sub-micromolar range with an IC50 value of 153.25 ± 1.58 nM (MAO-B selectivity index > 172), of 26.3 ± 1.03 μM to the MAO-A.
2. Results and Discussion
2.1. Molecular Docking of Coumarins in Clausena and Murraya
Through a previous literature collection, 141 known coumarins were found in Clausena plants and 177 coumarins found in Murraya plants. Molecular docking studies on coumarins from Clausena and Murraya were carried out, and the potential inhibitory activity of monoamine oxidase B of the two was compared. The docking results are shown in the following table (Table 1). A negative scoring value indicated that they could bind to the receptor protein. A smaller scoring value indicated better binding [14]. The results showed that only the compounds had better scores, those with scores below −85. The preliminary screening results showed that 40 of the coumarins in Clausena had the better scoring value, accounting for 28.4% of the total number, and 48 of the coumarins in Murraya had the better scoring value, accounting for 27.1% of the total number. The coumarins of Murraya and Clausena have similar molecular docking scores; there was no significant difference between the two. Therefore, Murraya could be a highly valuable research resource for its coumarins that have MAO-B inhibition potential.

Table 1.
Docking results of coumarins in Clausena and Murraya.
2.2. Structure Elucidation of New Coumarin 1
The powdered leaves and twigs of Murraya exotica L. (2.5 kg) were repeatedly extracted with 95% EtOH at room temperature. The extract was then concentrated under reduced pressure to give a brown syrup, which was suspended in water and successively partitioned with petroleum ether, ethyl acetate (EtOAc) and n-butanol (n-BuOH). The EtOAc fraction was subjected to a multi-step chromatography procedure to yield 32 coumarins (Figure 1), including a new coumarin (1).
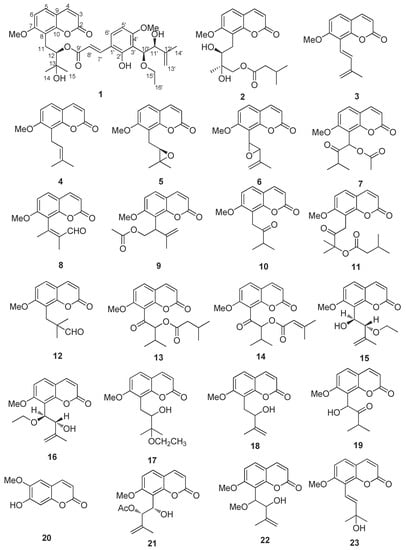
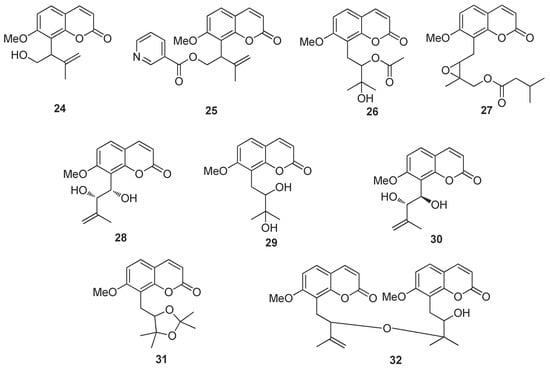
Figure 1.
The structures of coumarins from Murraya exotica L.
Compound (1) was obtained as a white solid. Its molecular formula was determined as C32H38O10 via the positive HRESIMS (m/z 605.2359 [M + Na]+, calcd for 605.2357) and 13C-NMR data, indicating 14 indices of hydrogen deficiency. Strong UV bands at λmax 322 nm showed the characteristic absorption of the coumarin skeleton [15], and the IR spectrum showed absorption bands for carbonyl (1732 cm−1) and aromatic ring (1600 cm−1 and 1465 cm−1) functionalities [15]. The 13C-NMR, DEPT, and HSQC spectra of (1) show the presence of 32 resonances comprised of 4 methyl carbons, 3 methylene carbons, 11 methylene carbons, 12 quaternary carbons and 2 methoxy carbons (see Supplementary Materials). The presence of an 8-prenylated-7-methoxycoumarin backbone as a common structural unit in (1) was further deduced by a methoxy singlet signal at δ 3.92, two sets of 1H AB doublets at δH 6.11 and 7.53 (each d, J = 9.4 Hz) and δH 7.29 and 6.79 (each d, J = 8.6 Hz), which were easily assignable to H-3 and H-4and to H-5 and H-6 on the coumarin skeleton, respectively, and a group of prenyl signals [δH 3.41 (1H, dd, J = 13.6, 10.8 Hz, H-11), 3.13 (1H, dd, J = 13.6, 2.4 Hz, H-11), 5.23 (1H, dd, J = 10.6, 2.5 Hz, H-12), 1.41 (3H, s, H-14), 1.35 (3H, s, H-15) (Table 2, see Supplementary Materials) [15,16]. The NMR spectroscopic data of (1) were similar to those of 10′-ethoxyexotimarin F [16], except that the absence of a methoxy group at C-5, which was deduced from its HMBC correlations of H-5 (δH 7.29) with C-4, C-7, C-9 and C-10 (Figure 2). The 1H-NMR spectrum exhibited a group of characteristic signals at δH 7.70, 6.28 (each 1H, d, J = 16.1 Hz), and 7.31, 6.38 (each 1H, d, J = 8.7 Hz)], confirmed the presence of a set of 2, 3, 4-trisubstituted cinnamoyl unit.

Table 2.
1H (400 MHz) and 13C (100 MHz) NMR data of (1) in CDCl3.
Figure 2.
Key 1H-1H COSY and HMBC of new compound (1).
In (1), a large coupling constant (J = 7.4 Hz) suggested the threo configuration of H-10′/H-11′ [16]. Recently, the circular dichroism exciton chirality method provided a powerful and rapid approach for assigning the absolute configuration of natural products [17]. The exciton chirality method was used to infer the configuration of compound (1) based on the sign of the excitonic couplet, using 10′-ethoxyexotimarin F as the reporter group. Thus, the absolute configuration of C-12 was deduced as R from a positive split chirality [298 nm (Δε −3.2), 344 nm (Δε +1.6)] determined from the ECD spectrum using the exciton chirality rule (Figure 3). The optical rotation of [α]26.7D −37.24 (c 0.14, MeOH) of (1) with the same negative sign of 10′-ethoxyexotimarin F supposed the same absolute configuration. Moreover, from a biosynthetic consideration, the absolute configurations of (1) were deduced to be identical to those of 10′-ethoxyexotimarin F. Thus, the absolute configuration of (1) was assigned as (12R, 10′S, 11′S). Hence, compound (1) was defined as 5-demethoxy-10′-ethoxyexotimarin F [(E)-(R)-3-hydroxy-1-(7-methoxy-2-oxo-2H-chromen-8-yl)-3-methylbutan-2-yl·3-(3-((1S, 2S)-1-ethoxy-2-hydroxy-3-methylbut-3-en-1-yl)-2-hydroxy-4-methoxyphenyl)acrylate].
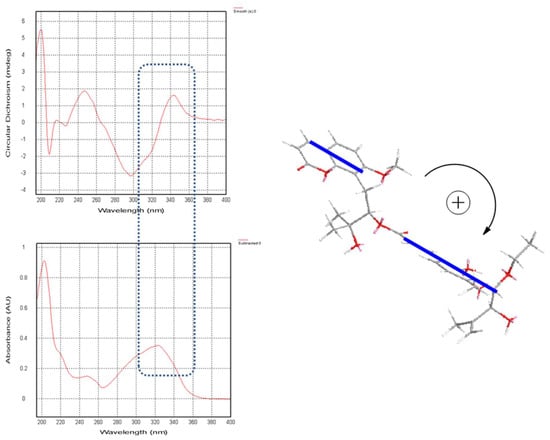
Figure 3.
CD spectrum, UV spectrum, and the exciton chirality of 1.
The 31 known coumarins (Figure 1) were identified by comparing their spectroscopic data with those reported in literature as exotimarin G (2) [7], trans-dehydroosthol (3) [7], osthol (4) [18], meranzin (5) [19], phebalosin (6) [19], hainanmurpanin (7) [20], murralonginal (8) [21], isomurralonginol acetate (9) [22], isomeranzin (10) [22], paniculonol isovalerate (11) [23], 7-methoxy-8-(2-formyl-2-methylpropyl)coumarin (12) [24], muralatin O (13) [19], isomurranganonsenecioate (14) [22], 2′-O-ethylmurrangatin (15) [25], muralatin K (16) [10], yuehgesin-C (17) [26], auraptenol (18) [22], murranganon (19) [22], scopoletin (20) [27], murrangatin acetate (21) [22], albiflorin-3 (22) [28], murraol (23) [22], isomurralonginol (24) [22], isomurralonginol nicotinate (25) [22], 2′-acetoxy-3′-dihydroxyl-osthole (26) [29], muralatin P (27) [19], murrangatin (28) [30], meranzin hydrate (29) [15], minumicrolin (30) [22], pranferin (31) [30], and murradimerin A (32) [31].
2.3. Virtual Screen of Coumarins Isolate from Murraya exotica L.
A total of 32 coumarin molecules from Murraya exotica L. were screened by molecular docking with the ligand C18_1503 in the target protein as the reference and the coumarin anisucoumaramide as the positive control. The results are shown in the following Table 3. The scores showed that nine coumarins were better, accounting for 28.1% of the total number, which was similar to the results in the previous study. In addition, the new coumarin 5-demethoxy-10′-ethoxyexotimarin F (1) had a higher scoring value and a higher docking advantage than the positive control anisucoumaramide and the original ligand C18_1503. Therefore, 2D diagrams of the interactions between the new coumarin (1), anisucoumaramide and C18_1503 and protein crystals were displayed, respectively (Figure 4). It could be found that all three had hydrogen bond interaction, spatial interaction with residue Ile 199, and hydrogen bond interaction with residue Tyr 326, which was consistent with the action reported in the literature [32,33]. Therefore, the new compound 5-demethoxy-10′-ethoxyexotimarin F (1) had a certain selective inhibition potential for MAO-B.

Table 3.
Docking scores of coumarins of Murraya exotica L.
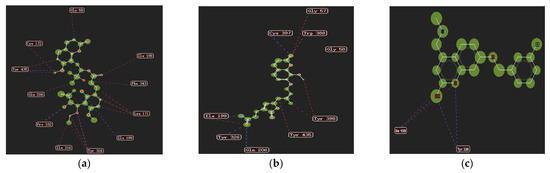
Figure 4.
Protein crystals 2D interactions with (1), anisucoumaramide, and C18_1503. (a) 5-demethoxy-10′-ethoxyexotimarin F (1); (b) anisucoumaramide; (c) C18_1503.
In the pharmacophore study of MAO-B selective inhibitor, the pharmacophore model 01 had the highest CAI value (see Table 4). Moreover, its other indicators were also high. Therefore, the pharmacophore model 01 was determined as that optimal pharmacophore. According to the obtained optimal pharmacophore, 32 coumarins isolated from Murraya exotica L. were virtually screened. The screening results are shown in Table 5. Overall, 27 molecules could be matched with the pharmacophore, indicating that the coumarins in Murraya exotica L. had high overall matching with the pharmacophore model of MAO-B selective inhibitor. The matching of each molecule was evaluated based on the Fit Value. A higher value indicated a higher degree of matching between the molecule and the pharmacophore model. As can be seen from the table, the new compound 5-demethoxy-10′-ethoxyexotimarin F (1) has a high matching property and, therefore, a high potential as a selective inhibitor of MAO-B.

Table 4.
Evaluation results of pharmacophore model *.

Table 5.
Pharmacophore model screening results.
2.4. Biological Activity of Coumarin 1
The potential inhibitory effects of the new coumarin (1) were evaluated on human recombinant monoamine oxidase (hMAO) isoforms. The inhibitory effects were assessed by measuring the production of H2O2 from p-tyramine using the Amplex Red MAO assay kit with selegiline and iproniazide as reference drugs. The IC50 values and MAO-B selectivity indices for the inhibitory effects of both the new compound and reference inhibitors were calculated (Table 6). Compound (1) inhibited MAO-B with an IC50 value of 153.25 ± 1.58 nM (MAO-B selectivity index > 172) but with an IC50 value of 26.3 ± 1.03 μM to the MAO-A, demonstrating that compound (1) shows selectivity for the MAO-B isoenzyme and inhibitory activity in the sub-micromolar range.

Table 6.
MAO-A and MAO-B inhibitory activity results.
The anti-inflammatory activity of (1) was evaluated by measuring the inhibition against LPS-induced NO production in RAW264.7 cells. Compound (1) displayed no inhibitory effect, showing only a 34.59% inhibition, less than 50%, compared with NG-monomethyl-l-arginine (l-NMMA) with an inhibition rate of 56.13%. Meanwhile, the acetylcholinesterase (AChE) and butyrylcholinesterase (BuChE) inhibitory activity of compound (1) was assayed. (1) did not show any inhibitory activity at a concentration of 50 μM. Tacrine (0.33 μM) was used as the positive control and showed 50.1% inhibition. The α-glucosidase enzymatic activity of compound (1) was investigated and found to be inactive since its percentage of inhibition was less than 50% at a concentration of 50 μM compared to 53.17 % inhibition of quercetin (positive control). Compound (1) did not lead to a significant inhibitory effect against four bacterial pathogens, Salmonella enterica (ATCC14028), Staphylococcus aureus (ATCC29213), Escherichia coli (ATCC25922), and Pseudomonas aeruginosa (ATCC27853), and fungal pathogen Candida albicans (ATCC10231) at a concentration of 100 μM.
3. Materials and Methods
3.1. General Experimental Procedures
The specific optical rotation data were acquired on a Rudolph Autopol III automatic polarimeter (Rudolph Research, Fairfield, NJ, USA). The UV spectra were recorded on a Shimadzu UV-2450 spectrophotometer (Shimadzu Corporation, Kyoto, Japan). IR spectra were recorded on a Thermo Nicolet Nexus 470 FT-IR spectrometer (Thermo Nicolet, Vernon Hills, IL, USA). The ECD data were acquired on a JASCO 810 CD spectrophotometer (Jasco Corporation, Tokyo, Japan). NMR spectra were recorded on a Brucker-400 and 600 NMR spectrometer (Bruker Corp. Billerica, MA, USA), with tetramethylsilane as an internal standard. HRESIMS experiments were measured on a Waters Xevo G2 Q-TOF mass spectrometer (Waters MS Technologies, Manchester, UK). Silica gel (100–200 mesh or 200–300 mesh, Qingdao Marine Chemical Co. Ltd., Qingdao, China) and Sephadex LH-20 (Amersham Biosciences, Uppsala, Sweden) were used for open CC. TLC analyses were carried out on the pre-coated silica gel GF254 plates (Qingdao Marine Chemical Co. Ltd., Qingdao, China). The spots were visualized under the UV lights (254 nm and 365 nm). All of the solvents were distilled prior to use.
3.2. Plant Materials
The leaves and twigs of Murraya exotica L. were collected in Hekou County, Honghe Prefecture, Yunnan Province in May 2019 and identified by Professor Chen Yu. The sample was deposited in the School of Chemical Science and Technology, Yunnan University.
3.3. Extraction and Isolation
Dried powdered leaves and twigs of Murraya exotica L. (2.5 kg) were extracted three times with 95% aqueous EtOH. The extract was evaporated under reduced pressure, and the residue (500 g) was suspended in H2O and partitioned successively with petroleum ether, EtOAc, and n-BuOH. The EtOAc extract (100 g) was subjected to silica gel CC and eluted with a stepwise gradient of petroleum ether-EtOAc (7:3, 3:2, 2:3, 1:4, and 0:1, v/v) to afford five fractions (F1–5), respectively. Then each part was subjected to a series of chromatographic techniques, such as silica gel column (mesh 200–300) and Sephadex LH-20. Fraction 1 (25 g) was separated on silica gel CC eluting with petroleum ether-EtOAc (4:1, v/v) and Sephadex LH-20 (CH2Cl2-MeOH, 1:1, v/v) to afford (3) (15.1 mg), (4) (3.5 mg), (5) (5.2 mg), (6) (10.1 mg), (7) (99.5 mg), (8) (11.4 mg), (9) (36 mg), (10) (16.0 mg), (11) (4.5 mg), (12) (5.5 mg). Fraction 2 (20 g) was chromatographed successively on silica gel CC and Sephadex LH-20 (CH2Cl2-MeOH, 1:1, v/v) to afford (13) (5.5 mg), (14) (10.3 mg), (15) (18 mg), (16) (5.4 mg), (17) (25.5 mg), (18) (7.7 mg), (19) (19.0 mg), (20) (7.1 mg). Fraction 3 (25 g) was chromatographed successively on silica gel CC and Sephadex LH-20 (CH2Cl2-MeOH, 1:1, v/v) to afford (1) (3 mg), (2) (5.3 mg), (21) (9.2 mg), (22) (1.5 mg), (23) (9.5 mg), (24) (5.0 mg), (25) (9.2 mg), (26) (3.1 mg), (27) (6.4 mg), (28) (18.0 mg), (29) (4.5 mg), (32) (12.1 mg). Fraction 4 (14 g) was chromatographed on silica gel CC eluting with petroleum ether-acetone (3:2, v/v) and Sephadex LH-20 (MeOH) to afford (30) (5 mg). Fraction 5 (16 g) was chromatographed on silica gel CC eluting with CH2Cl2-acetone (4:1, v/v) and Sephadex LH-20 (MeOH) to afford (31) (51.0 mg).
3.4. Structural Elucidation
5-demethoxy-10′-ethoxyexotimarin F (1): White solid; [α]26.7D -37.24 (c0.14, MeOH); UV (MeOH) λmax198, 246 and 322 nm; IR (KBr) νmax 3380, 2925, 2855, 1732, 1724, 1600, 1465 cm−1; 1H and 13C NMR data see Table 2; HRESIMS m/z 605.2359 [M + Na]+ (calcd for C32H37O10, 605.2357).
3.5. Molecular Docking
Molegro Virtual Docker 6.0 (MVD 6.0, 2013, Molexus, Odder, Denmark) semi-flexible docking software is mainly used, which is higher than the docking accuracy of most software on the market. The key is that it comes with a number of different scoring functions and search algorithms. The combination of the two can obtain the docking algorithm for the receptor protein. After repeated attempts using different scoring functions and search algorithms in the software, a docking method with good repeatability was finally obtained [34]. The RMSD value when reproducing the original protein-ligand was 1.066, less than 2, indicating that the docking method was available. In the finally determined docking method, the PLANTS Score [GRID] is used for the scoring function, and Iterated Simplex is used for the search algorithm. In this study [35], the protein crystal 2V61 obtained from the PDB database was combined with the selective inhibitor 7-(3-chlorobenzyloxy)-4-(methylamino) methyl-coumarin, providing more reference value for the docking analysis of coumarin compounds [36].
3.6. Pharmacophore Model
The pharmacophore model was constructed based on the common pharmacodynamic characteristics [37], and the active compounds related to MAO-B selective inhibitor were obtained by using the protein code P27338 of protein crystal 2V61 and the Binding Data Base database. The training set consisted of 17 active compounds. The specific structure and IC50 activity values are shown in Figure 5. The test set consisted of 50 active compounds and 100 inactive compounds [38,39,40,41]. Pharmacophore model evaluation is based on the CAI system, and the evaluation results are shown in Table 4 [42,43,44].
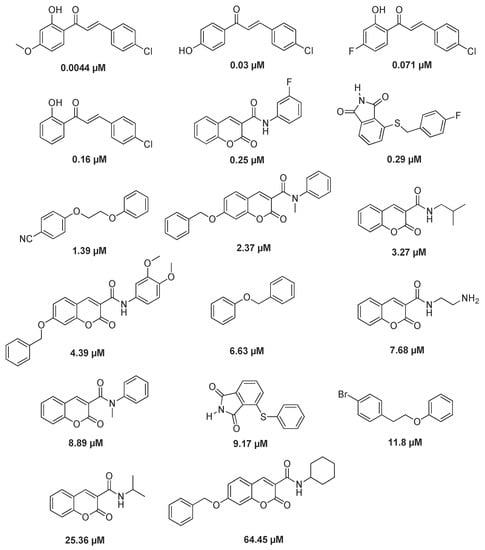
Figure 5.
Training set compounds and their activity value.
3.7. In Vitro Bioassay
3.7.1. In Vitro MAO Inhibitory Assay
The effects of (1) on the hMAO isoform enzymatic activity were evaluated by measuring the effects on the production of H2O2 from p-tyramine using a fluorimetric method following the experimental protocol previously described [6]. Selegiline and iproniazide served as reference inhibitors.
3.7.2. Anti-Inflammatory Assay
The Nitric oxide (NO) production inhibition of the compounds (1) was determined using a procedure described before [45].
3.7.3. Acetylcholinesterase/Butrylcholinesterase Inhibitory Activity
Acetylcholinesterase/butyrylcholinesterase (AChE/BuChE) inhibitory activity of the isolated compound (1) was assayed by the spectrophotometric method developed by Ellman’s method with slight modification [46]. S-Acetylthiocholine iodide, S-butyrylthiocholineiodide, 5,5′-dithio-bis-(2-nitrobenzoic) acid (DTNB, Ellman’s reagent), acetylcholinesterase and butyrylcholinesterase derived from human erythrocytes were purchased from Sigma Chemical (St. Louis, MO, USA). The compounds were dissolved in DMSO. The reaction mixture (totally 200 μL) containing phosphate buffer (pH 8.0), test compound (50 μM), and acetyl cholinesterase (0.02 U/mL) or butyrylcholinesterase (0.016 U/mL), was incubated for 20 min (37 °C). Then, the reaction was initiated by the addition of 40 μL of the solution containing DTNB (0.625 mM) and acetylthiocholine iodide (0.625 mM) or butyrylthiocholine iodide (0.625 mM) for AChE or BuChE inhibitory activity assay, respectively. The hydrolysis of acetylthiocholineorbutyrylthiocholine was monitored at 405 nm every 30 s for one hour. Tacrine was used as a positive control with a final concentration of 0.333 μM. All of the reactions were performed in triplicate. The percentage inhibition was calculated as follows: % inhibition = (E − S)/E × 100 (E is the activity of the enzyme without test compound, and S is the activity of the enzyme with test compound).
3.7.4. α-Glucosidase Inhibition Assay
An enzyme inhibitor screening model was chosen using 4-nitro-phenol-α-d-glucopyranoside (4-NPGP) and slightly modified [47]. Briefly, the test compound (50 μM), α-glucosidase solution (0.025 U/mL), phosphate buffer (pH 6.9), and (4-NPGP) (1 μM) were incubated in 96-well plates at 37 °C for 50 min. Absorbance at 405 nm was recorded on a microplate reader. Blank readings (no enzyme) were subtracted from each well, and the results were compared to the control. Quercetin was selected as the positive control. All of the reactions were repeated three times. The inhibition rate (%) was calculated as (1 − ODsample)/ODcontrol blank × 100%.
3.7.5. Antimicrobial Assay
The broth dilution method and Oxford cup method were used to detect antimicrobial activity against reference strains, including Salmonella enterica subsp. enterica (ATCC14028), Staphylococcus aureus subsp. aureus (ATCC29213), Escherichia coli (ATCC25922), Pseudomonas aeruginosa (ATCC27853), and Candida albicans (ATCC10231). Ceftazidime and penicillin G sodium and amphotericin B served as an antibacterial and antifungal positive control, respectively. The percentage inhibition of cell growth below 50% was regarded as inactive.
4. Conclusions
A new coumarin 5-demethoxy-10′-ethoxyexotimarin F (1) was obtained from Murraya exotica L. (1) could be a new potential MAO-B selective inhibitor, which showed better than the known positive control coumarin anisucoumaramide and the original ligand reference C18_1503 through molecular docking and pharmacophore model evaluation. Compound (1) showed selectivity for the MAO-B isoenzyme and inhibitory activity in the sub-micromolar range with an IC50 value of 153.25 ± 1.58 nM (MAO-B selectivity index > 172), of 26.3 ± 1.03 μM to the MAO-A. The exploration and discovery of bioactive components from medicinal herbs is one of the most important approaches for developing new drugs and improving the therapeutic properties in drug discovery. This study enriches the chemical diversity of coumarins in Murraya species and provides a theoretical basis for the traditional usage of Murraya exotica L. The results encourage us to further explore the potential of this family of derivatives as potential lead candidates for the treatment of neurodegenerative disorders.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/molecules27154950/s1, Figure S1. 1H NMR spectrum of 1 in CDCl3; Figure S2. 13C NMR and DEPT spectra of 1 in CDCl3; Figure S3. HSQC spectrum of 1 in CDCl3; Figure S4. HMBC spectrum of 1 in CDCl3; Figure S5. 1H-1H COSY spectrum of 1 in CDCl3; Figure S6. ROESY spectrum of 1 in CDCl3; Figure S7. HRESIMS spectrum of1; Figure S8. UV spectrum of 1; Figure S9. CD spectrum of 1.
Author Contributions
Conceived and designed the experiments, Y.-S.W. and J.-H.Y.; plant collection, Y.-S.W.; molecular docking, Z.-R.X.-H. and T.Y.; experiments, Y.-F.M., X.-F.F. and Y.-Y.Z.; data curation, Z.-R.X.-H.; writing—original draft preparation, Z.-R.X.-H., X.-F.F. and J.-H.Y.; writing—review and editing, Y.-S.W. and J.-H.Y.; supervision, Y.-S.W. and J.-H.Y.; project administration, Y.-Y.Z. and J.P.; funding acquisition, Y.-S.W. and J.-H.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by National Nature Science Foundation of China (No. 81960629, 82160661, and 21662040) and the Program for Yunnan Innovative Research Team (No. 202005AE160005).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
Sample Availability
Samples of the compounds are not available from the authors.
References
- Shih, J.C. Molecular basis of human MAO A and B. Neuropsychopharmacology 1991, 4, 1–7. [Google Scholar] [PubMed]
- Schwartz, T.L. A neuroscientific update on monoamine oxidase and its inhibitors. CNS Spectrums. 2013, 18 (Suppl. 1), 25–33. [Google Scholar] [CrossRef] [PubMed]
- Bonanno, L.; Ciurleo, R.; Marino, S.; Ruvolo, C.; Morabito, R.; Bramanti, A.; Corallo, F. Effect of MAO-B Inhibitors on Neurometabolic Profile of Patients Affected by Parkinson Disease: A Proton Magnetic Resonance Spectroscopy Study. J Clin. Med. 2022, 11, 1931. [Google Scholar] [CrossRef] [PubMed]
- Al Mamun, A.; Uddin, M.S. KDS2010: A Potent Highly Selective and Reversible MAO-B Inhibitor for Alzheimer’s Disease. Comb. Chem. High T. Scr. 2020, 23, 836–841. [Google Scholar] [CrossRef]
- Dhiman, P.; Malik, N.; Khatkar, A. 3D-QSAR and in-silico Studies of Natural Products and Related Derivatives as Monoamine Oxidase Inhibitors. Curr. Neuropharmacol. 2018, 16, 881–900. [Google Scholar] [CrossRef]
- Wang, Y.S.; Li, B.T.; Liu, S.X.; Wen, Z.Q.; Yang, J.H.; Zhang, H.B.; Hao, X.J. Anisucoumaramide, a Bioactive Coumarin from Clausena anisum-olens. J. Nat. Prod. 2017, 80, 798–804. [Google Scholar] [CrossRef]
- Liu, B.Y.; Zhang, C.; Zeng, K.W.; Li, J.; Guo, X.Y.; Zhao, M.B.; Tu, P.F.; Jiang, Y. Anti-inflammatory prenylated phenylpropenols and coumarin derivatives from Murraya exotica. J. Nat. Prod. 2018, 81, 22–33. [Google Scholar] [CrossRef]
- Liang, H.; Shi, Y.; Zeng, K.; Zhao, M.; Tu, P.; Jiang, Y. Coumarin derivatives from the leaves and twigs of Murraya exotica L. and their anti-inflammatory activities. Phytochemistry. 2020, 177, 112416. [Google Scholar] [CrossRef]
- Huang, L.; Feng, Z.L.; Wang, Y.T.; Lin, L.G. Anticancer carbazole alkaloids and coumarins from Clausena plants: A review. Chin. J. Nat. Med. 2017, 15, 881–888. [Google Scholar] [CrossRef]
- Lv, H.N.; Wang, S.; Zeng, K.W.; Li, J.; Guo, X.Y.; Ferreira, D.; Zjawiony, J.K.; Tu, P.F.; Jiang, Y. Anti-inflammatory coumarin and benzocoumarin derivatives from Murraya alata. J. Nat. Prod. 2015, 78, 279–285. [Google Scholar] [CrossRef]
- Wang, Y.S.; Huang, R.; Li, N.Z.; Yang, J.H. Coumarins from Clausena anisum-olens Merr. Biosci. Biotech. Bioch. 2010, 74, 1483–1484. [Google Scholar] [CrossRef]
- Takemura, Y.; Nakamura, K.; Hirusawa, T.; Ju-ichi, M.; Ito, C.; Furukawa, H. Four new furanone-coumarins from Clausena excavata. Chem. Pharm. Bull. 2000, 48, 582–584. [Google Scholar] [CrossRef]
- Zhang, D.X.; Hartley, T.G. Murraya J. Koenig ex Linnaeus. In Flora of China; Wu, Z.Y., Raven, P.H., Hong, D.Y., Eds.; Science Press: Beijing, China, 2008; Volume 11, pp. 85–87. [Google Scholar]
- Xu, S.K.; Peng, H.; Wang, N.; Zhao, M. Inhibition of TNF-α and IL-1 by compounds from selected plants for rheumatoid arthritis therapy: In vivo and in silico studies. Trop. J. Pharm. Res. 2018, 17, 277–285. [Google Scholar] [CrossRef]
- Raj, K.; Misra, S.C.; Kapil, R.S.; Popli, S.P. Coumarins from Murraya paniculata. Phytochemistry 1976, 15, 1787. [Google Scholar] [CrossRef]
- Wang, X.T.; Liang, H.Z.; Zeng, K.W.; Zhao, M.B.; Tu, P.F.; Li, J.; Jiang, Y. Panitins AG: Coumarin derivatives from Murraya paniculata from Guangxi Province, China shows variable NO inhibitory activity. Phytochemistry 2019, 162, 224–231. [Google Scholar] [CrossRef]
- Pescitelli, G. For a Correct Application of the CD Exciton Chirality Method: The Case of Laucysteinamide A. Mar. Drugs. 2018, 16, 388. [Google Scholar] [CrossRef]
- Steck, W. Paniculatin, a New Coumarin from Murraya paniculata (L.) Jack. Can. J. Chem. 1972, 50, 443–445. [Google Scholar] [CrossRef]
- You, C.X.; Guo, S.S.; Geng, Z.F.; Zhang, W.J.; Liang, J.Y.; Zhang, Z.; Wang, C.F.; Du, S.S.; Deng, Z.W. Repellent activity of compounds from Murraya alata Drake against Tribolium castaneum. Ind. Crop. Prod. 2017, 95, 460–466. [Google Scholar] [CrossRef]
- Yang, J.S.; Su, Y.L. Studeis on the constituents of Murraya paniculata (L.) Jack. Acta. Pharm. Sin. B. 1983, 18, 760–765. [Google Scholar]
- Saied, S.; Nizami, S.S.; Anis, I. Two new coumarins from Murraya paniculata. J. Asian. Nat. Prod. Res. 2008, 10, 515–519. [Google Scholar] [CrossRef]
- Ito, C.; Furukawa, H. Constituents of Murraya exotica L. Structure elucidation of new coumarins. Chem Pharm Bull. 1987, 35, 4277–4285. [Google Scholar] [CrossRef]
- Ito, C.; Furukawa, H. Two new coumarins from Murraya plants. Chem. Pharm. Bull. 1989, 37, 819–820. [Google Scholar] [CrossRef]
- Imai, F.; Kinoshita, T.; Sankawa, U. Constituents of the leaves of Murraya paniculata collected in Taiwan. Chem Pharm Bull. 1989, 37, 358–362. [Google Scholar] [CrossRef]
- Choudhary, M.I.; Azizuddin; Khalid, A.; Sultani, S.Z.; Atta-ur-Rahman. A New Coumarin from Murraya paniculate. Planta Med. 2002, 68, 81–83. [Google Scholar] [CrossRef]
- Lin, J.K.; Wu, T.S. Constituents of Flowers of Murraya Paniculata. J. Chin. Chem. Soc. Taip. 1994, 41, 213–216. [Google Scholar] [CrossRef]
- Wu, T.S.; Lin, C.N.; Yang, L.K.; Lin, S.T. Studies on the Constituents of Murraya Paniculata Jack (I). J. Chin. Chem. Soc. Taip. 1975, 22, 163–165. [Google Scholar] [CrossRef]
- Wu, T.S.; Liou, M.J.; Kuoh, C.S. Coumarins of the flowers of Murraya paniculate. Phytochemistry 1989, 28, 293–294. [Google Scholar] [CrossRef]
- Kikuchi, T.; Yokoi, T.; Umemoto, K.; Shingu, T. Constituents of Scaevola frutescens. Yaku gaku Zasshi. 1974, 94, 1616. [Google Scholar] [CrossRef][Green Version]
- Nikonov, G.K.; Saidkhodzhaev, A.I. Structure of pranferin, a new coumarin from Prangos ferulaceae roots. Khim. Prir. Soedin. 1971, 7, 255. [Google Scholar] [CrossRef]
- Teshima, N.; Tsugawa, M.; Tateishi, A.; Tokumaru, M.; Matsubara, R.; Kimachi, T.; Ju-ichi, M.; Ito, C.; Furukawa, H. Two new bicoumarins from the leaves of Murraya exotica. Heterocycles. 2004, 63, 2837–2843. [Google Scholar]
- Hubálek, F.; Binda, C.; Khalil, A.; Li, M.; Mattevi, A.; Castagnoli, N.; Edmondson, D.E. Demonstration of isoleucine 199 as a structural determinant for the selective inhibition of human monoamine oxidase B by specific reversible inhibitors. J. Biol. Chem. 2005, 280, 15761–15766. [Google Scholar] [CrossRef] [PubMed]
- Legoabe, L.J.; Petzer, A.; Petzer, J.P. Selected C7-substituted chromone derivatives as monoamine oxidase inhibitors. Bioorg Chem. 2012, 45, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.C.; Zhou, J.; Tang, Y.P.; Duan, J.A. Molecular Docking of COX-2 and Four Flavonoids from Scutellaria Baicalensis Georgi. Acta Chin. Med. Pharmacol. 2009, 37, 71–74. [Google Scholar]
- Rao, S.N.; Head, M.S.; Kulkarni, A.; Lalonde, J.M. Validation studies of the site-directed docking program LibDock. J. Chem. Inf. Model. 2007, 47, 2159–2171. [Google Scholar] [CrossRef]
- Binda, C.; Wang, J.; Pisani, L.; Caccia, C.; Carotti, A.; Salvati, P.; Edmondson, D.E.; Mattevi, A. Structures of human monoamine oxidase B complexes with selective noncovalent inhibitors: Safinamide and coumarin analogs. J. Med. Chem. 2007, 50, 5848–5852. [Google Scholar] [CrossRef]
- Suh, J.; Yum, E.K.; Cheon, H.G.; Cho, Y.S. Synthesis and biological evaluation of N-aryl-4-aryl-1,3-Thiazole-2-amine derivatives as direct 5-lipoxygenase inhibitors. Chem. Biol. Drug Des. 2012, 80, 90–99. [Google Scholar] [CrossRef]
- Chimenti, F.; Fioravanti, R.; Bolasco, A.; Chimenti, P.; Secci, D.; Rossi, F.; Yáñez, M.; Orallo, F.; Ortuso, F.; Alcaro, S. Chalcones: A valid scaffold for monoamine oxidases inhibitors. J. Med. Chem. 2009, 52, 2818–2824. [Google Scholar] [CrossRef]
- Chimenti, F.; Secci, D.; Bolasco, A.; Chimenti, P.; Bizzarri, B.; Granese, A.; Carradori, S.; Yáñez, M.; Orallo, F.; Ortuso, F.; et al. Synthesis, molecular modeling and selective inhibitory activity against human monoamine oxidases of 3-carboxamido-7-substituted coumarins. J. Med. Chem. 2009, 52, 1935–1942. [Google Scholar] [CrossRef]
- Manley-King, C.I.; Bergh, J.J.; Petzer, J.P. Monoamine oxidase inhibition by C4-substituted phthalonitriles. Bioorg. Chem. 2012, 40, 114–124. [Google Scholar] [CrossRef]
- Van der Walt, M.M.; Terre’Blanche, G.; Petzer, A.; Petzer, J.P. The adenosine receptor affinities and monoamine oxidase B inhibitory properties of sulfanylphthalimide analogues. Bioorg. Chem. 2015, 59, 117–123. [Google Scholar] [CrossRef]
- Yang, H. The research of constructing antivirus related pharmacophore and evaluating method. Beijing Univ. Chin. Med. 2010, 12, 83. [Google Scholar]
- Wang, X.; Xiang, Y.H.; Ren, Z.Z.; Zhang, Y.L.; Qiao, Y.J. Rational questing for inhibitors of endothelin converting enzyme-1 from Salvia miltiorrhiza by combining ligand and structure based virtual screening. Can. J. Chem. 2013, 91, 448–456. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, Y.L.; Wang, Y.; Ren, Z.Z.; Bao, H.J.; Qiao, Y.J. Study on relations between transient receptor potential vanilloid 1 and pungent property of traditional Chinese medicines. Chin. J. Chin. Mater. Med. 2014, 39, 2422–2427. [Google Scholar]
- Wang, D.S.; Nie, W.; Jiang, T.T.; Ding, L.F.; Song, L.D.; Wu, X.D.; Zhao, Q.S.; Caesalpanins, A.-C. three dimeric cassane diterpenoids from the seeds of Caesalpinia sappan L. Chem. Biodiversity 2020, 17, 2–9. [Google Scholar] [CrossRef]
- Ellman, G.L.; Courtney, K.D.; Andres, V.J.; Feather-Stone, R.M. A new and rapid colorimetric determination of acetylcholinesterase activity. Biochem. Pharmacol. 1961, 7, 88–95. [Google Scholar] [CrossRef]
- Zhang, L.; Tu, Z.C.; Yuan, T.; Wang, H.; Xie, X.; Fu, Z.F. Antioxidants and α-glucosidase inhibitors from Ipomoea batatas leaves identified by bioassay-guided approach and structure-activity relationships. Food Chem. 2016, 208, 61–67. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).