Abstract
Objective: This descriptive systematic review aimed to assess in the available literature on the current application and overall performance of Artificial Intelligence (AI) models in the diagnosis and classification of Rotator Cuff Tears (RCTs) using MRIs. Methods: The systematic review was performed by two of the authors from 2020 to November 2024. Only diagnostic studies involving AI application to MRI images of the rotator cuff were considered, including supraspinatus and biceps tears. Studies evaluating AI applications to Ultrasound or X-ray, or including only healthy rotator cuffs, were not analyzed in this paper. Results: The coronal plane in the T2 sequence emerged as the predominant imaging protocol, with the VGG network being the most widely utilized AI model. The studies included in this research exhibited a solid performance of the AI models with accuracy, ranging from 71.0% to 100%. The statistical analysis revealed no significant differences (p > 0.05) in accuracy, sensitivity, specificity, or precision between AI and human experts across studies that included such comparisons. Conclusions: While AI can significantly improve diagnostic efficiency and workflow optimization, future studies must focus on external validation, regulatory approval, and AI-human collaboration models to ensure safe and effective integration into orthopedic imaging.
1. Introduction
Rotator cuff disease is the most common cause of shoulder pain, affecting 6.8% to 22.4% of patients older than 40 [1].
Calcific tendinitis, tendinosis, and tendon tears are the main causes of rotator cuff pathology [2]. Specifically, the most prevalent condition in patients over 60 is Rotator Cuff Tears (RCTs), specifically of the supraspinatus muscle, which affects 61.9% of men and 38.1% of women. [3].
The diagnosis is primarily based on both patient-reported symptoms and imaging techniques, including ultrasound, X-ray, Computed Tomography (CT), and Magnetic Resonance Imaging (MRI). The current gold standard for RCT diagnosis, prognostic feature depiction, and surgical planning is MRI [4]. Advanced fatty infiltration of the muscle and reduced acromiohumeral distance have been shown to be associated with long-standing injury to the RC and are indicative of a pre-existing tear if detected shortly after a trauma [5]. Differences in tendon kinking, muscle edema, and the degree of muscle atrophy observed on MRI have been shown to help distinguish between different types and stages of rotator cuff injuries [6].
However, the possibility of misdiagnosis can be increased by a number of circumstances. These consist of deceptive image artifacts or the existence of other diseases that could obstacle the diagnosis. As a result, studies investigating the application of computer-aided diagnostic tools to improve diagnostic accuracy and clinical decision-making have significantly increased [7].
In particular, the application of Artificial Intelligence (AI) would be particularly valuable for pathology identification, improving the diagnostic performance of medical radiologists while minimizing subjectivity and mistakes caused by inattention and fatigue [8].
Various AI applications in musculoskeletal imaging have been reported in the literature, including fracture detection, bone age estimation, osteoarthritis grading, soft tissue tumor classification, and implant analysis. These models are trained on annotated imaging datasets to identify patterns, quantify structures, or predict clinical outcomes [9].
AI can be applied through various techniques, primarily Machine Learning (ML) and Deep Learning (DL). ML involves the creation of automated computer systems that predict outcomes using mathematical algorithms. These models are developed using two types of datasets: a training set for constructing the mathematical model and a testing set for evaluating its effectiveness [10].
In contrast, DL is a more sophisticated subset of ML that enables unsupervised learning from unstructured and unlabeled data, effectively filtering out irrelevant factors during the process [11].
These processes are trained on medical databases containing big data, most of which are generated by radiomics, a quantitative method in radiology that provides clinicians with additional information using advanced mathematical analysis. By analyzing patterns, intensity, shape, and pixel relationships, radiomics quantifies textural details, offering objective, data-driven insights that complement traditional image interpretation [12].
Previous systematic reviews, such as the ones published by Zhan et al. in 2024 [7] and Rodriguez et al. in 2023 [13], have analyzed AI applications in the identification of various rotator cuff pathologies across various imaging modalities.
Similarly, Garcia et al. in 2024 [14] evaluated the performance of both DL and ML models across various imaging techniques. However, there remains a gap in the literature specifically focused on MRI-based DL diagnosis of RCTs.
This review addresses that gap by offering a comprehensive and updated evaluation of DL models applied specifically to shoulder MRI for the diagnosis and classification of RCTs. In contrast to earlier work, this review emphasizes MRI as the gold standard in imaging and explores diagnostic performance in greater detail by reporting metrics, such as accuracy, sensitivity, specificity, precision, and Dice coefficient. It also includes tear stratification data, allowing for a deeper understanding of how well models differentiate between degrees of injury severity. Additionally, the review discusses comparisons between AI and human experts, critically evaluating the imaging protocols and model architectures employed. Finally, it outlines key limitations in the current literature and offers practical recommendations for standardization and future research directions. These contributions support clinical adoption and the design of more robust, reproducible AI tools in musculoskeletal radiology.
Hence, this systematic review aims to provide an updated analysis of the diagnostic performance of MRI-based AI models for detecting and classifying rotator cuff pathologies and to assess the impact of radiomics on improving diagnostic consistency and accuracy.
2. Materials and Methods
2.1. Eligibility Criteria
The present review includes retrospective diagnostic study designs published after December 2020. Considering the authors’ proficiency in various languages, articles in English and Italian were screened. Peer-reviewed articles of each level of evidence according to the Oxford classification were included.
Only studies involving the application of AI tools to MRI images of the rotator cuff were considered, including supraspinatus and biceps tears. Studies evaluating AI applications to Ultrasound or X-ray, or including only healthy rotator cuffs, were not analyzed in this study. Systematic reviews, technical notes, letters to editors, instructional courses, or studies including pathologies different from those were excluded. Studies lacking stratified results were not considered. In vitro, animal, cadaver, and biomechanical studies were excluded.
2.2. Information Sources
A systematic review was performed using the Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) guidelines [15]. The review protocol was not registered with PROSPERO, but it was performed in accordance with the PRISMA guidelines and registration information. Medline, EMBASE, Scopus, CINAHL, and CENTRAL bibliographic databases were searched using the following string:
((((((((((MRI) OR (magnetic AND resonance)) AND (shoulder AND joint)) AND (supraspinatus AND tendon)) OR (supraspinatus AND muscle)) AND (tear) OR (lesion)) AND (diagnostics) AND (segmentation)) AND (computer AND vision)) AND (artificial AND intelligence)) OR (deep AND learning)) AND (supraspinatus AND tear).
The search was performed by two authors from 2020 to November 2024, and articles from the inception of the database to November 2024 were searched.
Keywords were used both isolated and combined. Additional studies were searched among the reference lists of selected papers and systematic reviews.
2.3. Search Strategy and Data Collection Process
Data extraction was performed by two independent authors, and differences were reconciled by mutual agreement. In case of disagreement on the inclusion or exclusion of articles, a third reviewer was consulted. One author performed the review and organization of the titles in order to limit the bias.
The reviewers used the following screening approach: the title and abstract were reviewed first, then the full articles. The full text of papers not excluded was evaluated and eventually selected after a discussion between the reviewers. In case of disagreement, the third reviewer was consulted.
The initial search strategy was organized according to the PICO (Population, Intervention, Comparison, Outcome) structure. This systematic review aims to describe whether AI tools applied to shoulder MRIs of patients suspected of RCTs or other shoulder pathologies (P) performed automated diagnosis, segmentation, and classification (I) comparable to standard MRIs or radiologist diagnoses (C). The outcomes (O) assessed were: diagnostic accuracy, sensitivity, specificity, precision, and Dice coefficient.
The number of articles included or excluded was registered and reported in the PRISMA flowchart. Guidelines by Moher et al. were followed to design the PRISMA chart (Figure 1) [16].
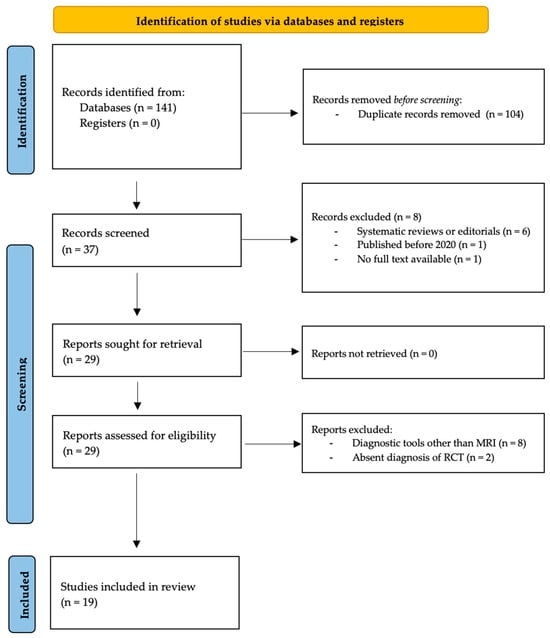
Figure 1.
PRISMA Flowchart.
2.4. Data Items
The extracted data included cohort demographics, such as author, year of publication, cohort sample size, mean age, and gender distribution, as well as study specifics such as study objective and rotator cuff pathology (Table 1). Tear specifics and classification were then reported in Table 2.

Table 1.
Study Objective and Cohort Characteristics.

Table 2.
Tear Stratification.
General study characteristics, including MRI imaging planes, MRI sequences, and number of slices, were also recorded (Table 3).

Table 3.
MRI Acquisition Parameters.
Moreover, information regarding the AI model application was summarized in Table 4, including the AI model, the number of slices, the training sets and test sets images, and ground truth references.

Table 4.
AI Model Specifics.
Finally, diagnostic performance outcomes evaluated included percentage of accuracy, sensitivity, specificity, precision, and Dice coefficient (Table 5).

Table 5.
AI Model Performance Analysis.
All results compatible with each outcome domain were sought, and any exclusions were based on predefined criteria related to our research questions.
2.5. Study Risk of Bias Assessment
Given the designs of the included studies, the quality of all included studies was assessed using the QUADAS-2 tool (https://mcguinlu.shinyapps.io/robvis/ accessed on 15 November 2024), which is designed to evaluate the accuracy of diagnostic studies [36]. Selected articles were independently evaluated by two reviewers and verified by a third in case of disagreement.
2.6. Synthesis Method
The synthesis of results was performed using a descriptive approach due to the high heterogeneity among the included studies, which precluded the possibility of conducting a meta-analysis. Data were extracted from each article and compiled in structured Excel spreadsheets to compare study characteristics across several domains, including cohort size, imaging protocols, AI model types, and diagnostic performance metrics (accuracy, sensitivity, specificity, precision, and Dice coefficient).
A qualitative synthesis was used to identify trends in AI model usage, commonly applied MRI sequences (e.g., T2-weighted coronal), and types of ground truth references.
When possible, a quantitative comparative analysis was also conducted for the studies that directly compared AI diagnostic performance with human experts. Paired t-tests were performed to compare the accuracy, sensitivity, specificity, and precision between AI and human diagnoses across those studies. A p-value less than 0.05 was considered statistically significant.
3. Results
3.1. Study Selection
This systematic review was conducted and reported in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) 2020 guidelines [15].
The literature search identified 141 articles published between 2020 and 2024. No additional studies were found in the grey literature, and no unpublished studies were retrieved. Duplicate removal resulted in the exclusion of 104 studies, leaving 37 articles for screening. Eight articles were excluded based on title and abstract (systematic reviews and editorials: n = 6; studies published before 2020: n = 1; no full text available: n = 1). Twenty-nine articles were screened by full text. Ten were excluded (US or X-ray-based studies: n = 8; absent diagnosis of RCT: n = 2). At the final screening, 19 articles met the selection criteria and were included in the review [17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35]. The PRISMA flowchart of the literature search is reported in Figure 1. Rules by Page et al. were followed in designing the PRISMA chart [15].
3.2. Quality of Evidence
The QUADAS-2 tool for diagnostic studies was used to assess the methodological quality of each article [36].
Out of the 19 included studies, four were identified as “low risk of bias” studies [19,24,28,30]; 13 were identified as “some concerns” studies [17,20,21,23,25,26,27,29,31,32,33,34,35], and two studies resulted in having a “high risk of bias” [18,22].
The risk of bias assessment is reported in Figure 2. Each study is evaluated across four domains: (1) Patient Selection, (2) Index Test, (3) Reference Standard, and (4) Flow and Timing. The color coding indicates the level of bias: green for low risk, yellow for some concerns, and red for high risk. This visual summary highlights the overall methodological quality and helps assess the reliability of diagnostic accuracy results reported in the included studies.
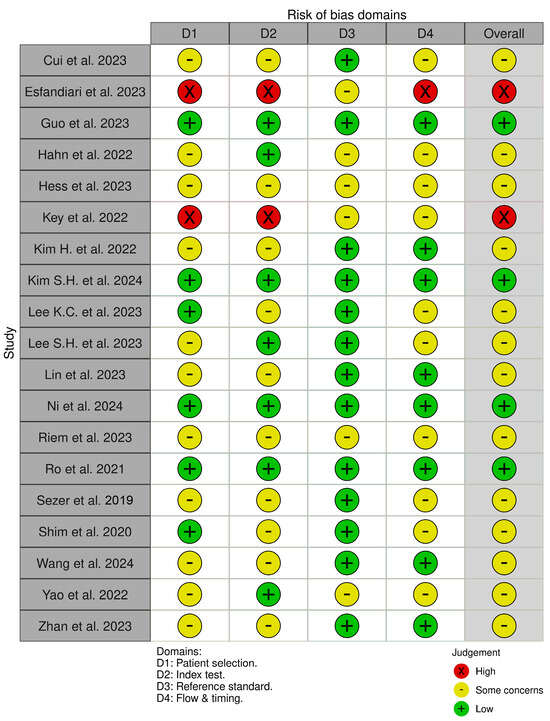
Figure 2.
QUADAS-2 Tool Results [17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35].
No formal assessment of reporting bias was conducted, as the review did not include a meta-analysis, and the small number of studies limited the applicability of publication bias detection tools.
3.3. Cohort Characteristics
All the selected studies correctly reported the number of patients. This review included 10,277 patients. The study by Shim et al. [32] reported the highest number of patients (n = 2124). The study by Kim et al. [23], on the other hand, reported the lowest number of patients (n = 56).
The mean age and gender were not specified by all articles; however, the lowest reported age was 47.2 ± 10.0, while the highest was 64.5 ± 8.2. Additionally, most of the studies included a predominantly female cohort, with only four studies reporting a majority of male participants [21,27,29,34].
A meta-analysis was not performed at the end of the review due to the heterogeneity of the data of the selected articles. The cohort characteristics are shown in Table 1.
3.4. Individual Study Objectives
All studies evaluated RCTs. Specifically, nine authors studied the Supraspinatus muscle [17,19,23,27,28,30,33,34,35], while two studies focused on the Biceps Muscle [20,22]. Three studies targeted RC segmentation [21,26,33].
Finally, 16 studies focused on the diagnosis of RCT by MRI imaging [17,18,19,20,22,23,24,25,27,28,29,30,31,32,34,35]. Of these, nine studies [19,23,27,28,29,30,31,32,35] further classified the tears between partial tear, full tear, or even small, medium, large, and massive. Across all papers reporting raw counts, the dataset comprises 6721 torn tendons versus 3123 intact tendons, indicating that the AI models were generally trained and tested on tear-heavy cohorts.
3.5. MRI Acquisition Parameters
The most common plane of acquisition for the MRI slices was the coronal plane, employed by 17 studies [17,18,19,20,21,23,24,25,26,27,28,30,31,32,33,34,35]. Ten studies used the sagittal plane [18,20,21,24,25,26,27,28,29,32], with only one of these utilizing it exclusively [29]. The least commonly used plane was the axial plane, which was employed in only nine studies [18,20,21,22,24,25,26,30,32], with only one of these utilizing it exclusively [22].
Regarding the MRI sequences, T2 and Proton Density (PD) were the most common, employed by nine [17,20,22,23,24,25,26,27,32,34,35] and seven studies [19,24,25,27,28,31,33], with six of these utilizing it exclusively in the first [17,20,22,23,34,35] or the second [19,28,31,33]. Finally, T1 was the least frequently utilized sequence, applied only in six articles [21,24,26,29,30,32], of which three used it solely [21,29,30].
Eight articles reported the number of slices analyzed [17,19,22,24,26,27,33,34]. The study by Lin et al. [27] reported the lowest number of slices (n = 32). The study by Yao et al. [34], on the other hand, reported the highest number of slices (n = 4287).
The MRI acquisition specifics are summarized in Table 3.
3.6. AI Models and Learning Data
The most commonly utilized AI models were U-Net and VGG, respectively, and were applied in five [17,26,29,33,34] and four studies [22,28,30,35]. These models were implemented either independently or in combination with other models. Additionally, ResNet [17,27,34] and nnU-Net [21,24] systems were employed to analyze MRIs in three studies each, while DenseNet was employed in two studies [17,23,35]. Finally, MobileNet [18], SqueezeNet [18], Xception [19], AIR Recon [20], INCA [22], YOLO [25], RC-MTL [28], CapsNet [31], and VRN [32] were each utilized in a single article.
To ensure the accuracy of the results provided by the AI models, all studies analyzed in this research established a ground truth reference, which was verified either before or after applying the AI tool to the MRI images. A musculoskeletal radiologist was consulted by eight authors [17,21,24,25,27,33,34,35], while in six studies [18,23,26,29,30,31], the comparison was performed by an orthopaedic surgeon. Lastly, the least commonly employed ground truth reference was represented by intra-operative arthroscopic findings in five studies [19,20,22,28,32].
All information regarding the employment of the models is reported in Table 4.
3.7. AI Model Performance Analysis
AI model accuracy was evaluated by 13 articles [17,18,19,20,22,25,27,28,30,31,32,34,35]. The lowest accuracy was obtained by Guo et al. [19] (71.0%), while the highest value was achieved by Key et al. [22] (100%). Sensitivity was reported in 14 articles [17,18,19,20,21,22,24,25,26,28,30,32,34,35]. The lowest sensitivity was registered by Hahn et al. [20] (72.7%), whereas both Hess et al. and Key et al. [21,22] reported the highest sensitivity (100%). Specificity was analyzed by 13 articles [17,18,19,20,21,22,25,26,28,30,32,34,35], of which the highest and lowest values were obtained, respectively, by Hahn et al. and Key et al. [20,22] (100%) and by Guo et al. [19] (69.6%). Eight studies analyzed the model precision [18,19,22,24,25,26,32,33], achieving values ranging from 54.0% [19] to 100% [22].
Finally, the Dice score was evaluated by eight studies [21,23,24,26,29,30,33,34], among which Ro et al. [30] reported the highest value (0.94) while Yao et al. [34] reported the lowest value (0.81).
Four studies compared the performance of the AI model with that of an orthopedic specialist [17,19,27,32]. The p-values for the evaluated metrics were calculated using a paired t-test, comparing AI and specialist results from the same studies in terms of accuracy, sensitivity, specificity, and precision (p = 0.87, 0.52, 0.68, and 0.63, respectively). No statistically significant differences were observed between AI and specialists.
The AI model results are reported in Table 5.
4. Discussion
This descriptive systematic review aimed to assess the available literature on the current application and overall performance of AI models in the diagnosis and classification of RCTs using MRIs.
The findings reveal that AI-based models exhibit high accuracy, sensitivity, and specificity, often approaching the performance of human specialists. The studies included in this research exhibited solid performance of the DL models, with accuracy, sensitivity, specificity, precision, and Dice ranging from 71.0% to 100%, 72.7% to 100%, 69.6% to 100%, 54.0% to 100%, and 0.94 to 0.81, respectively. Several studies exceeded 90% accuracy in classification tasks. The high values for sensitivity and specificity also showed that these models successfully detect both positive and negative cases.
Notably, the studies conducted by Key et al., Ni et al., and Ro et al. obtained the highest accuracy rates (100%, 98.0%, and 99.89%, respectively) [22,28,30]. These studies also employed the same AI model, VGG. This consistency in performance across different research groups may suggest a potential superiority of the VGG model in medical image analysis. This result has also been validated by the study conducted by Saavedra et al. [37] in 2023, who trained CNN models, including VGG-19, ResNet-50, and Inception-v3, to classify supraspinatus muscle fatty infiltration using shoulder T2-weighted MRI images. The VGG-19 model demonstrated exceptional performance, achieving an accuracy of 97.3%, a sensitivity of 94.7%, and a specificity of 97.5%.
The present review also reported a direct comparison between AI models and orthopedic specialists. Statistical analysis revealed no significant differences (p > 0.05) in accuracy, sensitivity, specificity, or precision between AI and human experts across studies that included such comparisons. This finding supports the notion that AI can serve as a reliable decision-support tool for radiologists and orthopedic surgeons. In particular, AI’s ability to rapidly analyze MRI scans and provide quantitative assessments holds significant promise for improving diagnostic efficiency.
However, it is important to note that the overall performance of AI models is not universally consistent across studies. The observed variability in AI results underscores the significant influence of factors such as dataset quality, annotation consistency, model training approaches, and ground truth reference. For instance, only five studies used arthroscopic confirmation of RCTs as the gold standard, which is currently identified as the gold standard for RCT diagnosis [38], while others relied solely on radiologist interpretations, which can introduce subjectivity. This discrepancy in ground truth labeling may lead to inconsistent AI training and variable performance metrics. However, not all rotator cuff tears necessitate surgical intervention. Therefore, arthroscopic confirmation is not always feasible [39]
Additionally, the MRI acquisition parameters varied significantly between the included studies, affecting model reproducibility. The coronal plane in the T2 sequence emerged as the most commonly employed imaging protocol across the studies analyzed. In particular, in the present literature, the coronal plane was found to be more appropriate for detecting tendon ruptures in the shoulder when using visual descriptors such as the mean intensity value of the supraspinatus tendon [40], as this plane offers enhanced sensitivity and specificity for identifying tendon ruptures, further confirmed by other research [7,34]. This consistency suggests that the coronal T2 sequence may offer particular advantages in visualizing the structures of interest, potentially contributing to more reliable assessments. It is recommended that further studies adopt this protocol to strengthen the validity of future research and enhance the comparability of results. Standardizing the imaging approach in this manner could promote greater homogeneity within study cohorts, thereby reducing variability and ensuring more robust and generalizable findings. Thus, while AI has shown strong performance in controlled environments, these sources of variability must be addressed before AI can be seamlessly integrated into clinical practice.
This review presents points of strength. Firstly, the articles selected for analysis were published between 2020 and 2024, ensuring that the included studies reflect the most recent advancements in the field. Additionally, the studies assessed using the QUADAS-2 tool demonstrated a relatively low risk of bias, further supporting the validity of the findings.
Nonetheless, this review is subject to certain limitations.
Despite the growing interest in AI applications for rotator cuff tear diagnosis, publicly available MRI datasets specifically labeled for this purpose remain limited. Most studies included in this review relied on private, institution-specific datasets, which restricts reproducibility and external validation.
Also, the results reached from this systematic review are limited in their generalizability due to the small number of included papers, which prevented the execution of a meta-analysis and conferred this review a predominantly descriptive character. Future research should seek to increase the dataset and implement more consistent procedures to improve comparability among studies. Furthermore, the studies analyzed exhibited a high degree of heterogeneity, encompassing variations in AI algorithms, evaluation criteria, and reference standards, as well as different MRI protocols in terms of plane and sequence of acquisition, number of slices, and ground truth method. In fact, the majority of studies had the MRIs first evaluated by either experienced orthopaedic surgeons or musculoskeletal radiologists, while only a minor subgroup utilized arthroscopic findings. The latter confers more objectivity to the findings, reducing potential error in the dataset. Adopting arthroscopy or surgical findings as the reference standard would be preferred for the design of more reliable and accurate AI models. Lastly, although this systematic review adhered to the PRISMA guidelines, the relatively small number of included studies limits the generalizability of the conclusions drawn.
Future research should aim to expand the size and diversity of datasets by incorporating multi-center, multi-population imaging sources, which would improve the robustness and generalizability of AI models. In addition, adopting more standardized methodologies, such as consistent MRI acquisition protocols, uniform tear classification systems, and clearly defined ground truth references, would greatly enhance the comparability of results across studies. These efforts would not only facilitate more reliable meta-analyses in the future but also support the development of clinically deployable AI tools that can perform accurately in varied real-world settings.
5. Conclusions
MRI is considered the gold standard for diagnosing supraspinatus muscle tears, with the T2-weighted coronal plane emerging as the most commonly and effectively used imaging sequence for this purpose. This review found that DL models, particularly VGG-based architectures, have shown promising results in automating the detection and classification of rotator cuff conditions. The studies included in this research exhibited solid performance of the DL models, with accuracy, sensitivity, specificity, precision, and Dice ranging from 71.0% to 100%, 72.7% to 100%, 69.6% to 100%, 54.0% to 100%, and 0.94 to 0.81, respectively. Moreover, the statistical analysis revealed no significant differences (p > 0.05) in accuracy, sensitivity, specificity, or precision between AI and human experts across studies that included such comparisons
However, methodological limitations, dataset variability, and lack of standardization remain key barriers to clinical implementation. While AI can significantly improve diagnostic efficiency and workflow optimization, future studies must focus on external validation, regulatory approval, and AI-human collaboration models to ensure safe and effective integration into orthopedic imaging.
Author Contributions
Conceptualization, U.G.L. and B.B.; methodology, L.P., A.C., E.S. and B.B.; software, A.d.S. and P.D.; validation, U.G.L. and M.M.; formal analysis, B.B., L.M. and M.M.; investigation, A.d.S., L.M. and A.C.; resources, L.P., A.C. and E.S.; data curation, M.M.; writing—original draft preparation, B.B.; writing—review and editing, U.G.L., M.M. and B.B.; visualization, P.D. and A.d.S.; supervision, M.M.; project administration, U.G.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available upon request from the corresponding author.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Hill, J.R.; Olson, J.J.; Sefko, J.A.; Steger-May, K.; Teefey, S.A.; Middleton, W.D.; Keener, J.D. Does surgical intervention alter the natural history of degenerative rotator cuff tears? Comparative analysis from a prospective longitudinal study. J. Shoulder Elb. Surg. 2024, 34, 430–440. [Google Scholar] [CrossRef] [PubMed]
- Via, A.G.; De Cupis, M.; Spoliti, M.; Oliva, F. Clinical and biological aspects of rotator cuff tears. Muscles Ligaments Tendons J. 2013, 3, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Redondo-Alonso, L.; Chamorro-Moriana, G.; Jiménez-Rejano, J.J.; López-Tarrida, P.; Ridao-Fernández, C. Relationship between chronic pathologies of the supraspinatus tendon and the long head of the biceps tendon: Systematic review. BMC Musculoskelet. Disord. 2014, 15, 377. [Google Scholar] [CrossRef]
- Calvo, E.; Guardado, C.R.; Morcillo, D.; Arce, G. Diagnosis and Classification of Rotator Cuff Tears. In Rotator Cuff Across the Life Span; Springer: Berlin/Heidelberg, Germany, 2019; pp. 3–10. [Google Scholar]
- Loew, M.; Magosch, P.; Lichtenberg, S.; Habermeyer, P.; Porschke, F. How to discriminate between acute traumatic and chronic degenerative rotator cuff lesions: An analysis of specific criteria on radiography and magnetic resonance imaging. J. Shoulder Elb. Surg. 2015, 24, 1685–1693. [Google Scholar] [CrossRef] [PubMed]
- Furrer, P.R.; Borbas, P.; Egli, R.J.; Zindel, C.; Wieser, K.; Bouaicha, S. MRI findings of traumatic and degenerative rotator cuff tears and introduction of the “cobra sign”. JSES Int. 2023, 7, 550–554. [Google Scholar] [CrossRef]
- Zhan, H.; Teng, F.; Liu, Z.; Yi, Z.; He, J.; Chen, Y.; Geng, B.; Xia, Y.; Wu, M.; Jiang, J. Artificial Intelligence Aids Detection of Rotator Cuff Pathology: A Systematic Review. Arthroscopy 2024, 40, 567–578. [Google Scholar] [CrossRef]
- Liu, F.; Guan, B.; Zhou, Z.; Samsonov, A.; Rosas, H.; Lian, K.; Sharma, R.; Kanarek, A.; Kim, J.; Guermazi, A.; et al. Fully Automated Diagnosis of Anterior Cruciate Ligament Tears on Knee MR Images by Using Deep Learning. Radiol. Artif. Intell. 2019, 1, 180091. [Google Scholar] [CrossRef]
- Gitto, S.; Serpi, F.; Albano, D.; Risoleo, G.; Fusco, S.; Messina, C.; Sconfienza, L.M. AI applications in musculoskeletal imaging: A narrative review. Eur. Radiol. Exp. 2024, 8, 22. [Google Scholar] [CrossRef]
- Lalehzarian, S.P.; Gowd, A.K.; Liu, J.N. Machine learning in orthopaedic surgery. World J. Orthop. 2021, 12, 685–699. [Google Scholar] [CrossRef]
- Lisacek-Kiosoglous, A.B.; Powling, A.S.; Fontalis, A.; Gabr, A.; Mazomenos, E.; Haddad, F.S. Artificial intelligence in orthopaedic surgery. Bone Jt. Res. 2023, 12, 447–454. [Google Scholar] [CrossRef]
- van Timmeren, J.E.; Cester, D.; Tanadini-Lang, S.; Alkadhi, H.; Baessler, B. Radiomics in medical imaging-”how-to” guide and critical reflection. Insights Imaging 2020, 11, 91. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, H.C.; Rust, B.; Hansen, P.Y.; Maffulli, N.; Gupta, M.; Potty, A.G.; Gupta, A. Artificial Intelligence and Machine Learning in Rotator Cuff Tears. Sports Med. Arthrosc. Rev. 2023, 31, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Velasquez Garcia, A.; Hsu, K.L.; Marinakis, K. Advancements in the diagnosis and management of rotator cuff tears. The role of artificial intelligence. J. Orthop. 2024, 47, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA Statement. Open Med. 2009, 3, e123–e130. [Google Scholar]
- Cui, J.; Xia, X.; Wang, J.; Li, X.; Huang, M.; Miao, S.; Hao, D.; Li, J. Fully Automated Approach for Diagnosis of Supraspinatus Tendon Tear on Shoulder MRI by Using Deep Learning. Acad. Radiol. 2023, 31, 994–1002. [Google Scholar] [CrossRef]
- Esfandiari, M.A.; Fallah Tafti, M.; Jafarnia Dabanloo, N.; Yousefirizi, F. Detection of the rotator cuff tears using a novel convolutional neural network from magnetic resonance image (MRI). Heliyon 2023, 9, e15804. [Google Scholar] [CrossRef]
- Guo, D.; Liu, X.; Wang, D.; Tang, X.; Qin, Y. Development and clinical validation of deep learning for auto-diagnosis of supraspinatus tears. J. Orthop. Surg. Res. 2023, 18, 426. [Google Scholar] [CrossRef]
- Hahn, S.; Yi, J.; Lee, H.J.; Lee, Y.; Lim, Y.J.; Bang, J.Y.; Kim, H.; Lee, J. Image Quality and Diagnostic Performance of Accelerated Shoulder MRI With Deep Learning-Based Reconstruction. AJR Am. J. Roentgenol. 2022, 218, 506–516. [Google Scholar] [CrossRef]
- Hess, H.; Ruckli, A.C.; Bürki, F.; Gerber, N.; Menzemer, J.; Burger, J.; Schär, M.; Zumstein, M.A.; Gerber, K. Deep-Learning-Based Segmentation of the Shoulder from MRI with Inference Accuracy Prediction. Diagnostics 2023, 13, 1668. [Google Scholar] [CrossRef]
- Key, S.; Demir, S.; Gurger, M.; Yilmaz, E.; Barua, P.D.; Dogan, S.; Tuncer, T.; Arunkumar, N.; Tan, R.S.; Acharya, U.R. ViVGG19: Novel exemplar deep feature extraction-based shoulder rotator cuff tear and biceps tendinosis detection using magnetic resonance images. Med. Eng. Phys. 2022, 110, 103864. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Shin, K.; Lee, E.S.; Chung, S.W.; Koh, K.H.; Kim, N. Can deep learning reduce the time and effort required for manual segmentation in 3D reconstruction of MRI in rotator cuff tears? PLoS ONE 2022, 17, e0274075. [Google Scholar] [CrossRef]
- Kim, S.H.; Yoo, H.J.; Yoon, S.H.; Kim, Y.T.; Park, S.J.; Chai, J.W.; Oh, J.; Chae, H.D. Development of a deep learning-based fully automated segmentation of rotator cuff muscles from clinical MR scans. Acta Radiol. 2024, 65, 1126–1132. [Google Scholar] [CrossRef]
- Lee, K.C.; Cho, Y.; Ahn, K.S.; Park, H.J.; Kang, Y.S.; Lee, S.; Kim, D.; Kang, C.H. Deep-Learning-Based Automated Rotator Cuff Tear Screening in Three Planes of Shoulder MRI. Diagnostics 2023, 13, 3254. [Google Scholar] [CrossRef]
- Lee, S.H.; Lee, J.; Oh, K.S.; Yoon, J.P.; Seo, A.; Jeong, Y.; Chung, S.W. Automated 3-dimensional MRI segmentation for the posterosuperior rotator cuff tear lesion using deep learning algorithm. PLoS ONE 2023, 18, e0284111. [Google Scholar] [CrossRef]
- Lin, D.J.; Schwier, M.; Geiger, B.; Raithel, E.; von Busch, H.; Fritz, J.; Kline, M.; Brooks, M.; Dunham, K.; Shukla, M.; et al. Deep Learning Diagnosis and Classification of Rotator Cuff Tears on Shoulder MRI. Investig. Radiol. 2023, 58, 405–412. [Google Scholar] [CrossRef]
- Ni, M.; Zhao, Y.; Zhang, L.; Chen, W.; Wang, Q.; Tian, C.; Yuan, H. MRI-based automated multitask deep learning system to evaluate supraspinatus tendon injuries. Eur. Radiol. 2024, 34, 3538–3551. [Google Scholar] [CrossRef]
- Riem, L.; Feng, X.; Cousins, M.; DuCharme, O.; Leitch, E.B.; Werner, B.C.; Sheean, A.J.; Hart, J.; Antosh, I.J.; Blemker, S.S. A Deep Learning Algorithm for Automatic 3D Segmentation of Rotator Cuff Muscle and Fat from Clinical MRI Scans. Radiol. Artif. Intell. 2023, 5, e220132. [Google Scholar] [CrossRef]
- Ro, K.; Kim, J.Y.; Park, H.; Cho, B.H.; Kim, I.Y.; Shim, S.B.; Choi, I.Y.; Yoo, J.C. Deep-learning framework and computer assisted fatty infiltration analysis for the supraspinatus muscle in MRI. Sci. Rep. 2021, 11, 15065. [Google Scholar] [CrossRef] [PubMed]
- Aysun Sezer and Hasan Basri, S. Capsule network-based classification of rotator cuff pathologies from MRI. Comput. Electr. Eng. 2019, 80, 106480. [Google Scholar] [CrossRef]
- Shim, E.; Kim, J.Y.; Yoon, J.P.; Ki, S.Y.; Lho, T.; Kim, Y.; Chung, S.W. Automated rotator cuff tear classification using 3D convolutional neural network. Sci. Rep. 2020, 10, 15632. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Liu, Y.; Zhou, Z. Supraspinatus extraction from MRI based on attention-dense spatial pyramid UNet network. J. Orthop. Surg. Res. 2024, 19, 60. [Google Scholar] [CrossRef]
- Yao, J.; Chepelev, L.; Nisha, Y.; Sathiadoss, P.; Rybicki, F.J.; Sheikh, A.M. Evaluation of a deep learning method for the automated detection of supraspinatus tears on MRI. Skelet. Radiol. 2022, 51, 1765–1775. [Google Scholar] [CrossRef] [PubMed]
- Zhan, J.; Liu, S.; Dong, C.; Ge, Y.; Xia, X.; Tian, N.; Xu, Q.; Jiang, G.; Xu, W.; Cui, J. Shoulder MRI-based radiomics for diagnosis and severity staging assessment of surgically treated supraspinatus tendon tears. Eur. Radiol. 2023, 33, 5587–5593. [Google Scholar] [CrossRef]
- Whiting, P.F.; Rutjes, A.W.; Westwood, M.E.; Mallett, S.; Deeks, J.J.; Reitsma, J.B.; Leeflang, M.M.; Sterne, J.A.; Bossuyt, P.M.; QUADAS-2 Group. QUADAS-2: A revised tool for the quality assessment of diagnostic accuracy studies. Ann. Intern. Med. 2011, 155, 529–536. [Google Scholar] [CrossRef]
- Saavedra, J.P.; Droppelmann, G.; García, N.; Jorquera, C.; Feijoo, F. High-accuracy detection of supraspinatus fatty infiltration in shoulder MRI using convolutional neural network algorithms. Front. Med. 2023, 10, 1070499. [Google Scholar] [CrossRef]
- Fazal Gafoor, H.; Jose, G.A.; Mampalli Narayanan, B. Role of Magnetic Resonance Imaging (MRI) in the Diagnosis of Rotator Cuff Injuries and Correlation With Arthroscopy Findings. Cureus 2023, 15, e50103. [Google Scholar] [CrossRef]
- Dickinson, R.N.; Kuhn, J.E. Nonoperative Treatment of Rotator Cuff Tears. Phys. Med. Rehabil. Clin. N. Am. 2023, 34, 335–355. [Google Scholar] [CrossRef]
- Ganal, E.; Ho, C.P.; Wilson, K.J.; Surowiec, R.K.; Smith, W.S.; Dornan, G.J.; Millett, P.J. Quantitative MRI characterization of arthroscopically verified supraspinatus pathology: Comparison of tendon tears, tendinosis and asymptomatic supraspinatus tendons with T2 mapping. Knee Surg. Sports Traumatol. Arthrosc. 2016, 24, 2216–2224. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).