Abstract
Plastid genome analysis of non-model organisms provides valuable information for basic research e.g., molecular evolutionary genomics, phylogeny and phylogeography. Deschampsia cespitosa is the most widespread species of the genus and it is a common grass that is found across Eurasia and North America. Scattered populations in regions of appropriate ecological conditions are also found in Australia, New Zealand and southern South America, where it is sympatric with D. antarctica. We analyzed the plastid genome of a sample of Deschampsia cespitosa of the Austrian Alps using high-throughput sequencing. The plastid (cp) genome shows the typical quadripartite structure with a length of 135,340 bp, comprising a large single-copy (LSC) region of 79,992 bp, a small single-copy (SSC) region of 12,572 bp and two inverted repeats (IR) regions of 21,388 bp each. It contains 115 genes, including 85 protein-coding genes, four ribosomal RNA genes and 30 transfer RNA genes. The GC content (%), number of repeats and microsatellites, RNA editing sites and codon usage were highly similar to those of D. antarctica. The results of this present study highlight the extremely conserved nature of the cp genome in this group, since the comparison involved individuals separated by about 13,000 km, from the Alps to Antarctica.
1. Introduction
The rapid decrease in costs of next-generation sequencing methods has resulted in an increase in the availability of completed plastid (cp) genomes [1], enabling comparisons at the genomic level even between closely related species [2,3]. The amount of cp genomes increased from 800 in 2016 [4] to about 2400 in 2018 [5], which allowing for close examination of features [6] and whole plastid-based phylogenetic relationships [2,7].
Plastid genomes of angiosperms range in size from 120 to 170 kb [8]. The cp genomes are highly conserved in a quadripartite organization, namely, a main large single-copy (LSC), a small single-copy (SSC) region and two inverted repeats (IRs) [9]; functional categories include (i) protein-coding genes, (ii) tRNA coding genes, (iii) introns and (iv) intergenic spacers. The number of genes varies between approximately 100 and 120 [10].
In the grass family (Poaceae), the cp genome size roughly varies from 134 kb in Oryza sativa [11] to 140 kb in Sorghum bicolor [12]. Until now, the Poaceae have been shown as divided in two major clades: one with three subfamilies (Bambusoideae, Ehrhartoideae and Pooideae; the BEP clade) and the other with seven subfamilies (Panicoideae, Arundinoideae, Chloridoideae, Centothecoideae, Micrairoideae, Aristidoideae and Danthonioideae; the PACCMAD clade) [7]. The BEP clade comprises the majority of the grasses of cold-temperate regions; plastid genome size ranges from 134,5 (Triticum aestivum) to 136,5 kb (Agrostis stolonifera) [12], with the Antarctic hair-grass (Deschampsia antarctica) having an intermediate plastid genome with a size of 135,362 bp [6]. As one of the only two flowering plants (and the only grass) native to Antarctica, D. antarctica has been much studied, and studies have focused on the mechanisms of adaptation of the species to the extreme environments of the Antarctic continent. These mechanisms include physiological and morphological attributes [13,14]. Study of genetical aspects has revealed that there is low genetic variation [15] and that Antarctic populations originated from populations in Patagonia [16]. Other studies on D. antarctica have included the search of genes expressed under cold stress [17], the later development of cold resistant transgenic rice plants [18] and the resistance to oxidative stress after exposure to UVB radiation [19]. Recently, the effect of AntartinaTM, which was developed from extracts of D. antarctica, has been evaluated as an inhibitor of colorectal carcinoma growth and liver metastasis [20].
A previous phylogenetic analysis [21] showed that Deschampsia antarctica to be closely related to D. cespitosa; these species are sympatric in Patagonia [22,23] and share many loci with repetitive DNA elements (L. González, unpublished data). Despite the many studies on D. antarctica, its relationships at the genomic level are not well understood. Available evidence points to D. cespitosa as its closest relative, and, therefore, we focus here on the common species D. cespitosa (tufted hair grass), a tussock-forming, wind-pollinated, self-incompatible species with wide morphological variation throughout its large, nearly cosmopolitan distribution. It is mainly distributed in temperate and cold-temperate zones of the Northern Hemisphere, but isolated populations are found in southern South America, Australasia and South Africa [22]. The aims of this study are to (1) present the complete cp genome of D. cespitosa, (2) to compare it with D. antarctica, and (3) to assess regions of sequence diversity to detect potential markers for genetic, phylogenetic and phylogeographic studies. The understanding of relationships could clarify the evolutionary processes acting between sympatric populations of the two putative species, that have led to one of them undergoing: (1) adaptation to extreme environments; (2) development of morphological differences; and (3) speciation. A comparative analysis of variation in features is presented as a step towards identifying similarities between these taxa. To provide an evolutionary framework, we selected some early-diverging Poaceae and some closely related BEP taxa to include in a phylogenetic reconstruction.
2. Results
2.1. Sequencing
Illumina paired-end sequencing yielded 11,058,892 raw reads with an average length of 350–400 bp (total 4,423,556,800 bp).
2.2. Genome Assembly and Characteristics
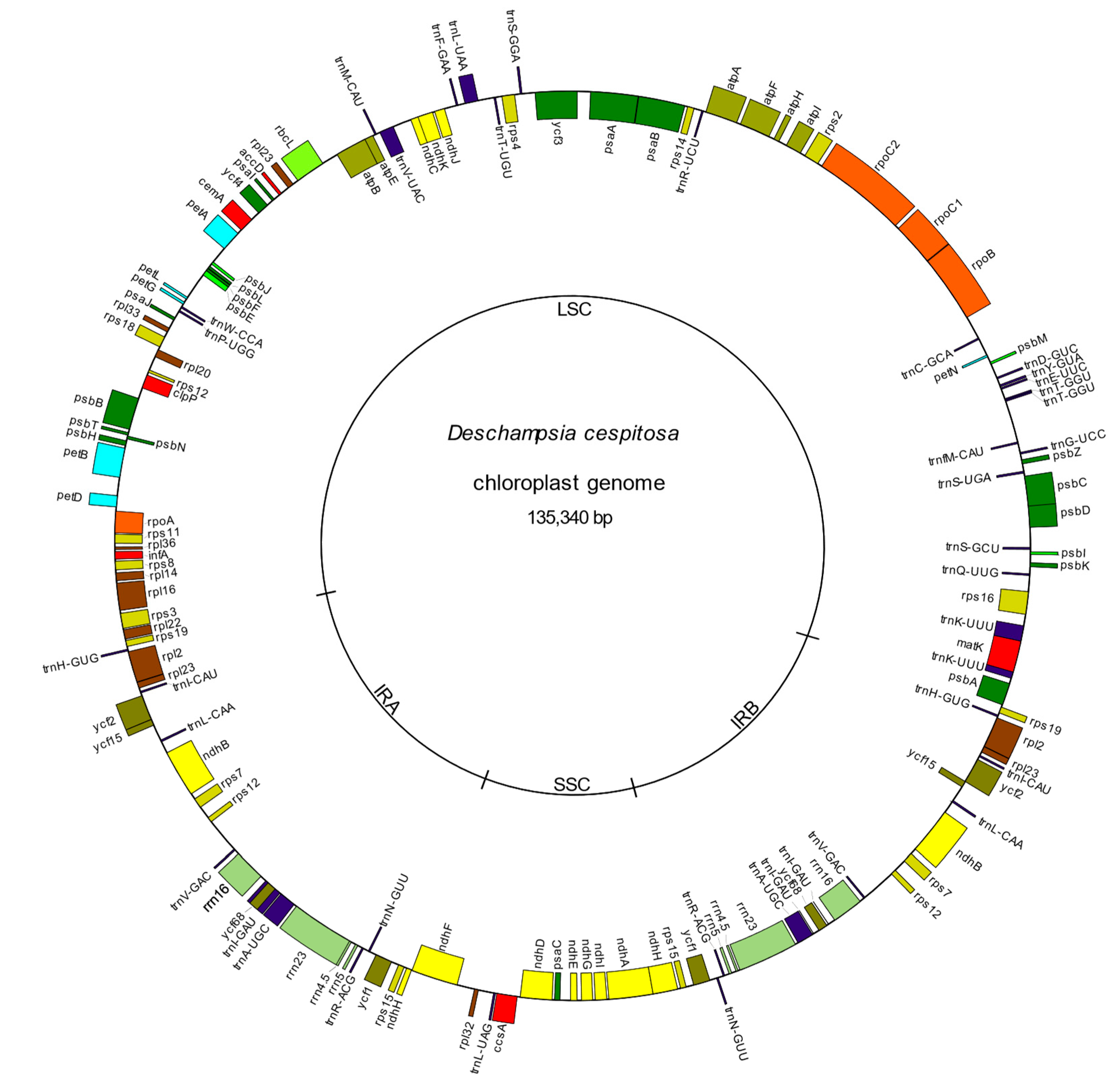
The cp genome of Deschampsia cespitosa resulted in a typical circular quadripartite structure of 135,340 bp length, with a LSC region of 79,992 bp, an SSC of 12,572 and two inverted repeats (IRa and IRb) each with a length of 21,388 bp. The average GC content was 38.27% (Table 1, Figure 1).

Table 1.
Plastid genomes of Deschampsia cespitosa and D. antarctica (from [5]).
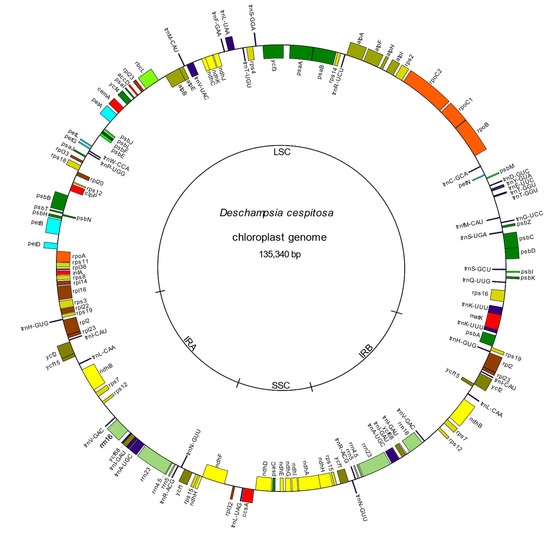
Figure 1.
The plastid genome of Deschampsia cespitosa. Genes outside and inside the circle are transcribed counterclockwise and clockwise, respectively.
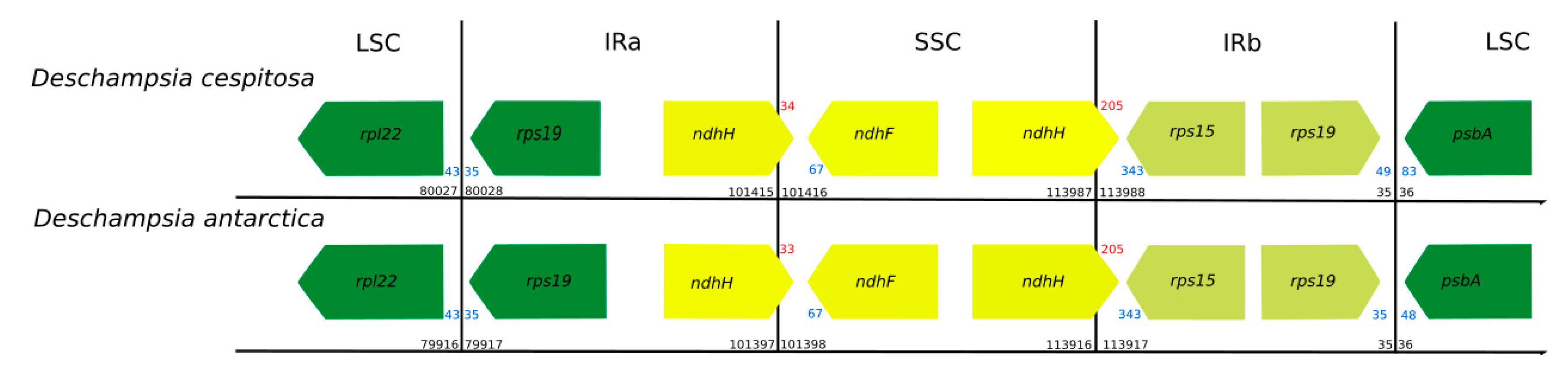
The cp genome of D. cespitosa contains 115 genes, including 81 protein-coding genes, four rRNA genes and 30 tRNA genes (Table S1). Twelve genes are duplicated in the IR and there are three open reading frames (ORF 188, ORF 56 and ORF 42). Gene order and size are nearly identical to those of D. antarctica. Both copies of the ndhH gene cross the boundaries between the regions: 34 bp in the IRa-SSC and 205 bp in the SSC-IRb (Figure 2). Of the 30 tRNA genes, 21 are located in the LSC, with trnT-GGU duplicated-. Furthermore, one gene is found in the SSC and the remaining eight are in the IRs. The four rNA genes are located in the inverted repeats, with two in each IR. Some functional groups are located in the LSC region, i.e., the small subunit of ribosome (rps genes, except rps15 in the SSC and rps7 and 12 in the IR), the large subunit (rpl genes except rpl32), the RNA polymerase (rpo genes), photosystems I (psa genes except psaC) and II (psb genes) and the cytochrome complex (pet genes). The NADH (nicotinamide adenine dinucleotide) dehydrogenase (ndh) genes are located in the SSC region, except for ndhB that is transpliced between the two IRs.
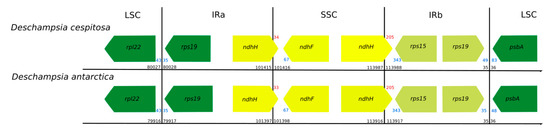
Figure 2.
Comparison of the boundaries between genome regions in Deschampsia cespitosa and D. antarctica (black numbers: region boundaries; red numbers: gene crossing region boundary; blue numbers: distance from gene end to region boundary. All numbers in bp. Direction of arrow indicates direction of transcription).
2.3. Repetitive Sequences
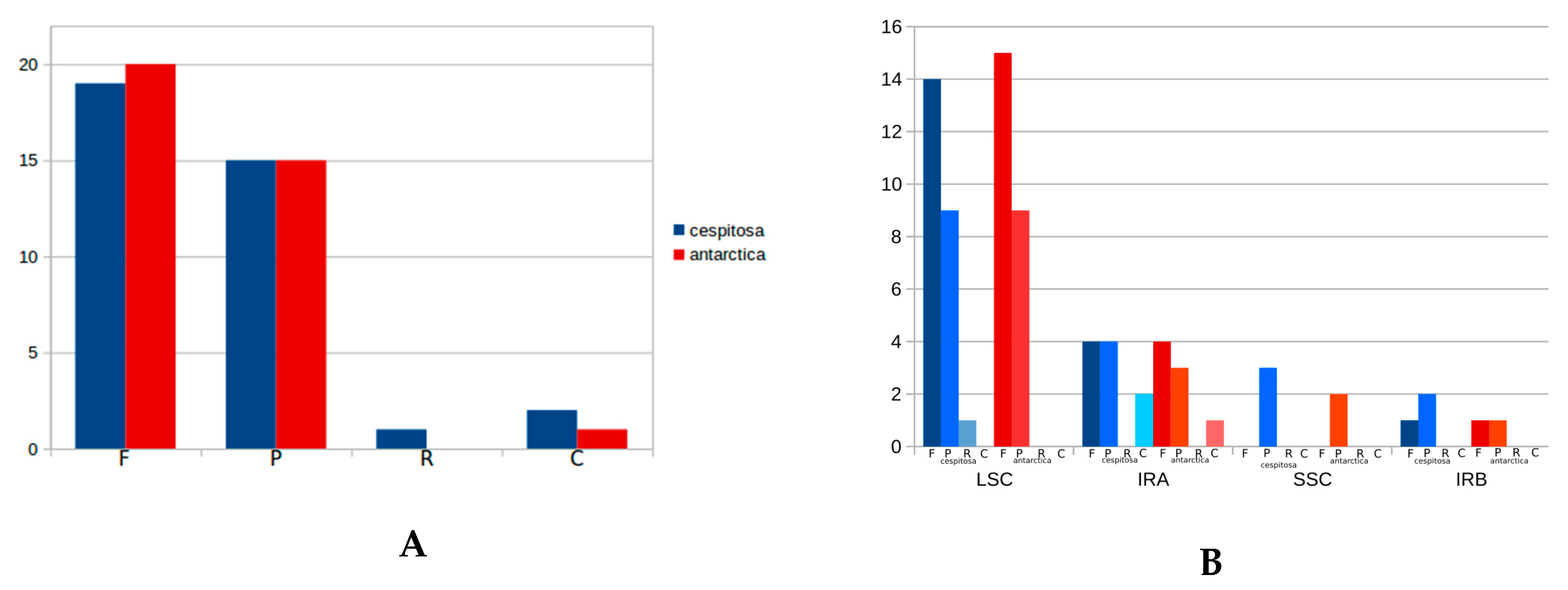
The cp genome of Deschampsia cespitosa contains 37 repeats (19 forward, 15 palindrome, two complementary and one reverse) (Table S2). Repeats were mostly located in intergenic spacers and coding sequences; a single repeat was found in non-coding sequences. A comparison with repeat sequences in D. antarctica is shown in Figure 3A,B. The length of repeats varies between 19 and 224.
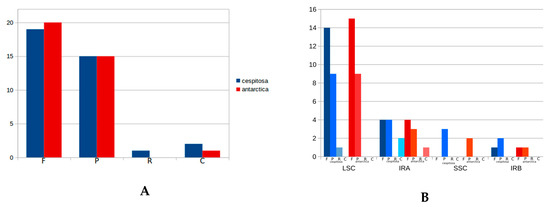
Figure 3.
(A) Type of repeat sequences in plastid genomes of Deschampsia cespitosa and D. antarctica (F: forward; P: palindrome; R: reverse; C: complementary); (B) Location of repeat sequences in genomic regions of plastid genomes in Deschampsia cespitosa and D. antarctica (LSC: Long single- copy; SSC: Short single-copy; IRA: Inverted repeat A; IRB: Inverted repeat B).
2.4. RNA Editing Sites
There are overall 41 RNA editing sites in 15 genes of the cp genome of D. cespitosa (Table S3). Most of the sites (18) were found in the ndhB gene, which is transpliced between the inverted repeats (nine sites each in IRa and IRb). Other genes with several editing sites include rpoC2 (LSC, five sites), matK, ndhA and rpoB (LSC, four sites).
2.5. Phylogenomic Comparison
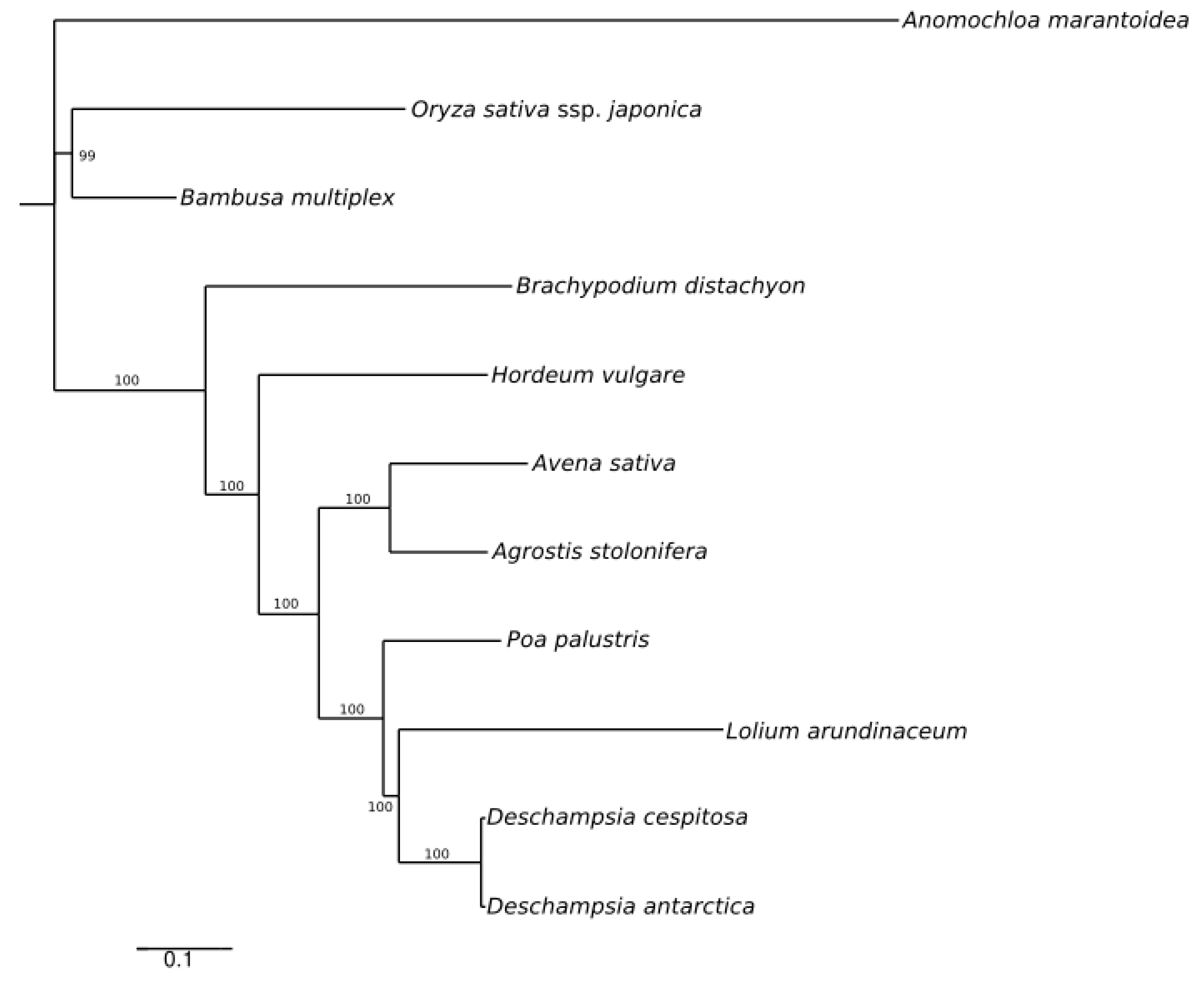
A maximum likelihood (ML) reconstruction with 11 selected whole cp genome sequences (Table S4) yielded a single tree with high support (Figure 4), depicting the low divergence between Deschampsia cespitosa and D. antarctica.
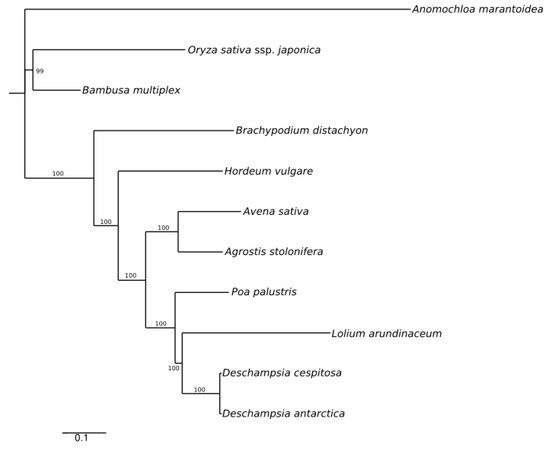
Figure 4.
ML phylogenetic tree based on plastid genomes of Deschampsia cespitosa and selected grass species.
2.6. Genomic Comparison of Deschampsia cespitosa with D. antarctica and other Poaceae
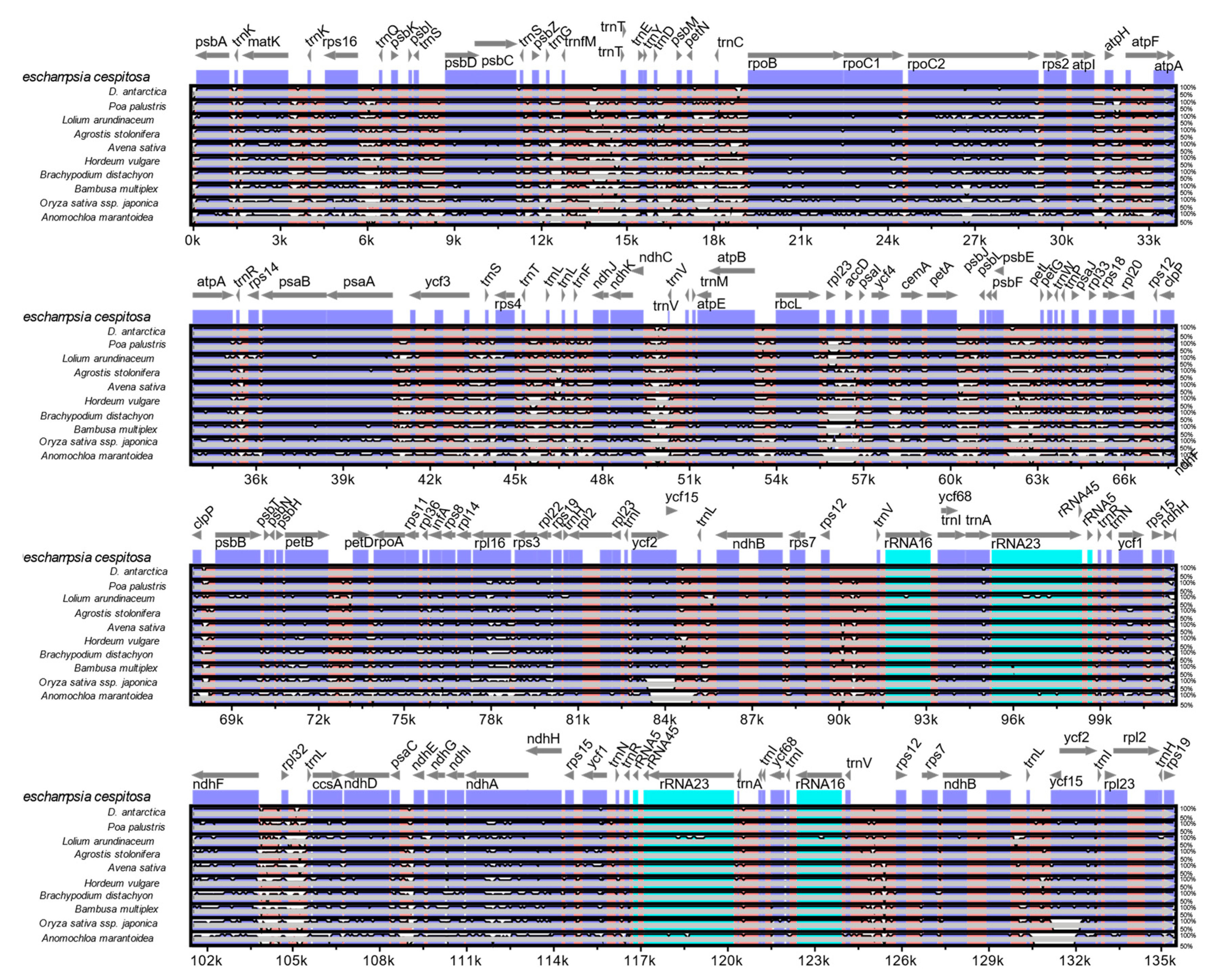
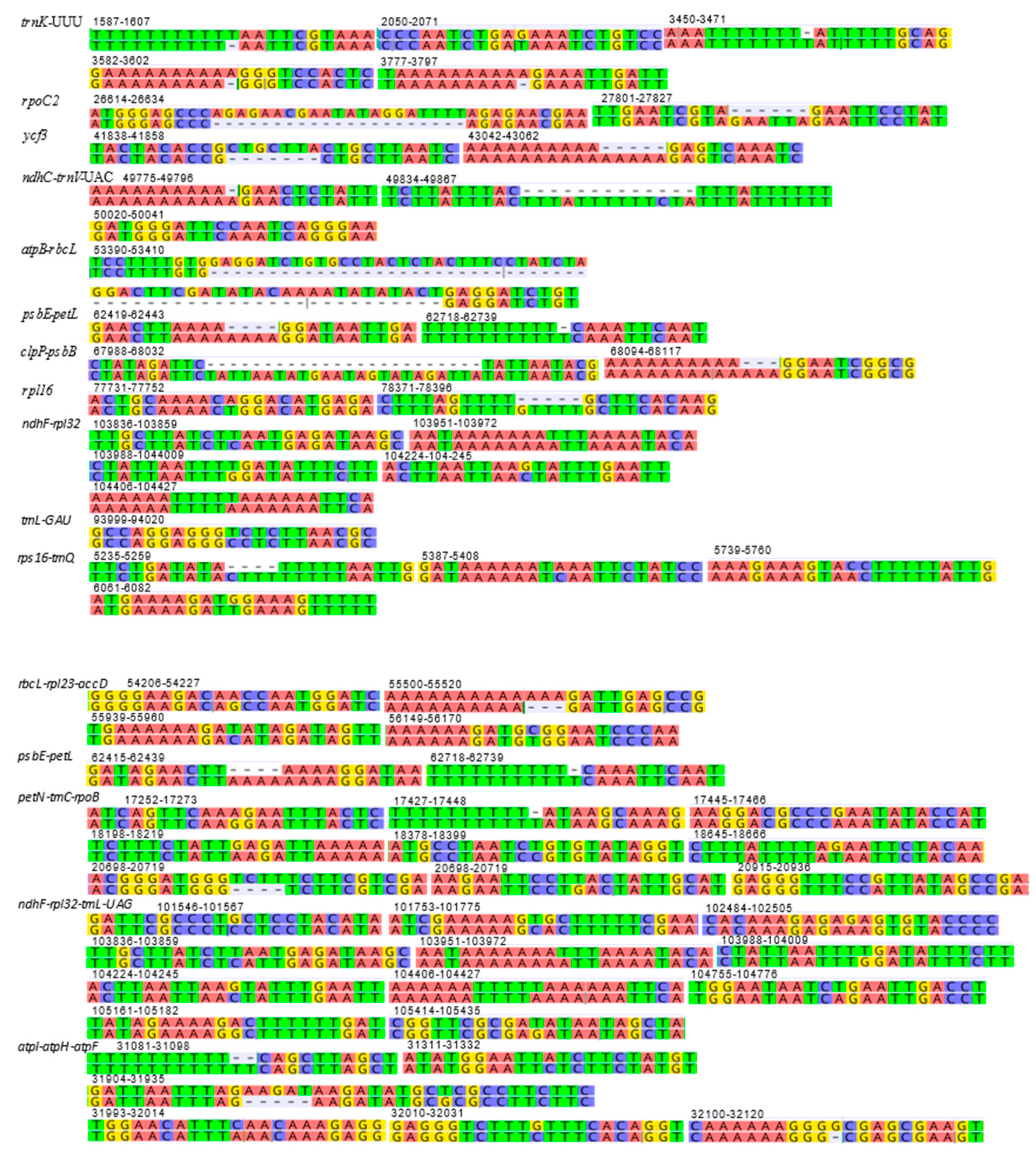
The mVISTA alignment of 11 plastid (details in Table S4) sequences shows that conservation decreases progressively among the BEP clade species towards the early-diverging Anomochloa maranthoidea (Figure 5; Figure S1). The mVISTA alignment shows potential variable sites between D. cespitosa and the other Poaceae in trnK, trnL, trnC rpoC2, rps14 and rpl16. Specifically focusing on differences in the plastid genome between Deschampsia cespitosa and D. antarctica, Figure 6 shows the structural differences (mutational differences, indels: insertions/deletions and inversions) of selected markers with both flanking regions in trnK-UUU, rpoC2, ycf3, ndhC-trnV-UAC, atpB-rbcL, psbE-petL, clpP-psbB, rpl16, ndhF-rpl32, trnI-GAU, rps16-trnQ, rbcL-rpl23-accD, psbE-petL, petN-trnC-rpoB, ndhF-rpl32-trnL-UAG and atpl-atpH-atpF. Most variations between Deschampsia cespitosa and D. antarctica are single mutations and small indels, which are more frequent in the intergenic spaces than in genes. Larger indels were found predominantly in intergenic spacers and to a lesser extent in coding regions, i.e., rpoC2 and ycf3.
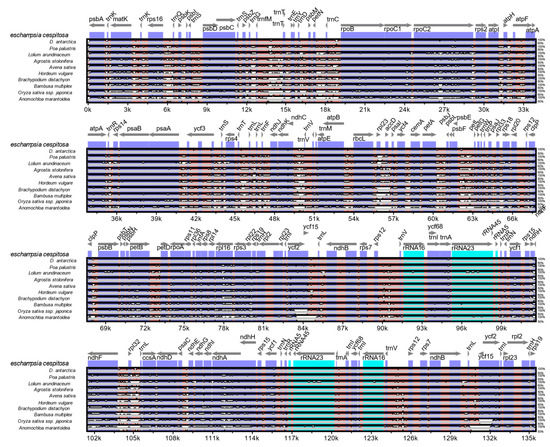
Figure 5.
Complete cp genome comparison of eleven species of Poaceae.
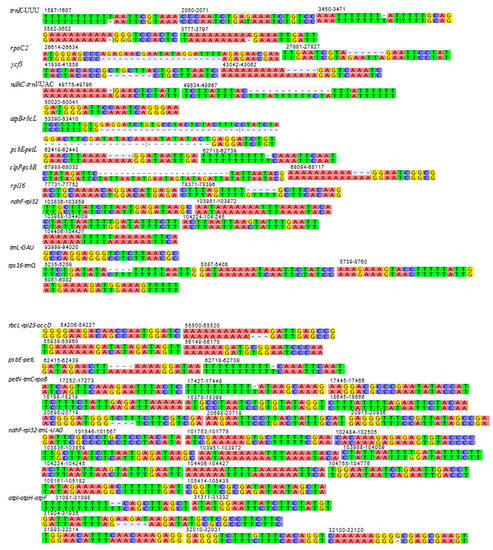
Figure 6.
Structural differences (mutational differences, insertions/deletions and inversions) of plastid genome between Deschampsia cespitosa and D. antarctica of selected regions of interest with an additional 10 bp at both flanking regions. Numbers indicate the position in the D. cespitosa plastid genome.
2.7. Microsatellites
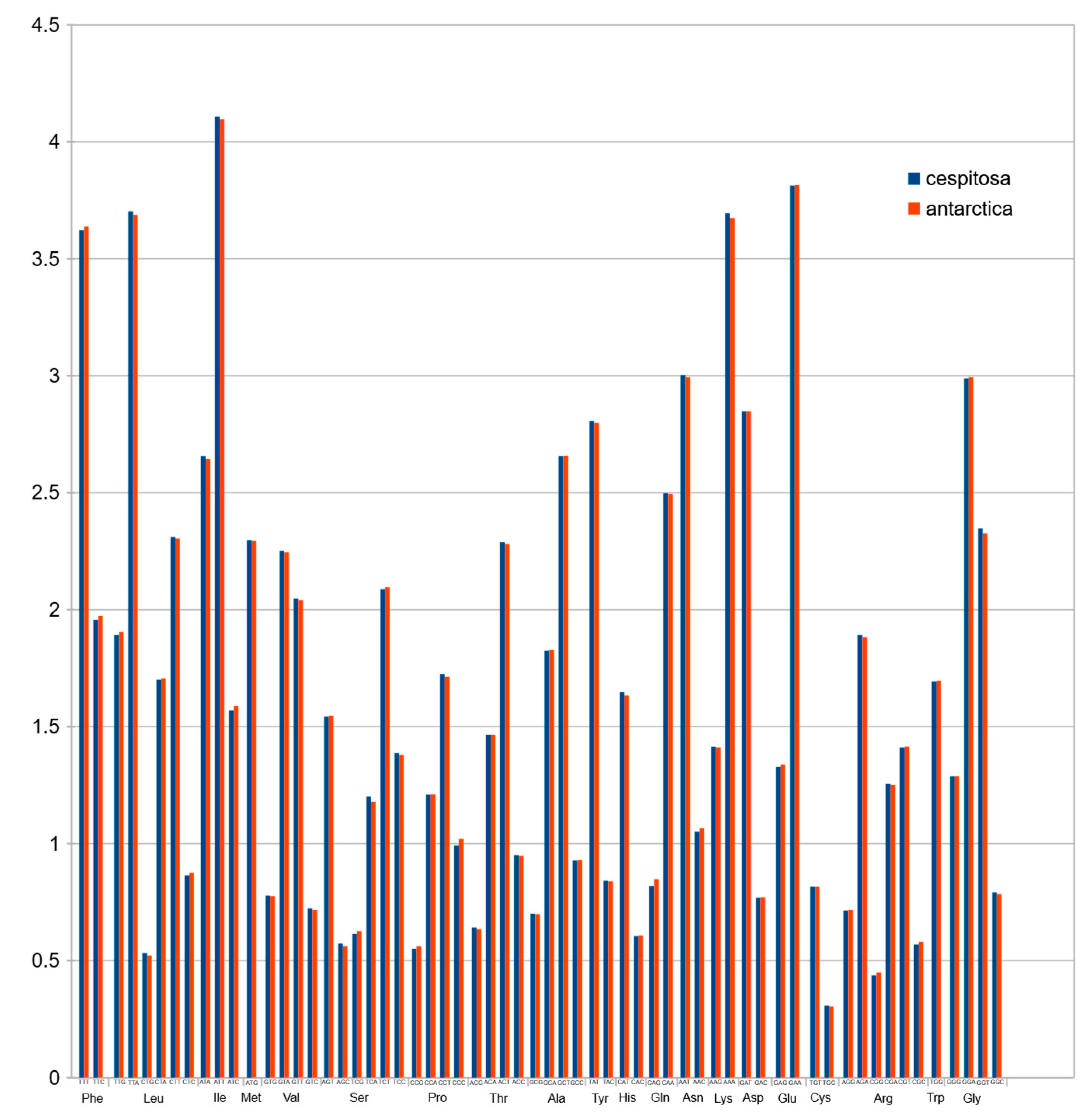
2.8. Codon Usage
Codon usage in protein-coding sequences Figure 6 showed a rather unimodal distribution, which is a common feature in grasses [24].
3. Discussion
Plastid genomes are uniparentally inherited [12], and the present study highlights its high conservation between two species of the genus Deschampsia. The comparison of genome size, gene order, repetitive sequences and codon usage between D. cespitosa and D. antarctica shows a high degree of similarity (Figure 7). Furthermore, the phylogenetic reconstruction shows little divergence between the two. A comparison per region of G-C content also shows essentially the same values (Table 1). The mVISTA alignment of the whole cp sequences (Figure 5; Figure S1) provides informative sites for phylogenetic and phylogeographic studies in the BEP clade. There are several mutations, indels and inversions that can be used for phylogeographic studies in the genus Deschampsia. Figure 6 shows structural differences in the plastid genome between Deschampsia cespitosa and D. antarctica of selected regions and their flanking regions. The main differences were detected in trnK-UUU, rpoC2, ycf3, ndhC-trnV-UAC, atpB-rbcL, psbE-petL, clpP-psbB, rpl16, ndhF-rpl32 and trnI-GAU. Some genes, i.e., rpoC2 and ycf3, have large indels differing between Deschampsia cespitosa and D. antarctica, which can be used in phylogenetic studies. Dispersed repeats of the forward, palindrome and reverse type are more common in the LSC region of both taxa, whereas only D. antarctica has a complementary repeat in the IRa region. The repeats are also more common in intergenic regions (Table S2). Codon usage in protein-coding sequences is rather similar between the two species and shows a bias to having an A in the third position [24]. The similarity of microsatellites between the two species (Table S3) allows for comparative mapping [25], especially in sympatric populations in Patagonia [23]. The major genomic region boundaries (Figure 2) are also highly similar and show few bp displacements between the two species.
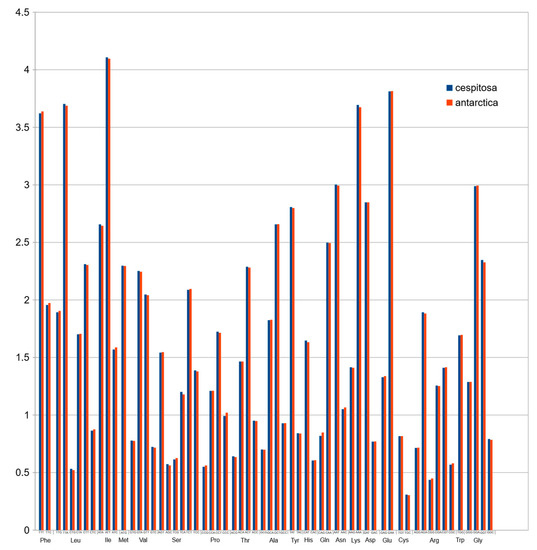
Figure 7.
Normalized codon usage of Deschampsia cespitosa and D. antarctica.
4. Materials and Methods
4.1. Sample Material, DNA Extraction and Sequencing
The individual Deschampsia cespitosa used for this analysis was collected in a clearing of a spruce forest in the Karawanken mountain range, Carinthia, Austria (a voucher specimen is deposited in the herbarium WU). Leaves were dried in silica gel and DNA was extracted from 20 mg of dry tissue with the DNeasy Plant Mini Kit (Qiagen, Hilden, Germany), following the manufacturer’s protocol. The total DNA was visualized with agarose gel electrophoresis on a transilluminator Gel Doc 2000 (Biorad, Vienna, Austria) while its quality and quantity were assessed using a Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, Vienna, Austria) and a Qubit 2.0 (Thermo Fisher Scientific). Whole cellular DNA (nuclear, mitochondrial and plastid DNA) was sheared with a Bioruptor® Pico sonication device (Diagenode, Liege, Belgium) using seven cycles of 15 s on and 90 s off at 4 °C in order to obtain fragments with an average size of 350 to 400 bp. Fragment size was checked afterwards with an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). The library was prepared with a TruSeq DNA PCR Free Library Kit (Illumina, San Diego, CA, USA), following the manufacturer’s protocol and barcodes provided with the kit. The library was sequenced in a 1/24 lane of an Illumina HiSeq 3000 at VBCF Vienna (https://www.vbcf.ac.at/facilities/next-generation-sequencing/).
4.2. Genome Assembly and Annotation
The BAM file with total genomic DNA sequences was downloaded from the facility and sorted with Bedtools [26] before fastq files were produced with Samtools [27]. Fastq files were processed with Fast-Plast [28] to assemble the cp genome. The Fast-Plast pipeline includes Trimmomatic [29], which performs an initial read cleaning; Bowtie2 [30], which reduces reads to only plastid-like reads; and SPAdes [31], which performs an initial assembly. The obtained genome was then annotated online using DOGMA [32] with default parameters, which used BLASTX and BLASTN searches to identify all genes by comparing them with a custom database of published cp genomes. The phylogenetically closest complete cp genome of D. antarctica was utilized to confirm the positions of start and stop codons and boundaries of exons or introns by alignment in the software Geneious 11.1.5 [33]. The annotated genome was finally uploaded in Genome VX [34] to produce the circular map (Figure 1). The complete cp genome was deposited in GenBank (GenBank accession MK262782).
4.3. Repeats Structure
Simple repetitive short sequences with repeated motifs of 1–10 bp are a common feature of plant genomes [35] that can be used in population genetics or to tag genes of interest [36]. Repeats (forward, palindrome, reverse and complement sequences) in the cp genome (Table S2) were detected with the REPuter server [37].
4.4. RNA Editing Sites
RNA editing sites are codon positions in mitochondrial and cp sequences, at which potential changes of C-to-U could result in changes in the encoded amino acid [38]. The editing sites in the cp genome of Deschampsia cespitosa (Table S3) were identified with the PREP (Predictive RNA Editor for Plants) suite, which is a set of web servers devoted to predicting such sites in plant organellar genes [38].
4.5. Phylogenetic Analysis
To verify the phylogenomic relationships of the newly obtained plastome of Deschampsia cespitosa in relation to other Poaceae, we selected early-diverging Poaceae and representatives of the BEP clade, especially members of the tribe Poeae ([7,39]; Agrostis stolonifera [12], Anomochloa maranthoidea [40], Avena sativa [41], Bambusa multiplex [42], Brachypodium distachyon [43], D. antarctica [6], Hordeum vulgare ssp. vulgare [12], Lolium arundinaceum [44], Oryza sativa ssp. japonica [11] and Poa palustris [41]) (Table S4). The data matrix was aligned with MAFFT [45] using standard settings. The alignment obtained with MAFFT was visually controlled with BioEdit [46]. The aligned matrix was uploaded in the IQ-TREE server [47] before a ML tree search with bootstrap support was run. The resulting tree (Figure 4) was edited with Inkscape [48].
4.6. Comparative Analyses
The comparative genomic analysis of Deschampsia cespitosa, early-diverging Poaceae and members of the BEP clade (Table S1) was conducted with mVISTA [49], which is a server for comparative analysis of genomic-level sequence. We used default parameters with the new sequence of D. cespitosa set as the reference (Figure 5; Figure S1). Comparisons between Deschampsia cespitosa and D. antarctica were made with Geneious [33].
4.7. Microsatellite Search
Microsatellites are ubiquitous components of all genomes [50] and are extremely variable [51]. We used Imperfect Microsatellite Extractor [52], which is an online server tool for finding microsatellites, Simple Sequence Repeats (SSRs) or Short Tandem Repeats (STRs) from genomic sequences, to detect microsatellites (Table S3). We followed a previous study [3] to create a similar comparison between two related species, setting minimum thresholds for search at seven for mononucleotide repeats, four for dinucleotide repeats and three for tri-, tetra-, penta- and hexanucleotide repeats.
4.8. Codon Usage
The codon usage percentage of protein coding sequences was estimated with Sequence Manipulation Suite, an online collection of Java Scripts for analyzing short DNA and protein sequences, using default settings. [53].
5. Conclusions
The results suggest a high similarity of the plastid genome of Deschampsia cespitosa and D. antarctica. The major genomic region boundaries (Figure 2) are highly similar and the few differences suggest a relatively stable cp genomic structure in the genus; however, the inclusion of further cp genomes of other species of the genus and D. cespitosa samples from other geographical regions is necessary to verify this. The similarity of the obtained cp genome may also highlight the possibility of using D. cespitosa for research in any of the fields mentioned in the introduction, which are currently restricted to D. antarctica. Available evidence [39] pointing to an origin of the genus in Eurasia and the worldwide distribution of D. cespitosa may reveal D. antarctica to be a cold-adapted form derived from the former once we can include more members of the genus in the analysis.
Supplementary Materials
The supplementary materials are available online.
Author Contributions
Conceptualization, J.O.C. and J.G.; Methodology J.O.C. and M.H.J.B.; Formal Analysis J.O.C., M.H.J.B. and Z.-Q.X.; Writing-Original Draft Preparation J.O.C.; Writing-Review & Editing J.O.C., M.H.J.B., Z.-Q.X. and J.G.; Project Administration J.G.; Funding Acquisition J.O.C. and J.G.
Funding
This research was funded by the Austrian Science Fund (FWF), Project P 30208-B29.
Acknowledgments
We thank O. Paun and M. R. McKain for assistance in data analysis. Comments by 3 reviewers helped to improve the text. M. F. Fay made a valuable contribution by improving the English.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
References
- Wysocki, W.P.; Clark, L.G.; Kelchner, S.A.; Burke, S.V.; Pires, J.C.; Edger, P.P.; Mayfield, D.R.; Triplett, J.K.; Columbus, J.T.; Ingram, A.L.; et al. A multi-step comparison of short-read full plastome sequence assembly methods in grasses. Taxon 2014, 63, 899–910. [Google Scholar] [CrossRef]
- Zhang, Y.J.; Ma, P.F.; Li, D.Z. High-throughput sequencing of six bamboo chloroplast genomes: Phylogenetic implications for temperate woody bamboos (Poaceae: Bambusoideae). PLoS ONE 2011, 6, e20596. [Google Scholar] [CrossRef] [PubMed]
- Menezes, A.P.; Resende-Moreira, L.C.; Buzatti, R.S.; Nazareno, A.G.; Carlsen, M.; Lobo, F.P.; Kalapothakis, E.; Lovato, M.B. Chloroplast genomes of Byrsonima species (Malpighiaceae): Comparative analysis and screening of high divergence sequences. Sci. Rep. 2018, 8, 2210. [Google Scholar] [CrossRef] [PubMed]
- Daniell, H.; Lin, C.-S.; Yu, M.; Chang, W.-J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016, 17, 134. [Google Scholar] [CrossRef] [PubMed]
- Magnoliophyta plastid genomes. Available online: https://www.ncbi.nlm.nih.gov/genome/browse#!/organelles/Magnoliophyta (accessed on 7 January 2019).
- Lee, J.; Kang, Y.; Shin, S.C.; Park, H.; Lee, H. Combined analysis of the chloroplast genome and transcriptome of the Antarctic vascular plant Deschampsia antarctica Desv. PLoS ONE 2014, 9, e92501. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.Q.; Ge, S. The phylogeny of the BEP clade in grasses revisited: Evidence from the whole-genome sequences of chloroplasts. Mol. Phylogenet. Evol. 2012, 62, 573–578. [Google Scholar] [CrossRef] [PubMed]
- Shaw, J.; Lickey, E.B.; Schilling, E.E.; Small, R.L. Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: The tortoise and the hare III. Am. J. Bot. 2007, 94, 275–288. [Google Scholar] [CrossRef]
- Palmer, J.D. Plastid chromosomes: Structure and evolution. In The Molecular Biology of Plastids: Cell Culture and Somatic Cell Genetics of Plants; Hermann, R.G., Ed.; Springer: Vienna, Austria, 1991; Volume 7A, pp. 5–53. [Google Scholar]
- Wicke, S.; Schneeweiss, G.M.; Müller, K.F.; dePamphilis, C.W.; Quandt, D. The evolution of the plastid chromosome in land plants: Gene content, gene order, gene function. Plant Mol. Biol. 2011, 76, 273–297. [Google Scholar] [CrossRef]
- Tang, J.; Xia, H.; Cao, M.; Zhang, X.; Zeng, W.; Hu, S.; Tong, W.; Wang, J.; Wang, J.; Yu, J.; et al. A comparison of rice chloroplast genomes. Plant Physiol. 2004, 135, 412–420. [Google Scholar] [CrossRef]
- Saski, C.; Lee, S.B.; Fjellheim, S.; Guda, C.; Jansen, R.K.; Luo, H.; Tomkins, J.; Rognli, O.A.; Daniell, H.; Clarke, J.L. Complete chloroplast genome sequences of Hordeum vulgare, Sorghum bicolor and Agrostis stolonifera, and comparative analyses with other grass genomes. Theor. Appl. Genet. 2007, 115, 571–590. [Google Scholar] [CrossRef]
- Alberdi, M.; Bravo, L.A.; Gutiérrez, A.; Gidekel, M.; Corcuera, L.J. Ecophysiology of Antarctic vascular plants. Physiol. Plant. 2002, 115, 479–486. [Google Scholar] [CrossRef] [PubMed]
- Romero, M.A.; Casanova, A.N.; Iturra, G.R.; Reyes, A.U.; Montenegro, G.L.; Alberdi, M.I. Leaf anatomy of Deschampsia antarctica (Poaceae) from the Maritime Antarctic and its plastic response to changes in the growth conditions. Rev. Chil. Hist. Nat. 1999, 72, 411–425. [Google Scholar]
- Holderegger, R.; Stehlik, I.; Lewis Smith, R.I.; Abbott, R.J. Populations of Antarctic hairgrass (Deschampsia antarctica) show low genetic diversity. Arct. Antarct. Alp. Res. 2003, 35, 214–217. [Google Scholar] [CrossRef]
- Fasanella, M.; Premoli, A.C.; Urdampilleta, J.D.; González, M.L.; Chiapella, J. How did a grass reach Antarctica? The Patagonian connection of Deschampsia antarctica (Poaceae). Bot. J. Linn. Soc. 2017, 185, 511–524. [Google Scholar] [CrossRef]
- Lee, H.; Kim, J.H.; Park, M.; Kim, I.C.; Yim, J.H.; Lee, H.K. Reference genes validation for qPCR normalization in Deschampsia antarctica during abiotic stresses. Antarct. Sci. 2010, 22, 477–484. [Google Scholar] [CrossRef]
- Byun, M.Y.; Lee, J.; Cui, L.H.; Kang, Y.; Oh, T.K.; Park, H.; Lee, H.; Kim, W.T. Constitutive expression of DaCBF7, an Antarctic vascular plant Deschampsia antarctica CBF homolog, resulted in improved cold tolerance in transgenic rice plants. Plant Sci. 2015, 236, 61–74. [Google Scholar] [CrossRef] [PubMed]
- Köhler, H.; Contreras, R.A.; Pizarro, M.; Cortés-Antíquera, R.; Zúñiga, G.E. Antioxidant Responses Induced by UVB Radiation in Deschampsia antarctica Desv. Front. Plant Sci. 2017, 8, 921. [Google Scholar] [CrossRef] [PubMed]
- Malvicini, M.; Gutierrez-Moraga, A.; Rodriguez, M.M.; Gomez-Bustillo, S.; Salazar, L.; Sunkel, C.; Nozal, L.; Salgado, A.; Hidalgo, M.; Lopez-Casas, P.P.; et al. A tricin derivative from Deschampsia antarctica Desv. inhibits colorectal carcinoma growth and liver metastasis through the induction of a specific immune response. Mol. Cancer Ther. 2018, 17, 966–976. [Google Scholar] [CrossRef] [PubMed]
- Chiapella, J. A molecular phylogenetic study of Deschampsia (Poaceae: Aveneae) inferred from nuclear ITS and plastid trnL sequence data: Support for the recognition of Avenella and Vahlodea. Taxon 2007, 56, 55–64. [Google Scholar]
- Chiapella, J.; Zuloaga, F.O. A Revision of Deschampsia, Avenella, and Vahlodea (Poaceae, Poeae, Airinae) in South America. Ann. Mo. Bot. Gard. 2010, 97, 141–162. [Google Scholar] [CrossRef]
- González, M.L.; Urdampilleta, J.D.; Fasanella, M.; Premoli, A.C.; Chiapella, J. Distribution of rDNA and polyploidy in Deschampsia antarctica E. Desv. in Antarctic and Patagonic populations. Polar Biol. 2016, 39, 1663–1677. [Google Scholar] [CrossRef]
- Sablok, G.; Nayak, K.C.; Vazquez, F.; Tatarinova, T.V. Synonymous codon usage, GC 3, and evolutionary patterns across plastomes of three pooid model species: Emerging grass genome models for monocots. Mol. Biotechnol. 2011, 49, 116–128. [Google Scholar] [CrossRef] [PubMed]
- Varshney, R.K.; Sigmund, R.; Börner, A.; Korzun, V.; Stein, N.; Sorrells, M.E.; Langridge, P.; Graner, A. Interspecific transferability and comparative mapping of barley EST-SSR markers in wheat, rye and rice. Plant Sci. 2005, 168, 195–202. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- McKain, M.R.; Wilson, M. Fast-Plast: Rapid de novo Assembly and Finishing for Whole Chloroplast Genomes. 2017. Available online: https://github.com/mrmckain/Fast-Plast (accessed on 20 November 2018).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S. Fast gapped-read alignment with Bowtie 2. Nature Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Wyman, S.K.; Jansen, R.K.; Boore, J.L. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 2004, 20, 3252–3255. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Conant, G.C.; Wolfe, K.H. GenomeVx: Simple web-based creation of editable circular chromosome maps. Bioinformatics 2008, 24, 861–862. [Google Scholar] [CrossRef] [PubMed]
- Levinson, G.; Gutman, G.A. Slipped-strand mispairing: A major mechanism for DNA sequence evolution. Mol. Biol. Evol. 1987, 4, 203–221. [Google Scholar] [CrossRef] [PubMed]
- Lauvergeat, V.; Barre, P.; Bonnet, M.; Ghesquiere, M. Sixty simple sequence repeat markers for use in the Festuca–Lolium complex of grasses. Mol. Ecol. Notes 2005, 5, 401–405. [Google Scholar] [CrossRef]
- Kurtz, S.; Choudhuri, J.V.; Ohlebusch, E.; Schleiermacher, C.; Stoye, J.; Giegerich, R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001, 29, 4633–4642. [Google Scholar] [CrossRef] [PubMed]
- Mower, J.P. The PREP Suite: Predictive RNA editors for plant mitochondrial genes, chloroplast genes and user-defined alignments. Nucleic Acids Res. 2009, 37, 253–259. [Google Scholar] [CrossRef] [PubMed]
- Soreng, R.J.; Peterson, P.M.; Romaschenko, K.; Davidse, G.; Zuloaga, F.O.; Judziewicz, E.J.; Filgueiras, T.S.; Davis, J.I.; Morrone, O. A worldwide phylogenetic classification of the Poaceae (Gramineae). J. Syst. Evol. 2015, 53, 117–137. [Google Scholar] [CrossRef]
- Morris, L.M.; Duvall, M.R. The chloroplast genome of Anomochloa marantoidea (Anomochlooideae; Poaceae) comprises a mixture of grass-like and unique features. Am. J. Bot. 2010, 97, 620–627. [Google Scholar] [CrossRef]
- Saarela, J.M.; Wysocki, W.P.; Barrett, C.F.; Soreng, R.J.; Davis, J.I.; Clark, L.G.; Kelchner, S.A.; Pires, J.C.; Edger, P.P.; Mayfield, D.R.; et al. Plastid phylogenomics of the cool-season grass subfamily: Clarification of relationships among early-diverging tribes. AoB Plants 2015, 7, plv046. [Google Scholar] [CrossRef]
- Gao, J.; Li, K.; Gao, L.Z. complete chloroplast genome sequence of Bambusa multiplex (Poaceae: Bambusoideae). Mitochondrial DNA A DNA Mapp. Seq. Anal. 2014, 27, 980–982. [Google Scholar] [CrossRef]
- Sancho, R.; Cantalapiedra, C.P.; López Alvarez, D.; Gordon, S.P.; Vogel, J.P.; Catalán, P.; Contreras Moreira, B. Comparative plastome genomics and phylogenomics of Brachypodium: Flowering time signatures, introgression and recombination in recently diverged ecotypes. New Phytol. 2017, 218, 1631–1644. [Google Scholar] [CrossRef]
- Cahoon, A.B.; Sharpe, R.M.; Mysayphonh, C.; Thompson, E.J.; Ward, A.D.; Lin, A. The complete chloroplast genome of tall fescue (Lolium arundinaceum; Poaceae) and comparison of whole plastomes from the family Poaceae. Am. J. Bot. 2010, 97, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2017, 9, bbx108. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Trifinopoulos, J.; Nguyen, L.T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, 232–235. [Google Scholar] [CrossRef] [PubMed]
- Free open-source vector graphics editor Version 0.92.3. Available online: https://gitlab.com/inkscape/inkscape (accessed on 20 November 2018).
- Frazer, K.A.; Pachter, L.; Poliakov, A.; Rubin, E.M.; Dubchak, I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004, 32, 273–279. [Google Scholar] [CrossRef] [PubMed]
- Tóth, G.; Gáspári, Z.; Jurka, J. Microsatellites in different eukaryotic genomes: Survey and analysis. Genome Res. 2000, 10, 967–981. [Google Scholar] [CrossRef]
- Ellegren, H. Microsatellites: Simple sequences with complex evolution. Nat. Rev. Genet. 2004, 5, 435–455. [Google Scholar] [CrossRef]
- Mudunuri, S.B.; Nagarajaram, H.A. IMEx: Imperfect microsatellite extractor. Bioinformatics 2007, 23, 1181–1187. [Google Scholar] [CrossRef]
- Stothard, P. The Sequence Manipulation Suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques 2000, 28, 1102–1104. [Google Scholar] [CrossRef]
Sample Availability: Samples are available from the authors. |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).