Characterization of Lysine Acetyltransferase Activity of Recombinant Human ARD1/NAA10
Abstract
1. Introduction
2. Results
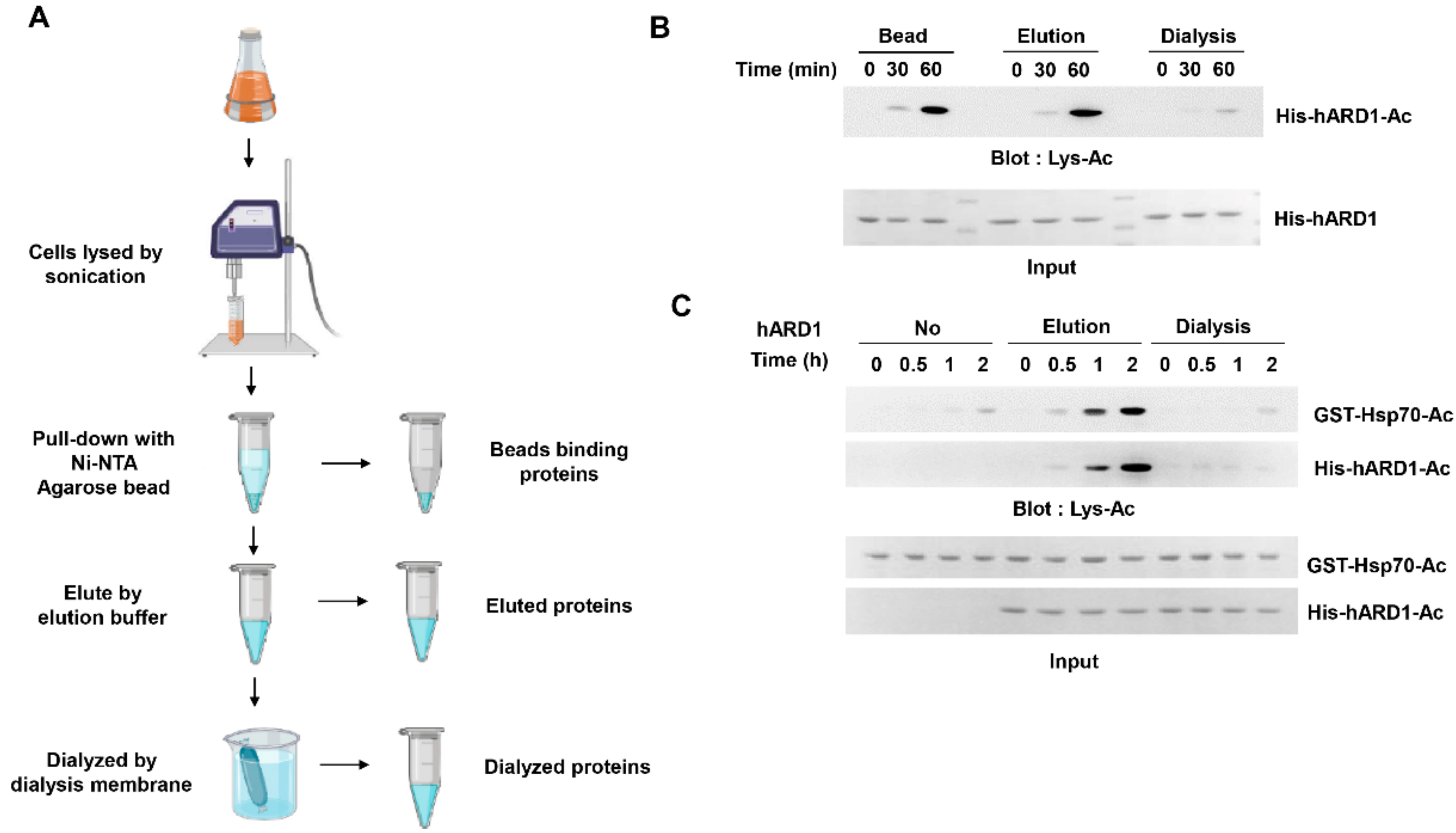
2.1. KAT Activity of rhARD1/NAA10 Is Lost During Dialysis
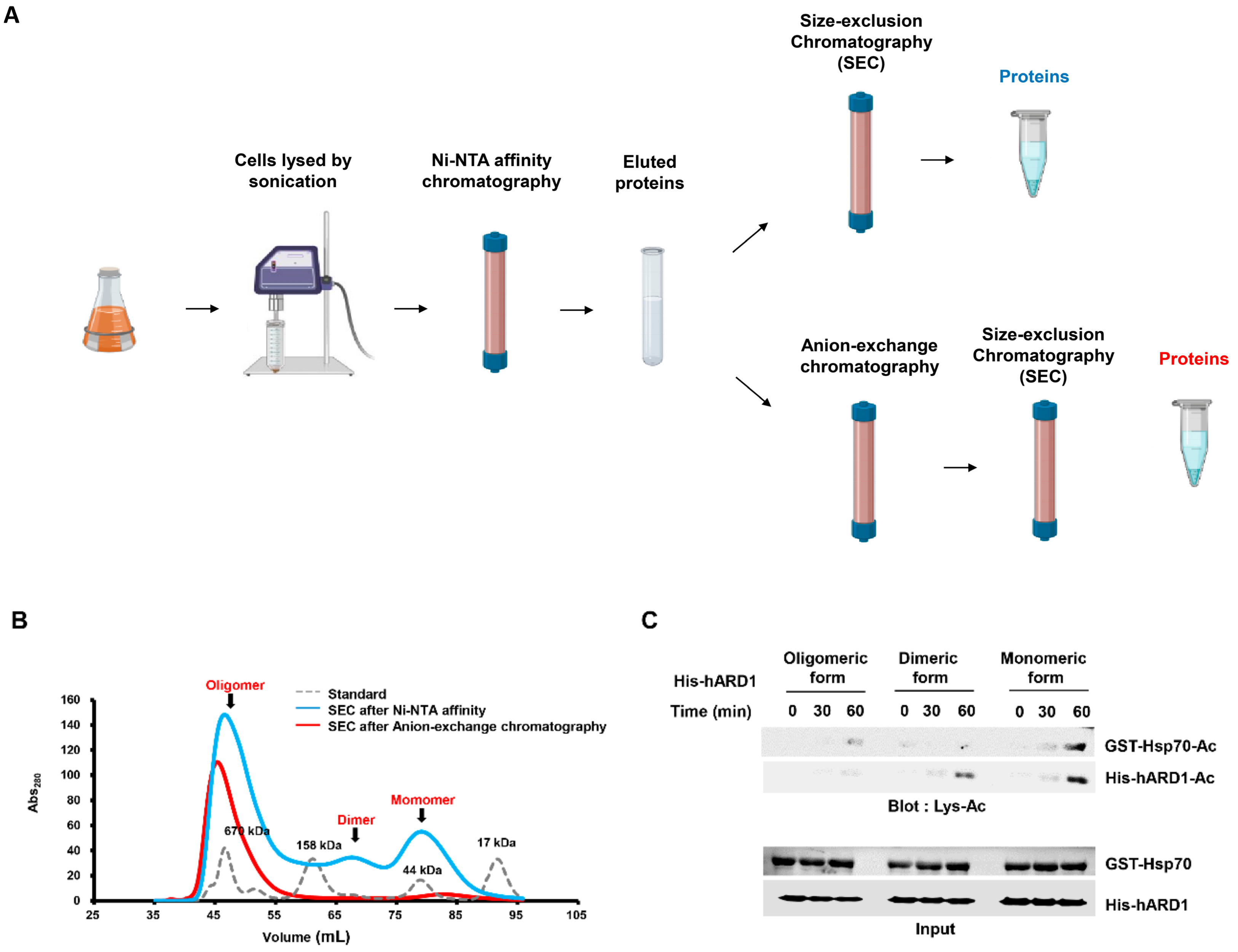
2.2. rhARD1/NAA10 Is Aggregated During Purification
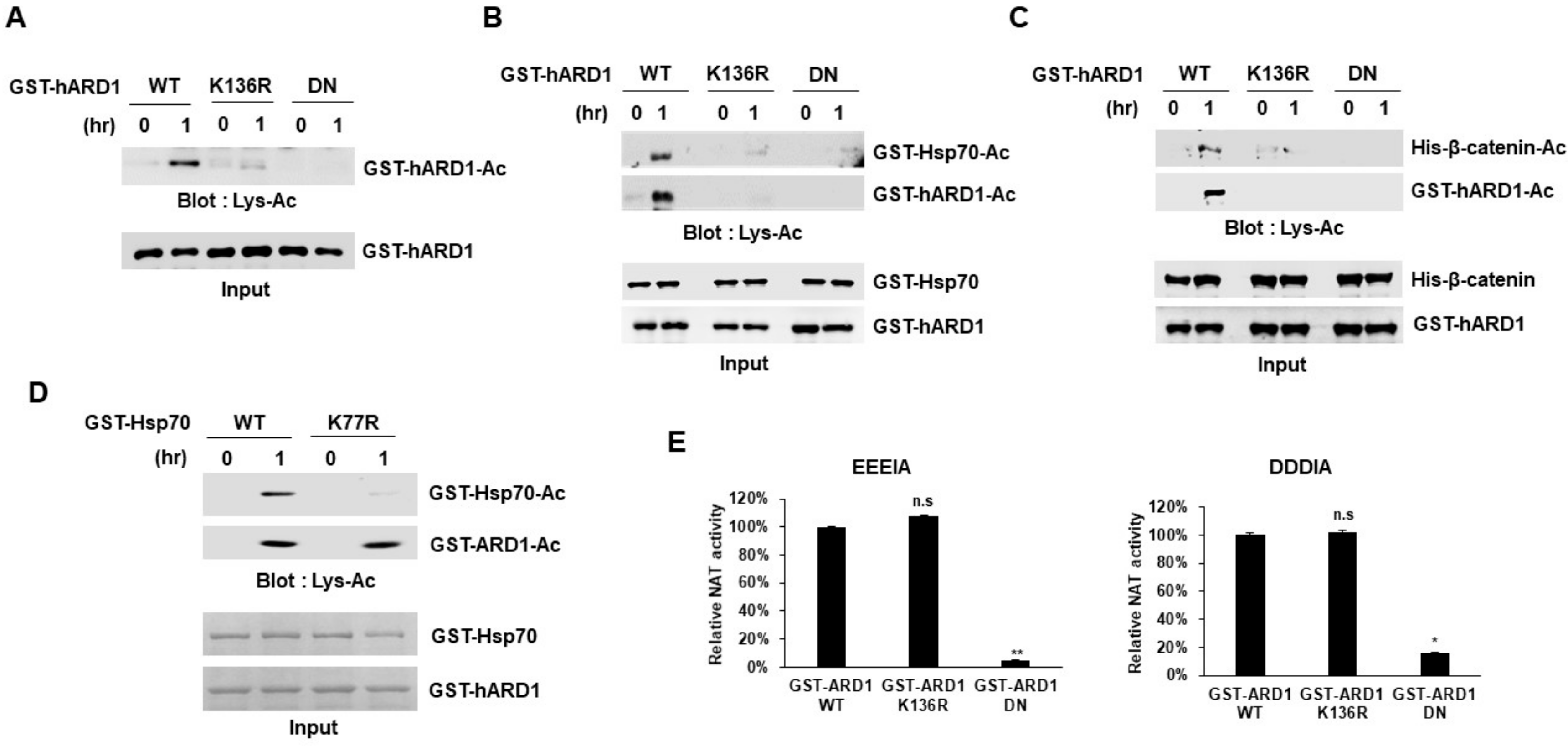
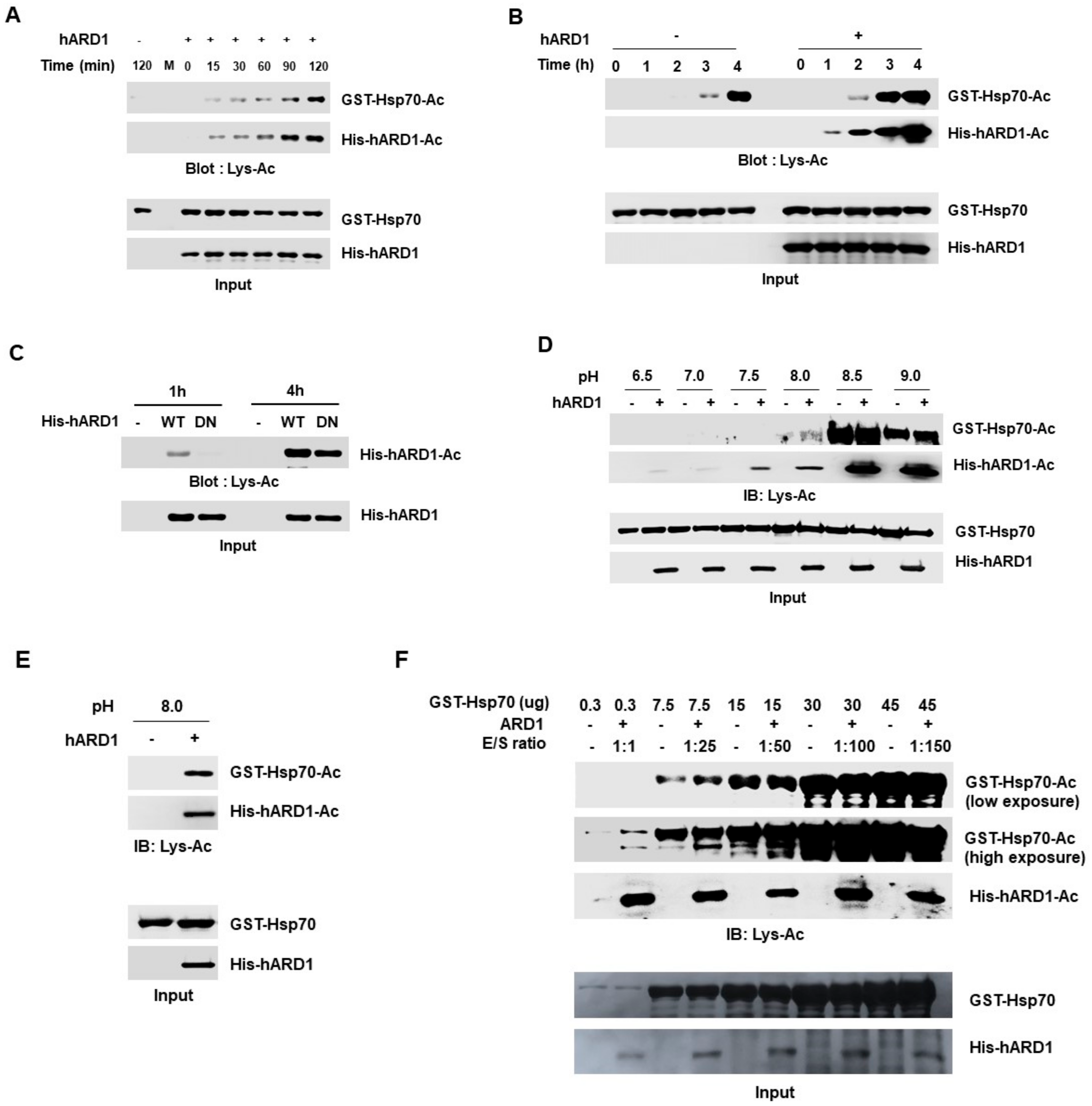
2.3. rhARD1/NAA10 Mediates Site-Specific Lysine Acetylation of Substrate That Is Distinctive from the Non-Specific Chemical Reaction
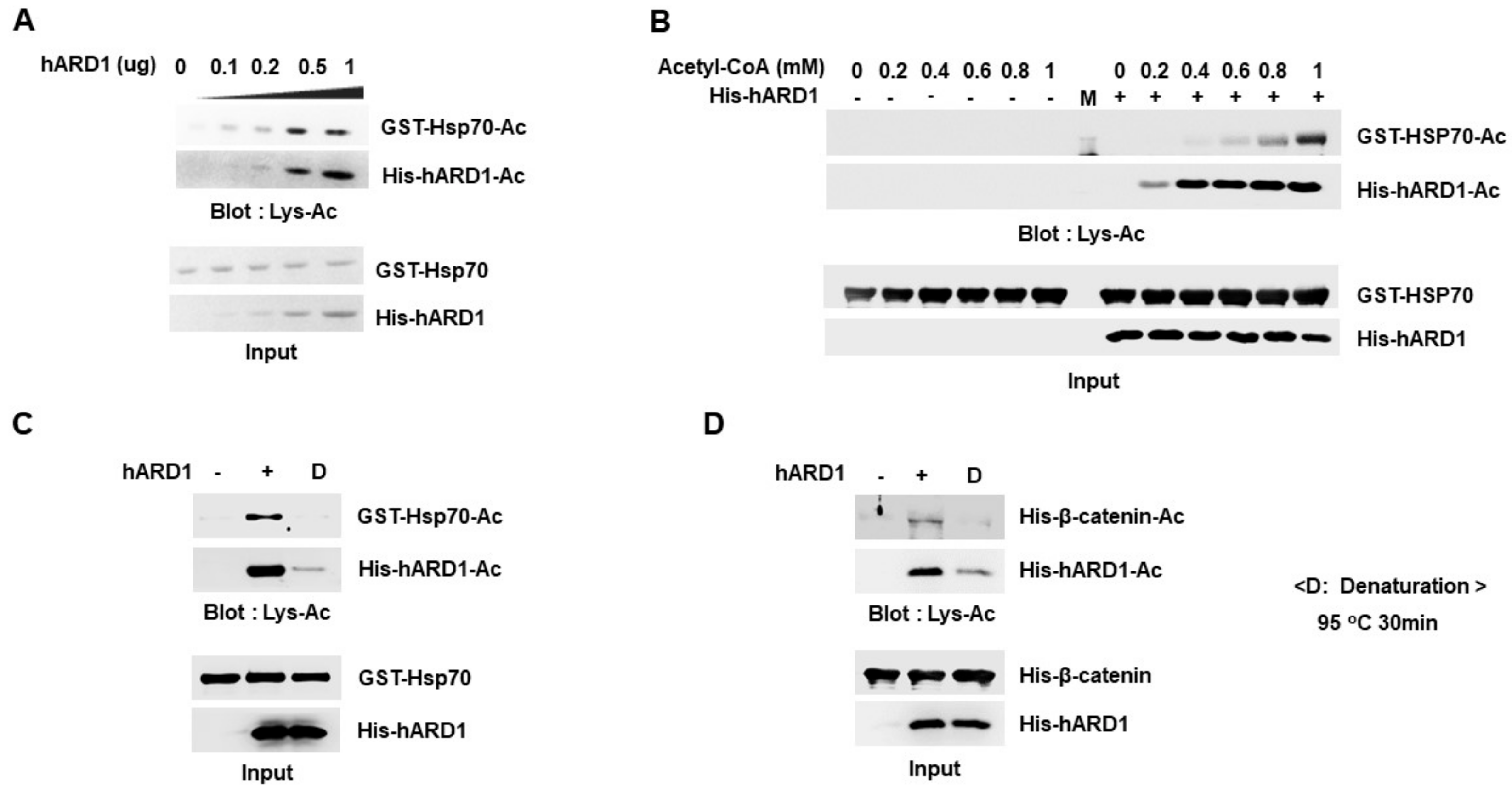
2.4. Lysine Acetylation Activity of hARD1/NAA10 Is Dependent on Concentration of Reactants
2.5. Limited Reaction Conditions Are Needed to Distinguish the hARD1/NAA10-Mediated Lysine Acetylation Reaction from Non-Specific Chemical Reaction
3. Discussion
4. Materials and Methods
4.1. Antibodies
4.2. Plasmid Construction
4.3. Expression and Purification of His-ARD1
4.4. Oligomeric Status Characterization by Size-Exclusion Chromatography
4.5. Expression and Purification of GST-ARD1/NAA10
4.6. Expression and Purification of GST-Hsp70
4.7. In Vitro Acetylation Assay
4.8. Immunoblotting
4.9. DTNB-Based Nt-Acetylation Assay
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Arnesen, T.; Van Damme, P.; Polevoda, B.; Helsens, K.; Evjenth, R.; Colaert, N.; Varhaug, J.E.; Vandekerckhove, J.; Lillehaug, J.R.; Sherman, F.; et al. Proteomics analyses reveal the evolutionary conservation and divergence of N-terminal acetyltransferases from yeast and humans. Proc. Natl. Acad. Sci. USA 2009, 106, 8157–8162. [Google Scholar] [CrossRef]
- Goetze, S.; Qeli, E.; Mosimann, C.; Staes, A.; Gerrits, B.; Roschitzki, B.; Mohanty, S.; Niederer, E.M.; Laczko, E.; Timmerman, E.; et al. Identification and Functional Characterization of N-Terminally Acetylated Proteins in Drosophila melanogaster. PLoS Biol. 2009, 7, e1000236. [Google Scholar] [CrossRef] [PubMed]
- Drazic, A.; Myklebust, L.M.; Ree, R.; Arnesen, T. The world of protein acetylation. Biochim. Et Biophys. Acta (Bba)-Proteins Proteom. 2016, 1864, 1372–1401. [Google Scholar] [CrossRef] [PubMed]
- Whiteway, M.; Szostak, J.W. The ARD1 gene of yeast functions in the switch between the mitotic cell cycle and alternative developmental pathways. Cell 1985, 43, 483–492. [Google Scholar] [CrossRef]
- Lee, M.-N.; Kweon, H.Y.; Oh, G.T. N-α-acetyltransferase 10 (NAA10) in development: The role of NAA10. Exp. Mol. Med. 2018, 50, 87. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.-W.; Bae, M.-K.; Ahn, M.-Y.; Kim, S.-H.; Sohn, T.-K.; Bae, M.-H.; Yoo, M.-A.; Song, E.J.; Lee, K.-J.; Kim, K.-W. Regulation and Destabilization of HIF-1α by ARD1-Mediated Acetylation. Cell 2002, 111, 709–720. [Google Scholar] [CrossRef]
- Lim, J.-H.; Park, J.-W.; Chun, Y.-S. Human Arrest Defective 1 Acetylates and Activates β-Catenin, Promoting Lung Cancer Cell Proliferation. Cancer Res. 2006, 66, 10677–10682. [Google Scholar] [CrossRef]
- Yoon, H.; Kim, H.-L.; Chun, Y.-S.; Shin, D.H.; Lee, K.-H.; Shin, C.S.; Lee, D.Y.; Kim, H.-H.; Lee, Z.H.; Ryoo, H.-M.; et al. NAA10 controls osteoblast differentiation and bone formation as a feedback regulator of Runx2. Nat. Commun. 2014, 5, 5176. [Google Scholar] [CrossRef]
- Shin, S.H.; Yoon, H.; Chun, Y.S.; Shin, H.W.; Lee, M.N.; Oh, G.T.; Park, J.W. Arrest defective 1 regulates the oxidative stress response in human cells and mice by acetylating methionine sulfoxide reductase A. Cell Death Dis. 2014, 5, e1490. [Google Scholar] [CrossRef]
- Shin, D.H.; Chun, Y.-S.; Lee, K.-H.; Shin, H.-W.; Park, J.-W. Arrest Defective-1 Controls Tumor Cell Behavior by Acetylating Myosin Light Chain Kinase. PLoS ONE 2009, 4, e7451. [Google Scholar] [CrossRef]
- DePaolo, J.S.; Wang, Z.; Guo, J.; Zhang, G.; Qian, C.; Zhang, H.; Zabaleta, J.; Liu, W. Acetylation of androgen receptor by ARD1 promotes dissociation from HSP90 complex and prostate tumorigenesis. Oncotarget 2016, 7, 71417–71428. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.H.; Park, J.-H.; Lee, E.J.; Vo, T.T.L.; Choi, H.; Kim, J.Y.; Jang, J.K.; Wee, H.-J.; Lee, H.S.; Jang, S.H.; et al. ARD1-mediated Hsp70 acetylation balances stress-induced protein refolding and degradation. Nat. Commun. 2016, 7, 12882. [Google Scholar] [CrossRef] [PubMed]
- Qian, X.; Li, X.; Cai, Q.; Zhang, C.; Yu, Q.; Jiang, Y.; Lee, J.-H.; Hawke, D.; Wang, Y.; Xia, Y.; et al. Phosphoglycerate Kinase 1 Phosphorylates Beclin1 to Induce Autophagy. Mol. Cell 2017, 65, 917–931.e916. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, P.; Ha, E.; Vo, T.T.L.; Seo, J.H. Diverse roles of arrest defective 1 in cancer development. Arch. Pharmacal. Res. 2019, 42, 1040–1051. [Google Scholar] [CrossRef] [PubMed]
- Arnesen, T.; Kong, X.; Evjenth, R.; Gromyko, D.; Varhaug, J.E.; Lin, Z.; Sang, N.; Caro, J.; Lillehaug, J.R. Interaction between HIF-1 alpha (ODD) and hARD1 does not induce acetylation and destabilization of HIF-1 alpha. Febs. Lett. 2005, 579, 6428–6432. [Google Scholar] [CrossRef]
- Magin, R.S.; March, Z.M.; Marmorstein, R. The N-terminal Acetyltransferase Naa10/ARD1 Does Not Acetylate Lysine Residues. J. Biol. Chem. 2016, 291, 5270–5277. [Google Scholar] [CrossRef]
- Liszczak, G.; Goldberg, J.M.; Foyn, H.; Petersson, E.J.; Arnesen, T.; Marmorstein, R. Molecular basis for N-terminal acetylation by the heterodimeric NatA complex. Nat. Struct. Mol. Biol. 2013, 20, 1098. [Google Scholar] [CrossRef]
- Gottlieb, L.; Marmorstein, R. Structure of Human NatA and Its Regulation by the Huntingtin Interacting Protein HYPK. Structure 2018, 26, 925–935.e928. [Google Scholar] [CrossRef]
- Aksnes, H.; Ree, R.; Arnesen, T. Co-translational, Post-translational, and Non-catalytic Roles of N-Terminal Acetyltransferases. Mol. Cell 2019, 73, 1097–1114. [Google Scholar] [CrossRef]
- Vo, T.T.L.; Jeong, C.-H.; Lee, S.; Kim, K.-W.; Ha, E.; Seo, J.H. Versatility of ARD1/NAA10-mediated protein lysine acetylation. Exp. Mol. Med. 2018, 50, 86. [Google Scholar] [CrossRef]
- Seo, J.H.; Cha, J.-H.; Park, J.-H.; Jeong, C.-H.; Park, Z.-Y.; Lee, H.-S.; Oh, S.H.; Kang, J.-H.; Suh, S.W.; Kim, K.H.; et al. Arrest Defective 1 Autoacetylation Is a Critical Step in Its Ability to Stimulate Cancer Cell Proliferation. Cancer Res. 2010, 70, 4422–4432. [Google Scholar] [CrossRef] [PubMed]
- Wagner, G.R.; Payne, R.M. Widespread and Enzyme-independent Nϵ-Acetylation and Nϵ-Succinylation of Proteins in the Chemical Conditions of the Mitochondrial Matrix. J. Biol. Chem. 2013, 288, 29036–29045. [Google Scholar] [CrossRef] [PubMed]
- Goss, G.; Grinstein, S. Chapter 7 Mechanisms of intracellular pH regulation. In Principles of Medical Biology; Bittar, E.E., Bittar, N., Eds.; Elsevier: London, UK, 1996; Volume 4, pp. 221–241. [Google Scholar]
- Casey, J.R.; Grinstein, S.; Orlowski, J. Sensors and regulators of intracellular pH. Nat. Rev. Mol. Cell Biol. 2010, 11, 50–61. [Google Scholar] [CrossRef] [PubMed]
- Arnesen, T.; Anderson, D.; Baldersheim, C.; Lanotte, M.; Varhaug, J.E.; Lillehaug, J.R. Identification and characterization of the human ARD1–NATH protein acetyltransferase complex. Biochem. J. 2005, 386, 433–443. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.H.; Park, J.-H.; Lee, E.J.; Kim, K.-W. Different subcellular localizations and functions of human ARD1 variants. Int. J. Oncol. 2015, 46, 701–707. [Google Scholar] [CrossRef] [PubMed]
- Van Damme, P.; Evjenth, R.; Foyn, H.; Demeyer, K.; De Bock, P.-J.; Lillehaug, J.R.; Vandekerckhove, J.; Arnesen, T.; Gevaert, K. Proteome-derived Peptide Libraries Allow Detailed Analysis of the Substrate Specificities of N(alpha)-acetyltransferases and Point to hNaa10p as the Post-translational Actin N(alpha)-acetyltransferase. Mol. Cell. Proteom. 2011, 10, M110.004580. [Google Scholar] [CrossRef]
- Park, J.-H.; Seo, J.H.; Wee, H.-J.; Vo, T.T.L.; Lee, E.J.; Choi, H.; Cha, J.-H.; Ahn, B.J.; Shin, M.W.; Bae, S.-J.; et al. Nuclear Translocation of hARD1 Contributes to Proper Cell Cycle Progression. PLoS ONE 2014, 9, e105185. [Google Scholar]
- Kang, J.; Chun, Y.-S.; Huh, J.; Park, J.-W. FIH permits NAA10 to catalyze the oxygen-dependent lysyl-acetylation of HIF-1α. Redox Biol. 2018, 19, 364–374. [Google Scholar] [CrossRef]
- Lee, E.J.; Seo, J.H.; Park, J.-H.; Vo, T.T.L.; An, S.; Bae, S.-J.; Le, H.; Lee, H.S.; Wee, H.-J.; Lee, D.; et al. SAMHD1 acetylation enhances its deoxynucleotide triphosphohydrolase activity and promotes cancer cell proliferation. Oncotarget 2017, 8, 68517–68529. [Google Scholar]
Sample Availability: Samples of the compounds are not available from the authors. |

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vo, T.T.L.; Park, J.-H.; Lee, E.J.; Nguyen, Y.T.K.; Han, B.W.; Nguyen, H.T.T.; Mun, K.C.; Ha, E.; Kwon, T.K.; Kim, K.-W.; et al. Characterization of Lysine Acetyltransferase Activity of Recombinant Human ARD1/NAA10. Molecules 2020, 25, 588. https://doi.org/10.3390/molecules25030588
Vo TTL, Park J-H, Lee EJ, Nguyen YTK, Han BW, Nguyen HTT, Mun KC, Ha E, Kwon TK, Kim K-W, et al. Characterization of Lysine Acetyltransferase Activity of Recombinant Human ARD1/NAA10. Molecules. 2020; 25(3):588. https://doi.org/10.3390/molecules25030588
Chicago/Turabian StyleVo, Tam Thuy Lu, Ji-Hyeon Park, Eun Ji Lee, Yen Thi Kim Nguyen, Byung Woo Han, Hien Thi Thu Nguyen, Kyo Cheol Mun, Eunyoung Ha, Taeg Kyu Kwon, Kyu-Won Kim, and et al. 2020. "Characterization of Lysine Acetyltransferase Activity of Recombinant Human ARD1/NAA10" Molecules 25, no. 3: 588. https://doi.org/10.3390/molecules25030588
APA StyleVo, T. T. L., Park, J.-H., Lee, E. J., Nguyen, Y. T. K., Han, B. W., Nguyen, H. T. T., Mun, K. C., Ha, E., Kwon, T. K., Kim, K.-W., Jeong, C.-H., & Seo, J. H. (2020). Characterization of Lysine Acetyltransferase Activity of Recombinant Human ARD1/NAA10. Molecules, 25(3), 588. https://doi.org/10.3390/molecules25030588







