Unraveling Synergism between Various GH Family Xylanases and Debranching Enzymes during Hetero-Xylan Degradation
Abstract
1. Introduction
2. Results and Discussion
2.1. Substrate Specificity Determination
2.2. Degradation Products of GH10 and 11 Xylanases on Xylans
2.3. Synergy Studies
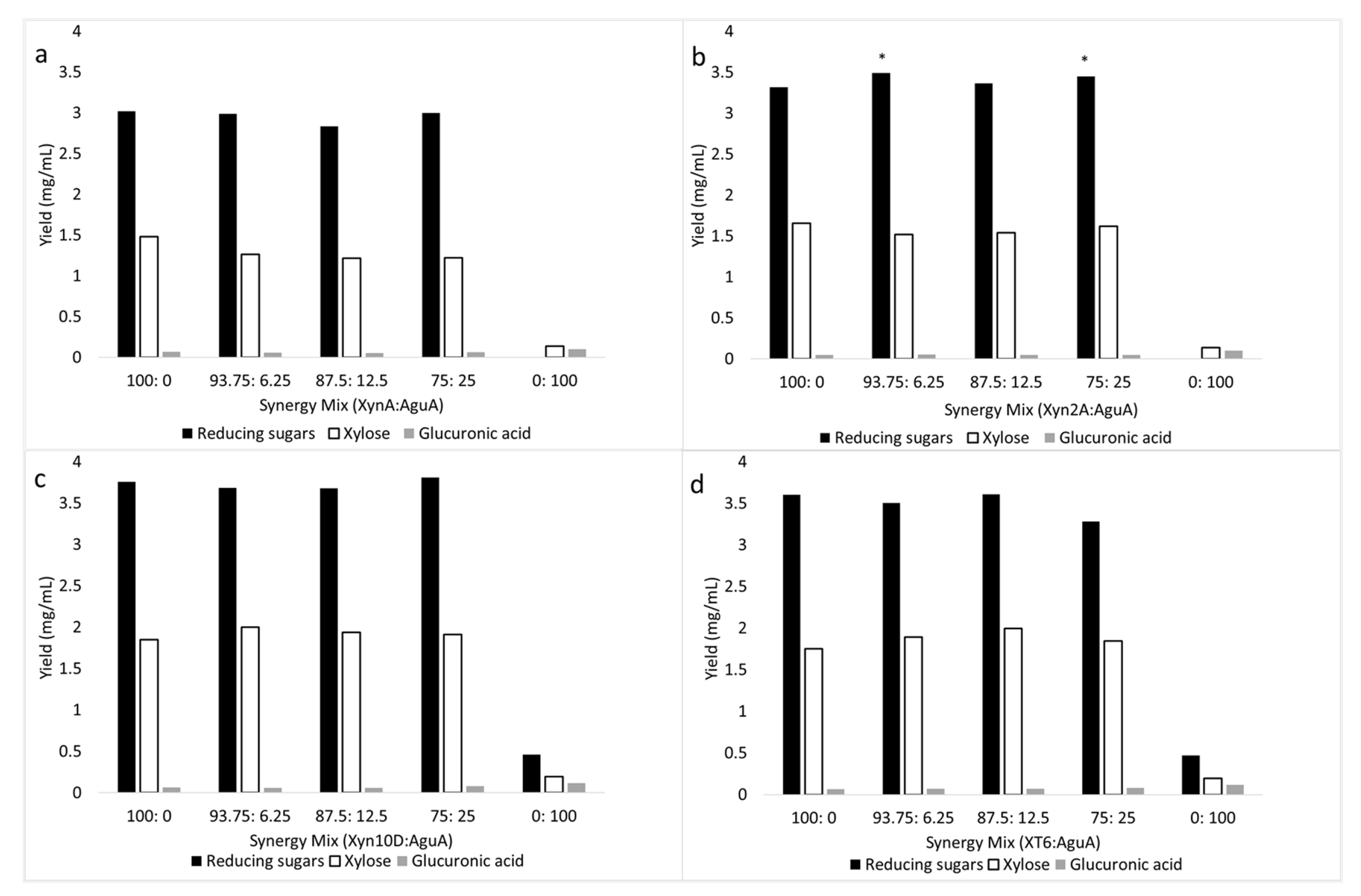
2.3.1. Synergism during Beechwood Glucuronoxylan Degradation
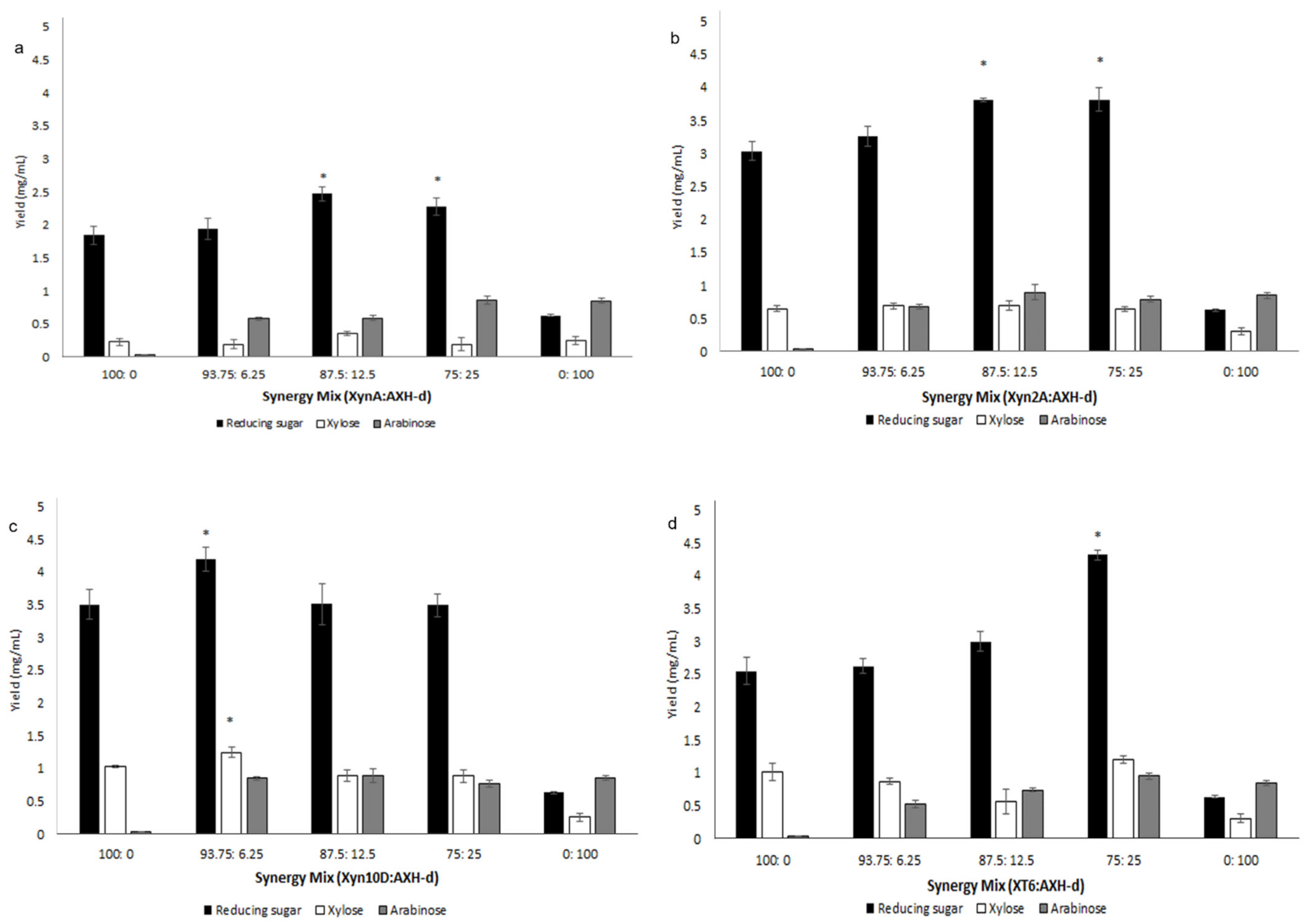
2.3.2. Synergism during Wheat Flour Arabinoxylan Degradation
3. Materials and Methods
3.1. Materials and Enzymes
3.2. Protein Determination
3.3. Substrate Specificity Determination
3.4. Determination of Degradation Products of Xylanases
3.5. Synergy Studies
3.6. Data Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Sweeney, M.D.; Xu, F. Biomass converting enzymes as industrial biocatalysts for fuels and chemicals: Recent developments. Catalysts 2012, 2, 244–263. [Google Scholar] [CrossRef]
- Subramaniyan, S.; Prema, P. Biotechnology of microbial xylanases: Enzymology, molecular biology, and application. Crit. Rev. Biotechnol. 2002, 22, 33–64. [Google Scholar] [CrossRef] [PubMed]
- Malgas, S.; Mafa, M.S.; Mkabayi, L.; Pletschke, B.I. A mini review of xylanolytic enzymes with regards to their synergistic interactions during hetero-xylan degradation. World J. Microbiol. Biotechnol. 2019, 35, 187. [Google Scholar] [CrossRef] [PubMed]
- Pinto, P.C.; Evtuguin, D.V.; Neto, C.P. Structure of hardwood glucuronoxylans: Modifications and impact on pulp retention during wood kraft pulping. Carbohydr. Polym. 2005, 60, 489–497. [Google Scholar] [CrossRef]
- Mafa, M.S.; Malgas, S.; Pletschke, B.I. Feruloyl esterase (FAE-1) sourced from a termite hindgut and GH10 xylanases synergy improves degradation of arabinoxylan. AMB Express 2021, 11, 21. [Google Scholar] [CrossRef]
- Kormelink, F.J.M.; Voragen, G.J. Degradation of different [(glucurono) arabino]xylans by a combination of purified xylan-degrading enzymes. Appl. Microbiol. Biotechnol. 1993, 38, 688–695. [Google Scholar] [CrossRef]
- York, W.S.; O’Neill, M.A. Biochemical control of xylan biosynthesis—which end is up? Curr. Opin. Plant Biol. 2008, 11, 258–265. [Google Scholar] [CrossRef]
- Peng, P.; She, D. Isolation, structural characterization, and potential applications of hemicelluloses from bamboo: A review. Carbohydr. Polym. 2014, 112, 701–720. [Google Scholar] [CrossRef]
- Revanappa, S.B.; Nandini, C.D.; Salimath, P.V. Structural variations of arabinoxylans extracted from different wheat (Triticum aestivum) cultivars in relation to chapati-quality. Food Hydrocoll. 2015, 43, 736–742. [Google Scholar] [CrossRef]
- Van Dyk, J.S.; Pletschke, B.I. A review of lignocellulose bioconversion using enzymatic hydrolysis and synergistic cooperation between enzymes-factors affecting enzymes, conversion and synergy. Biotechnol. Adv. 2012, 30, 1458–1480. [Google Scholar] [CrossRef]
- Malgas, S.; Thoresen, M.; van Dyk, S.J.; Pletschke, B.I. Time dependence of enzyme synergism during the degradation of model and natural lignocellulosic substrates. Enzyme Microb. Technol. 2017, 103, 1–11. [Google Scholar] [CrossRef]
- Biely, P.; Singh, S.; Puchart, V. Towards enzymatic breakdown of complex plant xylan structures: State of the art. Biotechnol. Adv. 2016, 34, 1260–1274. [Google Scholar] [CrossRef]
- Moreira, L.R.S.; Filho, E.X.F. Insights into the mechanism of enzymatic hydrolysis of xylan. Appl. Microbiol. Biotechnol. 2016, 100, 5205–5214. [Google Scholar] [CrossRef]
- Collins, T.; Gerday, C.; Feller, G. Xylanases, xylanase families and extremophilic xylanases. FEMS Microbiol. Rev. 2005, 29, 3–23. [Google Scholar] [CrossRef]
- Paës, G.; Berrin, J.G.; Beaugrand, J. GH11 xylanases: Structure/function/properties relationships and applications. Biotechnol. Adv. 2012, 30, 564–592. [Google Scholar] [CrossRef]
- Kumar, V.; Marin-Navarro, J.; Shukla, P. Thermostable microbial xylanases for pulp and paper industries: Trends, applications and further perspectives. World J. Microbiol. Biotechnol. 2016, 32, 34. [Google Scholar] [CrossRef]
- Mathibe, B.N.; Malgas, S.; Radosavljevic, L.; Kumar, V.; Shukla, P.; Pletschke, B.I. Tryptic mapping based structural insights of endo-1,4-β-xylanase from Thermomyces lanuginosus VAPS-24. Indian J. Microbiol. 2020, 60, 392–395. [Google Scholar] [CrossRef]
- Lagaert, S.; Pollet, A.; Courtin, C.M.; Volckaert, G. β-Xylosidases and α-l-arabinofuranosidases: Accessory enzymes for arabinoxylan degradation. Biotechnol. Adv. 2014, 32, 316–332. [Google Scholar] [CrossRef]
- Haltrich, D.; Nidetzky, B.; Kulbe, K.D.; Steiner, W.; Župančič, S. Production of fungal xylanases. Bioresour. Technol. 1996, 58, 137–161. [Google Scholar] [CrossRef]
- Polizeli, M.L.T.M.; Rizzatti, A.C.S.; Monti, R.; Terenzi, H.F.; Jorge, J.A.; Amorim, D.S. Xylanases from fungi: Properties and industrial applications. Appl. Microbiol. Biotechnol. 2005, 67, 577–591. [Google Scholar] [CrossRef]
- Nurizzo, D.; Nagy, T.; Gilbert, H.J.; Davies, G.J. The structural basis for catalysis and specificity of the Pseudomonas cellulosa alpha-glucuronidase, GlcA67A. Structure 2002, 10, 547–556. [Google Scholar] [CrossRef]
- Tenkanen, M.; Siika-Aho, M. An α-glucuronidase of Schizophyllum commune acting on polymeric xylan. J. Biotechnol. 2000, 78, 149–161. [Google Scholar] [CrossRef]
- McKee, L.S.; Sunner, H.; Anasontzis, G.E.; Toriz, G.; Gatenholm, P.; Bulone, V.; Vilaplana, F.; Olsson, L. A GH115 α-glucuronidase from Schizophyllum commune contributes to the synergistic enzymatic deconstruction of softwood glucuronoarabinoxylan. Biotechnol. Biofuels 2016, 9, 2. [Google Scholar] [CrossRef]
- Nagy, T.; Emami, K.; Fontes, C.M.G.; Ferreira, L.M.; Humphry, D.R.; Gilbert, H.J. The membrane-bound α-glucuronidase from Pseudomonas cellulosa hydrolyzes 4-O-methyl-d-glucuronoxylooligosaccharides but not 4-O-methyl-d-glucuronoxylan. J. Biotechnol. 2002, 184, 4925–4929. [Google Scholar] [CrossRef]
- Golan, G.; Shallom, D.; Teplitsky, A.; Zaide, G.; Shulami, S.; Baasov, T.; Stojanoff, V.; Thompson, A.; Shoham, Y.; Shoham, G. Crystal structures of Geobacillus stearothermophilus alpha-glucuronidase complexed with its substrate and products: Mechanistic implications. J. Biol. Chem. 2004, 279, 3014–3024. [Google Scholar] [CrossRef]
- Van Den Broek, L.A.M.; Lloyd, R.M.; Beldman, G.; Verdoes, J.C.; McCleary, B.V.; Voragen, A.G.V. Cloning and characterization of arabinoxylan arabinofuranohydrolase-D3 (AXHd3) from Bifidobacterium adolescentis DSM20083. Appl. Microbiol. Biotechnol. 2005, 67, 641–647. [Google Scholar] [CrossRef]
- Suresh, C.; Kitaoka, M.; Hayashi, K. A thermostable non-xylanolytic α-glucuronidase of Thermotoga maritima MSB8. Biosci. Biotechnol. Biochem. 2003, 67, 2359–2364. [Google Scholar] [CrossRef]
- Jordan, D.B.; Li, X.L. Variation in relative substrate specificity of bifunctional beta-d-xylosidase/alpha-l-arabinofuranosidase by single-site mutations: Roles of substrate distortion and recognition. Biochim. Biophys. Acta. Proteins Proteom. 2007, 1774, 1192–1198. [Google Scholar] [CrossRef]
- Malgas, S.; Pletschke, B.I. The effect of an oligosaccharide reducing-end xylanase, Bh Rex8A, on the synergistic degradation of xylan backbones by an optimised xylanolytic enzyme cocktail. Enzyme Microb. Technol. 2019, 122, 74–81. [Google Scholar] [CrossRef]
- Mendis, M.; Simsek, S. Production of structurally diverse wheat arabinoxylan hydrolyzates using combinations of xylanase and arabinofuranosidase. Carbohydr. Polym. 2015, 132, 452–459. [Google Scholar] [CrossRef]
- Rosa, L.; Ravanal, M.C.; Mardones, W.; Eyzaguirre, J. Characterization of a recombinant α-glucuronidase from Aspergillus fumigatus. Fungal Biol. 2013, 117, 380–387. [Google Scholar] [CrossRef] [PubMed]
- Gomes, J.; Gomes, I.; Kreiner, W.; Esterbauer, H.; Sinner, M.; Steiner, W. Production of high level of cellulase-free and thermostable xylanase by a wild strain of Thermomyces lanuginosus using beechwood xylan. J. Biotechnol. 1993, 30, 283–297. [Google Scholar] [CrossRef]
- Törrönen, A.; Rouvinen, J. Structural Comparison of Two Major endo-1,4-Xylanases from Trichoderma reesei. Biochemistry 1995, 34, 847–856. [Google Scholar] [CrossRef] [PubMed]
- Mi, S.; Jia, X.; Wang, J.; Qiao, W.; Peng, X.; Han, Y. Insight into glycoside hydrolases for debranched xylan degradation from extremely thermophilic bacterium Caldicellulosiruptor lactoaceticus. PLoS ONE 2014, 9, e106482. [Google Scholar] [CrossRef]
- Sydenham, R.; Zheng, Y.; Riemens, A.; Tsang, A.; Powlowski, J.; Storms, R. Cloning and enzymatic characterization of four thermostable fungal endo-1,4-β-xylanases. Appl. Microbiol. Biotechnol. 2014, 98, 3613–3628. [Google Scholar] [CrossRef]
- Mafa, M.S.; Malgas, S.; Rashamuse, K.; Pletschke, B.I. Delineating functional properties of a cello-oligosaccharide and β-glucan specific cellobiohydrolase (GH5_38): Its synergism with Cel6A and Cel7A for β-(1,3)-(1,4)-glucan degradation. Carbohydr. Res. 2020, 495, 108081. [Google Scholar] [CrossRef]
- Pletschke, B.I.; Malgas, S.; Bhattacharya, A.; Bhattacharya-Shrivastava, A.; Clarke, M.D.; Mafa, M.S.; Morake, S.; Thoresen, M. Enzyme synergism: A powerful tool for decreasing enzyme loading for efficient biomass conversion. In Proceedings of the 24th European Biomass Conference and Exhibition, Amsterdam, The Netherlands, 6–9 June 2016; Volume 2016. [Google Scholar]
- Ebringerová, A. Structural diversity and application potential of hemicelluloses. Macromol. Symp. 2006, 232, 1–12. [Google Scholar] [CrossRef]
- Cobucci-Ponzano, B.; Strazzulli, A.; Iacono, R.; Masturzo, G.; Giglio, R.; Rossi, M.; Moracci, M. Novel thermophilic hemicellulases for the conversion of lignocellulose for second generation biorefineries. Enzyme Microb. Technol. 2015, 78, 63–73. [Google Scholar] [CrossRef]
- Brumm, P.; Xie, D.; Allen, L.; Mead, D.A. Cloning, expression and characterization of the α-glucuronidase from the hyperthermophile Dictyoglomus turgidum DSM 6724Ô. J. Enzym. 2020, 1, 34–47. [Google Scholar] [CrossRef]
- Rhee, M.S.; Sawhney, N.; Kim, Y.S.; Rhee, H.J.; Hurlbert, J.C.; John, F.J.S.; Nong, G.; Rice, J.D.; Preston, J.F. GH115 α-glucuronidase and GH11 xylanase from Paenibacillus sp. JDR-2: Potential roles in processing glucuronoxylans. Appl. Microbiol. Biotechnol. 2017, 101, 1465–1476. [Google Scholar] [CrossRef]
- Ryabova, O.; Vršanská, M.; Kaneko, S.; van Zyl, W.H.; Biely, P. A novel family of hemicellulolytic α-glucuronidase. FEBS Lett. 2009, 583, 1457–1462. [Google Scholar] [CrossRef]
- Brüx, C.; Ben-David, A.; Shallom-Shezifi, D.; Leon, M.; Niefind, K.; Shoham, G.; Shoham, Y.; Schomburg, D. The structure of an inverting GH43 β-xylosidase from Geobacillus stearothermophilus with its substrate reveals the role of the three catalytic residues. J. Mol. Biol. 2006, 359, 97–109. [Google Scholar] [CrossRef]
- Rohman, A.; Dijkstra, B.W.; Puspaningsih, N.N.T. β-Xylosidases: Structural diversity, catalytic mechanism, and inhibition by monosaccharides. Int. J. Mol. Sci. 2019, 20, 5524. [Google Scholar] [CrossRef]
- Jordan, D.B.; Li, X.L.; Dunlap, C.A.; Whitehead, T.T.; Cotta, M.A. Structure-function relationships of a catalytically efficient β-d-xylosidase. Appl. Biochem. Biotechnol. 2007, 141, 51–76. [Google Scholar] [CrossRef]
- Sørensen, H.R.; Pedersen, S.; Jørgensen, C.T.; Meyer, A.S. Enzymatic hydrolysis of wheat arabinoxylan by a recombinant “minimal” enzyme cocktail containing β-xylosidase and novel endo-1,4-β-xylanase and α-l-arabinofuranosidase activities. Biotechnol. Prog. 2007, 23, 100–107. [Google Scholar] [CrossRef]
- Kumar, V.; Chhabra, D.; Shukla, P. Xylanase production from Thermomyces lanuginosus VAPS-24 using low cost agro-industrial residues via hybrid optimization tools and its potential use for saccharification. Bioresour. Technol. 2017, 243, 1009–1019. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Miller, G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Mkabayi, L.; Malgas, S.; Wilhelmi, B.S.; Pletschke, B.I. Evaluating feruloyl esterase—xylanase synergism for hydroxycinnamic acid and xylo-oligosaccharide production from untreated, hydrothermally pre-treated and dilute-acid pre-treated corn cobs. Agronomy 2020, 10, 688. [Google Scholar] [CrossRef]



| Property | Xylanolytic Enzyme Assessed | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Abf51A | AXH-d | AguA | Agu4B | SXA | XynA | Xyn2A | Xyn10D | XT6 | |
| GH family | 51 | 43 | 67 | 4 | 43 | 11 | 11 | 10 | 10 |
| Mass (kDa) | 58.5 | 59.4 | 93.2 | Nd | 61.9 | 20.9 | 25.1 | 39 | 43.8 |
| pH optimum | 5.5 | 6.0 | 7.0 | 7.5 | 5.0 | 7.0 | 6.0 | 5.0 | 6.0 |
| Temperature optimum | 50 | 70 | 70 | 40 | 40 | 50 | 50 | 60 | 70 |
| Substrate | Xylanolytic Enzyme Assessed | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Abf51A | AXH-d | AguA | Agu4B | SXA | XynA | Xyn2A | Xyn10D | XT6 | |
| pNPA | 161.9 | 2.6 | 2 | Nd | 19.8 | Nd | 2.1 | Nd | 2.1 |
| pNPX | 0.4 | 0.3 | 0.2 | Nd | 155.6 | Nd | 0.4 | Nd | 0.4 |
| Aldouronic acids | Nd | Nd | 5.7 | 0.68 | Nd | Nd | Nd | Nd | Nd |
| Beechwood xylan | 0 | 0.23 | 1.6 | 0.96 | 3.9 | 91 | 258.4 | 30.8 | 141.8 |
| Wheat flour xylan | 0 | 30.9 | 0.9 | 0 | 6.6 | 370.6 | 502.1 | 133.1 | 273.6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Malgas, S.; Mafa, M.S.; Mathibe, B.N.; Pletschke, B.I. Unraveling Synergism between Various GH Family Xylanases and Debranching Enzymes during Hetero-Xylan Degradation. Molecules 2021, 26, 6770. https://doi.org/10.3390/molecules26226770
Malgas S, Mafa MS, Mathibe BN, Pletschke BI. Unraveling Synergism between Various GH Family Xylanases and Debranching Enzymes during Hetero-Xylan Degradation. Molecules. 2021; 26(22):6770. https://doi.org/10.3390/molecules26226770
Chicago/Turabian StyleMalgas, Samkelo, Mpho S. Mafa, Brian N. Mathibe, and Brett I. Pletschke. 2021. "Unraveling Synergism between Various GH Family Xylanases and Debranching Enzymes during Hetero-Xylan Degradation" Molecules 26, no. 22: 6770. https://doi.org/10.3390/molecules26226770
APA StyleMalgas, S., Mafa, M. S., Mathibe, B. N., & Pletschke, B. I. (2021). Unraveling Synergism between Various GH Family Xylanases and Debranching Enzymes during Hetero-Xylan Degradation. Molecules, 26(22), 6770. https://doi.org/10.3390/molecules26226770








