Abstract
Pseudomonas aeruginosa is a major opportunistic pathogen in cystic fibrosis, wound and nosocomial infections, posing a serious burden to public health, due to its antibiotic resistance. The P. aeruginosa Pseudomonas Quinolone System (pqs) quorum sensing system, driven by the activation of the transcriptional regulator, PqsR (MvfR) by alkylquinolone (AQ) signal molecules, is a key player in the regulation of virulence and a potential target for the development of novel antibacterial agents. In this study, we performed in silico docking analysis, coupled with screening using a P. aeruginosa mCTX::PpqsA-lux chromosomal promoter fusion, to identify a series of new PqsR antagonists. The hit compounds inhibited pyocyanin and alkylquinolone signal molecule production in P. aeruginosa PAO1-L and PA14 strains. The inhibitor Ia, which showed the highest activity in PA14, reduced biofilm formation in PAO1-L and PA14, increasing their sensitivity to tobramycin. Furthermore, the hepatic and plasma stabilities for these compounds were determined in both rat and human in vitro microsomal assays, to gain a further understanding of their therapeutic potential. This work has uncovered a new class of P. aeruginosa PqsR antagonists with potential for hit to lead optimisation in the search for quorum sensing inhibitors for future anti-infective drug discovery programs.
1. Introduction
Antimicrobial resistance has emerged as a global threat to public health, driven by complex resistance mechanisms, a lack of new antibiotics and the misuse of clinically approved antibacterial agents [1,2]. In recent years, a novel approach to combat antimicrobial resistance has begun to attract attention, based on disarming bacterial virulence through the disruption of bacterial quorum sensing (QS)-mediated communication systems. QS employs diffusible signals, sometimes called autoinducers (AIs), to control bacterial community behaviour by co-ordinating gene expression at the population level, to promote pathogen survival and dissemination [3]. There are several approaches to target QS from a drug discovery point of view and these include inhibition of the biosynthesis of AIs, blocking the interaction of these signals with their receptors or through direct interference with the signals, using enzymes or antibodies [4,5,6]. Among the organisms most clinically resistant to antibiotics is Pseudomonas aeruginosa, an opportunistic gram-negative pathogen and leading cause of diverse nosocomial infections, mainly in immunocompromised patients and individuals with cystic fibrosis [7,8]. P. aeruginosa infections are commonly difficult to eradicate with conventional antibiotic therapy, particularly when established as biofilms.
P.aeruginosa possesses three integrated quorum sensing circuits, known as las, rhl and the Pseudomonas Quinolone System (pqs). The las and rhl systems are reliant on N-acylhomoserine lactones (AHLs), whereas the pqs system utilises alkylquinolone (AQ) derived signal molecules [9]. The latter is regulated by the interaction between the transcriptional regulator, PqsR (MvfR), and 2-heptyl-3-hydroxy-4(1H)-quinolone (PQS) or 2-heptyl-4-hydroxyquinoline (HHQ), which in turn induce the transcription of the pqsABCDEphnAB operon, leading to the production of more AQs and virulence factors, including pyocyanin as well as enhancing biofilm maturation [10,11]. It has been well documented that interfering with PqsR activity disrupts biofilm development and increases sensitivity to antibiotics, further validating its potential as a therapeutic target [12]. While the las and rhl systems have been studied extensively, with many medicinal chemistry approaches described for developing inhibitors that block binding of AHLs to their cognate receptors [13,14,15,16,17], the pqs system, as a therapeutic target, has received less attention. Amongst the limited number of PqsR inhibitors are quinazolinone, quinolinone, benzamide-benzimidazole and hydroxybenzamide derived molecules [18,19,20,21,22]. In this study, we employed an in silico virtual screening method to search for novel PqsR inhibitors in a library of quinoline-based molecules. Following that, the compounds were evaluated for their inhibitory activity, using a suite of in vitro assays, to reveal a new generation of PqsR inhibitors with low micromolar potency. The antagonists were then profiled for their metabolic stability in both humans and rats (plasma and liver microsomes), to gain a better understanding of their therapeutic potential.
2. Results
2.1. In Silico Virtual Screening for PqsR Antagonists
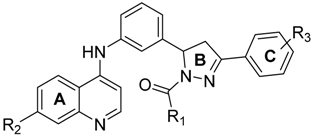
We employed a virtual screening approach, to identify new inhibitors of PqsR from a library of quinolone-derived molecules. The library was based on compounds, originally reported by Ramírez-Prada et al. [23] as antiprotozoal agents, as they share a 7-chloro-4-aminoquinoline core (Figure 1a) which closely resembles the native P. aeruginosa AQ co-inducer head group. The study was performed using the crystal structure of the PqsR ligand binding domain, complexed with the quinazolinone inhibitor, 3-NH2-7Cl-C9-QZN (abbreviated as QZN), reported by Ilangovan et al. [18] (Figure 1b). We speculated that 7-chloro-4-aminoquinoline could form a suitable isostere for the head group in the QZN antagonist, while the hydrophobic aromatic extension attached to the amino group could replace the C9 alkyl chain in QZN (Figure 1c). Hence the Glide module of the Schrödinger Suite was used to examine a total of 31 structures exploiting the extra-precision docking function [24].
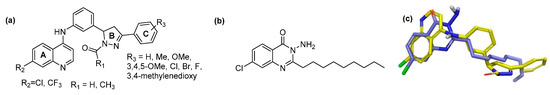
Figure 1.
Structures of PqsR antagonists. (a) Chemical structures of quinolone-based compound library; (b) Chemical structure of PqsR inhibitor 3-NH2-7-Cl-C9-QZN (QZN); (c) Overlay of a quinolone derivative (yellow) and QZN inhibitor (blue).
The ligand binding pocket in PqsR consists of an inner pocket (B pocket) which accommodates the quinoline head group and an elongated outer ‘A’ pocket, rich with hydrophobic residues that interact with the aliphatic chain of the co-inducers. The docking study confirmed that the favorable binding position for the quinoline library imitates the QZN inhibitor binding mode, where the 4-amino-quinoline core resides in the A pocket, while the aromatic tail extends to occupy the B pocket (Figure 2c). Moreover, the chlorine atom at the 7-position of the quinoline adopts the same orientation as the chlorine in the QZN and occupies the same small pocket formed by Thr265. The docking score, represented as XP GScore (Table 1), suggested that the best potential candidates are those with an unsubstituted ring C, while having a formyl or acetyl group on the pyrazoline ring B had no significant effect. The best fit compounds were Ia, Ii, IIa and IIi (Table 1).
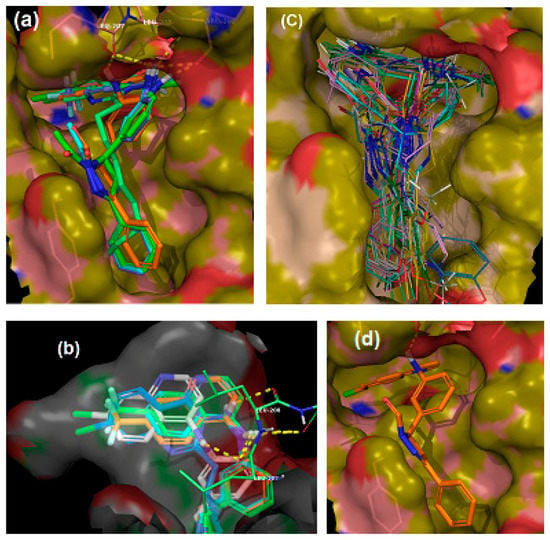
Figure 2.
In silico binding of putative inhibitors to the PqsR ligand binding domain. (a) Overlay of the highest scored compounds in the PqsR ligand binding domain, compared to QZN (solid green); (b) Overlay of quinoline core in the inner pocket of PqsR; (c) Overlay of the entire library of compounds in the PqsR binding domain; (d) Binding position for compound Ia.

Table 1.
Summary of docking scores and activity data for the library.
Interestingly, chlorine substitution at the 7-position of the quinoline ring resulted in a slightly higher GLIDE score, compared to the corresponding trifluoromethane-substituted compounds (e.g., compare Ia and Ii). The docking positions for these ligands indicated the presence of π-π interactions between ring C and Tyr258 as well as electrostatic contacts between the quinolone 4-amino substituent and the side chains of Leu207, Leu208 and Arg209 (Figure 2a,b,d).
2.2. Whole Cell Biosensor Reporter Screening for PqsR Inhibitors
To determine the ability of the quinolines to inhibit PqsR functionality, the P. aeruginosa biosensor strains, PAO1-L and PA14, incorporating a chromosomal mCTX::PpqsA-lux transcriptional fusion, were used. These report on the activation of the pqsA promoter, regulated by PqsR [25]. PAO1-L and PA14 were chosen because they are genetically amenable and belong to the two major P. aeruginosa genomic groups [26]. The quinolone compounds were incubated with the corresponding strain, and luminescence and optical density readings were recorded to monitor the effect of the quinolines on PqsR activity and bacterial growth, respectively. The compounds were screened at 10 µM concentration, and the activity was compared to a control of DMSO (0.1%) (Figure 3a). To be considered a hit, a threshold of at least 50% inhibition of the test compound, relative to the negative control had to be reached, without affecting bacterial growth. The library showed variable activity towards the reporters (Table 1, Figure 3a) and a total of four inhibitors (Ia, Ii, IIa and Iii) were identified with strong PqsR inhibitory activity for both P. aeruginosa strains, without interfering with bacterial growth (Figure 3a,d,e). Concentration-response experiments were then carried out to determine the IC50 values (Figure 3b,c), which are summarised in Table 1.
Figure 3.
Whole bacterial cell-based P. aeruginosa compound screen for both PqsR and growth inhibition (a) Activity against PA14 (green) and PAO1-L (purple) PpqsA-lux reporter strains at 10 µM test compound; (b) and (c) Dose-response curves for active compounds against PA14 (b) and PAO1-L (c); (d) and (e) growth curves for PA14 (d) and PAO1-L (e) in the presence of compound or DMSO control.
Ii and IIi had the same PqsR inhibitory activity against both strains (IC50 5 µM), while compound Ia was the most active inhibitor of PA14 (IC50 2.2 µM), but exhibited only modest activity against PAO1-L. In contrast, IIa was the most active PqsR antagonist for PAO1-L (IC50 1.5 µM), compared with PA14 (IC50 of 3.7 µM).
2.3. Impact of PqsR Antagonists on Pyocyanin Production
Pyocyanin is a blue redox-active toxin, produced by P. aeruginosa and strongly-controlled by PqsR via the regulation of pqsE expression [11,27,28]. Pyocyanin is essential for the pathogenicity of P. aeruginosa in lung infections [29]. The effect of the PqsR inhibitors on pyocyanin production by PA14 and PAO1-L strains, when grown in the presence or absence of the inhibitors, was investigated (Figure 4). All the compounds tested demonstrated significant inhibition when used at a concentration equivalent to three-fold their IC50 values. Compound Ia had the strongest effect on pyocyanin production in both strains, followed by Ii and IIi, while IIa had the weakest effect, with just over 25% reduction, despite its low micromolar potency in the reporter assay.
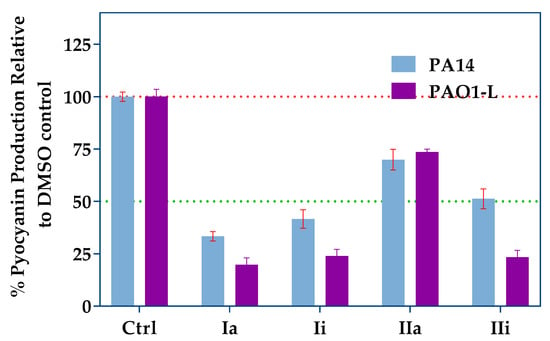
Figure 4.
Pyocyanin production assay using the top hits at concentrations equal to three times their IC50s. Blue bars represent PA14 and purple bars represent PAO1-L. Error bars represent standard deviation of n = 3 biological replicates.
2.4. Impact of Hit Compounds on Alkylquinolone Production
P. aeruginosa produces a broad range of alkylquinolones, via PqsABCDE, some of which (e.g., PQS and HHQ) act as quorum sensing signal molecules, while others (e.g., 2-heptyl-4-hydroxyquinoline N-oxide; HQNO) do not [11,30]. HQNO, can protect Staphylococcus aureus from vancomycin [31] and contributes to the environmental competitiveness of P. aeruginosa, through its activity as a potent inhibitor of the cytochrome bc1 complex [32]. AQ biosynthesis is tightly-regulated by PqsR, hence inhibition of this transcriptional regulator is reflected by a reduction in AQ production that can be used as a readout for PqsR inactivation. LC-MS/MS was used to quantify the major AQs HHQ, PQS and HQNO after incubation of the P. aeruginosa strains with the hit compounds, for 16 h. The assay was performed using both PAO1-L and PA14 strains which were treated with PqsR antagonists at concentrations three-fold higher than their IC50 values. The results obtained depended on the inhibitor and strain tested (Figure 5). Compound Ii showed the greatest inhibition of AQ production in PAO1-L; however, this was not observed for PA14. Furthermore, IIa had only a moderate effect, whereas Ia maintained a strongly-inhibited AQ production for both strains, in agreement with the pyocyanin data (compare Figure 4 and Figure 5).
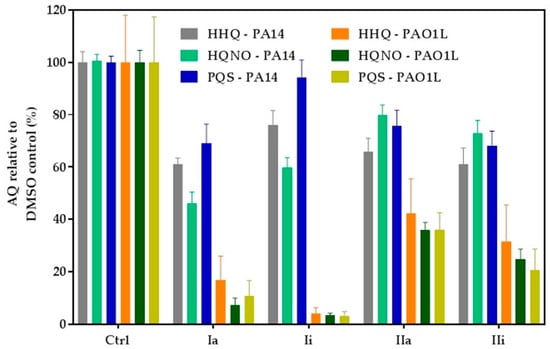
Figure 5.
Inhibition of alkylquinolone (AQ) production. PAO1-L and PA14 were treated with the corresponding PqsR inhibitors at 3× IC50 for 16 h and culture supernatants extracted for LC-MS/MS analysis. Each experiment was performed using three biological and three technical replicates. The bars show percentage levels in relation to a non-treated DMSO control.
2.5. Effect of PqsR Antagonists on Biofilms
One of the clinical challenges of P. aeruginosa infections is the difficulty associated with the effective treatment of biofilms due to their high resistance to antibiotics. The pqs system has been shown to regulate a number of genes required for biofilm development [27] and hence, inactivation of this system through antagonism of PqsR is a promising approach, as it can sensitize biofilms to conventional antibiotics [33]. The effect of Ia, with or without tobramycin, on PA14 and PAO1-L biofilms grown on glass coverslips, was determined. Ia was the compound of choice, as it demonstrated relatively strong inhibitory activity in each of the assays used in this study. The experiment was performed using green fluorescent labelled (GFP)-labelled PAO1-L and PA14 strains, grown under four different conditions: (i) no treatment (control); (ii) compound Ia at 8 µM in PA14 and 34 µM in PAO1-L (concentrations equal to 3× IC50); (iii) tobramycin at 100 µg/mL; (iv) Ia and tobramycin, at the same concentrations as (ii) and (iii).
Figure 6 shows that exposure to Ia had a substantial effect on biofilm development for both PAO1-L and PA14, reducing biomass with little impact on cell viability, as measured using propidium iodide (red bars) (Figure 6a,b,f,g,e,j). Treatment with tobramycin significantly reduced biofilm biomass, increasing the proportion of dead bacterial cells substantially (Figure 6c,h,e,j). Most importantly, the combined treatments had a synergistic effect with near complete biofilm eradication (Figure 6d,i,e,j). These data further validate the pqs system in Pseudomonas as a target that can aid the effectiveness of antibiotic treatment for biofilm-centred infections.
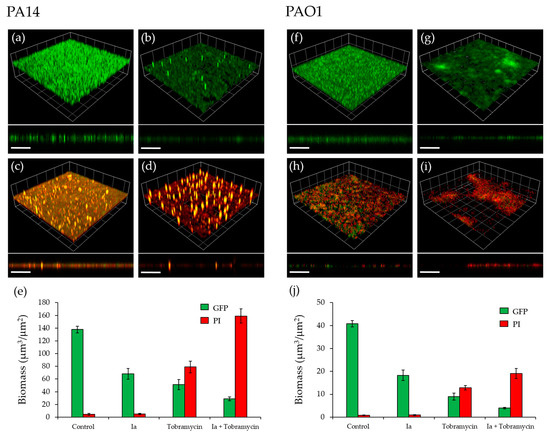
Figure 6.
Effect of Ia on P. aeruginosa biofilms. (a) Untreated GFP-labelled PA14 biofilm; (b) GFP- labelled PA14 biofilm grown with Ia at 8 µM; (c) GFP-labelled PA14 biofilm, treated with 100 µg/mL tobramycin for 4 h after 16 h of growth; (d) GFP-labelled PA14 biofilm grown with 8 µM Ia and treated with 100 µg/mL tobramycin for 4 h after 16 h of growth; (e) Biomass quantitation of PA14 biofilms; (f) Untreated GFP-labelled PAO1-L biofilm; (g) GFP-labelled PAO1-L biofilm grown with Ia at 34 µM; (h) GFP-labelled PAO1-L biofilm, treated with 100 µg/mL tobramycin for 4 h after 16 h of growth; (i) GFP-labelled PAO1-L biofilm grown with 34 µM Ia and treated with 100 µg/mL tobramycin for 4 h after 16 h of incubation; (j) Biomass quantitation of PAO1-L biofilms. Dead cells and extracellular DNA were stained with propidium iodide (PI). Three-dimensional (3D) sections and cross sections are shown. Scale bars represent 100 µm.
2.6. Determination of the Plasma and Hepatic Stability of Selected Quinolone Inhibitors
To gain a greater understanding of the potential of the quinolone inhibitors to progress towards a hit to lead optimization process, in vitro plasma and hepatic stability assays were performed. The results obtained (Table 2) show that each of the selected compounds possesses a high degree of stability in rat plasma and to a less extent in human plasma, where the half-lives varied, with the most stable compound being IIa (Table 2). Microsomal stability usually represents Phase 1 metabolism of a drug, although it does not incorporate factors such as protein binding, which can lead to enhanced in vivo stability. The majority of the compounds tested demonstrated moderate to high intrinsic clearance with relatively short half-lives of <30 min in both human and rat microsomes. Inhibitors Ia and Iii showed the greatest plasma stability.

Table 2.
Summary of plasma and microsomal stability of PqsR inhibitors.
3. Discussion
Antimicrobial resistance has become a global challenge with very slow progress in the development of novel therapeutics [2]. As a multi-antibiotic resistant Gram-negative pathogen, P. aeruginosa constitutes a major clinical threat [34,35]. There is a growing body of literature describing several approaches to attenuate bacterial virulence through interference with the expression or activity of virulence factors [36]. Despite some reports on the emergence of resistance to these agents, these approaches are becoming more popular for treating bacterial infections, as they pose less selective pressure on the bacterial pathogen [37,38]. Disruption of QS can provide an alternative approach to conventional antibacterial therapy, as it targets bacterial virulence, rather than bacterial viability [4,39,40,41]. The pqs system plays a critical role in regulating virulence and biofilm development, through the transcriptional regulator, PqsR [42]. In this study, we employed a molecular modelling approach using the Schrödinger Suite [24] to assess a series of 4-amino-quinoline-based compounds with three sites for amenable structural modifications. To our knowledge, this is the first report describing the use of this in silico approach to search for PqsR inhibitors. Our docking results highlighted four compounds with the potential to bind to PqsR, adopting similar conformations to the reported QZN inhibitor [15]. The interactions within the PqsR ligand-binding site and Ia, Ii, Ia, Ii were mainly dominated by a hydrophobic π-π interaction with Tyr258 and electrostatic interactions between the 4-amino and the side chains of Leu207, Leu208 and Arg209. Moreover, the chlorine and trifluoromethane groups at the 7-position adopted similar orientations towards Thr265, in agreement with the previously-reported binding orientation for QZN [18]. It is noteworthy that substitution at the para-position was not tolerated and resulted in a substantial reduction in the GLIDE docking score, as this may cause a change of conformation to the phenyl ring, affecting the π-π interaction with Tyr258. To confirm that these results can be translated to P. aeruginosa cells, we used biosensor reporter strains, in which a chromosomal transcriptional fusion of PpqsA-lux was introduced. This responds to endogenous AQ production by emitting light. In the presence of an antagonist, bioluminescence is reduced, providing a simple read-out [25]. Compounds active in this reporter assay may either be inhibitors of PqsR or of AQ biosynthetic enzymes. The results obtained with the quinolines confirmed that compounds (Ia, Ii, Ia, Ii) are active in both P. aeruginosa strains tested at low micromolar concentrations. In addition, there were some other hits with selective activity against one of the strains. These results highlight the importance of including multiple strains to validate the activity of an inhibitor, as its potency may vary in a strain-dependent manner. Interestingly, all reported PqsR inhibitors to date were examined using a single P. aeruginosa strain, highlighting the need for these compounds to be tested in different strains before further optimisation is carried out [19,22,43,44]. In fact, it is crucial that anti-Pseudomonas agents undergo a rigid assessment of their activity in representative strains and fresh clinical isolates before they can be considered candidates, as this would greatly reduce the chances of expensive failure at the clinical stage, due to lack of potency [26]. The reasons for such differences are not clear, as yet, but may be attributable to different cell envelope permeabilities, active efflux pumps or a secondary mode of action at a different target [45]. In the present study, only compounds that showed consistent potency in both PA14 and PAO1-L were further assessed using phenotypic analysis.
A pyocyanin quantitation assay upon treatment with the compounds revealed that Ia was the most effective at reducing phenazine production, followed by Ii. Surprisingly, IIa reduced pyocyanin modestly, despite being the most potent inhibitor in the bioreporter assay. In agreement with our findings for pyocyanin, Ia was the most potent inhibitor of AQ production, followed by IIa, the effect of which was substantial for PAO1-L. The remaining inhibitors showed only modest, variable effects on AQ production. Collectively, the inhibitory activity of the compounds, alongside the in silico modelling, is consistent with PqsR as their target. However, the PpqsA-lux reporter assay used does not distinguish between inhibitors of PqsR and AQ biosynthetic enzymes, and further work will be required to confirm that PqsR is their direct target [19,22,43,44]. Based on the results obtained, Ia was evaluated for anti-biofilm activity. Ia disrupted biofilm development and reduced biomass, without affecting bacterial viability. Furthermore, Ia sensitized the biofilm to tobramycin, as reflected by an improved potency in killing both PAO1-L and PA14 biofilms, a finding that supports previous reports that inhibition of pqs signalling disrupts biofilm integrity and its sensitivity to antibiotic treatment [33,46].
Before progressing a hit to lead optimisation process, it is important to determine the stability of the most active hits in plasma and hepatic microsome assays. The latter experiments are essential for predicting the metabolic stability of a putative drug and aid medicinal chemistry decisions [47,48]. The stability study showed that these inhibitors present a reasonable level of plasma stability; however, the intrinsic clearance values in hepatic microsomes were elevated and their half-lives were relatively short (less than 30 min). Thus, further chemical refinement to improve hepatic stability should be considered if these molecules are to be pursued as leads. Nevertheless, we have introduced a new class of low micromolar putative PqsR inhibitors that constitute a starting point for further medicinal chemistry studies aiming at optimising their potency and physicochemical properties, to generate clinically useful therapeutic agents particularly as adjuvants for antibiotics in the context of P. aeruginosa biofilm-centred infections.
4. Materials and Methods
4.1. Molecular Docking
4.1.1. Preparation of Protein and Receptor Grid Generation
The X-ray crystal structure of PqsR ligand binding domain in complex with the QZN inhibitor (PDB ID: 4JVI) was used as a protein template. The protein was prepared using the protein preparation wizard (Small-Molecule Drug Discovery Suite 2017-4, Schrödinger, LLC, New York, NY, USA), where hydrogen atoms were added, water molecules were removed and the correct bond order was assigned to the amino acid residues. Afterwards, a receptor grid generation was performed, based on defined residues around the ligand binding sites: (Ala102, Pro129, Ile149, Thr166, Ala 168, Val 170, Ile186, Ile189, Gln194, Ser196, Leu196, Leu197, Leu207, Leu208, Pro210, Val211, Trp234, Gly235, Pro238, Ser255). The inner grid box was set to 10 Å, while the outer box was 20 Å.
4.1.2. Ligand Preparation
The chemical structures of the inhibitors were sketched using ChemDraw (Version 16.0.1.4, PerkinElmer informatics) via an SDF file. LigPrep module (Small-Molecule Drug Discovery Suite 2017-4, Schrödinger, LLC, New York, NY, USA) was then used for final preparation of ligands into their lowest energy 3D conformations. The partial atomic charges were assigned to the molecular structures, using the 2005 implementation of the OPLS-AA force field. These energy-minimized structures were used for Glide) docking.
4.1.3. Molecular Docking
The “Extra Precision” (XP) mode of Glide docking (Small-Molecule Drug Discovery Suite 2017-4, Schrödinger, LLC, New York, NY, USA) was used to perform all docking calculations, using the OPLS-AA 2005 force field. The scale factor of 1.0 for van der Waals radii was applied to atoms of protein with absolute partial charges of less than or equal to 0.25. The number of position per ligand was set to five, after energy minimization. The best docked structures were chosen using an XP Glide Score (XP Gscore) function as well as visual observations.
4.2. Bacterial strains and growth conditions
The P. aeruginosa strains and plasmids used in this study are shown in Table 3. Bacteria were grown in lysogeny broth (LB) at 37 °C, unless stated otherwise. Where required, tetracycline (Tc) was added to the media at 125 µg/mL, to select for recombinants. Synthetic alkylquinolones were added at the concentrations indicated.

Table 3.
Bacterial strains and plasmids used in this study.
4.3. Biosensor Reporter Assay
Strains PA14 mCTX::PpqsA-lux and PAO1-L mCTX::PpqsA-lux were constructed using plasmid mini-CTX::PpqsA-lux, as previously described [50], and the assay was performed according to a published protocol [52]. For screening, the compounds were tested at a concentration of 10 µM, which was prepared from a 10 mM stock, in DMSO.
4.4. Pyocyanin Quantification
The experiment was performed following a published protocol with minor modifications [53]. P. aeruginosa strains were cultured into 5 mL fresh medium overnight. Compounds were assayed at 3× IC50s, for 16 h, at 37 °C (Kuhner LT W Shaker, Adolf Kühner AG, Basel, Switzerland). Cells were centrifuged at 10,000 RCF for 10 min (Allegra 64R centrifuge, Beckman Coulter, High Wycombe, UK) and the supernatant was transferred to 15 mL falcon tubes with a HSW 10 mL Soft-Ject Syringe and a 0.22 μM Sartorius syringe-driven filter (Fisher Brand, Loughborough, UK). Pyocyanin pigment was extracted into chloroform by mixing 7.5 mL of supernatant with 4.5 mL of chloroform. Pyocyanin was further extracted into 1.5 mL of 0.2 M HCL, which gave a pink/red solution, and the absorbance was measured at 520 nm.
4.5. LCMS-MS Alkyl Quinoline Quantification
For each test sample, 100 µL of sterile filtered supernatant (the same preparation as for pyocyanin) was spiked with 10 µL of an internal standard solution (10 µM d4-PQS in MeOH), and diluted with water, to a total volume of 500 µL. Samples were then extracted three times with an 0.5 mL aliquot of ethyl acetate, vortex mixing the aqueous/organic mix for 2 min, then removing the organic phase once the layers had successfully partitioned. For each sample, the combined organic extracts were dried under vacuum and re-dissolved in 100 µL MeOH prior to analysis. For the LC-MS/MS analysis of supernatant extracts, the chromatography was achieved using a Shimadzu series 10AD VP LC system (Columbia, MD, USA). The LC column, maintained at 40 °C, was a Phenomenex Gemini C18 (3.0 µm, 100 × 3.0 mm) (Macclesfield, Cheshire, UK) with an appropriate guard column. Mobile phase A was 0.1% (v/v) formic acid in water containing 2 mM 2-picolinic acid, and mobile phase B 0.1% (v/v) formic acid in methanol. The flow rate throughout the chromatographic separation was 450 µL/min. After an injection of a 2 µL/sample, a binary gradient, beginning initially at 30% B, increased linearly to 99% B over 5 min. The composition remained at 99% B for 3 min, decreased to 30% B over 1 min, and stayed at this composition for 4 min, to allow for column equilibration. The MS system used for analyte detection was an Applied Biosystems Qtrap 4000 hybrid triple-quadrupole linear ion trap mass spectrometer (Foster City, CA, USA), equipped with an electrospray ionisation (ESI) interface. Instrument control, data collection and analysis were conducted using Analyst software (Foster City, CA, USA). The MS analysis was achieved with positive electrospray (+ES) multiple reaction monitoring (MRM) screening of the LC eluent for specific AQ analytes. Where chromatographic peaks for HHQ, HQNO and PQS were detected, a peak area was determined, and analyte peak area/internal standard peak area calculated.
4.6. Biofilms
Biofilms were cultivated on borosilicate glass coverslips in petri dishes. P. aeruginosa strains, PAO1-L and PA14, were labelled by transformation with plasmid pMMG, which constitutively expresses GFP from the Ptac promoter [51]. Labelled strains were grown at 37 °C, for 16 h, in 2 mL RPMI-1640 (Lonza, Slough, UK), supplemented with 20 mM D-glucose (Sigma–Aldrich, Dorset, UK) and 2 µM FeCl3 (Sigma–Aldrich, Dorset, UK). Cultures were diluted 1:100 in fresh medium and allowed to grow for a further 4 h, or until an OD600 of 0.5 was reached. The mid-logarithmic cultures were diluted to an OD600 of 0.01 in 25 mL RPMI, supplemented with glucose and FeCl3, and inoculated into petri dishes containing UV sterilised borosilicate glass coverslips (22 × 22 mm, thickness no1) (VWR, Lutterworth, UK). Bacterial cells were seeded at 37 °C under static conditions for 1.5 h, and compound Ia was added to the culture at a concentration of 34 µM for P. aeruginosa PAO1-L and 8 µM for PA14 before dishes were moved to a shaker at 60 rpm and 37 °C for 15 h to form mature biofilms. Tobramycin and propidium iodide were added to the 15 h-old cultures at concentrations of 100 µg/mL and 2 µM, respectively, followed by further incubation for 4 h. Coverslips were examined under a Laser Scanning Fluorescent Microscope (LSM2, Zeiss, Oberkochen, Germany). Biofilms were visualised using egfp mode at an excitation wavelength of 488nm. Imaging was carried out using Zen 2011 imaging software (Zeiss, Oberkochen, Germany). A total of 5 Z-stacked images were collected per coverslip. Sampling was conducted at random from the central portion of each coverslip. Biomass was calculated using Image J (NIH, Bethesda, MD, USA) and Comstat 2.1. Software package (www.comstat.dk, lyngby, Denmark) [54].
4.7. Determination of Plasma Stablity
Dilutions of a 10 mM test compound DMSO stock solution were prepared so that the final DMSO concentration was 1% v/v and the final test compound concentration tested was 1 µM. Following the addition of the test compound to plasma, the samples were pre-incubated for 10 min prior to the start of the incubation by the addition of the test compound. The test compound was incubated (n = 2; 1 µM final concentration) at 37 °C with plasma for up to 2 h. Aliquots were sampled at several time points and mixed with acetonitrile (containing internal standard) to terminate the reaction and precipitate the proteins. All the samples were mixed, centrifuged and the supernatants analysed by UPLC-MSMS. Parent disappearance and half-life (t 1/2) values were determined from the slope of the parent depletion curve. Control compounds (imidapril, enalapril, tenofovir and/or propantheline), were respectively included.
4.8. Determination of Hepatic Stability
Dilutions of a 1 mM test compound DMSO stock solution were prepared in buffer (typically 0.01 M phosphate buffered saline (pH 7.4) so that the final DMSO concentration was 0.1% v/v and the final test compound concentration tested was 1 µM. Following addition of protein (0.5 mg/mL final) and NADPH, the samples were pre-incubated for 10 min prior to the start of the incubation by the addition of test compound. The test compounds were then incubated (n = 2; 1 µM final concentration) at 37 °C with tissue microsomes. Aliquots were sampled at several time points and mixed with acetonitrile (containing internal standard) to terminate the reaction and precipitate the proteins. All the samples were mixed, centrifuged and the supernatants analysed by UPLC-MS/MS. The intrinsic clearance (Clint) and half-life (t 1/2) values were determined from the slope of the parent depletion curve. Control compounds for low, moderate and high intrinsic clearance, respectively, for both human and rat microsomes, were included.
4.9. Data Analysis and Figure Preparation
Sigmoidal dose-response curves and the representation of all data were prepared using GraphPad Prism 7.
Acknowledgments
This work was supported by the following: SENBIOTAR grant funded by Medical Research Council grant ref MR/N501852/1, Bridging the Gaps project at The University of Nottingham, funded by the Engineering and Physical Sciences Research Council grant reference EP/M027333/1 in collaboration with Sygnature Discovery. Colciencias and Universidad del Valle for providing financial support for chemical synthesis.
Author Contributions
Fadi Soukarieh, Eduard Vico Oton, Jean-Frédéric Dubern, Nigel Halliday, Stephan Heeb, Michael J. Stocks, Miguel Cámara, Paul Williams conceived and designed the experiments; Fadi Soukarieh, Eduard Vico Oton, Janice Gomes, Jean-Frederic Dubern, Nigel Halliday performed the experiments and analyzed the data; Maria de Pilar Crespo, Jonathan Ramirez-Prada, Braulio Insuasty, Rodrigo Abonia, Jairo Quiroga contributed with compounds; Fadi Soukarieh, Miguel Cámara, Michael J. Stocks and Paul Williams wrote the paper.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Marston, H.D.; Dixon, D.M.; Knisely, J.M.; Palmore, T.N.; Fauci, A.S. Antimicrobial Resistance. JAMA 2016, 316, 1193–1204. [Google Scholar] [CrossRef] [PubMed]
- Ventola, C.L. The antibiotic resistance crisis: Part 1: Causes and threats. Pharm. Ther. 2015, 40, 277–283. [Google Scholar]
- Ng, W.L.; Bassler, B.L. Bacterial quorum-sensing network architectures. Annu. Rev. Genet. 2009, 43, 197–222. [Google Scholar]
- Reuter, K.; Steinbach, A.; Helms, V. Interfering with Bacterial Quorum Sensing. Perspect. Med. Chem. 2016, 8, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Williams, P. Strategies for inhibiting quorum sensing. Emerg. Top. Life Sci. 2017, 1, 23–30. [Google Scholar] [CrossRef]
- Rampioni, G.; Leoni, L.; Williams, P. The art of antibacterial warfare: Deception through interference with quorum sensing-mediated communication. Bioorg. Chem. 2014, 55, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Gellatly, S.L.; Hancock, R.E. Pseudomonas aeruginosa: New insights into pathogenesis and host defenses. Pathog. Dis. 2013, 67, 159–173. [Google Scholar] [CrossRef] [PubMed]
- Potron, A.; Poirel, L.; Nordmann, P. Emerging broad-spectrum resistance in Pseudomonas aeruginosa and Acinetobacter baumannii: Mechanisms and epidemiology. Int. J. Antimicrob. Agents 2015, 45, 568–585. [Google Scholar] [CrossRef] [PubMed]
- Welch, M.; Hodgkinson, J.T.; Gross, J.; Spring, D.R.; Sams, T. Ligand binding kinetics of the quorum sensing regulator PqsR. Biochemistry 2013, 52, 4433–4438. [Google Scholar] [CrossRef] [PubMed]
- Hodgkinson, J.T.; Gross, J.; Baker, Y.R.; Spring, D.R.; Welch, M. A new Pseudomonas quinolone signal (PQS) binding partner: MexG. Chem. Sci. 2016, 7, 2553–2562. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Krishnan, G.; Goumnerov, B.; Tsongalis, J.; Tompkins, R.; Rahme, L.G. A quorum sensing-associated virulence gene of Pseudomonas aeruginosa encodes a LysR-like transcription regulator with a unique self-regulatory mechanism. Proc. Natl. Acad. Sci. USA 2001, 98, 14613–14618. [Google Scholar] [CrossRef] [PubMed]
- Maura, D.; Rahme, L.G. Pharmacological Inhibition of the Pseudomonas aeruginosa MvfR Quorum Sensing System Interferes with Biofilm Formation and Potentiates Antibiotic-Mediated Biofilm Disruption. Antimicrob. Agents Chemother. 2017. [Google Scholar] [CrossRef] [PubMed]
- O'Loughlin, C.T.; Miller, L.C.; Siryaporn, A.; Drescher, K.; Semmelhack, M.; Bassler, B.L. A quorum-sensing inhibitor blocks Pseudomonas aeruginosa virulence and biofilm formation. Proc. Natl. Acad. Sci. USA 2013, 110, 17981–17986. [Google Scholar] [CrossRef] [PubMed]
- Amara, N.; Gregor, R.; Rayo, J.; Dandela, R.; Daniel, E.; Liubin, N.; Willems, H.M.E.; Ben-Zvi, A.; Krom, B.P.; Meijler, M.M. Fine-Tuning Covalent Inhibition of Bacterial Quorum Sensing. Chembiochem 2016, 17, 825–835. [Google Scholar] [CrossRef] [PubMed]
- Amara, N.; Mashiach, R.; Amar, D.; Krief, P.; Spieser, S.A.H.; Bottomley, M.J.; Aharoni, A.; Meijler, M.M. Covalent inhibition of bacterial quorum sensing. J. Am. Chem. Soc. 2009, 131, 10610–10619. [Google Scholar] [CrossRef] [PubMed]
- Hentzer, M.; Wu, H.; Andersen, J.B.; Riedel, K.; Rasmussen, T.B.; Bagge, N.; Kumar, N.; Schembri, M.A.; Song, Z.; Kristoffersen, P. Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J. 2003, 22, 3803–3815. [Google Scholar] [CrossRef] [PubMed]
- Moore, J.D.; Rossi, F.M.; Welsh, M.A.; Nyffeler, K.E.; Blackwell, H.E. A Comparative Analysis of Synthetic Quorum Sensing Modulators in Pseudomonas aeruginosa: New Insights into Mechanism, Active Efflux Susceptibility, Phenotypic Response, and Next-Generation Ligand Design. J. Am. Chem. Soc. 2015, 137, 14626–14639. [Google Scholar] [CrossRef] [PubMed]
- Ilangovan, A.; Fletcher, M.; Rampioni, G.; Pustelny, C.; Rumbaugh, K.; Heeb, S.; Camara, M.; Truman, A.; Chhabra, S.R.; Emsley, J.; et al. Structural basis for native agonist and synthetic inhibitor recognition by the Pseudomonas aeruginosa quorum sensing regulator PqsR (MvfR). PLoS Pathog. 2013, 9. [Google Scholar] [CrossRef] [PubMed]
- Starkey, M.; Lepine, F.; Maura, D.; Bandyopadhaya, A.; Lesic, B.; He, J.; Kitao, T.; Righi, V.; Milot, S.; Tzika, A. Identification of anti-virulence compounds that disrupt quorum-sensing regulated acute and persistent pathogenicity. PLoS Pathog. 2014, 10. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Kirsch, B.; Zimmer, C.; de Jong, J.C.; Henn, C.; Maurer, C.K.; Musken, M.; Haussler, S.; Steinbach, A.; Hartmann, R.W.; et al. Discovery of antagonists of PqsR, a key player in 2-alkyl-4-quinolone-dependent quorum sensing in Pseudomonas aeruginosa. Chem. Biol. 2012, 19, 381–390. [Google Scholar] [CrossRef] [PubMed]
- Klein, T.; Henn, C.; de Jong, J.C.; Zimmer, C.; Kirsch, B.; Maurer, C.K.; Pistorius, D.; Muller, R.; Steinbach, A.; Hartmann, R.W. Identification of small-molecule antagonists of the Pseudomonas aeruginosa transcriptional regulator PqsR: Biophysically guided hit discovery and optimization. ACS Chem. Biol. 2012, 7, 1496–1501. [Google Scholar] [CrossRef] [PubMed]
- Zender, M.; Klein, T.; Henn, C.; Kirsch, B.; Maurer, C.K.; Kail, D.; Ritter, C.; Dolazal, O.; Steinbach, A.; Hartmann, R.W. Discovery and biophysical characterization of 2-amino-oxadiazoles as novel antagonists of PqsR, an important regulator of Pseudomonas aeruginosa virulence. J. Med. Chem. 2013, 56, 6761–6774. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Prada, J.; Robledo, S.M.; Velez, I.D.; del Pilar Crespo, M.; Quiroga, J.; Abonia, R.; Montoya, A.; Svetaz, L.; Zacchino, S.; Insuasty, B. Synthesis of novel quinoline-based 4,5-dihydro-1H-pyrazoles as potential anticancer, antifungal, antibacterial and antiprotozoal agents. Eur. J. Med. Chem. 2017, 131, 237–254. [Google Scholar] [CrossRef] [PubMed]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 2006, 49, 6177–6196. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, M.P.; Diggle, S.P.; Crusz, S.A.; Chhabra, S.R.; Camara, M.; Williams, P. A dual biosensor for 2-alkyl-4-quinolone quorum-sensing signal molecules. Environ. Microbiol. 2007, 9, 2683–2693. [Google Scholar] [CrossRef] [PubMed]
- Freschi, L.; Jeukens, J.; Kukavica-Ibrulj, I.; Boyle, B.; Dupont, M.J.; Laroche, J.; Larose, S.; Maaroufi, H.; Fothergill, J.L.; Moore, M.; et al. Clinical utilization of genomics data produced by the international Pseudomonas aeruginosa consortium. Front. Microbiol. 2015, 6, 1036. [Google Scholar] [CrossRef] [PubMed]
- Rampioni, G.; Falcone, M.; Heeb, S.; Frangipani, E.; Fletcher, M.P.; Dubern, J.F.; Visca, P.; Leoni, L.; Cámara, M.; Williams, P. Unravelling the Genome-Wide Contributions of Specific 2-Alkyl-4-Quinolones and PqsE to Quorum Sensing in Pseudomonas aeruginosa. PLoS Pathog. 2016, 12. [Google Scholar] [CrossRef] [PubMed]
- Lau, G.W.; Hasset, D.J.; Ran, H.; Kong, F. The role of pyocyanin in Pseudomonas aeruginosa infection. Trends Mol. Med. 2004, 10, 599–606. [Google Scholar] [CrossRef] [PubMed]
- Lau, G.W.; Ran, H.; Kong, F.; Hassett, D.J.; Mavrodi, D. Pseudomonas aeruginosa pyocyanin is critical for lung infection in mice. Infect. Immun. 2004, 72, 4275–4278. [Google Scholar] [CrossRef] [PubMed]
- Williams, P.; Camara, M. Quorum sensing and environmental adaptation in Pseudomonas aeruginosa: A tale of regulatory networks and multifunctional signal molecules. Curr. Opin. Microbiol. 2009, 12, 182–191. [Google Scholar] [CrossRef] [PubMed]
- Orazi, G.; O’Toole, G.A. Pseudomonas aeruginosa Alters Staphylococcus aureus Sensitivity to Vancomycin in a Biofilm Model of Cystic Fibrosis Infection. mBio 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, L.A.; McKnight, S.L.; Kuznetsova, M.S.; Pesci, E.C.; Manoil1, C. Functions required for extracellular quinolone signaling by Pseudomonas aeruginosa. J. Bacteriol. 2002, 184, 6472–6480. [Google Scholar] [CrossRef] [PubMed]
- Maura, D.; Rahme, L.G. Pharmacological Inhibition of the Pseudomonas aeruginosa MvfR Quorum-Sensing System Interferes with Biofilm Formation and Potentiates Antibiotic-Mediated Biofilm Disruption. Antimicrob. Agents Chemother. 2017, 61. [Google Scholar] [CrossRef] [PubMed]
- Sonmezer, M.C.; Ertem, G.; Erdinc, F.S.; Kaya Kilic, E.; Tulek, N.; Adiloglu, A.; Hatipoglu, C. Evaluation of Risk Factors for Antibiotic Resistance in Patients with Nosocomial Infections Caused by Pseudomonas aeruginosa. Can. J. Infect. Dis. Med. Microbiol. 2016, 2016, 1321487. [Google Scholar] [CrossRef] [PubMed]
- Papenfort, K.; Bassler., B.L. Quorum sensing signal-response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 2016, 14, 576–588. [Google Scholar] [CrossRef] [PubMed]
- Muhlen, S.; Dersch, P. Anti-virulence Strategies to Target Bacterial Infections. Curr. Top. Microbiol. Immunol. 2016, 398, 147–183. [Google Scholar] [PubMed]
- Garcia-Contreras, R.; Maeda, T.; Wood, T.K. Resistance to quorum-quenching compounds. Appl. Environ. Microbiol. 2013, 79, 6840–6846. [Google Scholar] [CrossRef] [PubMed]
- Maeda, T.; García-Contreras, R.; Pu, M.; Sheng, L.; Garcia, L.R.; Tomás, M.; Wood, T.K. Quorum quenching quandary: Resistance to antivirulence compounds. ISME J. 2012, 6, 493–501. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, M.; Anjua, C.P.; Biswasa, L.; Kumarb, V.A.; Mohana, C.G.; Biswasa, R. Antibiotic resistance in Pseudomonas aeruginosa and alternative therapeutic options. Int. J. Med. Microbiol. 2016, 306, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Scutera, S.; Zucca, M.; Savoia, D. Novel approaches for the design and discovery of quorum-sensing inhibitors. Expert Opin. Drug Discov. 2014, 9, 353–366. [Google Scholar] [CrossRef] [PubMed]
- Defoirdt, T. Quorum-Sensing Systems as Targets for Antivirulence Therapy. Trends Microbiol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Tettmann, B.; Niewerth, C.; Kirschhöfer, F.; Neidig, A.; Dötsch, A.; Brenner-Weiss, G.; Fetzner, S.; Overhage, J. Enzyme-Mediated Quenching of the Pseudomonas Quinolone Signal (PQS) Promotes Biofilm Formation of Pseudomonas aeruginosa by Increasing Iron Availability. Front. Microbiol. 2016, 7, 1978. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Kirsch, B.; Maurer, C.K.; de Jong, J.C.; Braunshausen, A.; Steinbach, A.; Hartmann, R.W. Optimization of anti-virulence PqsR antagonists regarding aqueous solubility and biological properties resulting in new insights in structure-activity relationships. Eur. J. Med. Chem. 2014, 79, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Maurer, C.K.; Kirsch, B.; Steinbach, A.; Hartmann, R.W. Overcoming the unexpected functional inversion of a PqsR antagonist in Pseudomonas aeruginosa: An in vivo potent antivirulence agent targeting pqs quorum sensing. Angew. Chem. Int. Ed. Engl. 2014, 53, 1109–1112. [Google Scholar] [CrossRef] [PubMed]
- Masi, M.; Réfregiers, M.; Pos, K.M.; Pagès, J.M. Mechanisms of envelope permeability and antibiotic influx and efflux in Gram-negative bacteria. Nat. Microbiol. 2017, 2, 17001. [Google Scholar] [CrossRef] [PubMed]
- Boopathi, S.; Vashisth, R.; Manoharan, P.; Kandasamy, R.; Sivakumar, N. Stigmatellin Y—An anti-biofilm compound from Bacillus subtilis BR4 possibly interferes in PQS-PqsR mediated quorum sensing system in Pseudomonas aeruginosa. Bioorg. Med. Chem. Lett. 2017, 27, 2113–2118. [Google Scholar] [CrossRef] [PubMed]
- Hughes, J.P.; Rees, S.; Kalindjian, S.B.; Philpott, K.L. Principles of early drug discovery. Br. J. Pharmacol. 2011, 162, 1239–1249. [Google Scholar] [CrossRef] [PubMed]
- Katsuno, K.; Burrows, J.N.; Duncan, K.; Hooft van Huijsduijnen, R.; Kaneko, T.; Kita, K.; Mowbray, C.E.; Schmatz, D.; Warner, P.; Slingsby, B.T. Hit and lead criteria in drug discovery for infectious diseases of the developing world. Nat. Rev. Drug Discov. 2015, 14, 751–758. [Google Scholar] [CrossRef] [PubMed]
- Rahme, L.G.; Stevens, E.J.; Wolfort, S.F.; Shao, J.; Tompkins, R.G.; Ausubel, F.M. Common virulence factors for bacterial pathogenicity in plants and animals. Science 1995, 268, 1899–1902. [Google Scholar] [CrossRef] [PubMed]
- Diggle, S.P.; Matthijs, S.; Wright, V.J.; Fletcher, M.P.; Chhabra, S.R.; Lamont, I.L.; Kong, X.; Hider, R.C.; Cornelis, P.; Cámara, M.; et al. The Pseudomonas aeruginosa 4-quinolone signal molecules HHQ and PQS play multifunctional roles in quorum sensing and iron entrapment. Chem. Biol. 2007, 14, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Popat, R.; Crusz, S.A.; Messina, M.; Williams, P.; West, S.A.; Diggle, S.P. Quorum-sensing and cheating in bacterial biofilms. Proc. Biol. Sci. 2012, 279, 4765–4771. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, M.P.; Diggle, S.P.; Cámara, M.; Williams, P. Biosensor-based assays for PQS, HHQ and related 2-alkyl-4-quinolone quorum sensing signal molecules. Nat. Protoc. 2007, 2, 1254–1262. [Google Scholar] [CrossRef] [PubMed]
- Essar, D.W.; Eberly, L.; Hadero, A.; Crawford, I.P. Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: Interchangeability of the two anthranilate synthases and evolutionary implications. J. Bacteriol. 1990, 172, 884–900. [Google Scholar] [CrossRef] [PubMed]
- Heydorn, A.; Nielsen, A.T.; Hentzer, M.; Sternberg, C.; Givskov, M.; Ersbøll, B.K.; Molin, S. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 2000, 146, 2395–2407. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).