Fatty Acid Synthase: An Emerging Target in Cancer
Abstract
1. Introduction
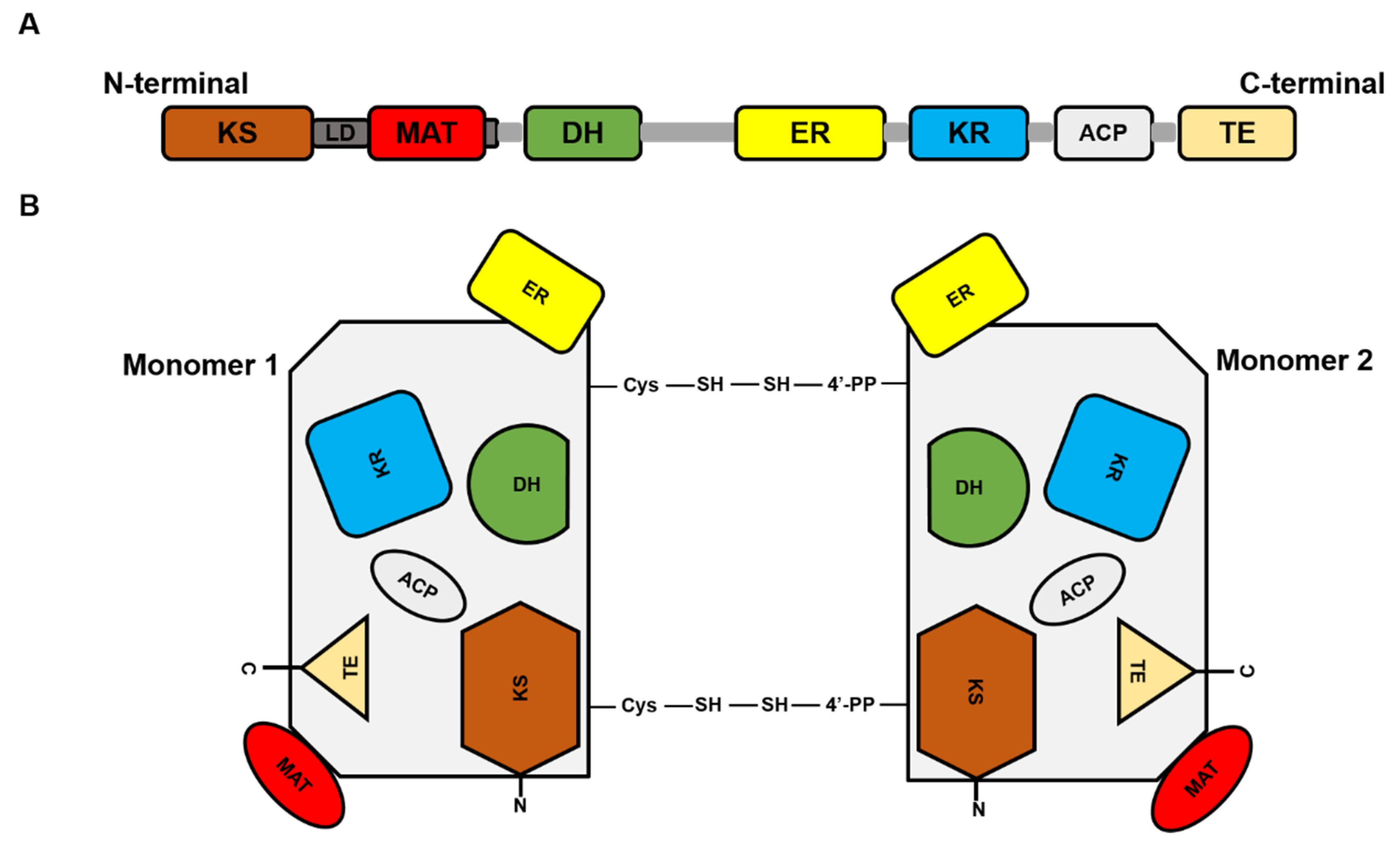
2. FASN in Normal Physiology
3. FASN Regulation
4. FASN and Cancer
5. FASN Influences Organelle Activity in Tumor Cells
5.1. Mitochondria
5.2. Peroxisomes
5.3. Nucleus
5.4. Endoplasmic Reticulum
6. FASN Is Functionally Linked to Metabolic Pathways
7. Targeting FASN in Cancer
8. Conclusions
Funding
Conflicts of Interest
References
- Hanahan, D.; Weinberg, R.A. The Hallmarks of Cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Warburg, O. On the Origin of Cancer Cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Ventura, R.; Mordec, K.; Waszczuk, J.; Wang, Z.; Lai, J.; Fridlib, M.; Buckley, D.; Kemble, G.; Heuer, T.S. Inhibition of de novo Palmitate Synthesis by Fatty Acid Synthase Induces Apoptosis in Tumor Cells by Remodeling Cell Membranes, Inhibiting Signaling Pathways, and Reprogramming Gene Expression. EBioMedicine 2015, 2, 808–824. [Google Scholar] [CrossRef]
- Chirala, S.S.; Wakil, S.J. Structure and function of animal fatty acid synthase. Lipids 2004, 39, 1045–1053. [Google Scholar] [CrossRef] [PubMed]
- Chirala, S.S.; Chang, H.; Matzuk, M.; Abu-Elheiga, L.; Mao, J.; Mahon, K.; Finegold, M.; Wakil, S.J. Fatty acid synthesis is essential in embryonic development: Fatty acid synthase null mutants and most of the heterozygotes die in utero. Proc. Natl. Acad. Sci. USA 2003, 100, 6358–6363. [Google Scholar] [CrossRef]
- Rooney, S.A. Fatty acid biosynthesis in developing fetal lung. Am. J. Physiol. Cell. Mol. Physiol. 1989, 257, L195–L201. [Google Scholar] [CrossRef]
- Semenkovich, C.F.; Coleman, T.; Fiedorek, F.T. Human fatty acid synthase mRNA: Tissue distribution, genetic mapping, and kinetics of decay after glucose deprivation. J. Lipid Res. 1995, 36, 1507–1521. [Google Scholar]
- Chung, K.-P.; Hsu, C.-L.; Fan, L.-C.; Huang, Z.; Bhatia, D.; Chen, Y.-J.; Hisata, S.; Cho, S.J.; Nakahira, K.; Imamura, M.; et al. Mitofusins regulate lipid metabolism to mediate the development of lung fibrosis. Nat. Commun. 2019, 10, 3390. [Google Scholar] [CrossRef]
- Rudolph, M.C.; McManaman, J.L.; Phang, T.; Russell, T.; Kominsky, U.J.; Serkova, N.J.; Stein, T.; Anderson, S.M.; Neville, M.C. Metabolic regulation in the lactating mammary gland: A lipid synthesizing machine. Physiol. Genom. 2007, 28, 323–336. [Google Scholar] [CrossRef]
- Smith, S.; Dils, R. Factors affecting the chain length of fatty acids synthesised by lactating-rabbit mammary glands. Biochim. Biophys. Acta 1966, 116, 23–40. [Google Scholar] [CrossRef]
- Smith, S.; Gagné, H.T.; Pitelka, D.R.; Abraham, S. The effect of dietary fat on lipogenesis in mammary gland and liver from lactating and virgin mice. Biochem. J. 1969, 115, 807–815. [Google Scholar] [CrossRef] [PubMed]
- Suburu, J.; Shi, L.; Wu, J.; Wang, S.; Samuel, M.; Thomas, M.J.; Kock, N.D.; Yang, G.; Kridel, S.; Chen, Y.Q. Fatty acid synthase is required for mammary gland development and milk production during lactation. Am. J. Physiol. Metab. 2014, 306, E1132–E1143. [Google Scholar] [CrossRef]
- Hudgins, L.C.; Hellerstein, M.; Seidman, C.; Neese, R.; Diakun, J.; Hirsch, J. Human fatty acid synthesis is stimulated by a eucaloric low fat, high carbohydrate diet. J. Clin. Investig. 1996, 97, 2081–2091. [Google Scholar] [CrossRef] [PubMed]
- Jensen-Urstad, A.P.; Semenkovich, C.F. Fatty acid synthase and liver triglyceride metabolism: Housekeeper or messenger? Biochim. Biophys. Acta 2012, 1821, 747–753. [Google Scholar] [CrossRef]
- Kersten, S.; Seydoux, J.; Peters, J.M.; Gonzalez, F.J.; Desvergne, B.; Wahli, W. Peroxisome proliferator-activated receptor alpha mediates the adaptive response to fasting. J. Clin. Investig. 1999, 103, 1489–1498. [Google Scholar] [CrossRef]
- Bowers, M.; Liang, T.; Gonzalez-Bohorquez, D.; Zocher, S.; Jaeger, B.N.; Kovacs, W.J.; Röhrl, C.; Cramb, K.M.; Winterer, J.; Kruse, M.; et al. FASN-Dependent Lipid Metabolism Links Neurogenic Stem/Progenitor Cell Activity to Learning and Memory Deficits. Cell Stem Cell 2020. [Google Scholar] [CrossRef]
- Bian, Y.; Yu, Y.; Wang, S.; Li, L. Up-regulation of fatty acid synthase induced by EGFR/ERK activation promotes tumor growth in pancreatic cancer. Biochem. Biophys. Res. Commun. 2015, 463, 612–617. [Google Scholar] [CrossRef]
- Kumar-Sinha, C.; Ignatoski, K.W.; Lippman, M.E.; Ethier, S.P.; Chinnaiyan, A.M. Transcriptome analysis of HER2 reveals a molecular connection to fatty acid synthesis. Cancer Res. 2003, 63, 132–139. [Google Scholar]
- Ravacci, G.R.; Brentani, M.M.; Tortelli, T.C.; Torrinhas, R.S.M.M.; Santos, J.R.; Logullo, A.F.; Waitzberg, D.L. Docosahexaenoic Acid Modulates a HER2-Associated Lipogenic Phenotype, Induces Apoptosis, and Increases Trastuzumab Action in HER2-Overexpressing Breast Carcinoma Cells. Biomed. Res. Int. 2015, 2015, 838652. [Google Scholar] [CrossRef]
- Demoulin, J.B.; Ericsson, J.; Kallin, A.; Rorsman, C.; Rönnstrand, L.; Heldin, C.H. Platelet-derived growth factor stimulates membrane lipid synthesis through activation of phosphatidylinositol 3-kinase and sterol regulatory element-binding proteins. J. Biol. Chem. 2004, 279, 35392–35402. [Google Scholar] [CrossRef] [PubMed]
- Eberlé, D.; Hegarty, B.; Bossard, P.; Ferré, P.; Foufelle, F. SREBP transcription factors: Master regulators of lipid homeostasis. Biochimie 2004, 86, 839–848. [Google Scholar] [CrossRef] [PubMed]
- Liang, G.; Yang, J.; Horton, J.D.; Hammer, R.E.; Goldstein, J.L.; Brown, M.S. Diminished hepatic response to fasting/refeeding and liver X receptor agonists in mice with selective deficiency of sterol regulatory element-binding protein-1c. J. Biol. Chem. 2002, 277, 9520–9528. [Google Scholar] [CrossRef] [PubMed]
- Horton, J.D.; Shimomura, I.; Brown, M.S.; Hammer, R.E.; Goldstein, J.L.; Shimano, H. Activation of cholesterol synthesis in preference to fatty acid synthesis in liver and adipose tissue of transgenic mice overproducing sterol regulatory element-binding protein-2. J. Clin. Investig. 1998, 101, 2331–2339. [Google Scholar] [CrossRef]
- Shimano, H.; Horton, J.D.; Hammer, R.E.; Shimomura, I.; Brown, M.S.; Goldstein, J.L. Overproduction of cholesterol and fatty acids causes massive liver enlargement in transgenic mice expressing truncated SREBP-1a. J. Clin. Investig. 1996, 98, 1575–1584. [Google Scholar] [CrossRef] [PubMed]
- Guo, D.; Bell, E.H.; Mischel, P.; Chakravarti, A. Targeting SREBP-1-driven lipid metabolism to treat cancer. Curr. Pharm. Des. 2014, 20, 2619–2626. [Google Scholar] [CrossRef]
- Horton, J.D.; Bashmakov, Y.; Shimomura, I.; Shimano, H. Regulation of sterol regulatory element binding proteins in livers of fasted and refed mice. Proc. Natl. Acad. Sci. USA 1998, 95, 5987–5992. [Google Scholar] [CrossRef]
- Shimomura, I.; Bashmakov, Y.; Ikemoto, S.; Horton, J.D.; Brown, M.S.; Goldstein, J.L. Insulin selectively increases SREBP-1c mRNA in the livers of rats with streptozotocin-induced diabetes. Proc. Natl. Acad. Sci. USA 1999, 96, 13656–13661. [Google Scholar] [CrossRef]
- Guo, D.; Prins, R.M.; Dang, J.; Kuga, D.; Iwanami, A.; Soto, H.; Lin, K.Y.; Huang, T.T.; Akhavan, D.; Hock, M.B.; et al. EGFR signaling through an Akt-SREBP-1-dependent, rapamycin-resistant pathway sensitizes glioblastomas to antilipogenic therapy. Sci. Signal. 2009, 2, ra82. [Google Scholar] [CrossRef]
- Heemers, H.V.; Verhoeven, G.; Swinnen, J.V. Androgen activation of the sterol regulatory element-binding protein pathway: Current insights. Mol. Endocrinol. 2006, 20, 2265–2277. [Google Scholar] [CrossRef]
- Heemers, H.; Maes, B.; Foufelle, F.; Heyns, W.; Verhoeven, G.; Swinnen, J.V. Androgens stimulate lipogenic gene expression in prostate cancer cells by activation of the sterol regulatory element-binding protein cleavage activating protein/sterol regulatory element-binding protein pathway. Mol. Endocrinol. 2001, 15, 1817–1828. [Google Scholar] [CrossRef] [PubMed]
- Lupu, R.; Menendez, J.A. Targeting fatty acid synthase in breast and endometrial cancer: An alternative to selective estrogen receptor modulators? Endocrinology 2006, 147, 4056–4066. [Google Scholar] [CrossRef]
- Lacasa, D.; Le Liepvre, X.; Ferre, P.; Dugail, I. Progesterone stimulates adipocyte determination and differentiation 1/sterol regulatory element-binding protein 1c gene expression. potential mechanism for the lipogenic effect of progesterone in adipose tissue. J. Biol. Chem. 2001, 276, 11512–11516. [Google Scholar] [CrossRef] [PubMed]
- Graner, E.; Tang, D.; Rossi, S.; Baron, A.; Migita, T.; Weinstein, L.J.; Lechpammer, M.; Huesken, D.; Zimmermann, J.; Signoretti, S.; et al. The isopeptidase USP2a regulates the stability of fatty acid synthase in prostate cancer. Cancer Cell 2004, 5, 253–261. [Google Scholar] [CrossRef]
- Lin, H.-P.; Cheng, Z.-L.; He, R.-Y.; Song, L.; Tian, M.-X.; Zhou, L.-S.; Groh, B.S.; Liu, W.-R.; Ji, M.; Ding, C.; et al. Destabilization of Fatty Acid Synthase by Acetylation Inhibits De Novo Lipogenesis and Tumor Cell Growth. Cancer Res. 2016, 76, 6924–6936. [Google Scholar] [CrossRef] [PubMed]
- Floris, A.; Mazarei, M.; Yang, X.; Robinson, A.E.; Zhou, J.; Barberis, A.; D’Hallewin, G.; Azara, E.; Spissu, Y.; Iglesias-Ara, A.; et al. SUMOylation Protects FASN Against Proteasomal Degradation in Breast Cancer Cells Treated with Grape Leaf Extract. Biomolecules 2020, 10, 529. [Google Scholar] [CrossRef]
- Gu, L.; Zhu, Y.; Lin, X.; Tan, X.; Lu, B.; Li, Y. Stabilization of FASN by ACAT1-mediated GNPAT acetylation promotes lipid metabolism and hepatocarcinogenesis. Oncogene 2020, 39, 2437–2449. [Google Scholar] [CrossRef]
- Humbert, M.; Seiler, K.; Mosimann, S.; Rentsch, V.; McKenna, S.L.; Tschan, M.P. Autophagy-mediated degradation of Fatty Acid Synthase (FASN) facilitates ATRA-induced granulocytic differentiation of acute myeloid leukemia (AML) cells. bioRxiv 2020, 924555. [Google Scholar] [CrossRef]
- Wang, H.; Luo, Q.F.; Peng, A.; Long, X.H.; Wang, T.F.; Liu, Z.L.; Zhang, G.M.; Zhou, R.P.; Gao, S.; Zhou, Y.; et al. Positive feedback regulation between Akt phosphorylation and fatty acid synthase expression in osteosarcoma. Int. J. Mol. Med. 2013, 33, 633–639. [Google Scholar] [CrossRef][Green Version]
- Hu, N.; Li, Y.; Zhao, Y.; Wang, Q.; You, J.-C.; Zhang, S.; Ye, L.-H. A novel positive feedback loop involving FASN/p-ERK1/2/5-LOX/LTB4/FASN sustains high growth of breast cancer cells. Acta Pharmacol. Sin. 2011, 32, 921–929. [Google Scholar] [CrossRef]
- Tanaka, T.; Nakata, T.; Oka, T.; Ogawa, T.; Okamoto, F.; Kusaka, Y.; Sohmiya, K.; Shimamoto, K.; Itakura, K. Defect in human myocardial long-chain fatty acid uptake is caused by FAT/CD36 mutations. J. Lipid Res. 2001, 42, 751–759. [Google Scholar] [PubMed]
- Endemann, G.; Stanton, L.W.; Madden, K.S.; Bryant, C.M.; White, R.T.; Protter, A.A. CD36 is a receptor for oxidized low density lipoprotein. J. Boil. Chem. 1993, 268, 11811–11816. [Google Scholar]
- Baranova, I.N.; Bocharov, A.V.; Vishnyakova, T.G.; Kurlander, R.; Chen, Z.; Fu, D.; Arias, I.M.; Csako, G.; Patterson, A.P.; Eggerman, T.L. CD36 Is a Novel Serum Amyloid A (SAA) Receptor Mediating SAA Binding and SAA-induced Signaling in Human and Rodent Cells. J. Boil. Chem. 2010, 285, 8492–8506. [Google Scholar] [CrossRef] [PubMed]
- Seimon, T.A.; Nadolski, M.J.; Liao, X.; Magallon, J.; Nguyen, M.; Feric, N.T.; Koschinsky, M.L.; Harkewicz, R.; Witztum, J.L.; Tsimikas, S.; et al. Atherogenic Lipids and Lipoproteins Trigger CD36-TLR2-Dependent Apoptosis in Macrophages Undergoing Endoplasmic Reticulum Stress. Cell Metab. 2010, 12, 467–482. [Google Scholar] [CrossRef]
- Ladányi, A.; Mukherjee, A.; Kenny, H.A.; Johnson, A.; Mitra, A.K.; Sundaresan, S.; Nieman, K.M.; Pascual, G.; Benitah, S.A.; Montag, A.; et al. Adipocyte-induced CD36 expression drives ovarian cancer progression and metastasis. Oncogene 2018, 37, 2285–2301. [Google Scholar] [CrossRef]
- Pan, J.; Fan, Z.; Wang, Z.; Dai, Q.; Xiang, Z.; Yuan, F.; Yan, M.; Zhu, Z.; Liu, B.-Y.; Li, C. CD36 mediates palmitate acid-induced metastasis of gastric cancer via AKT/GSK-3β/β-catenin pathway. J. Exp. Clin. Cancer Res. 2019, 38, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Han, H.; Liu, L.; Duan, Y.; Yang, X.; Ma, C.; Zhu, Y.; Han, J.; Li, X.; Chen, Y. CD36 plays a critical role in proliferation, migration and tamoxifen-inhibited growth of ER-positive breast cancer cells. Oncogenesis 2018, 7, 98. [Google Scholar] [CrossRef] [PubMed]
- Nath, A.; Li, I.; Roberts, L.R.; Chan, C. Elevated free fatty acid uptake via CD36 promotes epithelial-mesenchymal transition in hepatocellular carcinoma. Sci. Rep. 2015, 5, 14752. [Google Scholar] [CrossRef]
- Nath, A.; Chan, C. Genetic alterations in fatty acid transport and metabolism genes are associated with metastatic progression and poor prognosis of human cancers. Sci. Rep. 2016, 6, 18669. [Google Scholar] [CrossRef]
- Yen, M.C.; Chou, S.K.; Kan, J.Y.; Kuo, P.L.; Hou, M.F.; Hsu, Y.L. New Insight on Solute Carrier Family 27 Member 6 (SLC27A6) in Tumoral and Non-Tumoral Breast Cells. Int. J. Med. Sci. 2019, 16, 366–375. [Google Scholar] [CrossRef]
- Yen, M.C.; Chou, S.K.; Kan, J.Y.; Kuo, P.L.; Hou, M.F.; Hsu, Y.L. Solute Carrier Family 27 Member 4 (SLC27A4) Enhances Cell Growth, Migration, and Invasion in Breast Cancer Cells. Int. J. Mol. Sci. 2018, 19, 3434. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Di Martino, J.S.; Bowman, R.M.; Campbell, N.; Baksh, S.C.; Simon-Vermot, T.; Kim, I.S.; Haldeman, P.; Mondal, C.; Yong-Gonzales, V.; et al. Adipocyte-Derived Lipids Mediate Melanoma Progression via FATP Proteins. Cancer Discov. 2018, 8, 1006–1025. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-S.; Jung, J.; Jeong, H.; Lee, J.H.; Oh, H.E.; Lee, E.S.; Choi, J.W. High Membranous Expression of Fatty Acid Transport Protein 4 Is Associated with Tumorigenesis and Tumor Progression in Clear Cell Renal Cell Carcinoma. Dis. Markers 2019, 2019, 5702026. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Rao, E.; Zeng, J.; Hao, J.; Sun, Y.; Liu, S.; Sauter, E.R.; Bernlohr, D.A.; Cleary, M.P.; Suttles, J.; et al. Adipose Fatty Acid Binding Protein Promotes Saturated Fatty Acid-Induced Macrophage Cell Death through Enhancing Ceramide Production. J. Immunol. 2016, 198, 798–807. [Google Scholar] [CrossRef]
- Zimmerman, A.W.; Veerkamp, J.H. New insights into the structure and function of fatty acid-binding proteins. Cell Mol. Life Sci. 2002, 59, 1096–1116. [Google Scholar] [CrossRef]
- Coe, N.R.; Bernlohr, D.A. Physiological properties and functions of intracellular fatty acid-binding proteins. Biochim. Biophys. Acta 1998, 1391, 287–306. [Google Scholar] [CrossRef]
- Amiri, M.; Yousefnia, S.; Seyed Forootan, F.; Peymani, M.; Ghaedi, K.; Nasr Esfahani, M.H. Diverse roles of fatty acid binding proteins (FABPs) in development and pathogenesis of cancers. Gene 2018, 676, 171–183. [Google Scholar] [CrossRef]
- Furuhashi, M.; Hotamisligil, G.S. Fatty acid-binding proteins: Role in metabolic diseases and potential as drug targets. Nat. Rev. Drug Discov. 2008, 7, 489–503. [Google Scholar] [CrossRef]
- Kawaguchi, K.; Senga, S.; Kubota, C.; Kawamura, Y.; Ke, Y.; Fujii, H. High expression of Fatty Acid-Binding Protein 5 promotes cell growth and metastatic potential of colorectal cancer cells. FEBS Open Biol. 2016, 6, 190–199. [Google Scholar] [CrossRef]
- Tölle, A.; Suhail, S.; Jung, M.; Jung, K.; Stephan, C. Fatty acid binding proteins (FABPs) in prostate, bladder and kidney cancer cell lines and the use of IL-FABP as survival predictor in patients with renal cell carcinoma. BMC Cancer 2011, 11, 302. [Google Scholar] [CrossRef]
- Uehara, H.; Takahashi, T.; Oha, M.; Ogawa, H.; Izumi, K. Exogenous fatty acid binding protein 4 promotes human prostate cancer cell progression. Int. J. Cancer 2014, 135, 2558–2568. [Google Scholar] [CrossRef] [PubMed]
- Medes, G.; Thomas, A.; Weinhouse, S. Metabolism of neoplastic tissue. IV. A study of lipid synthesis in neoplastic tissue slices in vitro. Cancer Res. 1953, 13, 27–29. [Google Scholar] [PubMed]
- Szutowicz, A.; Kwiatkowski, J.; Angielski, S. Lipogenetic and glycolytic enzyme activities in carcinoma and nonmalignant diseases of the human breast. Br. J. Cancer 1979, 39, 681–687. [Google Scholar] [CrossRef] [PubMed]
- Kuhajda, F.P.; Jenner, K.; Wood, F.D.; Hennigar, R.A.; Jacobs, L.B.; Dick, J.D.; Pasternack, G.R. Fatty acid synthesis: A potential selective target for antineoplastic therapy. Proc. Natl. Acad. Sci. USA 1994, 91, 6379–6383. [Google Scholar] [CrossRef]
- Kastaniotis, A.J.; Autio, K.J.; Kerätär, J.M.; Monteuuis, G.; Mäkelä, A.M.; Nair, R.R.; Pietikäinen, L.P.; Shvetsova, A.; Chen, Z.; Hiltunen, K. Mitochondrial fatty acid synthesis, fatty acids and mitochondrial physiology. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2017, 1862, 39–48. [Google Scholar] [CrossRef]
- Hiltunen, K.; Autio, K.J.; Schonauer, M.S.; Kursu, V.S.; Dieckmann, C.L.; Kastaniotis, A.J. Mitochondrial fatty acid synthesis and respiration. Biochim. Biophys. Acta 2010, 1797, 1195–1202. [Google Scholar] [CrossRef]
- Madigan, A.A.; Rycyna, K.J.; Parwani, A.V.; Datiri, Y.J.; Basudan, A.M.; Sobek, K.M.; Cummings, J.L.; Basse, P.H.; Bacich, D.J.; O’Keefe, D.S. Novel Nuclear Localization of Fatty Acid Synthase Correlates with Prostate Cancer Aggressiveness. Am. J. Pathol. 2014, 184, 2156–2162. [Google Scholar] [CrossRef]
- Hillebrand, M.; Gersting, S.W.; Lotz-Havla, A.S.; Schäfer, A.; Rosewich, H.; Valerius, O.; Muntau, A.C.; Gärtner, J. Identification of a New Fatty Acid Synthesis-Transport Machinery at the Peroxisomal Membrane. J. Boil. Chem. 2011, 287, 210–221. [Google Scholar] [CrossRef]
- Jensen-Urstad, A.P.L.; Song, H.; Lodhi, I.J.; Funai, K.; Yin, L.; Coleman, T.; Semenkovich, C.F. Nutrient-dependent phosphorylation channels lipid synthesis to regulate PPARα. J. Lipid Res. 2013, 54, 1848–1859. [Google Scholar] [CrossRef]
- Jin, Q.; Yuan, L.X.; Boulbes, D.R.; Baek, J.M.; Wang, Y.-N.; Gomez-Cabello, D.; Hawke, D.; Yeung, S.-C.J.; Lee, M.-H.; Hortobágyi, G.N.; et al. Fatty acid synthase phosphorylation: A novel therapeutic target in HER2-overexpressing breast cancer cells. Breast Cancer Res. 2010, 12, R96. [Google Scholar] [CrossRef]
- Di Vizio, D.; Adam, R.M.; Kim, J.; Kim, R.; Sotgia, F.; Williams, T.; Demichelis, F.; Solomon, K.R.; Loda, M.; Rubin, M.A.; et al. Caveolin-1 interacts with a lipid raft-associated population of fatty acid synthase. Cell Cycle 2008, 7, 2257–2267. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Lu, J.; Su, H.; Yang, J.; Zhou, D. Fatty acid synthase cooperates with protrudin to facilitate membrane outgrowth of cellular protrusions. Sci. Rep. 2017, 7, 46569. [Google Scholar] [CrossRef] [PubMed]
- Momcilovic, M.; Jones, A.; Bailey, S.T.; Waldmann, C.M.; Li, R.; Lee, J.T.; Abdelhady, G.; Gomez, A.; Holloway, T.; Schmid, E.; et al. In vivo imaging of mitochondrial membrane potential in non-small-cell lung cancer. Nature 2019, 575, 380–384. [Google Scholar] [CrossRef] [PubMed]
- LeBleu, V.S.; O’Connell, J.T.; Herrera, K.N.G.; Wikman, H.; Pantel, K.; Haigis, M.C.; De Carvalho, F.M.; Damascena, A.; Chinen, L.T.D.; Rocha, R.M.; et al. PGC-1α mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nature 2014, 16, 992–1003. [Google Scholar] [CrossRef]
- Bin Ali, A.; Levantini, E.; Teo, J.T.; Goggi, J.; Clohessy, J.G.; Wu, C.S.; Chen, L.; Yang, H.; Krishnan, I.; Kocher, O.; et al. Fatty acid synthase mediates EGFR palmitoylation in EGFR mutated non-small cell lung cancer. EMBO Mol. Med. 2018, 10, e8313. [Google Scholar] [CrossRef]
- Zaytseva, Y.Y.; Rychahou, P.; Le, A.-T.; Scott, T.L.; Flight, R.M.; Kim, J.T.; Harris, J.; Liu, J.; Wang, C.; Morris, A.J.; et al. Preclinical evaluation of novel fatty acid synthase inhibitors in primary colorectal cancer cells and a patient-derived xenograft model of colorectal cancer. Oncotarget 2018, 9, 24787–24800. [Google Scholar] [CrossRef]
- Zaytseva, Y.Y.; Harris, J.W.; Mitov, M.I.; Kim, J.T.; Butterfield, D.A.; Lee, E.Y.; Weiss, H.L.; Gao, T.; Evers, B.M. Increased expression of fatty acid synthase provides a survival advantage to colorectal cancer cells via upregulation of cellular respiration. Oncotarget 2015, 6, 18891–18904. [Google Scholar] [CrossRef]
- Lee, C.-K.; Jeong, S.-H.; Jang, C.; Bae, H.; Kim, Y.H.; Park, I.; Kim, S.K.; Koh, G.Y. Tumor metastasis to lymph nodes requires YAP-dependent metabolic adaptation. Science 2019, 363, 644–649. [Google Scholar] [CrossRef]
- Park, J.H.; Vithayathil, S.; Kumar, S.; Sung, P.-L.; Dobrolecki, L.E.; Putluri, V.; Bhat, V.B.; Bhowmik, S.K.; Gupta, V.; Arora, K.; et al. Fatty Acid Oxidation-Driven Src Links Mitochondrial Energy Reprogramming and Oncogenic Properties in Triple-Negative Breast Cancer. Cell Rep. 2016, 14, 2154–2165. [Google Scholar] [CrossRef]
- Wang, Y.-N.; Zeng, Z.-L.; Lu, J.; Wang, Y.; Liu, Z.-X.; He, M.-M.; Zhao, Q.; Wang, Z.-X.; Li, T.; Lu, Y.-X.; et al. CPT1A-mediated fatty acid oxidation promotes colorectal cancer cell metastasis by inhibiting anoikis. Oncogene 2018, 37, 6025–6040. [Google Scholar] [CrossRef]
- Bollu, L.R.; Ren, J.; Blessing, A.M.; Katreddy, R.R.; Gao, G.; Xu, L.; Wang, J.; Su, F.; Weihua, Z. Involvement of de novo synthesized palmitate and mitochondrial EGFR in EGF induced mitochondrial fusion of cancer cells. Cell Cycle 2014, 13, 2415–2430. [Google Scholar] [CrossRef] [PubMed]
- Chandran, K.; Goswami, S.; Sharma-Walia, N. Implications of a peroxisome proliferator-activated receptor alpha (PPARα) ligand clofibrate in breast cancer. Oncotarget 2015, 7, 15577–15599. [Google Scholar] [CrossRef]
- Wu, X.; Dong, Z.; Wang, C.J.; Barlow, L.J.; Fako, V.; Serrano, M.A.; Zou, Y.; Liu, J.-Y.; Zhang, J.-T. FASN regulates cellular response to genotoxic treatments by increasing PARP-1 expression and DNA repair activity via NF-κB and SP1. Proc. Natl. Acad. Sci. USA 2016, 113, E6965–E6973. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Jiang, S.; Fu, Q.; Smith, K.D.; Tu, K.; Li, H.; Zhao, Y. FASN, ErbB2-mediated glycolysis is required for breast cancer cell migration. Oncol. Rep. 2016, 35, 2715–2722. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, A.P.; Jacobs, S.R.; Freemerman, A.J.; Makowski, L.; Rathmell, J.C.; Dittmer, D.P.; Damania, B. Dysregulation of fatty acid synthesis and glycolysis in non-Hodgkin lymphoma. Proc. Natl. Acad. Sci. USA 2012, 109, 11818–11823. [Google Scholar] [CrossRef]
- Zhan, N.; Li, B.; Xu, X.; Xu, J.; Hu, S. Inhibition of FASN expression enhances radiosensitivity in human non-small cell lung cancer. Oncol. Lett. 2018, 15, 4578–4584. [Google Scholar] [CrossRef]
- Little, J.L.; Wheeler, F.B.; Fels, D.R.; Koumenis, C.; Kridel, S.J. Inhibition of fatty acid synthase induces endoplasmic reticulum stress in tumor cells. Cancer Res. 2007, 67, 1262–1269. [Google Scholar] [CrossRef]
- Pizer, E.S.; Jackisch, C.; Wood, F.D.; Pasternack, G.R.; Davidson, N.E.; Kuhajda, F.P. Inhibition of fatty acid synthesis induces programmed cell death in human breast cancer cells. Cancer Res. 1996, 56, 2745–2747. [Google Scholar]
- Sokolowska, E.; Presler, M.; Goyke, E.; Milczarek, R.; Swierczynski, J.; Sledzinski, T. Orlistat Reduces Proliferation and Enhances Apoptosis in Human Pancreatic Cancer Cells (PANC-1). Anticancer Res. 2017, 37, 6321–6327. [Google Scholar] [CrossRef]
- Voss, K.; Luthers, C.R.; Pohida, K.; Snow, A.L. Fatty Acid Synthase Contributes to Restimulation-Induced Cell Death of Human CD4 T Cells. Front. Mol. Biosci. 2019, 6, 106. [Google Scholar] [CrossRef]
- Jia, D.; Park, J.H.; Jung, K.H.; Levine, H.; Kaipparettu, B.A. Elucidating the Metabolic Plasticity of Cancer: Mitochondrial Reprogramming and Hybrid Metabolic States. Cells 2018, 7, 21. [Google Scholar] [CrossRef]
- Labora, J.A.F.; Carpintero-Fernández, P.; Jordan, S.J.D.; Shikh-Bahaei, T.; Abdullah, S.M.; Mahenthiran, M.; Rodríguez-Navarro, J.A.; Niklison-Chirou, M.V.; O’Loghlen, A. FASN activity is important for the initial stages of the induction of senescence. Cell Death Dis. 2019, 10, 318. [Google Scholar] [CrossRef]
- Cerveny, K.L.; Tamura, Y.; Zhang, Z.; Jensen, R.E.; Sesaki, H. Regulation of mitochondrial fusion and division. Trends Cell Biol. 2007, 17, 563–569. [Google Scholar] [CrossRef]
- Tyagi, S.; Gupta, P.; Saini, A.S.; Kaushal, C.; Sharma, S. The peroxisome proliferator-activated receptor: A family of nuclear receptors role in various diseases. J. Adv. Pharm Technol. Res. 2011, 2, 236–240. [Google Scholar] [CrossRef] [PubMed]
- Dreyer, C.; Krey, G.; Keller, H.; Givel, F.; Helftenbein, G.; Wahli, W. Control of the peroxisomal beta-oxidation pathway by a novel family of nuclear hormone receptors. Cell 1992, 68, 879–887. [Google Scholar] [CrossRef]
- Chakravarthy, M.V.; Pan, Z.; Zhu, Y.; Tordjman, K.; Schneider, J.G.; Coleman, T.; Turk, J.; Semenkovich, C.F. “New” hepatic fat activates PPARα to maintain glucose, lipid, and cholesterol homeostasis. Cell Metab. 2005, 1, 309–322. [Google Scholar] [CrossRef] [PubMed]
- Chakravarthy, M.V.; Zhu, Y.; López, M.; Yin, L.; Wozniak, D.F.; Coleman, T.; Hu, Z.; Wolfgang, M.; Vidal-Puig, A.; Lane, M.D.; et al. Brain fatty acid synthase activates PPARα to maintain energy homeostasis. J. Clin. Investig. 2007, 117, 2539–2552. [Google Scholar] [CrossRef] [PubMed]
- Pawlak, M.; Lefebvre, P.; Staels, B. Molecular mechanism of PPARα action and its impact on lipid metabolism, inflammation and fibrosis in non-alcoholic fatty liver disease. J. Hepatol. 2015, 62, 720–733. [Google Scholar] [CrossRef] [PubMed]
- Wierzbicki, M.; Chabowski, A.; Zendzian-Piotrowska, M.; Gorski, J. Differential effects of in vivo PPAR alpha and gamma activation on fatty acid transport proteins expression and lipid content in rat liver. J. Physiol. Pharmacol. 2009, 60, 99–106. [Google Scholar]
- Ahmed, W.; Ziouzenkova, O.; Brown, J.; Devchand, P.; Francis, S.; Kadakia, M.; Kanda, T.; Orasanu, G.; Sharlach, M.; Zandbergen, F.; et al. PPARs and their metabolic modulation: New mechanisms for transcriptional regulation? J. Intern. Med. 2007, 262, 184–198. [Google Scholar] [CrossRef]
- Li, M.-Y.; Yuan, H.; Ma, L.T.; Kong, A.W.Y.; Hsin, M.K.Y.; Yip, J.H.Y.; Underwood, M.J.; Chen, G.G. Roles of Peroxisome Proliferator–Activated Receptor–α and –γ in the Development of Non–Small Cell Lung Cancer. Am. J. Respir. Cell Mol. Boil. 2010, 43, 674–683. [Google Scholar] [CrossRef] [PubMed]
- Ventura-Clapier, R.; Garnier, A.; Veksler, V. Transcriptional control of mitochondrial biogenesis: The central role of PGC-1. Cardiovasc. Res. 2008, 79, 208–217. [Google Scholar] [CrossRef] [PubMed]
- Billi, A.M.; Cappellini, A.; Manzoli, L.; Cocco, L.; Martelli, A.M.; Faenza, I. Metabolism and signaling activities of nuclear lipids. Cell. Mol. Life Sci. 2004, 61, 1143–1156. [Google Scholar] [CrossRef]
- Scaglia, N.; Tyekucheva, S.; Zadra, G.; Photopoulos, C.; Loda, M. De novo fatty acid synthesis at the mitotic exit is required to complete cellular division. Cell Cycle 2014, 13, 859–868. [Google Scholar] [CrossRef]
- Rodriguez Sawicki, L.; Garcia, K.A.; Corsico, B.; Scaglia, N. lipogenesis at the mitotic exit is used for nuclear envelope reassembly/expansion. Implications for combined chemotherapy. Cell Cycle 2019, 18, 1646–1659. [Google Scholar] [CrossRef]
- Han, J.; Kaufman, R.J. The role of ER stress in lipid metabolism and lipotoxicity. J. Lipid Res. 2016, 57, 1329–1338. [Google Scholar] [CrossRef]
- Rutkowski, D.T.; Wu, J.; Back, S.-H.; Callaghan, M.U.; Ferris, S.P.; Iqbal, J.; Clark, R.; Miao, H.; Hassler, J.R.; Fornek, J.; et al. UPR Pathways Combine to Prevent Hepatic Steatosis Caused by ER Stress-Mediated Suppression of Transcriptional Master Regulators. Dev. Cell 2008, 15, 829–840. [Google Scholar] [CrossRef]
- Sankaranarayanapillai, M.; Zhang, N.; Baggerly, K.A.; Gelovani, J.G. Metabolic shifts induced by fatty acid synthase inhibitor orlistat in non-small cell lung carcinoma cells provide novel pharmacodynamic biomarkers for positron emission tomography and magnetic resonance spectroscopy. Mol. Imaging Biol. 2013, 15, 136–147. [Google Scholar] [CrossRef]
- Knowles, L.M.; Smith, J.W. Genome-wide changes accompanying knockdown of fatty acid synthase in breast cancer. BMC Genom. 2007, 8, 168. [Google Scholar] [CrossRef]
- Flavin, R.; Peluso, S.; Nguyen, P.L.; Loda, M. Fatty acid synthase as a potential therapeutic target in cancer. Future Oncol. 2010, 6, 551–562. [Google Scholar] [CrossRef]
- Pizer, E.S.; Wood, F.D.; Heine, H.S.; Romantsev, F.E.; Pasternack, G.R.; Kuhajda, F.P. Inhibition of fatty acid synthesis delays disease progression in a xenograft model of ovarian cancer. Cancer Res. 1996, 56, 1189–1193. [Google Scholar] [PubMed]
- Angeles, T.S.; Hudkins, R.L. Recent advances in targeting the fatty acid biosynthetic pathway using fatty acid synthase inhibitors. Expert. Opin. Drug Discov. 2016, 11, 1187–1199. [Google Scholar] [CrossRef] [PubMed]
- Corominas-Faja, B.; Vellon, L.; Cuyàs, E.; Buxó, M.; Martin-Castilló, B.; Serra, L.; García, J.; Lupu, R.; Menendez, J.A. Clinical and therapeutic relevance of the metabolic oncogene fatty acid synthase in HER2+ breast cancer. Histol. Histopathol. 2016, 32, 687–698. [Google Scholar] [CrossRef] [PubMed]
- Relat, J.; Blancafort, A.; Oliveras, G.; Cufí, S.; Haro, D.; Marrero, P.F.; Puig, T. Different fatty acid metabolism effects of (−)-Epigallocatechin-3-Gallate and C75 in Adenocarcinoma lung cancer. BMC Cancer 2012, 12, 280. [Google Scholar] [CrossRef] [PubMed]
- Loftus, T.M.; Jaworsky, D.E.; Frehywot, G.L.; Townsend, C.A.; Ronnett, G.V.; Lane, M.D.; Kuhajda, F.P. Reduced Food Intake and Body Weight in Mice Treated with Fatty Acid Synthase Inhibitors. Science 2000, 288, 2379–2381. [Google Scholar] [CrossRef]
- Landree, L.E. C75, a Fatty Acid Synthase Inhibitor, Modulates AMP-activated Protein Kinase to Alter Neuronal Energy Metabolism. J. Boil. Chem. 2003, 279, 3817–3827. [Google Scholar] [CrossRef]
- Cheng, X.; Li, L.; Uttamchandani, M.; Yao, S.Q. In situ proteome profiling of C75, a covalent bioactive compound with potential anticancer activities. Org. Lett. 2014, 16, 1414–1417. [Google Scholar] [CrossRef]
- Sternby, B.; Hartmann, D.; Borgström, B.; Nilsson, A. Degree of in vivo inhibition of human gastric and pancreatic lipases by Orlistat (Tetrahydrolipstatin, THL) in the stomach and small intestine. Clin. Nutr. 2002, 21, 395–402. [Google Scholar] [CrossRef]
- Pemble, C.W.; Johnson, L.C.; Kridel, S.J.; Lowther, W.T. Crystal structure of the thioesterase domain of human fatty acid synthase inhibited by Orlistat. Nat. Struct. Mol. Biol. 2007, 14, 704–709. [Google Scholar] [CrossRef]
- Zhi, J.; Melia, A.T.; Eggers, H.; Joly, R.; Patel, I.H. Review of limited systemic absorption of orlistat, a lipase inhibitor, in healthy human volunteers. J. Clin. Pharmacol. 1995, 35, 1103–1108. [Google Scholar] [CrossRef]
- Brenner, A.J.; Von Hoff, D.D.; Infante, J.R.; Patel, M.R.; Jones, S.F.; Burris, H.A.; Rubino, C.; McCulloch, W.; Zhukova-Harrill, V.; Kemble, G. First-in-human investigation of the oral first-in-class fatty acid synthase (FASN) inhibitor, TVB-2640. J. Clin. Oncol. 2015, 33, TPS2615. [Google Scholar] [CrossRef]
- Daemen, A.; Peterson, D.; Sahu, N.; Mccord, R.; Du, X.; Liu, B.; Kowanetz, K.; Hong, R.; Moffat, J.; Gao, M.; et al. Metabolite profiling stratifies pancreatic ductal adenocarcinomas into subtypes with distinct sensitivities to metabolic inhibitors. Proc. Natl. Acad. Sci. USA 2015, 112, E4410–E4417. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Zhou, L.; Li, H.; Tian, Y.; Li, J.; Dong, L.; Zhao, Y.; Wei, D. Fatty acid synthase affects expression of ErbB receptors in epithelial to mesenchymal transition of breast cancer cells and invasive ductal carcinoma. Oncol. Lett. 2017, 14, 5934–5946. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yan, C.; Wei, H.; Minjuan, Z.; Yan, X.; Jingyue, Y.; Wenchao, L.; Sheng, H. The mTOR Inhibitor Rapamycin Synergizes with a Fatty Acid Synthase Inhibitor to Induce Cytotoxicity in ER/HER2-Positive Breast Cancer Cells. PLoS ONE 2014, 9, e97697. [Google Scholar] [CrossRef]
- Shiragami, R.; Murata, S.; Kosugi, C.; Tezuka, T.; Yamazaki, M.; Hirano, A.; Yoshimura, Y.; Suzuki, M.; Shuto, K.; Koda, K. Enhanced antitumor activity of cerulenin combined with oxaliplatin in human colon cancer cells. Int. J. Oncol. 2013, 43, 431–438. [Google Scholar] [CrossRef]
- Gouw, A.M.; Eberlin, L.S.; Margulis, K.; Sullivan, D.K.; Toal, G.G.; Tong, L.; Zare, R.N.; Felsher, D.W. Oncogene KRAS activates fatty acid synthase, resulting in specific ERK and lipid signatures associated with lung adenocarcinoma. Proc. Natl. Acad. Sci. USA 2017, 114, 4300–4305. [Google Scholar] [CrossRef]
- Chang, L.; Wu, P.; Senthilkumar, R.; Tian, X.; Liu, H.; Shen, X.; Tao, Z.; Huang, P. Loss of fatty acid synthase suppresses the malignant phenotype of colorectal cancer cells by down-regulating energy metabolism and mTOR signaling pathway. J. Cancer Res. Clin. Oncol. 2015, 142, 59–72. [Google Scholar] [CrossRef]
- Wang, T.F.; Wang, H.; Peng, A.; Luo, Q.F.; Liu, Z.L.; Zhou, R.P.; Gao, S.; Zhou, Y.; Chen, W.Z. Inhibition of fatty acid synthase suppresses U-2 OS cell invasion and migration via downregulating the activity of HER2/PI3K/AKT signaling pathway in vitro. Biochem. Biophys. Res. Commun. 2013, 440, 229–234. [Google Scholar] [CrossRef]
- Deepa, P.R.; Vandhana, S.; Krishnakumar, S. Fatty Acid Synthase Inhibition Induces Differential Expression of Genes Involved in Apoptosis and Cell Proliferation in Ocular Cancer Cells. Nutr. Cancer 2013, 65, 311–316. [Google Scholar] [CrossRef]
- Jiang, B.; Li, E.-H.; Lu, Y.-Y.; Jiang, Q.; Cui, D.; Jing, Y.-F.; Xia, S.-J. Inhibition of Fatty-acid Synthase Suppresses P-AKT and Induces Apoptosis in Bladder Cancer. Urology 2012, 80, 484.e9–484.e15. [Google Scholar] [CrossRef]
- Zecchin, K.G.; Rossato, F.A.; Raposo, H.F.; Melo, D.R.; Alberici, L.C.; Oliveira, H.C.F.; Castilho, R.F.; Coletta, R.D.; Vercesi, A.E.; Graner, E. Inhibition of fatty acid synthase in melanoma cells activates the intrinsic pathway of apoptosis. Lab. Investig. 2010, 91, 232–240. [Google Scholar] [CrossRef]
- Menendez, J.A.; Lupu, R. Fatty acid synthase regulates estrogen receptor-α signaling in breast cancer cells. Oncogenesis 2017, 6, e299. [Google Scholar] [CrossRef] [PubMed]
- Menendez, J.A.; Vazquez-Martin, A.; Fernandez-Real, J.M.; Oliveras-Ferraros, C.; Navarrete, J.M.; Martin-Castilló, B.; Del Barco, S.; Brunet, J. Fatty acid synthase activity regulates HER2 extracellular domain shedding into the circulation of HER2-positive metastatic breast cancer patients. Int. J. Oncol. 2009, 35, 1369–1376. [Google Scholar] [CrossRef] [PubMed]
- Vazquez-Martin, A.; Colomer, R.; Brunet, J.; Lupu, R.; Menendez, J.A. Overexpression of fatty acid synthase gene activates HER1/HER2 tyrosine kinase receptors in human breast epithelial cells. Cell Prolif. 2008, 41, 59–85. [Google Scholar] [CrossRef] [PubMed]
- Puig, T.; Vázquez-Martín, A.; Relat, J.; Petriz, J.; Menendez, J.A.; Porta, R.; Casals, G.; Marrero, P.F.; Haro, D.; Brunet, J.; et al. Fatty acid metabolism in breast cancer cells: Differential inhibitory effects of epigallocatechin gallate (EGCG) and C75. Breast Cancer Res. Treat. 2007, 109, 471–479. [Google Scholar] [CrossRef]
- Duan, J.; Chen, L.; Zhou, M.; Zhang, J.; Sun, L.; Huang, N.; Bin, J.; Liao, Y.; Liao, W. MACC1 decreases the chemosensitivity of gastric cancer cells to oxaliplatin by regulating FASN expression. Oncol. Rep. 2017, 37, 2583–2592. [Google Scholar] [CrossRef]
- Rahman, M.T.; Nakayama, K.; Ishikawa, M.; Rahman, M.; Katagiri, H.; Katagiri, A.; Ishibashi, T.; Iida, K.; Miyazaki, K. Fatty Acid Synthase Is a Potential Therapeutic Target in Estrogen Receptor-/Progesterone Receptor-Positive Endometrioid Endometrial Cancer. Oncology 2013, 84, 166–173. [Google Scholar] [CrossRef]
- Uddin, S.; Hussain, A.R.; Ahmed, M.; Bu, R.; Ahmed, S.O.; Ajarim, D.; Al-Dayel, F.; Bavi, P.; Al-Kuraya, K.S. Inhibition of Fatty Acid Synthase Suppresses c-Met Receptor Kinase and Induces Apoptosis in Diffuse Large B-Cell Lymphoma. Mol. Cancer Ther. 2010, 9, 1244–1255. [Google Scholar] [CrossRef]
- Chen, H.-W.; Chang, Y.-F.; Chuang, H.-Y.; Tai, W.-T.; Hwang, J.-J. Targeted therapy with fatty acid synthase inhibitors in a human prostate carcinoma LNCaP/tk-luc-bearing animal model. Prostate Cancer Prostatic Dis. 2012, 15, 260–264. [Google Scholar] [CrossRef][Green Version]
- Chang, L.; Fang, S.; Chen, Y.; Yang, Z.; Yuan, Y.; Zhang, J.; Ye, L.; Gu, W. Inhibition of FASN suppresses the malignant biological behavior of non-small cell lung cancer cells via deregulating glucose metabolism and AKT/ERK pathway. Lipids Health Dis. 2019, 18, 118. [Google Scholar] [CrossRef]
- Gélébart, P.; Zak, Z.; Anand, M.; Belch, A.; Lai, R. Blockade of Fatty Acid Synthase Triggers Significant Apoptosis in Mantle Cell Lymphoma. PLoS ONE 2012, 7, e33738. [Google Scholar] [CrossRef]
- Cioccoloni, G.; Aquino, A.; Notarnicola, M.; Caruso, M.G.; Bonmassar, E.; Zonfrillo, M.; Caporali, S.; Faraoni, I.; Villivà, C.; Fuggetta, M.P.; et al. Fatty acid synthase inhibitor orlistat impairs cell growth and down-regulates PD-L1 expression of a human T-cell leukemia line. J. Chemother. 2019, 32, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Chuang, H.-Y.; Lee, Y.-P.; Lin, W.-C.; Lin, Y.-H.; Hwang, J.-J. Fatty Acid Inhibition Sensitizes Androgen-Dependent and -Independent Prostate Cancer to Radiotherapy via FASN/NF-κB Pathway. Sci. Rep. 2019, 9, 13284. [Google Scholar] [CrossRef] [PubMed]
- Wright, C.; Iyer, A.K.V.; Kaushik, V.; Azad, N. Anti-Tumorigenic Potential of a Novel Orlistat-AICAR Combination in Prostate Cancer Cells. J. Cell. Biochem. 2017, 118, 3834–3845. [Google Scholar] [CrossRef] [PubMed]
- Seguin, F.; Carvalho, M.A.; Bastos, D.C.; Agostini, M.; Zecchin, K.G.; Alvarez-Flores, M.; Chudzinski-Tavassi, A.M.; Coletta, R.D.; Graner, E. The fatty acid synthase inhibitor orlistat reduces experimental metastases and angiogenesis in B16-F10 melanomas. Br. J. Cancer 2012, 107, 977–987. [Google Scholar] [CrossRef]
- You, B.-J.; Chen, L.-Y.; Hsu, P.-H.; Sung, P.-H.; Hung, Y.-C.; Lee, H.-Z. Orlistat Displays Antitumor Activity and Enhances the Efficacy of Paclitaxel in Human Hepatoma Hep3B Cells. Chem. Res. Toxicol. 2019, 32, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Agostini, M.; Almeida, L.Y.; Bastos, D.C.; Ortega, R.M.; Moreira, F.S.; Seguin, F.; Zecchin, K.G.; Raposo, H.F.; Oliveira, H.C.F.; Salo, T.; et al. The Fatty Acid Synthase Inhibitor Orlistat Reduces the Growth and Metastasis of Orthotopic Tongue Oral Squamous Cell Carcinomas. Mol. Cancer Ther. 2013, 13, 585–595. [Google Scholar] [CrossRef]
- Papaevangelou, E.; Almeida, G.S.; Box, C.; DeSouza, N.M.; Chung, Y.-L. The effect of FASN inhibition on the growth and metabolism of a cisplatin-resistant ovarian carcinoma model. Int. J. Cancer 2018, 143, 992–1002. [Google Scholar] [CrossRef]
- Mims, J.; Bansal, N.; Bharadwaj, M.S.; Chen, X.; Molina, A.J.; Tsang, A.W.; Furdui, C.M. Energy metabolism in a matched model of radiation resistance for head and neck squamous cell cancer. Radiat. Res. 2015, 183, 291–304. [Google Scholar] [CrossRef]
- Grube, S.; Dünisch, P.; Freitag, D.; Klausnitzer, M.; Sakr, Y.; Walter, J.; Kalff, R.; Ewald, C. Overexpression of fatty acid synthase in human gliomas correlates with the WHO tumor grade and inhibition with Orlistat reduces cell viability and triggers apoptosis. J. Neurooncol. 2014, 118, 277–287. [Google Scholar] [CrossRef]
- Li, C.-F.; Fang, F.-M.; Chen, Y.-Y.; Liu, T.-T.; Chan, T.-C.; Yu, S.-C.; Chen, L.-T.; Huang, H.-Y. Overexpressed Fatty Acid Synthase in Gastrointestinal Stromal Tumors: Targeting a Progression-Associated Metabolic Driver Enhances the Antitumor Effect of Imatinib. Clin. Cancer Res. 2017, 23, 4908–4918. [Google Scholar] [CrossRef] [PubMed]
- Kant, S.; Kumar, A.; Singh, S.M. Myelopoietic Efficacy of Orlistat in Murine Hosts Bearing T Cell Lymphoma: Implication in Macrophage Differentiation and Activation. PLoS ONE 2013, 8, e82396. [Google Scholar] [CrossRef] [PubMed]
- Kant, S.; Kumar, A.; Singh, S.M. Fatty acid synthase inhibitor orlistat induces apoptosis in T cell lymphoma: Role of cell survival regulatory molecules. Biochim. Biophys. Acta 2012, 1820, 1764–1773. [Google Scholar] [CrossRef] [PubMed]
- Czumaj, A.; Zabielska, J.; Pakiet, A.; Mika, A.; Rostkowska, O.; Makarewicz, W.; Kobiela, J.; Sledzinski, T.; Stelmanska, E. In Vivo Effectiveness of Orlistat in the Suppression of Human Colorectal Cancer Cell Proliferation. Anticancer Res. 2019, 39, 3815–3822. [Google Scholar] [CrossRef]
- Carvalho, M.A.; Zecchin, K.G.; Seguin, F.; Bastos, D.C.; Agostini, M.; Rangel, A.L.C.; Veiga, S.S.; Raposo, H.F.; Oliveira, H.C.F.; Loda, M.; et al. Fatty acid synthase inhibition with Orlistat promotes apoptosis and reduces cell growth and lymph node metastasis in a mouse melanoma model. Int. J. Cancer 2008, 123, 2557–2565. [Google Scholar] [CrossRef]
- Sagimet Biosciences. Available online: https://sagimet.com/programs/pipeline/ (accessed on 17 August 2020).
- De Aquino, I.G.; Bastos, D.C.; Cuadra-Zelaya, F.J.M.; Teixeira, I.F.; Salo, T.; Della Coletta, R.; Graner, E. Anticancer properties of the fatty acid synthase inhibitor TVB-3166 on oral squamous cell carcinoma cell lines. Arch. Oral Boil. 2020, 113, 104707. [Google Scholar] [CrossRef]
- Tao, T.; Su, Q.; Xu, S.; Deng, J.; Zhou, S.; Zhuang, Y.; Huang, Y.; He, C.; He, S.; Peng, M.; et al. Down-regulation of PKM2 decreases FASN expression in bladder cancer cells through AKT/mTOR/SREBP-1c axis. J. Cell. Physiol. 2018, 234, 3088–3104. [Google Scholar] [CrossRef]

| Role of FASN | Function | Test Model | Tumor Type | Reference |
|---|---|---|---|---|
| Promote | Lipogenesis | In vitro | Breast | [70] |
| Signal transduction | In vitro, Mouse | Breast, Prostate, Colorectal, Non-small Lung | [40,71,75,76] | |
| Cytoskeleton remodeling | In vitro | Cervical | [72] | |
| Mitochondria respiration | In vitro, Mouse | Colorectal, Melanoma, Breast | [77,78,79] | |
| Fatty Acid Oxidation | In vitro, Mouse | Melanoma, Breast, Colorectal | [78,79,80] | |
| Mitochondria fusion | In vitro | Prostate, Breast | [81] | |
| Lipolysis | In vitro | Breast | [82] | |
| DNA repair | In vitro | Breast, Pancreatic | [83] | |
| Cell cycle | In vitro | Cervical | [9] | |
| Glycolysis | In vitro | Colorectal, Breast, Non-Hodgkin Lymphoma | [77,84,85] | |
| Cell migration | In vitro | Breast | [84] | |
| Repress | DNA repair | In vitro | Non-small Lung | [86] |
| Unfolded Protein Response | In vitro | Colorectal | [87] | |
| Programmed cell death | In vitro | Breast, Pancreatic | [88,89] |
| Inhibitor | Drug Development Stage | Test Model | Disease Type | Reference |
|---|---|---|---|---|
| Cerulenin | Preclinical | In vitro | Breast Cancer | [88,123,124] |
| Colon Cancer | [125] | |||
| Adenocarcinoma Lung Cancer | [126] | |||
| Colorectal Cancer | [127] | |||
| Retinoblastoma | [128] | |||
| Ocular Cancer | [129] | |||
| Bladder Cancer | [130] | |||
| Melanoma | [131] | |||
| Mouse | Ovarian Cancer | [111] | ||
| Adenocarcinoma Lung Cancer | [126] | |||
| C75 | Preclinical | In vitro | Breast Cancer | [113,132,133,134,135] |
| Adenocarcinoma Lung Cancer | [114] | |||
| Gastric Cancer | [136] | |||
| Endometrial Cancer | [137] | |||
| B-cell Lymphoma | [138] | |||
| Mouse | Adenocarcinoma Lung Cancer | [114] | ||
| Prostate Cancer | [139] | |||
| Orlistat | FDA Approved | Human | Obesity Management | [118] |
| Preclinical | In vitro | Non-Small Cell Lung Cancer | [75,140] | |
| B-cell Lymphoid Cancer | [141] | |||
| Pancreatic Cancer | [89] | |||
| T-cell Leukemia | [142] | |||
| Prostate Cancer | [143,144] | |||
| Melanoma | [131,145] | |||
| Retinoblastoma | [128] | |||
| Hepatocellular Carcinoma | [146] | |||
| Oral Squamous Cell Carcinoma | [147] | |||
| Ovarian Cancer | [148] | |||
| Head and Neck Squamous Cell Cancer | [149] | |||
| Brain Cancer | [150] | |||
| Mouse | Gastrointestinal Cancer | [151] | ||
| Oral Squamous Cell Cancer | [148] | |||
| T-cell Lymphoma | [152,153] | |||
| Ovarian Cancer | [149] | |||
| Colorectal Cancer | [154] | |||
| Melanoma | [145,155] | |||
| Prostate Cancer | [143] | |||
| Non-Small Cell Lung Cancer | [75,140] | |||
| TVB-2640 | Phase 1 Clinical Trial (ongoing) | Human | Colon Cancer and other cancer type that can be removed by surgery (ID: NCT02980029) | [156] |
| Phase II Clinical Trial (ongoing) | Human | Breast Cancer (ID: NCT03179904) | [156] | |
| KRAS mutated Non-Small Cell Lung Cancer (ID: NCT03808558) | [156] | |||
| Astrocytoma (ID: NCT03032484) | [156] | |||
| TVB-3664 | Preclinical | In vitro | Colon Cancer | [76] |
| Mouse | Colon Cancer | [76] | ||
| TVB-3166 | Preclinical | In vitro | Oral Squamous Cell Cancer | [157] |
| Bladder Cancer | [158] | |||
| Colorectal Cancer | [4] | |||
| Non-Small Cell Lung Cancer | [4] | |||
| Prostate Cancer | [4] | |||
| Breast Cancer | [4] | |||
| Ovarian Cancer | [4] | |||
| Mouse | Pancreatic Cancer | [4] | ||
| Ovarian Cancer | [4] | |||
| Non-Small Cell Lung Cancer | [4] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fhu, C.W.; Ali, A. Fatty Acid Synthase: An Emerging Target in Cancer. Molecules 2020, 25, 3935. https://doi.org/10.3390/molecules25173935
Fhu CW, Ali A. Fatty Acid Synthase: An Emerging Target in Cancer. Molecules. 2020; 25(17):3935. https://doi.org/10.3390/molecules25173935
Chicago/Turabian StyleFhu, Chee Wai, and Azhar Ali. 2020. "Fatty Acid Synthase: An Emerging Target in Cancer" Molecules 25, no. 17: 3935. https://doi.org/10.3390/molecules25173935
APA StyleFhu, C. W., & Ali, A. (2020). Fatty Acid Synthase: An Emerging Target in Cancer. Molecules, 25(17), 3935. https://doi.org/10.3390/molecules25173935






