Abstract
In this continuing work, we have updated our recently proposed Multi-fingerprint Similarity Search algorithm (MuSSel) by enabling the generation of dominant ionized species at a physiological pH and the exploration of a larger data domain, which included more than half a million high-quality small molecules extracted from the latest release of ChEMBL (version 24.1, at the time of writing). Provided with a high biological assay confidence score, these selected compounds explored up to 2822 protein drug targets. To improve the data accuracy, samples marked as prodrugs or with equivocal biological annotations were not considered. Notably, MuSSel performances were overall improved by using an object-relational database management system based on PostgreSQL. In order to challenge the real effectiveness of MuSSel in predicting relevant therapeutic drug targets, we analyzed a pool of 36 external bioactive compounds published in the Journal of Medicinal Chemistry from October to December 2018. This study demonstrates that the use of highly curated chemical and biological experimental data on one side, and a powerful multi-fingerprint search algorithm on the other, can be of the utmost importance in addressing the fate of newly conceived small molecules, by strongly reducing the attrition of early phases of drug discovery programs.
1. Introduction
Conceiving a drug to bias a specific target is a challenging and risky bet. As is well known, drug discovery often ends in costly flops, with about 12 years being necessary to obtain a marketable drug, and research and development investments often exceeding US$ 1 billion. It goes without saying that reducing attrition in early development is by far more important than filling a pipeline with poorly chosen late-stage products, which are likely to fail and fail expensively [1]. The fiasco in the clinic is often due to sloppy early target validation as drugs do not work or are unsafe. More often than not, the key to success is the quick and accurate identification of drug targets with real-life potential.
In this scenario, we have recently developed a Multi-fingerprint Similarity Search algorithm (MuSSel) in an attempt to better approach protein drug target and bioactivity prediction [2]. By exploiting a large collection of high-quality experimental bioactivity data available from ChEMBL (version 22.1), our first release of MuSSel made use of a pool of 13 selected molecular fingerprints (FPs) to return an informed prediction of therapeutically relevant protein drug targets based on a consensus scheme for a given user query. In addition, MuSSel was also effective in mining ChEMBL data relevant to bioactivity prediction and, more specifically, to quantitatively assess the Ki or IC50 values provided that a relevant similarity threshold was found and that activity cliffs were not experienced. The interested reader is refereed elsewhere [2] for a comprehensive description of the MuSSel architecture.
In this continuing work, our efforts were mainly directed to improving the selection of ChEMBL experimental data in an attempt to increase the overall reliability of the MuSSel results as far as protein target prediction is concerned. Building on this idea, we used the latest release of ChEMBL (version 24.1, at the time of writing) [3] as a training set, which encompassed a wider data landscape (i.e., 862,311 biologically-annotated records) compared to the previous version of ChEMBL (version 22.1) (694,532 biologically-annotated records) [4]. On the other hand, we exploited a high number of upstream molecular options to raise the level of data curation of our training set, which, for instance, applied salt stripping and SMILES standardization [5]. Great attention was then paid to ionizable compounds; these included about 250,000 compounds, thus amounting to half of the data taken from the latest release of ChEMBL (version 24.1, at the time of writing). In the present work, we generated the dominant state for each ionizable compound at pH 7.4 [6], thus allowing the user the chance to gain more realistic results after a screening campaign. For the sake of completeness, a parallel investigation was also carried out to prove the effectiveness of the 13 selected FPs in discerning neutral from ionized pairs. Interestingly, we observed that five out of 13 FPs returned similarity values that were likely to be pH dependent. The prediction power of this refined version of MuSSel was challenged by employing a more severe validation strategy, resulting in encouraging results with a significant improvement compared to our initial approach. Moreover, the predictive strength of this revised version of MuSSel was further and successfully tested on an external set of 36 properly selected bioactive drug-like compounds published in the Journal of Medicinal Chemistry in the previous three months (from October to December 2018) and thus not included in the latest release of ChEMBL (version 24.1, at the time of writing). Interestingly, we observed that MuSSel returned reliable results, being able to properly predict the reported protein drug target for 18 out of 36 bioactive drug-like compounds. This retrospective exercise gave us the valuable chance to infer some general predictive trends and, more importantly, to gain a wealth of preliminary information about some specific therapeutic classes [7]. The main aim of this study is to describe an advanced drug discovery tool, which relates newly designed small drug-like molecules to the most probable protein drug targets and unveils new potentially clinical uses for known drugs for apparently unrelated diseases.
2. Results and Discussion
2.1. A Multi-Fingerprints Similarity Analysis Comparing Ionized and Neutral Molecular Pairs
Based on our previous works [2,8], 13 different types of FPs were calculated by means of the RDKit [9] and Pybel [10] python packages and the CDK Java library [11,12]. The calculated FPs are summarized in Table 1.

Table 1.
Fingerprint notations along with the open-source software packages used for their calculation.
As recently explained elsewhere [2,19], these FPs were properly selected after conducting a correlation analysis of the Tanimoto similarity coefficients (Tc) calculated for one million pairs that were randomly selected. To make a fair comparison of the above selected 13 FPs, we randomly generated 10 million pairs according to the approach suggested by Maggiora et al. [20]. In this respect, we calculated the Tc distributions of the 13 different FPs to designate a statistically significant similarity threshold Tcm%, which indicated, for each considered FP, the value of Tc, which met or exceeded the percentage of comparison m%. For a more comprehensive view, the interested reader is referred to our recent work [2]. We then carried out a preliminary investigation to assess the sensitivity of the 13 selected FPs when dealing with ionized compounds at a physiological pH compared to the corresponding neutral species. To this end, we defined two groups containing the same pool of one million pairs of compounds that were first ionized at a physiological pH and in then in a neutral condition, irrespective of pH. This pool of one million pairs of compounds was obtained by random generation from the ensemble of about 250,000 ionizable entries taken from ChEMBL (version 24.1) and then made available in MuSSel. For each pair, the molecular similarity was measured considering that the partners were both ionized on one side and neutral on the other. These similarity measures were thus repeated by using all the 13 FPs implemented in MuSSel. Of course, identical similarity values were expected for those FPs unable to discern a given pair where the partners were both ionized or both neutral. Likewise, different similarity values should occur in the case of FPs distinguishing a given pair if the partners are both charged or both neutral. Based on this idea, we investigated the similarity values calculated by using the 13 FPs implemented in MuSSel for the same pool of one million pairs existing as ionized and neutral forms. Interestingly, our analysis revealed that a pH-dependent similarity was found in five out of the 13 FPs, including klekota_roth, cdk_maccs, pubchem, substructure, and FeatMFP1. For the sake of clarity, we plotted their calculated similarity values in the case of ionized (i.e., y axis) and neutral (i.e., x axis) partners for each pair of compounds randomly generated. As shown in Figure 1, each graph can be split into four areas. The first and the second areas collected pairs, colored in orange and purple, respectively, whose similarity values were always under and over their calculated statistically significant similarity threshold [2,20], irrespective of the ionization state. The third area collected pairs colored in green whose similarity values exceeded their calculated statistically significant similarity threshold, having been awarded in terms of molecular similarity on the basis of their ionized state. Finally, the fourth area collected pairs colored in red whose similarity values deteriorated following the ionization. Bearing this in mind, we could observe that klekota_roth, cdk_maccs, pubchem, and substructure FPs tended to move pairs towards the green rather than red areas. This could likely indicate that such FPs could have a major role in dealing with ionized pairs. On the other hand, a higher number of pairs was in the red zone when using the FeatMFP1, likely suggesting that this FP, although pH-dependent, was not very successful at screening ionized queries. For the sake of comparison, the interested reader can find the same graph generated for all the other remaining 8 FPs in Figure S1.
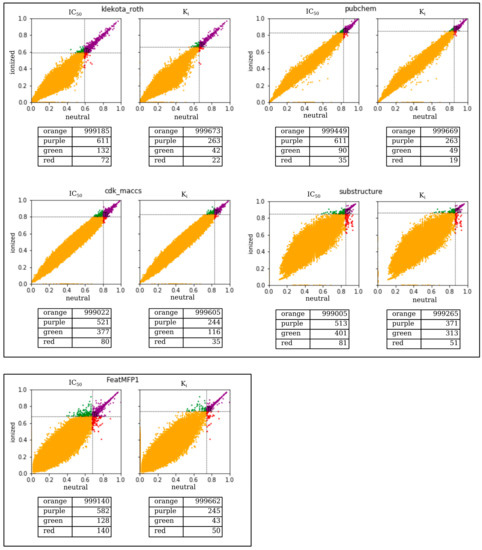
Figure 1.
Similarity comparisons of one million neutral vs. ionized pairs of compounds by using klekota_roth, cdk_maccs, pubchem and substructure, and FeatMFP1 FPs. Orange/purple pairs have similarity values always under/over the threshold, respectively, irrespective of the ionization state. Green/red pairs have similarity values awarded/penalized after ionization, respectively.
For the sake of completeness, we also assessed the overall effect of ionization on predictions. To this end, we used two copies of our entire MuSSel database. The first contained all the compounds in a neutral state and the second contained compounds as ionized species depending on the physiological pH. These two copies were thus used to predict, separately for both Ki and IC50 pools, an external set made of 5000 compounds ionized at a physiological pH extracted from the relative pool. As illustrated in Table 2, the results show that the statistics are slightly improved in the case of predictions based on the ionized database. Although the improvements are small, the results reflect a more realistic picture based on the effect of a physiological pH on ionizable species.

Table 2.
For both the Ki and IC50 pools, predictions are based on first using the neutral database and then the ionized database on the same external dataset comprised of 5000 ionized compounds at a physiological pH randomly discarded by the training set based on ChEMBL (version 24.1). Using both Ki and IC50 protein drug target data, the predictions were considered successful if a match was found as the top-one (p1) or within the top-five (p5).
2.2. Ki and IC50 based Protein Drug Target Predictions
In the present investigation, our attention was mostly directed to assessing the impact on the prediction of ionized dominant species calculated for a larger basis of data (that is about 862,311 vs. 694,532 of our previous analysis) provided with a higher biological assay confidence score. The same setting of calibration parameters already tuned in our previous work was used [2]. For the ease of comparison, we preferred to first predict the same three external sets discussed in our previous work. Basically, a prediction was flagged as correct if a match was found as the top-one (that is p1) or within the top-five (that is p5) calculated protein drug targets after selecting, by chance, one experimental biological annotation for each external set compound. In this respect, each of these three external sets contained 300 compounds that were randomly selected considering the difference between ChEMBL (version 23) and ChEMBL (version 22.1) [21,22,23]. Of course, these external set compounds were excluded from our new training set before their prediction. However, due to this difference in the collection of congeneric series recently published in the scientific literature, this widely employed validation strategy could not ensure that these external sets could really reflect the same proportion of protein drug targets existing in the training data set. In this respect, the usage of the latest release of ChEMBL (version 24.1, at the time of writing), which encompassed the previous ones, could have had the effect of resulting in an impressive improvement of statistics for both the Ki and IC50 pools in comparison with our previous work.
To avoid the above mentioned risk of misrepresentation of data in the external sets, we herein carried out a further validation analysis based on the prediction of a pool of 1000 compounds blindly extracted from the latest release of ChEMBL (version 24.1, at the time of writing) and left out from our new training set. In addition, we also considered the chance of having multiple experimental biological annotations for a ligand. To deal with this more complex but indeed more realistic description of data, a prediction was herein considered successful if a match was found as the top-one (that is p1) or within the top-five (that is p5) calculated protein drug targets after scanning all the available and experimentally measured biological annotations. Based on this counting approach, this new validation test returned encouraging statistics, reported in Table 3, with top-one and top-five protein drug targets ranging from 90.77% to 94.32% in the case of Ki and from 90.1% to 93.2% in the case of IC50, respectively. The interested reader can inspect all the external set data by browsing the content of the File S1 enclosed as Supplementary Materials.

Table 3.
Each of Ext1, Ext2, and Ext3 comprised 300 molecules randomly taken from the difference between ChEMBL (version 23) and ChEMBL (version 22.1). Ext4 comprised 1000 compounds randomly discarded from the training set based on ChEMBL (version 24.1). Using both Ki and IC50 protein drug target data, the predictions were considered successful if a match was found as the top-one (p1) or within the top-five (p5) targets.
2.3. Case Studies
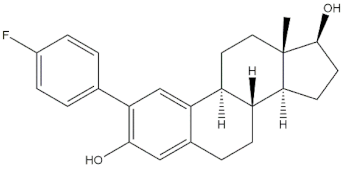
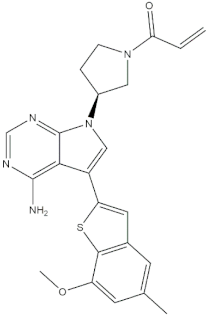
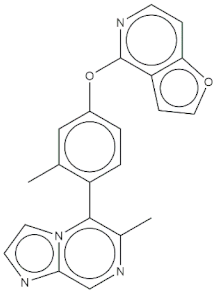
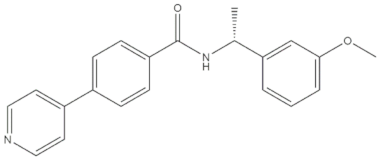
The real predictive strength of MuSSel was finally challenged by conducting a retrospective exercise on a pool of drug-like small molecules whose experimentally determined protein drug targets have just been published in the Journal of Medicinal Chemistry and are therefore not yet covered in the latest release of ChEMBL (version 24.1, at the time of writing). More specifically, we only selected research papers published from October to December of the year 2018 including specific keywords (that were one of the following: discovery, synthesis, identification, design, or optimization) in the title and provided with SMILES notations. The selection was further limited to only small molecules, apart from radioligands, having experimentally established information and whose protein drug target was included among the 2822 explored by MuSSel. In this way, we collected 36 small molecules whose chemical structures were thus submitted to MuSSel to test its potential in properly pairing the real protein drug targets. We observed that the target of 18 small molecules out of 36 was properly ranked in the top-five and, very satisfactorily, in 16 out of 18 compounds, the right target was the top-one. For the sake of comparison, our previous MuSSel release was able to correctly match 15 out of 36 protein drug targets. A comprehensive view of the chemical structures of the 18 entries whose protein drug targets were successfully predicted is given in Table 4. A closer look revealed that MuSSel was effective in associating the real and predicted protein drug target, apart from compounds 2 and 10. For the former, the Heat shock protein 90 kDa beta member 1 was predicted in place of its real isoform, which is actually the Heat shock protein 90 alpha [24], while for the latter, the fibroblast growth factor receptor 1 was predicted in place of its real isoform, which is actually the fibroblast growth factor receptor 2 [25]. The same analysis was carried out for unsuccessfully predicted protein drug targets and a comprehensive list is enclosed as Table S1. By analyzing the number of entries per targets selected in MuSSel, we observed that properly predicted targets were significantly more populated than those unsuccessfully predicted. For the sake of completeness, the same pool of 36 small molecules was also challenged by using the SwissTargetPrediction [26] and the Polypharmacology Browser 2 webserver [27]. Interestingly, a nice overlap was observed by comparing the results obtained by MuSSel with those of the other two platforms. More specifically, MuSSel, SwissTargetPrediction, and Polypharmacology Browser 2 were successful in properly matching the actual protein drug target of 18, 12, and 13 out of 36 cases, respectively. Interestingly, the combined use of the three platforms had the effect of increasing the overall accuracy to 21 out of 36 initial queries, as described in Table S2 of the Supplementary Materials. The approach herein adopted was based on a scheme implying that a given protein drug target, reported in the articles published by the Journal of Medicinal Chemistry, was successfully predicted if at least one of the three platforms returned the right answer [28]. For a more informed view, the interested reader is referred to the Supplementary Materials enclosed in the File S2, which also contains full, detailed reports provided by SwissTargetPrediction and by the Polypharmacology Browser 2 webserver.

Table 4.
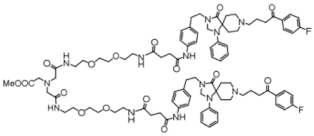
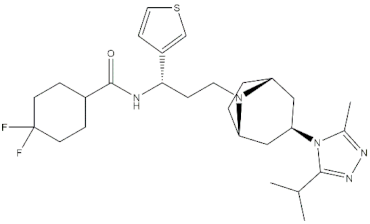
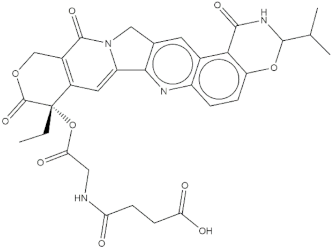
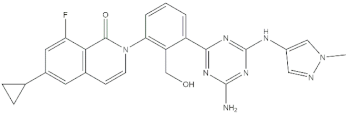
Chemical structures of the 18 entries selected from the Journal of Medicinal Chemistry (i.e., inspecting papers published from October to December 2018) whose protein drug targets were successfully predicted. For each entry, the name of the protein drug target with the corresponding number of associated compounds, as well as the ChEMBL ID available in MuSSel, are reported. A parallel table with the unsuccessful cases is enclosed in the Supplementary Materials.
3. Materials and Methods
3.1. Construction of the Ki and IC50 Database
ChEMBL (version 24.1) was downloaded as a PostgreSql cartridge format and stored in a local machine. MuSSel-related tables were associated with three ChEMBL macroareas: ‘COMPOUND INFORMATION’ (tables molecule_dictionary and compound_records); ‘EXPERIMENTAL DATA’ (tables activities and assays); and ‘TARGET INFORMATION’ (tables target_dictionary, target_components, component_sequences, component_class, and protein_classification). As is usual in relational databases, the table associations were based on links between primary keys and foreign keys, as described in ChEMBL [45]. A materialized view named MuSSel_data was thus built by querying the ChEMBL database for retrieving entries matching the following criteria: only ‘small molecule’ in the molecule_type field was chosen; molecules marked in the database as prodrugs were removed (about 300 records); a target was set if the target_type was indicated as ‘SINGLE_PROTEIN’ or ‘PROTEIN COMPLEX’ no restriction was applied on organism to avoid missing therapeutically-relevant targets, such as those typical of antiviral and antibiotic drugs; all records containing any warning on the data assay were removed (data_validity_comment field); only records with an assay confidence_score greater than 5 were considered on a scale from 1 to 9; only activity records containing ‘IC50’, ‘Ki’, ‘Kd’, and ‘EC50’ as standard_type were maintained; and ambiguous biological data with standard_relation equal to ‘>’ in the activity table were removed. Based on this filtering process, 1,280,553 records were selected for MuSSel. The whole pool of MuSSel_data was split into four main groups by standard_type activity values. The interested reader can find the SQL query used for MuSSel_data generation in Figure S2 of the Supplementary Materials and a formal scheme as Figure S3 of the Supplementary Materials. In this study, only IC50 and Ki groups were analyzed according to our previous investigation [2]. Within each group, the records were split into protein drug target ensembles contacting at least 10 compounds or data were otherwise not further considered. In the case of multiple ligand annotations within the same protein drug target, the lowest activity value (i.e., the best measure for Ki or IC50) was retained [2]. The Ki pool contained 288,178 annotated entries covering 1265 targets, while the IC50 pool included 522,594 covering 2441 targets. The interested reader is referred to the File S3 of the Supplementary Materials for a complete list of the protein drug targets explored in MuSSel.
3.2. Canonicalization and Correction of Chemical Structures
Validation control was performed in a canonical SMILES format stored in MuSSel_data, using an in-house python script based on RDkit and the MolVS library [9]. The main steps were the fragment strip and the standardization. The fragment strip (also termed as salt strip) was based on the identification of all the fragments in SMILES strings in order to maintain only the largest one. In the case of enantiomers, only one was randomly picked. The standardization implied a sequence of structure optimization steps consisting of SMILES sanitization (i.e., valence error correction), metal disconnection, the application of normalization rules, and stereochemistry recalculation [46,47].
3.3. Generation of Dominant Ionized Species at a Physiological pH and Neutral Forms
Compounds were ionized at pH 7.4 by using the ChemAxon cxcalc majormicrospecies plugin [48]. Neutral forms were regenerated from standardized SMILES using the Rdkit package [9].
3.4. Fingerprints Generation
For each molecule included in the MuSSel_data table, 13 different types of FPs were calculated by means of the RDKit [9], Pybel [10], and CDK [11] packages and loaded as tables in the MuSSel database. The calculated FPs are summarized in Table 1. Two parallel sets of FPs were generated when a molecule could exist in the ionized and neutral form based on the physiological pH of 7.4. In this respect, a first set of 246,959 FPs was generated in the neutral form and a second parallel set of 509,058 FPs was derived to account for the possible ionization states. This heavy computational task was carried out by using the ReCaS-Bari system [49].
3.5. Construction of the External Sets
Based on the same procedures described in the previous paragraphs, three external sets were generated using the ionized SMILES strings exploiting the same external sets already challenged in our previous work and a corresponding set of FPs was computed [2]. Each external set contained 300 compounds that were randomly selected considering the difference between ChEMBL (version 23) and ChEMBL (version 22.1). The compounds of each external set were excluded by MuSSel_data before their prediction. In addition, an external set of 1000 compounds and one made of 5000 ionized compounds were randomly extracted from both the Ki and IC50 pools and left out from our training set before their prediction. All the external sets are included in the File S1 enclosed as Supplementary Materials.
3.6. Selection of Prospective Queries From Recently Published Scientific Articles
A systematic PubMed search of scientific articles recently published in the Journal of Medicinal covering the period from October to December 2018 was performed. Papers containing at least one of the following keywords: discovery, synthesis, identification, design or optimization, in the titles and provided with SMILES notations in the related Supporting Information were considered. For the sake of completeness, we only selected those small molecules, apart from radioligands, having experimentally established information and whose protein drug targets were included in the MuSSel database. Finally, a pool of 36 queries was generated. Additional details are reported in Table 4 and in Table S1.
3.7. Protein Drug Target Multi-FPs Similarity Search Algorithm
The protein drug target multi-FPs similarity search algorithm returned an overall score on the basis of the similarity values measured by using the pool of selected FPs listed in Table 1. More specifically, the Tc value between the query compound and all the entries associated with each protein drug target available in MuSSel was calculated by using each of the 13 FPs. A given query was paired to a protein drug target if there was at least one entry having a Tc no lower than the pre-calculated similarity thresholds Tcm% for a minimum number of FPs. If this condition held true, a score (SC) was thus measured for the protein drug target:
where was the maximum Tc value, based on the i-th FP type, between the query and the molecules associated with the drug target provided that . Finally, the selected protein drug targets were ranked according to the assigned SC values. For additional details, the interested reader is referred to our previous work [2].
4. Conclusions
In this study, we improved our multi-FPs similarity search algorithm, named MuSSel, by considering a wider and even higher quality pool of about half a million small drug-like molecules extracted by the latest release of ChEMBL (version 24.1, at the time of writing). This refined larger pool included biological data not limited to Homo Sapiens as the organism and covered 2440 experimentally determined biological targets. In addition, the multi-FPs similarity search was carried out considering the calculated dominant ionized species at a physiological pH for each small molecule taken from ChEMBL. Compared to our previous investigation, the net effect of these changes was a significant improvement of the external set statistics. Interestingly, this trend was substantially confirmed by a new second validation set and even by the retrospective exercise based on a pool of 36 selected molecules just published in the Journal of Medicinal Chemistry. In this work, we have launched a private platform, accessible on demand, which allows the interested users to screen single or even multiple queries at a time, as normally requested in reverse screening campaigns, which have become an integral part of drug discovery pipelines [50]. The herein proposed method showed very promising performances and can offer a useful and easy-to-run tool capable of pairing novel compounds to putative protein drug targets, as well as repurposing known drugs to apparently unrelated diseases, explicitly accounting for their potential toxicity and/or unwanted side effects. It is noteworthy that the multi-fingerprint search algorithm also demonstrated a great potential for the prediction of acute oral toxicity [51].
Supplementary Materials
The supplementary materials are available online.
Author Contributions
Software, M.M., D.A, and F.C.; validation, M.M., D.A., and F.C.; formal analysis, D.T., A.R.T., and G.F.M.; data curation, M.M.; draft preparation, O.N.; supervision, O.N.
Funding
This research received no external funding.
Acknowledgments
We thank ChemAxon for the academic use of the Marvin and Cxcalc plugin and ReCaS-Bari for the availability of high-performance computing resources and support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Smith, C. Drug target validation: Hitting the target. Nature 2003, 422, 341–347. [Google Scholar] [CrossRef] [PubMed]
- Alberga, D.; Trisciuzzi, D.; Montaruli, M.; Leonetti, F.; Mangiatordi, G.F.; Nicolotti, O. A new approach for drug target and bioactivity prediction: The Multifingerprint Similarity Search Algorithm (MuSSeL). J. Chem. Inf. Model. 2019, 59, 586–596. [Google Scholar] [CrossRef] [PubMed]
- ChEMBL v.24.1. Available online: http://ftp.ebi.ac.uk/pub/databases/chembl/ChEMBLdb/latest/ (accessed on 11 February 2019).
- ChEMBL v.22.1. Available online: http://ftp.ebi.ac.uk/pub/databases/chembl/ChEMBLdb/releases/chembl_22_1/ (accessed on 11 February 2019).
- Xia, J.; Tilahun, E.L.; Reid, T.E.; Zhang, L.; Wang, X.S. Benchmarking methods and data sets for ligand enrichment assessment in virtual screening. Methods San. Diego. Calif. 2015, 71, 146–157. [Google Scholar] [CrossRef] [PubMed]
- Kenny, P.W.; Sadowski, J. Structure Modification in Chemical Databases. In Chemoinformatics in Drug Discovery; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2005; pp. 271–285. [Google Scholar]
- Nicolotti, O.; Miscioscia, T.F.; Leonetti, F.; Muncipinto, G.; Carotti, A. Screening of matrix metalloproteinases available from the protein data bank: Insights into biological functions, domain organization, and zinc binding groups. J. Chem. Inf. Model. 2007, 47, 2439–2448. [Google Scholar] [CrossRef] [PubMed]
- Floris, M.; Manganaro, A.; Nicolotti, O.; Medda, R.; Mangiatordi, G.F.; Benfenati, E. A generalizable definition of chemical similarity for read-across. J. Cheminform. 2014, 6, 39. [Google Scholar] [CrossRef] [PubMed]
- Landrum, G. RDKit: Open-Source Cheminformatics. 2006. Available online: https://www.rdkit.org/ (accessed on 11 February 2019).
- O’Boyle, N.M.; Morley, C.; Hutchison, G.R. Pybel: A Python wrapper for the OpenBabel cheminformatics toolkit. Chem. Cent. J. 2008, 2, 5. [Google Scholar] [CrossRef] [PubMed]
- Steinbeck, C.; Han, Y.; Kuhn, S.; Horlacher, O.; Luttmann, E.; Willighagen, E. The Chemistry Development Kit (CDK): An open-source java library for chemo- and bioinformatics. J. Chem. Inf. Comput. Sci. 2003, 43, 493–500. [Google Scholar] [CrossRef]
- Steinbeck, C.; Hoppe, C.; Kuhn, S.; Floris, M.; Guha, R.; Willighagen, E.L. Recent developments of the Chemistry Development Kit (CDK)—An open-source java library for chemo- and bioinformatics. Curr. Pharm. Des. 2006, 12, 2111–2120. [Google Scholar] [CrossRef]
- Rogers, D.; Hahn, M. Extended-connectivity fingerprints. J. Chem. Inf. Model. 2010, 50, 742–754. [Google Scholar] [CrossRef]
- Gobbi, A.; Poppinger, D. Genetic optimization of combinatorial libraries. Biotechnol. Bioeng. 1998, 61, 47–54. [Google Scholar] [CrossRef]
- Carhart, R.E.; Smith, D.H.; Venkataraghavan, R. Atom pairs as molecular features in structure-activity studies: Definition and applications. J. Chem. Inf. Comput. Sci. 1985, 25, 64–73. [Google Scholar] [CrossRef]
- Nilakantan, R.; Bauman, N.; Dixon, J.S.; Venkataraghavan, R. Topological torsion: A new molecular descriptor for SAR applications. Comparison with other descriptors. J. Chem. Inf. Comput. Sci. 1987, 27, 82–85. [Google Scholar] [CrossRef]
- PubChem Substructure Fingerprint v1.3. Available online: ftp://ftp.ncbi.nlm.nih.gov/pubchem/specifications/pubchem_fingerprints.txt (accessed on 11 February 2019).
- Klekota, J.; Roth, F.P. Chemical substructures that enrich for biological activity. Bioinforma. Oxf. Engl. 2008, 24, 2518–2525. [Google Scholar] [CrossRef] [PubMed]
- Riniker, S.; Landrum, G.A. Open-source platform to benchmark fingerprints for ligand-based virtual screening. J. Cheminform. 2013, 5, 26. [Google Scholar] [CrossRef] [PubMed]
- Maggiora, G.; Vogt, M.; Stumpfe, D.; Bajorath, J. Molecular similarity in medicinal chemistry. J. Med. Chem. 2014, 57, 3186–3204. [Google Scholar] [CrossRef] [PubMed]
- Gaulton, A.; Bellis, L.J.; Bento, A.P.; Chambers, J.; Davies, M.; Hersey, A.; Light, Y.; McGlinchey, S.; Michalovich, D.; Al-Lazikani, B.; et al. ChEMBL: A large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2012, 40, D1100–D1107. [Google Scholar] [CrossRef] [PubMed]
- Gaulton, A.; Hersey, A.; Nowotka, M.; Bento, A.P.; Chambers, J.; Mendez, D.; Mutowo, P.; Atkinson, F.; Bellis, L.J.; Cibrián-Uhalte, E.; et al. The ChEMBL database in 2017. Nucleic Acids Res. 2017, 45, D945–D954. [Google Scholar] [CrossRef]
- Bento, A.P.; Gaulton, A.; Hersey, A.; Bellis, L.J.; Chambers, J.; Davies, M.; Krüger, F.A.; Light, Y.; Mak, L.; McGlinchey, S.; et al. The ChEMBL bioactivity database: An update. Nucleic Acids Res. 2014, 42, D1083–D1090. [Google Scholar] [CrossRef]
- Jiang, F.; Guo, A.; Xu, J.; You, Q.D.; Xu, X.L. Discovery of a Potent Grp94 Selective inhibitor with anti-inflammatory efficacy in a mouse model of ulcerative colitis. J. Med. Chem. 2018, 61, 9513–9533. [Google Scholar] [CrossRef]
- Wang, Y.; Li, L.; Fan, J.; Dai, Y.; Jiang, A.; Geng, M.; Ai, J.; Duan, W. Discovery of potent irreversible pan-fibroblast growth factor receptor (FGFR) inhibitors. J. Med. Chem. 2018, 61, 9085–9104. [Google Scholar] [CrossRef]
- Gfeller, D.; Grosdidier, A.; Wirth, M.; Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: A web server for target prediction of bioactive small molecules. Nucleic Acids Res. 2014, 42, W32–W38. [Google Scholar] [CrossRef] [PubMed]
- Awale, M.; Reymond, J.L. Polypharmacology Browser PPB2: Target prediction combining nearest neighbors with machine learning. J. Chem. Inf. Model. 2019, 59, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Milosevic, Z.; Hutle, M.; Schiper, A. Unifying byzantine consensus algorithms with weak interactive consistency. In Proceedings of the Principles of Distributed Systems; Springer: Berlin/Heidelberg, Germany, 2009; pp. 300–314. [Google Scholar]
- Ghosh, A.K.; Williams, J.N.; Ho, R.Y.; Simpson, H.M.; Hattori, S.; Hayashi, H.; Agniswamy, J.; Wang, Y.-F.; Weber, I.T.; Mitsuya, H. Design and synthesis of potent HIV-1 protease inhibitors containing bicyclic oxazolidinone scaffold as the P2 ligands: Structure–activity studies and biological and X-ray structural studies. J. Med. Chem. 2018, 61, 9722–9737. [Google Scholar] [CrossRef] [PubMed]
- Bergkemper, M.; Kronenberg, E.; Thum, S.; Börgel, F.; Daniliuc, C.; Schepmann, D.; Nieto, F.R.; Brust, P.; Reinoso, R.F.; Alvarez, I.; et al. Synthesis, receptor affinity, and antiallodynic activity of spirocyclic σ receptor ligands with exocyclic amino moiety. J. Med. Chem. 2018, 61, 9666–9690. [Google Scholar] [CrossRef] [PubMed]
- Brnardic, E.J.; Ye, G.; Brooks, C.; Donatelli, C.; Barton, L.; McAtee, J.; Sanchez, R.M.; Shu, A.; Erhard, K.; Terrell, L.; et al. Discovery of pyrrolidine sulfonamides as selective and orally bioavailable antagonists of transient receptor potential vanilloid-4 (TRPV4). J. Med. Chem. 2018, 61, 9738–9755. [Google Scholar] [CrossRef]
- Pulido, D.; Casadó-Anguera, V.; Pérez-Benito, L.; Moreno, E.; Cordomí, A.; López, L.; Cortés, A.; Ferré, S.; Pardo, L.; Casadó, V.; et al. Design of a true bivalent ligand with picomolar binding affinity for a G protein-coupled receptor homodimer. J. Med. Chem. 2018, 61, 9335–9346. [Google Scholar] [CrossRef]
- Peng, P.; Chen, H.; Zhu, Y.; Wang, Z.; Li, J.; Luo, R.-H.; Wang, J.; Chen, L.; Yang, L.M.; Jiang, H.; et al. Structure-based design of 1-Heteroaryl-1,3-propanediamine derivatives as a novel series of CC-chemokine receptor 5 antagonists. J. Med. Chem. 2018, 61, 9621–9636. [Google Scholar] [CrossRef]
- Pan, P.; Chen, J.; Li, X.; Li, M.; Yu, H.; Zhao, J.J.; Ni, J.; Wang, X.; Sun, H.; Tian, S.; et al. Structure-based drug design and identification of H2O-soluble and low toxic hexacyclic camptothecin derivatives with improved efficacy in cancer and lethal inflammation models in vivo. J. Med. Chem. 2018, 61, 8613–8624. [Google Scholar] [CrossRef]
- Kawahata, W.; Asami, T.; Kiyoi, T.; Irie, T.; Taniguchi, H.; Asamitsu, Y.; Inoue, T.; Miyake, T.; Sawa, M. Design and synthesis of novel amino-triazine analogues as selective Bruton’s tyrosine kinase inhibitors for treatment of rheumatoid arthritis. J. Med. Chem. 2018, 61, 8917–8933. [Google Scholar] [CrossRef]
- Dutour, R.; Roy, J.; Cortés-Benítez, F.; Maltais, R.; Poirier, D. Targeting Cytochrome P450 (CYP) 1B1 Enzyme with four series of a-ring substituted estrane derivatives: Design, synthesis, inhibitory activity, and selectivity. J. Med. Chem. 2018, 61, 9229–9245. [Google Scholar] [CrossRef]
- Davoren, J.E.; Nason, D.; Coe, J.; Dlugolenski, K.; Helal, C.; Harris, A.R.; LaChapelle, E.; Liang, S.; Liu, Y.; O’Connor, R.; et al. Discovery and lead optimization of atropisomer D1 agonists with reduced desensitization. J. Med. Chem. 2018, 61, 11384–11397. [Google Scholar] [CrossRef] [PubMed]
- Hobson, A.D.; Judge, R.A.; Aguirre, A.L.; Brown, B.S.; Cui, Y.; Ding, P.; Dominguez, E.; DiGiammarino, E.; Egan, D.A.; Freiberg, G.M.; et al. Identification of selective dual ROCK1 and ROCK2 inhibitors using structure-based drug design. J. Med. Chem. 2018, 61, 11074–11100. [Google Scholar] [CrossRef]
- Cao, X.; Zhang, Y.; Chen, Y.; Qiu, Y.; Yu, M.; Xu, X.; Liu, X.; Liu, B.F.; Zhang, L.; Zhang, G. Synthesis and biological evaluation of fused tricyclic heterocycle piperazine (piperidine) derivatives as potential multireceptor atypical antipsychotics. J. Med. Chem. 2018, 61, 10017–10039. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Murugan, N.A.; Tian, Y.; Bertagnin, C.; Fang, Z.; Kang, D.; Kong, X.; Jia, H.; Sun, Z.; Jia, R.; et al. Structure-based optimization of N-substituted oseltamivir derivatives as potent anti-Influenza A virus agents with significantly improved potency against oseltamivir-resistant N1-H274Y variant. J. Med. Chem. 2018, 61, 9976–9999. [Google Scholar] [CrossRef] [PubMed]
- Rageot, D.; Bohnacker, T.; Melone, A.; Langlois, J.-B.; Borsari, C.; Hillmann, P.; Sele, A.M.; Beaufils, F.; Zvelebil, M.; Hebeisen, P.; et al. Discovery and preclinical characterization of 5-[4,6-Bis({3-oxa-8-azabicyclo[3.2.1]octan-8-yl})-1,3,5-triazin-2-yl]-4-(difluoromethyl)pyridin-2-amine (PQR620), a highly potent and selective mTORC1/2 inhibitor for cancer and neurological disorders. J. Med. Chem. 2018, 61, 10084–10105. [Google Scholar] [CrossRef] [PubMed]
- Damalanka, V.C.; Han, Z.; Karmakar, P.; O’Donoghue, A.J.; La Greca, F.; Kim, T.; Pant, S.M.; Helander, J.; Klefström, J.; Craik, C.S.; et al. Discovery of selective matriptase and hepsin Serine protease inhibitors: Useful chemical tools for cancer cell biology. J. Med. Chem. 2019, 62, 480–490. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yang, J.; Aguilar, A.; McEachern, D.; Przybranowski, S.; Liu, L.; Yang, C.-Y.; Wang, M.; Han, X.; Wang, S. Discovery of MD-224 as a first-in-class, highly potent, and efficacious proteolysis targeting chimera murine double minute 2 degrader capable of achieving complete and durable tumor regression. J. Med. Chem. 2019, 62, 448–466. [Google Scholar] [CrossRef] [PubMed]
- Ju, Y.; Wu, J.; Yuan, X.; Zhao, L.; Zhang, G.; Li, C.; Qiao, R. Design and evaluation of potent EGFR inhibitors through the incorporation of macrocyclic polyamine moieties into the 4-anilinoquinazoline scaffold. J. Med. Chem. 2018, 61, 11372–11383. [Google Scholar] [CrossRef]
- ChEMBL. Available online: https://www.ebi.ac.uk/chembl/faq (accessed on 11 February 2019).
- Glaab, E. Building a virtual ligand screening pipeline using free software: A survey. Brief. Bioinform. 2016, 17, 352–366. [Google Scholar] [CrossRef]
- González-Medina, M.; Naveja, J.J.; Sánchez-Cruz, N.; Medina-Franco, J.L. Open chemoinformatic resources to explore the structure, properties and chemical space of molecules. RSC Adv. 2017, 7, 54153–54163. [Google Scholar] [CrossRef]
- Chemaxon. Available online: https://chemaxon.com/ (accessed on 11 February 2019).
- Antonacci, M.; Bellotti, R.; Cafagna, F.; de Palma, M.; Diacono, D.; Donvito, G.; Italiano, A.; Gervasoni, R.; Maggi, G.; Miniello, G.; et al. The ReCaS Project: The Bari Infrastructure. In High Performance Scientific Computing Using Distributed Infrastructures; World Scientific: Singapore, 2017; pp. 17–33. ISBN 978-981-4759-70-0. [Google Scholar]
- Ziegler, S.; Pries, V.; Hedberg, C.; Waldmann, H. Target identification for small bioactive molecules: Finding the needle in the haystack. Angew. Chem. Int. Ed. Engl. 2013, 52, 2744–2792. [Google Scholar] [CrossRef] [PubMed]
- Alberga, D.; Trisciuzzi, D.; Mansouri, K.; Mangiatordi, G.F.; Nicolotti, O. Prediction of acute oral systemic toxicity using a multifingerprint similarity approach. Toxicol. Sci. Off. J. Soc. Toxicol. 2019, 167, 484–495. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).