The Human Vulvar Microbiome: A Systematic Review
Abstract
1. Introduction
2. Materials and Methods
2.1. Search Strategy
2.2. Inclusion Criteria
2.3. Exclusion Criteria
2.4. Study Selection
2.5. Data Extraction
2.6. Assessment of Risk of Bias and Level of Evidence
3. Results
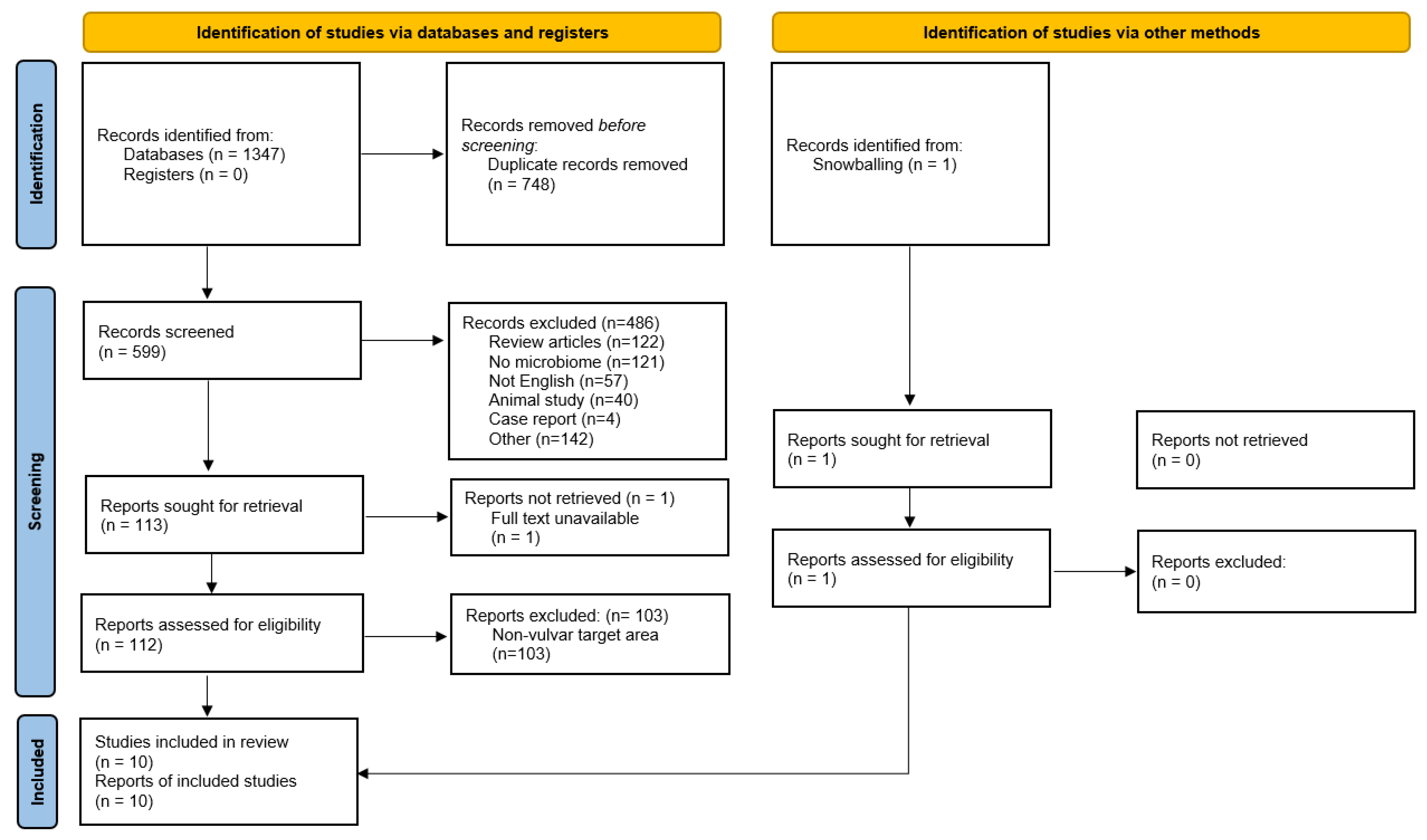
3.1. Number of Retrieved Papers
3.2. The Healthy Vulvar Microbiome
3.3. The Association of the Menstrual Cycle and Obesity and the Vulvar Microbiome
3.4. The Microbiome in Vulvovaginal Disease
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Byrd, A.L.; Belkaid, Y.; Segre, J.A. The human skin microbiome. Nat. Rev. Microbiol. 2018, 16, 143–155. [Google Scholar] [CrossRef]
- Feng, Q.; Liang, S.; Jia, H.; Stadlmayr, A.; Tang, L.; Lan, Z.; Zhang, D.; Xia, H.; Xu, X.; Jie, Z.; et al. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat. Commun. 2015, 6, 6528. [Google Scholar] [CrossRef] [PubMed]
- Niemeyer-van der Kolk, T.; van der Wall, H.E.C.; Balmforth, C.; Van Doorn, M.B.A.; Rissmann, R. A systematic literature review of the human skin microbiome as biomarker for dermatological drug development. Br. J. Clin. Pharmacol. 2018, 84, 2178–2193. [Google Scholar] [CrossRef] [PubMed]
- Hale, V.L.; Chen, J.; Johnson, S.; Harrington, S.C.; Yab, T.C.; Smyrk, T.C.; Nelson, H.; Boardman, L.A.; Druliner, B.R.; Levin, T.R.; et al. Shifts in the Fecal Microbiota Associated with Adenomatous Polyps. Cancer Epidemiol. Biomark. Prev. 2017, 26, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Rajagopala, S.V.; Vashee, S.; Oldfield, L.M.; Suzuki, Y.; Venter, J.C.; Telenti, A.; Nelson, K.E. The Human Microbiome and Cancer. Cancer Prev. Res. 2017, 10, 226–234. [Google Scholar] [CrossRef]
- Chen, J.; Douglass, J.; Prasath, V.; Neace, M.; Atrchian, S.; Manjili, M.H.; Shokouhi, S.; Habibi, M. The microbiome and breast cancer: A review. Breast Cancer Res. Treat. 2019, 178, 493–496. [Google Scholar] [CrossRef]
- Schmidt, B.L.; Kuczynski, J.; Bhattacharya, A.; Huey, B.; Corby, P.M.; Queiroz, E.L.S.; Nightingale, K.; Kerr, A.R.; DeLacure, M.D.; Veeramachaneni, R.; et al. Changes in Abundance of Oral Microbiota Associated with Oral Cancer. PLoS ONE 2014, 9, e98741. [Google Scholar] [CrossRef]
- Parsonnet, J.; Friedman, G.D.; Vandersteen, D.P.; Chang, Y.; Vogelman, J.H.; Orentreich, N.; Sibley, R.K. Helicobacter pylori Infection and the Risk of Gastric Carcinoma. N. Engl. J. Med. 1991, 325, 1127–1131. [Google Scholar] [CrossRef] [PubMed]
- Cubie, H.A. Diseases associated with human papillomavirus infection. Virology 2013, 445, 21–34. [Google Scholar] [CrossRef]
- Matson, V.; Chervin, C.S.; Gajewski, T.F. Cancer and the Microbiome—Influence of the Commensal Microbiota on Cancer, Immune Responses, and Immunotherapy. Gastroenterology 2021, 160, 600–613. [Google Scholar] [CrossRef]
- Helmink, B.A.; Khan, M.A.W.; Hermann, A.; Gopalakrishnan, V.; Wargo, J.A. The microbiome, cancer, and cancer therapy. Nat. Med. 2019, 25, 377–388. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, D.J.; Rebeck, O.N.; Dantas, G. Complex interactions between the microbiome and cancer immune therapy. Crit. Rev. Clin. Lab. Sci. 2019, 56, 567–585. [Google Scholar] [CrossRef]
- Hacker, N.F.; Eifel, P.J.; van der Velden, J. Cancer of the vulva. Int. J. Gynecol. Obstet. 2012, 119, S90–S96. [Google Scholar] [CrossRef]
- Nitecki, R.; Feltmate, C.M. Human papillomavirus and nonhuman papillomavirus pathways to vulvar squamous cell carcinoma: A review. Curr. Probl. Cancer 2018, 42, 476–485. [Google Scholar] [CrossRef] [PubMed]
- de Sanjosé, S.; Alemany, L.; Ordi, J.; Tous, S.; Alejo, M.; Bigby, S.M.; Joura, E.A.; Maldonado, P.; Laco, J.; Bravo, I.G.; et al. Worldwide human papillomavirus genotype attribution in over 2000 cases of intraepithelial and invasive lesions of the vulva. Eur. J. Cancer 2013, 49, 3450–3461. [Google Scholar] [CrossRef]
- Van De Nieuwenhof, H.P.; Van Kempen, L.C.L.T.; De Hullu, J.A.; Bekkers, R.L.M.; Bulten, J.; Melchers, W.J.G.; Massuger, L.F.A.G. The etiologic role of HPV in vulvar squamous cell carcinoma fine tuned. Cancer Epidemiol. Biomark. Prev. 2009, 18, 2061–2067. [Google Scholar] [CrossRef]
- Thuijs, N.B.; van Beurden, M.; Bruggink, A.H.; Steenbergen, R.D.M.M.; Berkhof, J.; Bleeker, M.C.G.G. Vulvar intraepithelial neoplasia: Incidence and long-term risk of vulvar squamous cell carcinoma. Int. J. Cancer 2021, 148, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Lockhart, J.; Gray, N.M.; Cruickshank, M.E. The development and evaluation of a questionnaire to assess the impact of vulval intraepithelial neoplasia: A questionnaire study. BJOG An Int. J. Obstet. Gynaecol. 2013, 120, 1133–1142. [Google Scholar] [CrossRef]
- Mullen, M.M.; Merfeld, E.C.; Palisoul, M.L.; Massad, L.S.; Woolfolk, C.; Powell, M.A.; Mutch, D.G.; Thaker, P.H.; Hagemann, A.R.; Kuroki, L.M. Wound Complication Rates After Vulvar Excisions for Premalignant Lesions. Obstet. Gynecol. 2019, 133, 658–665. [Google Scholar] [CrossRef]
- Grimm, D.; Eulenburg, C.; Brummer, O.; Schliedermann, A.-K.; Trillsch, F.; Prieske, K.; Gieseking, F.; Selka, E.; Mahner, S.; Woelber, L. Sexual activity and function after surgical treatment in patients with (pre)invasive vulvar lesions. Support. Care Cancer 2016, 24, 419–428. [Google Scholar] [CrossRef]
- Pérez-López, F.R.; Vieira-Baptista, P. Lichen sclerosus in women: A review. Climacteric 2017, 20, 339–347. [Google Scholar] [CrossRef] [PubMed]
- Norenhag, J.; Du, J.; Olovsson, M.; Verstraelen, H.; Engstrand, L.; Brusselaers, N. The vaginal microbiota, human papillomavirus and cervical dysplasia: A systematic review and network meta-analysis. BJOG An Int. J. Obstet. Gynaecol. 2020, 127, 171–180. [Google Scholar] [CrossRef]
- Łaniewski, P.; Cui, H.; Roe, D.J.; Barnes, D.; Goulder, A.; Monk, B.J.; Greenspan, D.L.; Chase, D.M.; Herbst-Kralovetz, M.M. Features of the cervicovaginal microenvironment drive cancer biomarker signatures in patients across cervical carcinogenesis. Sci. Rep. 2019, 9, 7333. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.; Kouzy, R.; Jaoude, J.A.; Noticewala, S.S.; Delgado Medrano, A.Y.; Klopp, A.H.; Taniguchi, C.M.; Colbert, L.E. Microbiome factors in HPV-driven carcinogenesis and cancers. PLoS Pathog. 2020, 16, e1008524. [Google Scholar] [CrossRef] [PubMed]
- ALY, R.; BRITZ, M.B.; MAIBACH, H.I. Quantitative microbiology of human vulva. Br. J. Dermatol. 1979, 101, 445–448. [Google Scholar] [CrossRef]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. PLoS Med. 2009, 6, e1000097. [Google Scholar] [CrossRef]
- Booth, A.; Clarke, M.; Dooley, G.; Ghersi, D.; Moher, D.; Petticrew, M.; Stewart, L. The nuts and bolts of PROSPERO: An international prospective register of systematic reviews. Syst. Rev. 2012, 1, 2. [Google Scholar] [CrossRef]
- Moher, D.; Booth, A.; Stewart, L. How to reduce unnecessary duplication: Use PROSPERO. BJOG An Int. J. Obstet. Gynaecol. 2014, 121, 784–786. [Google Scholar] [CrossRef]
- Browne, H.P.; Forster, S.C.; Anonye, B.O.; Kumar, N.; Neville, B.A.; Stares, M.D.; Goulding, D.; Lawley, T.D. Culturing of “unculturable” human microbiota reveals novel taxa and extensive sporulation. Nature 2016, 533, 543–546. [Google Scholar] [CrossRef]
- Fettweis, J.M.; Serrano, M.G.; Girerd, P.H.; Jefferson, K.K.; Buck, G.A. A new era of the vaginal microbiome: Advances using next-generation sequencing. Chem. Biodivers. 2012, 9, 965–976. [Google Scholar] [CrossRef]
- Walker, A.W.; Duncan, S.H.; Louis, P.; Flint, H.J. Phylogeny, culturing, and metagenomics of the human gut microbiota. Trends Microbiol. 2014, 22, 267–274. [Google Scholar] [CrossRef]
- Peterson, J.; Garges, S.; Giovanni, M.; McInnes, P.; Wang, L.; Schloss, J.A.; Bonazzi, V.; McEwen, J.E.; Wetterstrand, K.A.; Deal, C.; et al. The NIH Human Microbiome Project. Genome Res. 2009, 19, 2317–2323. [Google Scholar] [CrossRef] [PubMed]
- Ouzzani, M.; Hammady, H.; Fedorowicz, Z.; Elmagarmid, A. Rayyan-a web and mobile app for systematic reviews. Syst. Rev. 2016, 5, 210. [Google Scholar] [CrossRef]
- Porritt, K.; Gomersall, J.; Lockwood, C. JBI’s Systematic Reviews: Study selection and critical appraisal. Am. J. Nurs. 2014, 114, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Guyatt, G.H.; Oxman, A.D.; Vist, G.E.; Kunz, R.; Falck-Ytter, Y.; Alonso-Coello, P.; Schünemann, H.J. GRADE: An emerging consensus on rating quality of evidence and strength of recommendations. BMJ 2008, 336, 924–926. [Google Scholar] [CrossRef]
- Park, S.Y.; Lee, E.S.; Lee, S.R.; Kim, S.H.; Chae, H.D. Vaginal Microbiome Is Associated With Vulvodynia, Vulvar Pain Syndrome: A Case-Control Study. Sex. Med. 2021, 9, 100314. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, T.; Akiba, S.; Sato, N.; Fujimura, T.; Takagi, Y.; Kitahara, T.; Takema, Y.; Iizuka, H.; Sengoku, K. Study of the vulvar skin in healthy Japanese women: Components of the stratum corneum and microbes. Int. J. Dermatol. 2013, 52, 1500–1505. [Google Scholar] [CrossRef]
- Bruning, E.; Chen, Y.; McCue, K.A.; Rubino, J.R.; Wilkinson, J.E.; Brown, A.D.G. A 28 Day Clinical Assessment of a Lactic Acid-containing Antimicrobial Intimate Gel Wash Formulation on Skin Tolerance and Impact on the Vulvar Microbiome. Antibiotics 2020, 9, 55. [Google Scholar] [CrossRef]
- Brown, C.J.; Wong, M.; Davis, C.C.; Kanti, A.; Zhou, X.; Forney, L.J. Preliminary characterization of the normal microbiota of the human vulva using cultivation-independent methods. J. Med. Microbiol. 2007, 56, 271–276. [Google Scholar] [CrossRef]
- Zhou, X.; Bent, S.J.; Schneider, M.G.; Davis, C.C.; Islam, M.R.; Forney, L.J. Characterization of vaginal microbial communities in adult healthy women using cultivation-independent methods. Microbiology 2004, 150, 2565–2573. [Google Scholar] [CrossRef]
- Costello, E.K.; Lauber, C.L.; Hamady, M.; Fierer, N.; Gordon, J.I.; Knight, R. Bacterial community variation in human body habitats across space and time. Science 2009, 326, 1694–1697. [Google Scholar] [CrossRef] [PubMed]
- Shiraishi, T.; Fukuda, K.; Morotomi, N.; Imamura, Y.; Mishima, J.; Imai, S.; Miyazawa, K.; Taniguchi, H. Influence of menstruation on the microbiota of healthy women’s labia minora as analyzed using a 16s rRNA gene-based clone library method. Jpn. J. Infect. Dis. 2011, 64, 76–80. [Google Scholar] [PubMed]
- Hickey, R.J.; Zhou, X.; Settles, M.L.; Erb, J.; Malone, K.; Hansmann, M.A.; Shew, M.L.; Van Der Pol, B.; Fortenberry, J.D.; Forney, L.J. Vaginal microbiota of adolescent girls prior to the onset of menarche resemble those of reproductive-age women. MBio 2015, 6, e00097-15. [Google Scholar] [CrossRef]
- Vongsa, R.; Hoffman, D.; Shepard, K.; Koenig, D. Comparative study of vulva and abdominal skin microbiota of healthy females with high and average BMI. BMC Microbiol. 2019, 19, 16. [Google Scholar] [CrossRef] [PubMed]
- Murina, F.; Caimi, C.; Di Pierro, F.; Di Francesco, S.; Cetin, I. Features of the Vaginal and Vestibular Microbioma in Patients With Vestibulodynia: A Case-Control Study. J. Low. Genit. Tract Dis. 2020, 24, 290–294. [Google Scholar] [CrossRef]
- Jayaram, A.; Witkin, S.S.; Zhou, X.; Brown, C.J.; Rey, G.E.; Linhares, I.M.; Ledger, W.J.; Forney, L.J. The bacterial microbiome in paired vaginal and vestibular samples from women with vulvar vestibulitis syndrome. Pathog. Dis. 2014, 72, 161–166. [Google Scholar] [CrossRef][Green Version]
- Chattopadhyay, S.; Arnold, J.D.; Malayil, L.; Hittle, L.; Mongodin, E.F.; Marathe, K.S.; Gomez-Lobo, V.; Sapkota, A.R. Potential role of the skin and gut microbiota in premenarchal vulvar lichen sclerosus: A pilot case-control study. PLoS ONE 2021, 16, e0245243. [Google Scholar] [CrossRef]
- Yang, X.; Da, M.; Zhang, W.; Qi, Q.; Zhang, C.; Han, S. Role of Lactobacillus in cervical cancer. Cancer Manag. Res. 2018, 10, 1219–1229. [Google Scholar] [CrossRef]
- Wang, H.; Ma, Y.; Li, R.; Chen, X.; Wan, L.; Zhao, W. Associations of cervicovaginal lactobacilli with high-risk human papillomavirus infection, cervical intraepithelial neoplasia, and cancer: A systematic review and meta-analysis. J. Infect. Dis. 2019, 220, 1243–1254. [Google Scholar] [CrossRef] [PubMed]
- Grice, E.A.; Segre, J.A. The skin microbiome. Nat. Rev. Microbiol. 2011, 9, 244–253. [Google Scholar] [CrossRef]
- Gajer, P.; Brotman, R.M.; Bai, G.; Sakamoto, J.; Schütte, U.M.E.; Zhong, X.; Koenig, S.S.K.; Fu, L.; Ma, Z.; Zhou, X.; et al. Temporal dynamics of the human vaginal microbiota. Sci. Transl. Med. 2012, 4, 132ra52. [Google Scholar] [CrossRef] [PubMed]
- Guinan, M.; Dan, B.B.; Guidotti, R.J.; Reingold, A.L.; Schmid, G.P.; Guinan, M.; Dan, B.B.; Guidotti, R.J.; Reingold, A.L.; Schmid, G.P.; et al. Vaginal Colonization with Staphylococcus aureus in Healthy Vaginal Colonization with Staphylococcus aureus in Healthy Women: A Review of Four Studies Women: A Review of Four Studies Repository Citation Repository Citation. Available online: https://digitalscholarship.unlv.edu/community_health_sciences_fac_articles/11ThisresponseorcommentisavailableatDigitalScholarship@UNLV:https://digitalscholarship.unlv.edu/community_health_sciences_fac_articles/11 (accessed on 16 April 2021).
- Linnemann, C.C.; Staneck, J.L.; Hornstein, S.; Barden, T.P.; Rauh, J.L.; Bonventre, P.F.; Buncher, C.R.; Beiting, A. The epidemiology of genital colonization with Staphylococcus aureus. Ann. Intern. Med. 1982, 96, 940–944. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, Y.; Takeshita, T. The oral microbiome and human health. J. Oral Sci. 2017, 59, 201–206. [Google Scholar] [CrossRef]
- Randis, T.M.; Ratner, A.J. Gardnerella and Prevotella: Co-conspirators in the Pathogenesis of Bacterial Vaginosis. J. Infect. Dis. 2019, 220, 1085–1088. [Google Scholar] [CrossRef] [PubMed]
- Grice, E.A.; Segre, J.A. The human microbiome: Our second genome. Annu. Rev. Genomics Hum. Genet. 2012, 13, 151–170. [Google Scholar] [CrossRef]
- Oh, J.; Byrd, A.L.; Deming, C.; Conlan, S.; Kong, H.H.; Segre, J.A.; Barnabas, B.; Blakesley, R.; Bouffard, G.; Brooks, S.; et al. Biogeography and individuality shape function in the human skin metagenome. Nature 2014, 514, 59–64. [Google Scholar] [CrossRef]
- Farage, M.A.; Miller, K.W.; Elsner, P.; Maibach, H.I. Intrinsic and extrinsic factors in skin ageing: A review. Int. J. Cosmet. Sci. 2008, 30, 87–95. [Google Scholar] [CrossRef]
- Chen, Y.; Bruning, E.; Rubino, J.; Eder, S.E. Role of female intimate hygiene in vulvovaginal health: Global hygiene practices and product usage. Womens Health 2017, 13, 58–67. [Google Scholar] [CrossRef]
- Langan, E.A.; Künstner, A.; Miodovnik, M.; Zillikens, D.; Thaçi, D.; Baines, J.F.; Ibrahim, S.M.; Solbach, W.; Knobloch, J.K. Combined culture and metagenomic analyses reveal significant shifts in the composition of the cutaneous microbiome in psoriasis. Br. J. Dermatol. 2019, 181, 1254–1264. [Google Scholar] [CrossRef]
- Ring, H.C.; Thorsen, J.; Saunte, D.M.; Lilje, B.; Bay, L.; Riis, P.T.; Larsen, N.; Andersen, L.O.; Nielsen, H.V.; Miller, I.M.; et al. The follicular skin microbiome in patients with hidradenitis suppurativa and healthy controls. JAMA Dermatol. 2017, 153, 897–905. [Google Scholar] [CrossRef]
- Mitra, A.; MacIntyre, D.A.; Lee, Y.S.; Smith, A.; Marchesi, J.R.; Lehne, B.; Bhatia, R.; Lyons, D.; Paraskevaidis, E.; Li, J.V.; et al. Cervical intraepithelial neoplasia disease progression is associated with increased vaginal microbiome diversity. Sci. Rep. 2015, 5, 16865. [Google Scholar] [CrossRef]
- Lee, J.E.; Lee, S.; Lee, H.; Song, Y.M.; Lee, K.; Han, M.J.; Sung, J.; Ko, G.P. Association of the Vaginal Microbiota with Human Papillomavirus Infection in a Korean Twin Cohort. PLoS ONE 2013, 8, e63514. [Google Scholar] [CrossRef]
- Audirac-Chalifour, A.; Torres-Poveda, K.; Bahena-Román, M.; Téllez-Sosa, J.; Martínez-Barnetche, J.; Cortina-Ceballos, B.; López-Estrada, G.; Delgado-Romero, K.; Burguete-García, A.I.; Cantú, D.; et al. Cervical microbiome and cytokine profile at various stages of cervical cancer: A pilot study. PLoS ONE 2016, 11, e0153274. [Google Scholar] [CrossRef]
- Adebamowo, S.N.; Ma, B.; Zella, D.; Famooto, A.; Ravel, J.; Adebamowo, C. Mycoplasma hominis and mycoplasma genitalium in the vaginal microbiota and persistent high-risk human papillomavirus infection. Front. Public Health 2017, 5, 140. [Google Scholar] [CrossRef]
- Nowak, R.G.; Bentzen, S.M.; Ravel, J.; Crowell, T.A.; Dauda, W.; Ma, B.; Liu, H.; Blattner, W.A.; Baral, S.D.; Charurat, M.E. Anal Microbial Patterns and Oncogenic Human Papillomavirus in a Pilot Study of Nigerian Men Who Have Sex with Men at Risk for or Living with HIV. AIDS Res. Hum. Retrovir. 2019, 35, 267–275. [Google Scholar] [CrossRef] [PubMed]
- Onywera, H.; Williamson, A.-L.L.; Cozzuto, L.; Bonnin, S.; Mbulawa, Z.Z.A.A.; Coetzee, D.; Ponomarenko, J.; Meiring, T.L. The penile microbiota of Black South African men: Relationship with human papillomavirus and HIV infection. BMC Microbiol. 2020, 20, 78. [Google Scholar] [CrossRef] [PubMed]
- Cohen, A.J.; Gaither, T.W.; Srirangapatanam, S.; Castellanos, E.R.; Enriquez, A.; Fergus, K.B.; Fadrosh, D.; Lynch, S.; Mmonu, N.A.; Breyer, B.N. Synchronous genitourinary lichen sclerosus signals a distinct urinary microbiome profile in men with urethral stricture disease. World J. Urol. 2020, 39, 605–611. [Google Scholar] [CrossRef]
- Levy, A.; Browne, B.; Fredrick, A.; Stensland, K.; Bennett, J.; Sullivan, T.; Rieger-Christ, K.M.; Vanni, A.J. Insights into the Pathophysiology of Urethral Stricture Disease due to Lichen Sclerosus: Comparison of Pathological Markers in Lichen Sclerosus Induced Strictures vs Nonlichen Sclerosus Induced Strictures. J. Urol. 2019, 201, 1158–1162. [Google Scholar] [CrossRef]
- Aidé, S.; Lattario, F.R.; Almeida, G.; Do Val, I.C.; Da Costa Carvalho, M. Epstein-barr virus and human papillomavirus infection in vulvar lichen sclerosus. J. Low. Genit. Tract Dis. 2010, 14, 319–322. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Fu, Q.; Zhang, X. The presence of human papillomavirus and Epstein-Barr virus in male Chinese lichen sclerosus patients: A single center study. Asian J. Androl. 2016, 18, 650–653. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, S.; Al-Maweri, S.A.; Alaizari, N.; Umair, A.; Ariffin, Z.; Alhajj, M.N.; Kassim, S.; Awan, K.H. The association between Epstein-Barr virus and oral lichen planus: A systematic review and meta-analysis. J. Oral Pathol. Med. 2020, 49, 969–976. [Google Scholar] [CrossRef]
- Brady, G.; MacArthur, G.J.; Farrell, P.J. Epstein-Barr virus and Burkitt lymphoma. Postgrad. Med. J. 2008, 84, 372–377. [Google Scholar] [CrossRef]
- Tsang, C.M.; Tsao, S.W. The role of Epstein-Barr virus infection in the pathogenesis of nasopharyngeal carcinoma. Virol. Sin. 2015, 30, 107–121. [Google Scholar] [CrossRef] [PubMed]
- Crombie, J.L.; LaCasce, A.S. Epstein Barr virus associated B-cell lymphomas and iatrogenic lymphoproliferative disorders. Front. Oncol. 2019, 9, 109. [Google Scholar] [CrossRef] [PubMed]
- Ravel, J.; Gajer, P.; Abdo, Z.; Schneider, G.M.; Koenig, S.S.K.; McCulle, S.L.; Karlebach, S.; Gorle, R.; Russell, J.; Tacket, C.O.; et al. Vaginal microbiome of reproductive-age women. Proc. Natl. Acad. Sci. USA 2011, 108, 4680–4687. [Google Scholar] [CrossRef]
- Borgdorff, H.; van der Veer, C.; van Houdt, R.; Alberts, C.J.; de Vries, H.J.; Bruisten, S.M.; Snijder, M.B.; Prins, M.; Geerlings, S.E.; Schim van der Loeff, M.F.; et al. The association between ethnicity and vaginal microbiota composition in Amsterdam, the Netherlands. PLoS ONE 2017, 12, e0181135. [Google Scholar] [CrossRef]
- Zhou, X.; Hansmann, M.A.; Davis, C.C.; Suzuki, H.; Brown, C.J.; Schütte, U.; Pierson, J.D.; Forney, L.J. The vaginal bacterial communities of Japanese women resemble those of women in other racial groups. FEMS Immunol. Med. Microbiol. 2010, 58, 169–181. [Google Scholar] [CrossRef] [PubMed]
- Huttenhower, C.; Gevers, D.; Knight, R.; Abubucker, S.; Badger, J.H.; Chinwalla, A.T.; Creasy, H.H.; Earl, A.M.; Fitzgerald, M.G.; Fulton, R.S.; et al. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef]
- Zhou, X.; Brown, C.J.; Abdo, Z.; Davis, C.C.; Hansmann, M.A.; Joyce, P.; Foster, J.A.; Forney, L.J. Differences in the composition of vaginal microbial communities found in healthy Caucasian and black women. ISME J. 2007, 1, 121–133. [Google Scholar] [CrossRef]
- Coolen, M.J.L.; Post, E.; Davis, C.C.; Forney, L.J. Characterization of microbial communities found in the human vagina by analysis of terminal restriction fragment length polymorphisms of 16S rRNA genes. Appl. Environ. Microbiol. 2005, 71, 8729–8737. [Google Scholar] [CrossRef]
- Graspeuntner, S.; Loeper, N.; Künzel, S.; Baines, J.F.; Rupp, J. Selection of validated hypervariable regions is crucial in 16S-based microbiota studies of the female genital tract. Sci. Rep. 2018, 8, 9678. [Google Scholar] [CrossRef] [PubMed]
- Meisel, J.S.; Hannigan, G.D.; Tyldsley, A.S.; SanMiguel, A.J.; Hodkinson, B.P.; Zheng, Q.; Grice, E.A. Skin Microbiome Surveys Are Strongly Influenced by Experimental Design. J. Investig. Dermatol. 2016, 136, 947–956. [Google Scholar] [CrossRef] [PubMed]
- Kong, H.H.; Andersson, B.; Clavel, T.; Common, J.E.; Jackson, S.A.; Olson, N.D.; Segre, J.A.; Traidl-Hoffmann, C. Performing Skin Microbiome Research: A Method to the Madness. J. Investig. Dermatol. 2017, 137, 561–568. [Google Scholar] [CrossRef] [PubMed]

| Author, Year | Focus | Study Subjects | Subject Age | Ethnicity + Country | Sample Locations | Sample Type | Microbial Analysis | Key Findings | Limitations | Risk of Bias | Level of Evidence | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Health and the influence of patient factors | Brown et al, 2007 | Healthy | 4 HV | 28–44 years | Caucasian, USA | Labia minora, labia majora, vagina | Scrape samples | 16S rRNA amplification of unspecified region | Bacteriome vulva ≈ vagina. Dominant vulvar species: 3/4 L. crispatus or L. iners, 1/4 L. iners, Atopobium vaginae and Megaspheara elsdenii. | Small sample size. No longitudinal sampling. No specification of sequencing region. | Low | Very low |
| Bruning et al, 2020 | Healthy | 36 HV | 18–55 years | Caucasian, USA | Labia majora | Modified liquid cup scrub method | 16S rRNA amplification of the V1–V3 region and fungal ITS sequencing | Bacterial relative abundance at baseline: 27–47% Corynebacterium, 12–18% Lactobacillus, 4–10% Staphylococcus, 3–12% Prevotella, 1–13% Propionibacterium and 3–5% Finegoldia. Fungal relative abundance at baseline: 20–50% Cryptococcus, 0.3–21% Malassezia, 1–12% Cladosporium and 2% Rhodoturula | Focus reporting on effects investigational product, not microbiome. 34/36 bacterial, 16/36 fungal samples showed amplification for analysis. | Low | Low | |
| Miyamoto et al, 2013 | Healthy | 40 HV | 20–40 years | Japanese, Japan | Labia majora, groin, mons pubis, inner thigh | Saline wetted sterile swabs | qPCR for specific genera (S. epidermidis, S. aureus, P. acnes, Lactobacilli spp., Prevotella spp.) | Labia + groin vs. mons pubis/inner thigh: ↑ Lactobacilli and S. aureus. Prevotella spp. on labia + groin only. | No extensive sequencing data due to employed procedure. No longitudinal sampling. | Low | Very low | |
| Costello et al, 2009 | Healthy | 3 HV | 30–35 years | Unknown, USA | Labia minora | NaCl + Tween wetted sterile swabs | 16S rRNA amplification of the V2 region | Predominant taxa: Lactobacillus (48.6%)¸ Prevotella (16%) and Finegoldia (8.9%) | Small sample size. Focus not on vulvar microbiome, but on other body sites. | Low | Very low | |
| Shiraishi et al, 2010 | Menstruation | 10 HV | 31–43 years | Japanese, Japan | Labia minora, vagina (3/10) | Scrape samples | 16S rRNA amplification of the V3–V4 region | No species consistently changed abundance before or during menstruation. 7/10 showed predominance of L. crispatus or L. iners | Small sample size. Only 3/10 vaginal cross–reference samples. No longitudinal sampling. | Low | Very low | |
| Hickey et al, 2015 | Menarche | 32 HV | 10–12.9 years | Mixed Black, Caucasian, Native American, Hispanic. USA | Labia minora, vagina | Dry, sterile flocked swabs | 16S rRNA amplification of the V1–V3 region | Bacteriome vulva ≈ vagina (mean more similarities before menarche. Greater variety bacterial taxa on vulva compared to the vagina. Abundance lactic acid producing bacteria increases with puberty on vulva and vagina | Focus on vaginal microbiome. No report comparing the pre- and post-menarchal vulvar microbiome | Low | Low | |
| Vongsa et al, 2019 | Obesity | 20 obese (BMI >30) 20 HV (BMI 18-25) | 18–35 years | Unknown, USA | Labia majora, labia minora | Swab | 16S rRNA amplification of unspecified region | Obese vs. HV: ↑ Corynebacerium spp. and Anaerococcus spp., ↓ Lactobacillus spp. Labia majora more diverse than labia minora. | No longitudinal sampling. No ethnicity data disclosure. No specification of sequencing region. | Low | Low | |
| Disease | Jayaram et al, 2014 | Vulvar vestibulitis syndrome (VVS) | 20 VVS 15 HV | Mean 30.8 (VVS) and 32.6 (HV) years | Caucasian, USA | Vestibulum, vagina | Swab | 16S rRNA amplification of the V1–V3 region | No differences vulvar or vaginal bacteriome composition cases vs. controls. Bacteriome vestibulum ≈ vagina. Most prevalent VVS: Lactobacillus, Gardnerella, Atopobium. Most prevalent HV: Lactobacillus, Streptococcus and Gardnerella | No longitudinal sampling. | Low | Low |
| Murina et al, 2020 | Provoked vestibulodynia (PVD) | 20 PVD 18 HV | 23–48 years | Caucasian, Italy | Vestibulum, vagina | Swab | 16S rRNA amplification of the V3 region | L. gasseri only dominant in PVD Most prevalent genera PVD: Lactobacillus, Gardnerella and Atopobium. Most prevalent genera HV: Lactobacillus, Gardnerella and Bifidobacetrium. | No longitudinal sampling. | Low | Low | |
| Chattopadhyay et al, 2021 | Pre-menarchal lichen sclerosus | 5 LS 5 NSVV 3 HV | Mean 6 years | Mixed Caucasian, Black and Hispanic, USA | Labial fold, perineum, feces | Dry flocked swabs | 16S rRNA amplification of the V3–V4 region | LS vs. HV: vulvar bacteriome ↑ Porphyromonas spp., Parvimonas spp., Peptoniphilus spp. Prevotella spp., Dialister spp. and ↓ Peptostreptococcus spp. Corynebacterium spp. Faecal bacteriome LS: ↑ Dialister spp. | Small sample size No longitudinal sampling Only premenarchal girls No species level determination due to employed procedure. | Low | Very low |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pagan, L.; Ederveen, R.A.M.; Huisman, B.W.; Schoones, J.W.; Zwittink, R.D.; Schuren, F.H.J.; Rissmann, R.; Piek, J.M.J.; van Poelgeest, M.I.E. The Human Vulvar Microbiome: A Systematic Review. Microorganisms 2021, 9, 2568. https://doi.org/10.3390/microorganisms9122568
Pagan L, Ederveen RAM, Huisman BW, Schoones JW, Zwittink RD, Schuren FHJ, Rissmann R, Piek JMJ, van Poelgeest MIE. The Human Vulvar Microbiome: A Systematic Review. Microorganisms. 2021; 9(12):2568. https://doi.org/10.3390/microorganisms9122568
Chicago/Turabian StylePagan, Lisa, Roos A. M. Ederveen, Bertine W. Huisman, Jan W. Schoones, Romy D. Zwittink, Frank H. J. Schuren, Robert Rissmann, Jurgen M. J. Piek, and Mariëtte I. E. van Poelgeest. 2021. "The Human Vulvar Microbiome: A Systematic Review" Microorganisms 9, no. 12: 2568. https://doi.org/10.3390/microorganisms9122568
APA StylePagan, L., Ederveen, R. A. M., Huisman, B. W., Schoones, J. W., Zwittink, R. D., Schuren, F. H. J., Rissmann, R., Piek, J. M. J., & van Poelgeest, M. I. E. (2021). The Human Vulvar Microbiome: A Systematic Review. Microorganisms, 9(12), 2568. https://doi.org/10.3390/microorganisms9122568






