Abstract
This study represents the first survey studying the occurrence, genetic diversity, and pathogenicity of Botryosphaeriaceae species associated with symptomatic citrus species in citrus-production areas in five European countries. Based on morphological features and phylogenetic analyses of internal transcribed spacer (ITS) of nuclear ribosomal DNA (nrDNA), translation elongation factor 1-alpha (TEF1) and β-tubulin (TUB2) genes, nine species were identified as belonging to the genera Diplodia, Dothiorella, Lasiodiplodia, and Neofusicoccum. Isolates of Neofusicoccum parvum and Diplodia pseudoseriata were the most frequently detected, while Dothiorella viticola had the widest distribution, occurring in four of the five countries sampled. Representative isolates of the nine Botryosphaeriaceae species used in the pathogenicity tests caused similar symptoms to those observed in nature. Isolates assayed were all re-isolated, thereby fulfilling Koch’s postulates. Isolates of Diplodia pseudoseriata and Diplodia olivarum are recorded for the first time on citrus and all species found in our study, except N. parvum, are reported for the first time on citrus in Europe.
1. Introduction
Citrus production represents one of the most important fruit industries worldwide in terms of total yield. Greece, Italy, Portugal, and Spain are the most important European producers of citrus fruit []. In 2019, nearly 11 million tons of citrus was produced in Europe on approximately 515,000 ha []. Most canker diseases of citrus, as well as further fruit-tree crops, are caused by a broad range of fungal species that infect the wood mainly through winter pruning wounds and a subsequent colonization of vascular tissues []. Several abiotic and biotic factors are considered responsible for rots and gumming on the trunk and main branches in citrus. Frost damage, sunscald, or water distribution can promote the infection of numerous ascomycetes and basidiomycetes []. Several fungal infections involving twigs, branches and trunks of citrus caused by Colletotrichum and Diaporthe species were reported in different continents [,,,,]. Guarnaccia and Crous [] reported serious cankers developing in woody tissues of lemon trees caused by Diaporthe spp., often with a gummose exudate, causing serious blight and dieback. Canker diseases of citrus are also caused by other fungal genera such as Fusarium and Neocosmospora [], Peroneutypa [,], and Phaeoacremonium []. Recently, significant attention has been dedicated to revising species and genera of Botryosphaeriaceae, which encompass species with a cosmopolitan distribution that are able to cause diseases of numerous plant species worldwide [,].
Botryosphaeriaceae (Botryosphaeriales) include several species reported as endophytes, latent, and woody plant pathogens on a broad range of host [,,]. This family has undergone significant revision after the adoption of molecular tools to resolve its taxonomy [,,,,,,,]. Recently, the taxonomy of Botryosphaeriaceae (and other families in Botryosphaeriales) has been reviewed by Phillips et al. [] based on morphology of the sexual morphs, phylogenetic relationships on internal transcribed spacer (ITS) and 28S large subunit (LSU) of nuclear ribosomal DNA (nrDNA) sequences and evolutionary divergence times. The authors highlighted the main findings made by Yang et al. [] who included new families, genera, and species in Botryosphaeriales based on morphology and multi-marker phylogenetic analyses of a large collection of isolates. Currently, six families are accepted in Botryosphaeriales and 22 genera have been included in Botryosphaeriaceae [,,].
The most common symptoms observed in association with species of Botryosphaeriaceae are twig, branch and trunk cankers, die-back, collar rot, root cankers, gummosis, decline and, in severe cases, plant death [,]. Plant infections mainly occur through natural openings or wounds, but these fungal species could also survive in latency. This ability could lead to their spread worldwide through asymptomatic plant material, seedlings and fruit, frequently circumventing the adopted quarantine measures []. Moreover, stress and non-optimal plant growth conditions consistently induce the expression of diseases associated with Botryosphaeriaceae species. Thus, global warming could increase plant stress and induce favourable conditions for the development of Botryosphaeriaceae diseases [,,]. Species within the Botryosphaeriaceae represent a serious threat to different crops including major fruit, berry fruit and nut crops cultivated in sub-tropical, tropical, or temperate areas [,,,].
Several species of Diplodia (Di.), Dothiorella (Do.), Lasiodiplodia, Neofusicoccum, and Neoscytalidium (Ne.) have been previously reported to affect Citrus species [,,,]. For example, Ne. dimidiatum has been reported causing citrus branch canker in California [] and Italy []; Do. viticola, L. citricola, L. theobromae, and Ne. dimidiatum have been described in association with branch and trunk dieback of citrus trees in Iran [,] and Dothiorella spp. have been detected as causal agents of citrus gummosis in Tunisia []. Moreover, Di. seriata, Di. mutila, Do. viticola, L. mediterranea and L. mitidjana, have been recovered from symptomatic citrus trees in Algeria [].
Considering the important economic value of Citrus spp., a large survey of Botryosphaeriaceae affecting plants cultivated in the major citrus production areas of Europe was considered imperative. Identification in light of modern taxonomic concepts via morphological characterization and multi-marker DNA sequence data was necessary to adopt efficient control strategies against the pathogens that could affect these crops. Thus, several surveys have been conducted in Greece, Italy, Portugal, Spain, and Malta during 2015 and 2016. In particular, the aims of this study were to (1) conduct extensive surveys for sampling symptomatic plant materials; (2) obtain a broad collection of Botryosphaeriaceae isolates; (3) subject those isolates to DNA multi-marker sequence analyses combined with morphological characterization, and (4) evaluate the pathogenicity of the isolated species to citrus plants.
2. Results
2.1. Field Sampling and Fungal Isolation
In this study, the sampling focused on symptomatic plants of Citrus limon, C. reticulata, C. sinensis, C. sinensis × Poncirus trifoliata, and Microcitrus australasica. Samples were collected in 19 orchards (Table 1). Citrus trees showed various external disease symptoms, including partial or complete yellowing, wilting leaves and twigs, and dieback of branch tips, but also defoliation and branch decline. Canker and cracking of the bark associated with gummose exudate occurred on trunks and branches. Internal observation of infected branches revealed black to brown wood discoloration in cross-sections, wedge-shaped necrosis or irregular wood discoloration. Twigs were wilted and occasionally presenting sporocarps (Figure 1). Symptoms were detected in all the orchards and regions investigated. A total of 63 fungal isolates were collected and were found to be characterized by dark green to grey, fast-growing mycelium on MEA. Moreover, the isolates produced pycnidia on pine needles within 40 days, containing pigmented or hyaline conidia. According to these characteristics, the fungal isolates were classified as Botryosphaeriaceae spp. based on comparison with the previous generic descriptions []. Among the collected isolates, 18 were obtained from trunk cankers, 10 were associated with branch infections, and 35 from twig dieback (Table 2).

Table 1.
Geographical sites investigated and sampled.

Figure 1.
Symptoms on citrus tissues with associated Botryosphaeriacae species. (A) Branch decline in commercial lemon orchard. (B) Trunk canker and bark cracking of C. sinensis. (C,D) Trunk and branch canker with gummosis of C. sinensis plants. (E,F) External cracking with gummosis and internal wood discoloration of the same affected branch of C. reticulata plant. (G,H) Internal wood discoloration and branch blight of C. limon. (I) Twig dieback of young C. sinensis × P. trifoliata and M. australasica (J) plants.

Table 2.
GenBank accession numbers of sequences of Diplodia, Dothiorella, Lasiodiplodia, and Neofusicoccum species used in the phylogenetic analyses. Isolates and sequences obtained in this study are given in bold.
2.2. Phylogenetic Analyses
A combined multi-marker (ITS, TEF1, and TUB2) phylogenetic tree was inferred for each genus (Diplodia, Dothiorella, Lasiodiplodia, and Neofusicoccum) obtained in this study (Figure 2, Figure 3, Figure 4 and Figure 5). The best nucleotide models for the Bayesian Inference analysis of each dataset were as follows: SYM (symmetrical model) + I (proportion of invariable sites) + G (gamma distribution) (Diplodia, Dothiorella, Lasiodiplodia, and Neofusicoccum) for ITS; GTR (generalized time-reversible model) + G (Diplodia, Dothiorella and Neofusicoccum) and HKY (Hasegawa–Kishino–Yano) + I + G (Lasiodiplodia) for TEF1 and GTR + G (Diplodia, Lasiodiplodia and Neofusicoccum) and GTR + I + G (Dothiorella) for TUB2. The Diplodia phylogenetic analysis revealed the isolates as belonging to Di. pseudoseriata (15 isolates, BPP = 1 and ML-BS = 100), Di. seriata (9 isolates, BPP = 1 and ML-BS = 95), Di. olivarum (2 isolates, Bayesian posterior probabilities (BPP) = 1 and maximum likelihood bootstrapped (ML-BS) = 99), and Di. mutila (1 isolate, BPP = 0.99 and ML-BS = 87) (Figure 2). The Dothiorella phylogeny (Figure 3) grouped the isolates together within Do. viticola (9 isolates, BPP = 1 and ML-BS = 99). The Lasiodiplodia phylogenetic analysis placed five isolates as L. theobromae (BPP = 1 and ML-BS = 98) (Figure 4). The Neofusicoccum phylogeny (Figure 5) grouped sequences from our isolates as belonging to N. luteum (2 isolates, BPP = 1 and ML-BS = 94), N. parvum (16 isolates) and N. mediterraneum (4 isolates, BPP = 1 and ML-BS = 98).
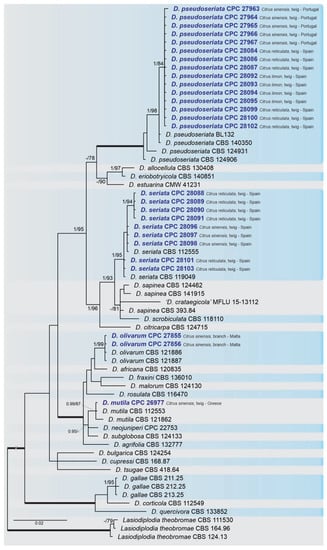
Figure 2.
Bayesian inference analysis of Diplodia species using ITS rDNA, TEF1 and TUB2 sequences. Isolates obtained in this study are in bold and blue. Bayesian posterior probability (BPP) and maximum likelihood-bootstrap (ML-BS) values equal or greater than 0.95 and 70%, respectively, are shown near nodes. Thickened branches represent clades with ML-BS = 100% and a BPP = 1.0. The tree was rooted to L. theobormae (CBS 111530, CBS 164.96 and CBS 124.13).
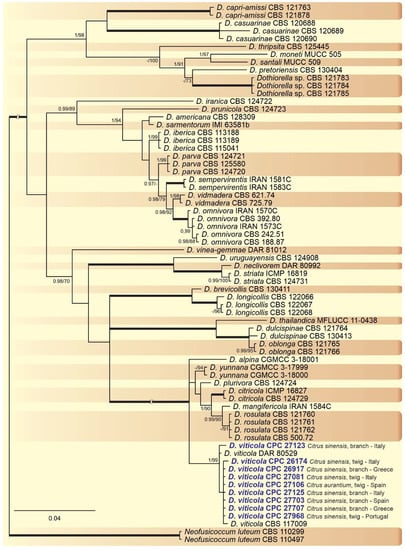
Figure 3.
Bayesian inference analysis of Dothiorella species using ITS rDNA, TEF1, and TUB2 sequences. Isolates obtained in this study are in bold and blue. Bayesian posterior probability (BPP) and ML bootstrap (ML-BS) values equal or greater than 0.95 and 70%, respectively, are shown near nodes. Thickened branches represent clades with ML-BS = 100% and a BPP = 1.0. The tree was rooted to N. luteum (CBS 110299 and CBS 110497).
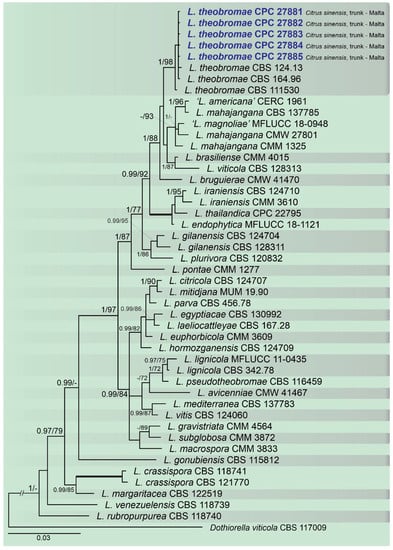
Figure 4.
Bayesian inference analysis of Lasiodiplodia species using ITS rDNA, TEF1, and TUB2 sequences. Isolates obtained in this study are in bold and blue. Bayesian posterior probability (BPP) and ML bootstrap (ML-BS) values equal or greater than 0.95 and 70%, respectively, are shown near nodes. Thickened branches represent clades with ML-BS = 100% and a BPP = 1.0. The tree was rooted to Do. viticola (CBS 117009).
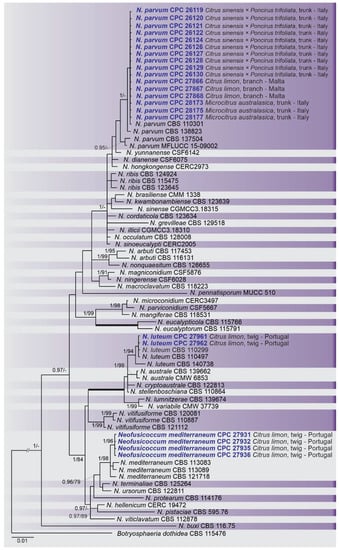
Figure 5.
Bayesian inference analysis of species Neofusicoccum using ITS rDNA, TEF1, and TUB2 sequences. Isolates obtained in this study are in bold and blue. Bayesian posterior probability (BPP) and ML bootstrap (ML-BS) values equal or greater than 0.95 and 70%, respectively, are shown near nodes. Thickened branches represent clades with ML-BS = 100% and a BPP = 1.0. The tree was rooted to B. dothidea (CBS 115476).
2.3. Occurrence of Botryosphaeriaceae among Countries and Citrus Species
Among countries, Do. viticola was found in Greece, Italy, Portugal, and Spain; N. parvum in Italy and Malta, and Di. pseudoseriata in Portugal and Spain. In addition, Di. mutila and Di. seriata were exclusively isolated in Greece and Spain, respectively; L. theobromae and Di. olivarum were only found in Malta, and N. luteum and N. mediterraneum were exclusively found in Portugal. Based on the citrus species, N. parvum (25.4%) and Di. pseudoseriata (23.8%) were the most frequently detected Botryosphaeriaceae spp. on C. sinensis × P. trifoliata, C. limon, C. reticulata, C. sinensis, and/or M. australasica; Di. seriata (on C. reticulata and C. sinensis); and Do. viticola (on C. aurantium and C. sinensis) had an equal percentage of frequency (14.3%); Di. mutila (exclusively found on C. sinensis), N. luteum and N. mediterraneum (only found on C. limon) and Di. olivarum and L. theobromae (exclusively found on C. sinensis) had low frequency values varying from 1.6% to 7.9%.
2.4. Pathogenicity Tests
All isolates caused lesions on wood of inoculated plants 60 d after inoculation (Figure 6) and the fungi were successfully re-isolated. No lesions were observed on control plants. The frequency of re-isolation was between 90% and 95%. The identities of the respective inoculated and re-isolated species were confirmed using culture and molecular analysis as described above, fulfilling Koch’s postulates. Lesions and internal discolouration were observed in correspondence to the inoculation points (Figure 7). The inoculated species that showed high aggressiveness on C. sinensis, C. limon, and C. reticulata were Di. seriata, Di. olivarum, L. theobromae, N. mediterraneum, N. luteum, and N. parvum (with mean lesion length (MLL) ranged from 5.25 to 6.96 cm). Weak symptoms were caused by Di. pseudoseriata, Di. mutila, and Do. viticola on the same species (with MLL ranged from 0.17 to 0.58 cm).

Figure 6.
Pathogenicity tests of selected Botryosphaeriacae isolates on citrus plants 60 d after inoculation. (A,B) Shoot blight of C. reticulata and C. sinensis plants inoculated with N. mediterraneum. (C) Internal lesion with abundant gummosis of C. sinensis plant caused by N. parvum. (D,E) Internal discoloration of C. sinensis and C. reticulata twigs inoculated with L. theobromae.
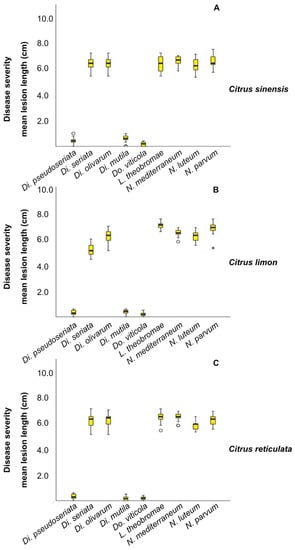
Figure 7.
Box plot showing the results of the pathogenicity tests. Boxes represent the interquartile range, while the horizontal line within each box indicates the average value. The Kruskal–Wallis test was carried out to compare the mean lesion lengths (cm) from inoculation with nine Botryosphaeriaceae representative isolates on C. sinensis (A), C. limon (B) and C. reticulata (C). p < 0.05 was taken to indicate a significant difference. °: Outliers. *: Extreme values.
For each tested host species, the pairwise comparison, obtained from the Kruskal–Wallis test, showed significant differences (p < 0.05) between the species Di. seriata, Di. olivarum, L. theobromae, N. mediterraneum, N. luteum, and N. parvum and the remaining pathogens Di. pseudoseriata, Di. mutila and Do. viticola (Supplementary Tables S1–S3). No significant differences were observed within the group composed by Di. seriata, Di. olivarum, L. theobromae, N. mediterraneum, N. luteum, and N. parvum. Moreover, N. parvum revealed to be highly aggressive on M. australasica and C. sinensis x P. trifoliata with similar level of aggressiveness (Figure 8). The tested strain developed a MLL = 7.83 cm on M. australasica and a MLL = 7.45 cm on C. sinensis × P. trifoliata.
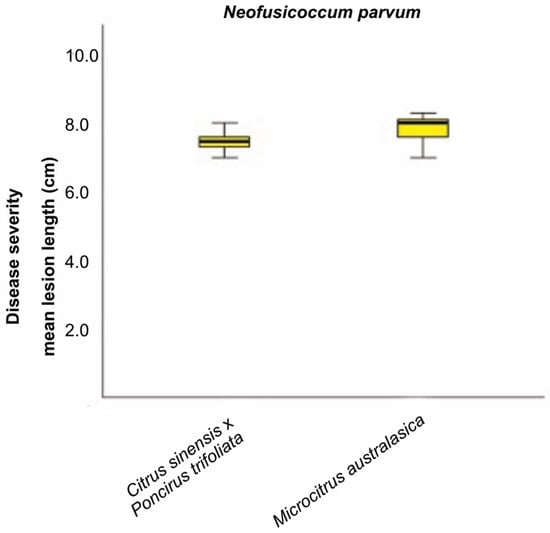
Figure 8.
The Kruskal–Wallis test was carried out to compare the mean lesion lengths (cm) from inoculation with one N. parvum representative isolate on C. sinensis × P. trifoliata and M. australasica. Significant difference was accepted for p < 0.05.
3. Discussion
Several Botryosphaeriaceae spp. have been detected in association with citrus cankers worldwide. Diplodia seriata, Di. mutila, Do. iberica, Do. viticola, L. parva, N. australe, N. luteum, N. mediterraneum, N. parvum, and Ne. dimidiatum have been recovered from necrotic tissues of branch canker and rootstock citrus samples in California [,,]. Recently, Di. citricarpa was described for a fungus on twigs of Citrus sp. in Iran [] and L. mitidjana was introduced for a fungus causing branch canker and dieback of C. sinensis in Algeria []. Botryosphaeriaceae spp. causing disease on citrus are known in European countries, where N. parvum and Ne. dimidiatum were reported on C. reticulata in Greece and on C. sinensis in Italy, respectively [,].
This study represents the first large survey aimed at studying the occurrence, genetic diversity, and pathogenicity of Botryosphaeriaceae species associated with symptomatic citrus species of citrus-producing areas in Greece, Italy, Portugal, Malta, and Spain [,]. Results obtained during our study have added new information about the pathogenicity of Botryosphaeriaceae spp. in citrus-producing areas of these European countries. Symptomatic plants were observed during fieldwork in all the citrus orchards and regions investigated and all isolates used in the pathogenicity test caused lesions on wood of inoculated citrus plants. Phylogenetic multi-marker analyses recognized botryosphaeriaceous isolates in four Diplodia species, with Di. pseudoseriata (15 isolates) being the most common; followed by three Neofusicoccum species, with N. parvum (16 isolates) as dominant species, Do. viticola (9 isolates), and L. theobromae (5 isolates). All species found in this study, except Di. pseudoseriata and Di. olivarum, which are reported for the first time on Citrus spp., have been found in citrus-producing areas of California (USA) [,,].
Diplodia and Neofusicoccum species were dominant in this study. Different species of Neofusicoccum and Diplodia were the most frequently detected pathogens causing gummosis on citrus in California [] and Di. citricarpa was a new species isolated from Citrus sp. in Iran []. Species of Diplodia, Dothiorella, Lasiodiplodia, and Neofusicoccum detected in our study are widely reported as pathogens of other host plants in Algeria and Tunisia [,], Australia [], Brazil [], China [,], Chile [], Italy, Portugal [,,,], South Africa [], and the USA [,,]. The results obtained in our study provide valuable information related to the richness, occurrence, and pathogenicity of Botryosphaeriaceae species in association with citrus species. This study is also the first major survey for Botryosphaeriaceae species associated with symptomatic citrus species in citrus-producing areas of five European countries, providing essential information for future monitoring. Moreover, while previous reports of canker diseases of citrus were based exclusively on morphological observations, the current study aimed to investigate the fungi affecting the major citrus production areas in Europe by large-scale sampling, morphology, and DNA phylogeny. The information achieved with this study about Botryosphaeriaceae population and citrus canker etiology provide fundamental knowledge to start further studies aimed to improve the disease management.
4. Materials and Methods
4.1. Field Sampling and Fungal Isolation
During 2015 and 2016 more than 90 sites in the most important citrus-producing areas of Europe were investigated. The surveys were conducted in Andalusia, Valencia, and the Balearic Islands (Spain); Apulia, Calabria, Sicily, and Aeolian Islands (Italy); Algarve (Portugal); Arta, Crete, Missolonghi, and Nafplio (Greece); Malta and Gozo (Malta) [,]. Twig, branch and trunk portions showing cankers and dieback were collected. Investigated species of Citrus and allied genera of the Rutaceae family such as Microcitrus included: C. limon, C. reticulata, C. sinensis, M. australasica, and C. sinensis × P. trifoliata.
Wood fragments (5 × 5 mm) were collected from the margin between necrotic and healthy tissues. Then, each fragment was disinfected by immersion in 70% ethanol for 5 s, 4% sodium hypochlorite for 90 s, sterilised distilled water for 60 s and then dried on sterile filter paper. The fragments were placed into Petri dishes containing malt extract agar (MEA) [] supplemented with penicillin (100 μg/mL) and streptomycin (100 μg/mL) (MEA-PS) and incubated at 25 °C until characteristic Botryosphaeriaceae colonies were observed. A second procedure was used with plant material incubated in moist chambers at 20 ± 3 °C for up to 10 d and inspected daily for fungal sporulation. The conidia obtained through both procedures were collected and crushed in a drop of sterile water and then spread over the surface of MEA-PS plates. After 24 h, germinating spores were individually transferred onto MEA plates. The isolates used in this study are maintained in the working collection of Pedro Crous (CPC), housed at the Westerdijk Fungal Biodiversity Institute (CBS), Utrecht, The Netherlands.
The occurrence of botryosphaeriaceous fungi among countries and citrus species was evaluated as the number of isolates from each fungal species against the total number of isolates and expressed as a percentage.
4.2. DNA Extraction, Polymerase Chain Reaction (PCR) Amplification and Sequencing
Colonies grown on MEA for 7 days were used to perform total DNA extraction using the Wizard®® Genomic DNA Purification Kit (Promega, Madison, WI, USA) standard protocol. The primer pair ITS4/ITS5 [] was used to amplify the ITS. The primer sets EF1-728F/EF2 [,] and Bt2a/Bt2b [] were used to amplify partial fragments of the TEF1 and TUB2 genes, respectively. Amplification by PCR was conducted as described by Yang et al. []. The PCR products were sequenced in both directions using the BigDye®® Terminator v. 3.1 Cycle Sequencing Kit (Applied Biosystems Life Technologies, Carlsbad, CA, USA), after which amplicons were purified through Sephadex G-50 Fine columns (GE Healthcare, Freiburg, Germany) in MultiScreen HV plates (Millipore, Billerica, MA, USA). Purified sequence reactions were analyzed on an Applied Biosystems 3730xl DNA Analyzer (Life Technologies, Carlsbad, CA, USA). The DNA sequences generated were analyzed and consensus sequences were computed using SeqMan Pro (DNASTAR, Madison, WI, USA). Sequences obtained in this study were deposited in GenBank https://academic.oup.com/nar/article/49/D1/D92/5983623 (accessed on 30 January 2021) (Table 2).
4.3. Phylogenetic Analyses
The phylogenetic analyses included DNA sequences generated in this study along with DNA sequences retrieved from GenBank (Table 2) and represent 124 Botryosphaeriaceae species (Diplodia = 23; Dothiorella = 31; Lasiodiplodia = 31; Neofusicoccum = 39) following recent studies [,,]. Alignments were first made using MAFFT v. 7 [] and manually checked and edited using MEGA v.7 []. Maximum Likelihood (ML) and Bayesian Inference (BI) analyses were conducted using RAxML-HPC BlackBox v.8.2.8 [] and MrBayes v.3.2.7a on XSEDE, respectively, at the CIPRES Science Gateway. The best nucleotide models for the BI analysis were calculated using MrModelTest v.2.3 [] while GTR + I + G was used for ML analysis. Clade stability of the ML phylogeny was assessed with 1000 bootstrap replicates. The BI analysis lasted for one million generations, a burning value of 25% and chains were sampled every 1000 generations. Values of ML bootstrap (ML-BS) and BI posterior probability (BPP) equal or greater than 70% and 0.95, respectively, were considered significant. Individual gene phylogenies were visually inspected and compared for topological incongruences before combining into a multi-marker sequence alignment. The combined alignments used to perform the phylogenetic inferences were deposited in TreeBASE (study ID S27709).
4.4. Pathogenicity Tests
Pathogenicity tests with nine Botryosphaeriaceae species isolated from the European citrus samples were performed to satisfy Koch’s postulates.
One isolate of Di. pseudoseriata (CPC 28084), Di. seriata (CPC 28091), Di. olivarum (CPC 27855), Di. mutila (CPC 26977), Do. viticola (CPC 27125), L. theobromae (CPC 27881), N. mediterraneum (CPC 27931), N. luteum (CPC 27961), and N. parvum (CPC 28175) were respectively inoculated onto potted 2-y-old healthy plants of lemon (C. limon), mandarin (C. reticulata) and sweet orange (C. sinensis). The strain of N. parvum was also inoculated onto potted 2-y-old healthy plants of Australasian lime (M. australasica) and Carrizo citrange (C. sinensis × P. trifoliata).
Three plants for each isolate were inoculated, each having five wounds on twigs made using a sterile blade. Mycelial plugs (5 mm diam.), taken from the margin of actively growing colonies on MEA, were placed on the wound sites on each plant. An equivalent number of plants and inoculation sites were inoculated with sterile MEA plugs and served as controls. The inoculation sites were covered with Parafilm®® (American National Can, Chicago, IL, USA). The inoculated plants were incubated with a 16 h photoperiod in a growth chamber at 100% relative humidity and 25 ± 1 °C. After 2 months external symptoms were assessed. Twigs were cut and the bark peeled off to check for any internal discolouration and the total, upward and downward lesion length was taken to evaluate the MLL. Small sections (0.5 cm) of symptomatic tissue from the edge of twig lesions were placed on MEA to re-isolate the fungal species and were identified based on TEF1 sequencing to fulfil Koch’s postulates. The experiment was conducted twice and each trial was considered a replicate. Because no normal distribution was observed in the lesion dimension data, the Kruskal–Wallis non-parametric test (at P = 0.05) was performed to determine significant differences among isolates. The data analysis was conducted using SPSS software 26 (IBM Corporate).
Supplementary Materials
The following are available online at https://www.mdpi.com/2223-7747/10/3/492/s1, Tables S1–S3. Kruskal-Wallis test results with multiple comparisons for disease severity between different Botryosphaeriaceae spp. on artificially inoculated twigs of C. sinensis (Table S1), C. limon (Table S2) and C. reticulata (Table S3).
Author Contributions
Conceptualization, V.G., G.P. and P.W.C.; methodology, J.D.P.B., D.A. and V.G.; software, J.D.P.B., V.G. and D.A.; validation, G.P., J.D.P.B. and V.G.; formal analysis, J.D.P.B., D.A. and V.G.; investigation, V.G. and D.A.; resources, P.W.C.; data curation, J.D.P.B. and V.G.; writing—original draft preparation, J.D.P.B. and V.G.; writing—review and editing, G.P., M.L.G. and P.W.C.; supervision, G.P., M.L.G. and P.W.C.; project administration, P.W.C.; funding acquisition, P.W.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Acknowledgments
The authors thank G. Gilardi (AGROINNOVA—University of Torino) and A. Azzaro for technical support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- FAOSTAT. Food and Agriculture Organization of the United Nations. 2019. Available online: http://www.fao.org/faostat/en/#home (accessed on 26 February 2020).
- Eurostat. Citrus Fruit Statistics; Eurostat: Luxembourg, 2020. [Google Scholar]
- Crous, P.W.; Wingfield, M.J. Fungi infecting woody plants: Emerging frontiers. Persoonia 2018, 40, i–iii. [Google Scholar] [CrossRef]
- Fawcett, H.S. Citrus Diseases and Their Control; McGraw-Hill: New York, NY, USA, 1936. [Google Scholar]
- Huang, F.; Chen, G.Q.; Hou, X.; Fu, Y.S.; Cai, L.; Hyde, K.D.; Li, H.Y. Colletotrichum species associated with cultivated citrus in China. Fungal Divers. 2013, 61, 61–74. [Google Scholar] [CrossRef]
- Huang, F.; Hou, X.; Dewdney, M.M.; Fu, Y.S.; Chen, G.Q.; Hyde, K.D.; Li, H. Diaporthe species occurring on citrus in China. Fungal Divers. 2013, 61, 237–250. [Google Scholar] [CrossRef]
- Mahadevakumar, S.; Yadav, V.; Tejaswini, G.S.; Sandeep, S.N.; Janardhana, G.R. First report of Phomopsis citri associated with dieback of Citrus lemon in India. Plant Dis. 2014, 98, 1281. [Google Scholar] [CrossRef]
- Guarnaccia, V.; Crous, P.W. Species of Diaporthe on Camellia and Citrus in the Azores Islands. Phytopathol. Mediterr. 2018, 57, 307–319. [Google Scholar]
- Mayorquin, J.S.; Nouri, M.T.; Peacock, B.B.; Trouillas, F.P.; Douhan, G.W.; Kallsen, C.; Eskalen, A. Identification, Pathogenicity, and Spore Trapping of Colletotrichum karstii Associated with Twig and Shoot Dieback in California. Plant Dis. 2019, 103, 1464–1473. [Google Scholar] [CrossRef] [PubMed]
- Guarnaccia, V.; Crous, P.W. Emerging citrus diseases in Europe caused by species of Diaporthe. IMA Fungus 2017, 8, 317–334. [Google Scholar] [CrossRef] [PubMed]
- Sandoval-Denis, M.; Guarnaccia, V.; Polizzi, G.; Crous, P.W. Symptomatic Citrus trees reveal a new pathogenic lineage in Fusarium and two new Neocosmospora species. Persoonia 2018, 40, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Timmer, L.W.; Garnsey, S.M.; Graham, J.H. Compendium of Citrus Diseases, 2nd ed.; American Phytopathological Society: Saint Paul, MN, USA, 2000. [Google Scholar]
- Mayorquin, J.S.; Wang, D.H.; Twizeyimana, M.; Eskalen, A. Identification, distribution, and pathogenicity of Diatrypaceae and Botryosphaeriaceae associated with Citrus branch canker in the southern California desert. Plant Dis. 2016, 100, 2402–2413. [Google Scholar] [CrossRef] [PubMed]
- Espargham, N.; Mohammadi, H.; Gramaje, D.A. Survey of Trunk Disease Pathogens within Citrus Trees in Iran. Plants 2020, 9, 754. [Google Scholar] [CrossRef]
- Phillips, A.J.L.; Alves, A.; Abdollahzadeh, J.; Slippers, B.; Wingfield, M.J.; Groenewald, J.Z.; Crous, P.W. The Botryosphaeriaceae: Genera and species known from culture. Stud. Mycol. 2013, 76, 51–167. [Google Scholar] [CrossRef]
- Yang, T.; Groenewald, J.Z.; Cheewangkoon, R.; Jami, F.; Abdollahzadeh, J.; Lombard, L.; Crous, P.W. Families, genera, and species of Botryosphaeriales. Fungal Biol. 2017, 121, 322–346. [Google Scholar] [CrossRef] [PubMed]
- Slippers, B.; Wingfield, M.J. Botryosphaeriaceae as endophytes and latent pathogens of woody plants: Diversity, ecology and impact. Fungal Biol. Rev. 2007, 21, 90–106. [Google Scholar] [CrossRef]
- Crous, P.W.; Slippers, B.; Wingfield, M.J.; Rheeder, J.; Marasas, W.F.O.; Philips, A.J.L.; Alves, A.; Burgess, T.; Barber, P.; Groenewald, J.Z. Phylogenetic lineages in the Botryosphaeriaceae. Stud. Mycol. 2006, 55, 235–253. [Google Scholar] [CrossRef] [PubMed]
- Crous, P.W.; Giraldo, A.; Hawksworth, D.L.; Robert, V.; Kirk, P.M.; Guarro, J.; Robbertse, B.; Schoch, C.L.; Damm, U.; Trakunyingcharoen, T.; et al. The genera of fungi: Fixing the application of type species of generic names. IMA Fungus 2014, 5, 141–160. [Google Scholar] [CrossRef] [PubMed]
- Crous, P.W.; Muller, M.M.; Sanchez, R.M.; Giordano, L.; Bianchinotti, M.V.; Anderson, F.E.; Groenewald, J.Z. Resolving Tiarosporella spp. allied to Botryosphaeriaceae and Phacidiaceae. Phytotaxa 2015, 202, 73–93. [Google Scholar] [CrossRef]
- Schoch, C.L.; Shoemaker, R.A.; Seifert, K.A.; Hambleton, S.; Spatafora, J.W.; Crous, P.W. A multigene phylogeny of the Dothideomycetes using four nuclear loci. Mycologia 2006, 98, 1041–1052. [Google Scholar] [CrossRef]
- Slippers, B.; Boissin, E.; Phillips, A.J.L.; Groenewald, J.Z.; Lombard, L.; Wingfield, M.J.; Postma, A.; Burgess, T.; Crous, P.W. Phylogenetic lineages in the Botryosphaeriales: A systematic and evolutionary framework. Stud. Mycol. 2013, 76, 31–49. [Google Scholar] [CrossRef] [PubMed]
- Phillips, A.J.L.; Hyde, K.D.; Alves, A.; Liu, J. Families in Botryosphaeriales: A phylogenetic, morphological and evolutionary perspective. Fungal Divers. 2019, 94, 1–22. [Google Scholar] [CrossRef]
- Wijayawardene, N.N.; Hyde, K.D.; Al-Ani, L.K.T.; Tedersoo, L.; Haelewaters, D.; Rajeshkumar, K.C.; Zhao, R.L.; Aptroot, A.; Leontyev, D.V.; Saxena, R.K.; et al. Outline of Fungi and fungus-like taxa. Mycosphere 2020, 11, 1060–1456. [Google Scholar] [CrossRef]
- Zhang, W.; Groenewald, J.Z.; Lombard, L.; Schumacher, R.K.; Phillips, A.J.L.; Crous, P.W. Evaluating species in Botryosphaeriales. Persoonia 2021, 46, 63–115. [Google Scholar]
- Mehl, J.W.M.; Slippers, B.; Roux, J.; Wingfield, M.J. Cankers and other diseases caused by the Botryosphaeriaceae. In Infectious Forest Diseases; Gonthier, P., Nicolotti, G., Eds.; CAB International: Boston, MA, USA, 2013; pp. 298–317. [Google Scholar]
- Pour, F.N.; Ferreira, V.; Félix, C.; Serôdio, J.; Alves, A.; Duarte, A.S.; Esteves, A.C. Effect of temperature on the phytotoxicity and cytotoxicity of Botryosphaeriaceae fungi. Fungal Biol. 2020, 124, 571–578. [Google Scholar] [CrossRef]
- Guarnaccia, V.; Martino, I.; Tabone, G.; Brondino, L.; Gullino, M.L. Fungal pathogens associated with stem blight and dieback of blueberry in northern Italy. Phytopathol. Mediterr. 2020, 59, 229–245. [Google Scholar]
- Marsberg, A.; Kemler, M.; Jami, F.; Nagel, J.H.; Postma-Smidt, A.; Naidoo, S.; Wingfield, M.J.; Crous, P.W.; Spatafora, J.W.; Hesse, C.N.; et al. Botryosphaeria dothidea: A latent pathogen of global importance to woody plant health. Mol. Plant Pathol. 2017, 18, 477–488. [Google Scholar] [CrossRef] [PubMed]
- Aiello, D.; Gusella, G.; Fiorenza, A.; Guarnaccia, V.; Polizzi, G. Identification of Neofusicoccum parvum causing canker and twig blight on Ficus carica in Italy. Phytopathol. Mediterr. 2020, 59, 213–218. [Google Scholar]
- Adesemoye, A.O.; Eskalen, A. First Report of Spencermartinsia viticola, Neofusicoccum australe, and N. parvum causing branch canker of citrus in California. Plant Dis. 2011, 95, 770. [Google Scholar] [CrossRef] [PubMed]
- Polizzi, G.; Aiello, D.; Vitale, A.; Giuffrida, F.; Groenewald, J.; Crous, P.W. First report of shoot blight, canker, and gummosis caused by Neoscytalidium dimidiatum on citrus in Italy. Plant Dis. 2009, 93, 1215. [Google Scholar] [CrossRef]
- Berraf-Tebbal, A.; Mahamedi, A.E.; Aigoun-Mouhous, W.; Špetík, M.; Čechová, J.; Pokluda, R.; Baránek, M.; Eichmeier, A.; Alves, A. Lasiodiplodia mitidjana sp. nov. and other Botryosphaeriaceae species causing branch canker and dieback of Citrus sinensis in Algeria. PLoS ONE 2020, 15, e0232448. [Google Scholar] [CrossRef]
- Abdollahzadeh, J.; Javadi, A.; Goltapeh, E.M.; Zare, R.; Phillips, A.J. Phylogeny and morphology of four new species of Lasiodiplodia from Iran. Persoonia 2010, 25, 1–10. [Google Scholar] [CrossRef]
- Hamrouni, N.; Nouri, M.; Trouillas, F.; Said, A.; Sadfi-Zouaoui, N.; Hajlaoui, M. Dothiorella gummosis caused by Dothiorella viticola, first record from citrus in Tunisia. New Dis. Rep. 2018, 38, 10. [Google Scholar] [CrossRef]
- Adesemoye, A.O.; Mayorquin, J.S.; Wang, D.H.; Twizeyimana, M.; Lynch, S.C.; Eskalen, A. Identification of species of Botryosphaeriaceae causing bot gummosis in citrus in California. Plant Dis. 2014, 98, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Vakalounakis, D.J.; Ntougias, S.; Kavroulakis, N.; Protopapadakis, E. Neofusicoccum parvum and Diaporthe foeniculina associated with twig and shoot blight and branch canker of citrus in Greece. J. Phytopathol. 2019, 167, 527–537. [Google Scholar] [CrossRef]
- Guarnaccia, V.; Groenewald, J.Z.; Li, H.; Glienke, C.; Carstens, E.; Hattingh, V.; Crous, P.W. First report of Phyllosticta citricarpa and description of two new species, P. paracapitalensis and P. paracitricarpa, from citrus in Europe. Stud. Mycol. 2017, 87, 161–185. [Google Scholar] [CrossRef]
- Linaldeddu, B.T.; Deidda, A.; Scanu, B.; Franceschini, A.; Serra, S.; Berraf-Tebbal, A.; Boutiti, M.Z.; Jamâa, M.L.B.; Phillips, A.J.L. Diversity of Botryosphaeriaceae species associated with grapevine and other woody hosts in Italy, Algeria and Tunisia, with descriptions of Lasiodiplodia exigua and Lasiodiplodia mediterranea sp. nov. Fungal Divers. 2015, 71, 201–214. [Google Scholar] [CrossRef]
- Mahamedi, A.E.; Phillips, A.J.L.; Lopes, A.; Djellid, Y.; Arkam, M.; Eichmeier, A.; Zitouni, A.; Alves, A.; Berraf-Tebbal, A. Diversity, distribution and host association of Botryosphaeriaceae species causing oak decline across different forest ecosystems in Algeria. Eur. J. Plant Pathol. 2020, 158, 745–765. [Google Scholar] [CrossRef]
- Burgess, T.I.; Tan, Y.P.; Garnas, J.; Edwards, J.; Scarlett, K.A.; Shuttleworth, L.A.; Daniel, R.; Dann, E.K.; Parkinson, L.E.; Dinh, Q. Current status of the Botryosphaeriaceae in Australia. Australas. Plant Pathol. 2018, 48, 35–44. [Google Scholar] [CrossRef]
- Machado, A.R.; Custódio, F.A.; Cabral, P.G.C.; Capucho, A.S.; Pereira, O.L. Botryosphaeriaceae species causing dieback on Annonaceae in Brazil. Plant Pathol. 2019, 68, 1394–1406. [Google Scholar] [CrossRef]
- Xu, C.; Zhang, H.; Zhou, Z.; Hu, T.; Wang, S.; Wang, Y.; Cao, K. Identification and distribution of Botryosphaeriaceae species associated with blueberry stem blight in China. Eur. J. Plant Pathol. 2015, 143, 737–752. [Google Scholar] [CrossRef]
- Li, G.; Slippers, B.; Wingfield, M.J.; Chen, S. Variation in Botryosphaeriaceae from Eucalyptus plantations in YunNan Province in southwestern China across a climatic gradient. IMA Fungus 2020, 11, 22. [Google Scholar] [CrossRef]
- Valencia, A.L.; Pilar, M.; Gil, B.A.; Latorre, I.; Rosales, M. Characterization and Pathogenicity of Botryosphaeriaceae Species Obtained from Avocado Trees with Branch Canker and Dieback and from Avocado Fruit with Stem End Rot in Chile. Plant Dis. 2019, 103, 996–1005. [Google Scholar] [CrossRef]
- Gusella, G.; Aiello, D.; Polizzi, G. First report of leaf and twig blight of Indian hawthorn (Rhaphiolepis indica) caused by Neofusicoccum parvum in Italy. J. Plant Pathol. 2020, 102, 275. [Google Scholar] [CrossRef]
- Alves, A.; Linaldeddu, B.T.; Deidda, A.; Scanu, B.; Phillips, A.J.L. The complex of Diplodia species associated with Fraxinus and some other woody hosts in Italy and Portugal. Fungal Divers. 2014, 67, 143–156. [Google Scholar] [CrossRef]
- Dissanayake, A.J.; Camporesi, E.; Hyde, K.D.; Yan, J.Y.; Li, X.H. Saprobic Botryosphaeriaceae, including Dothiorella italica sp nov., associated with urban and forest trees in Italy. Mycosphere 2017, 8, 1157–1176. [Google Scholar] [CrossRef]
- Jami, F.; Slippers, B.; Wingfield, M.J.; Loots, M.T.; Gryzenhout, M. Temporal and spatial variation of Botryosphaeriaceae associated with Acacia karroo in South Africa. Fungal Ecol. 2015, 15, 51–62. [Google Scholar] [CrossRef]
- Crous, P.W.; Verkley, G.J.M.; Groenewald, J.Z.; Samson, R.A. (Eds.) Fungal Biodiversity; CBS Laboratory Manual Series 1; Centraalbureau Voor Schimmelcultures: Utrecht, The Netherlands, 2009; pp. 1–269. [Google Scholar]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, A.M., Gelfard, D.H., Snindky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Carbone, I.; Kohn, L.M. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 1999, 91, 553–556. [Google Scholar] [CrossRef]
- O’Donnell, K.; Kistler, H.C.; Cigelnik, E.; Ploetz, R.C. Multiple evolutionary origins of the fungus causing Panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Natl. Acad. Sci. USA 1998, 95, 2044–2049. [Google Scholar] [CrossRef] [PubMed]
- Glass, N.L.; Donaldson, G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 1995, 61, 1323–1330. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Nylander, J. MrModeltest v. 2 Program Distributed by the Author; Evolutionary Biology Centre, Uppsala University: Uppsala, Sweden, 2004. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).