Abstract
Carbapenemase-producing Klebsiella pneumoniae (CPKP) emerged in Greece in 2002 and became endemic thereafter. Driven by a notable variability in the phenotypic testing results for carbapenemase production in K. pneumoniae isolates from the intensive care units (ICUs) of our hospital, we performed a study to assess the molecular epidemiology of CPKP isolated between 2016 and 2019 using pulse-field gel electrophoresis (PFGE) including isolates recovered from 165 single patients. We investigated the molecular relatedness among strains recovered from rectal surveillance cultures and from respective subsequent infections due to CPKP in the same individual (48/165 cases). For the optimal interpretation of our findings, we carried out a systematic review regarding the clonality of CPKP isolated from clinical samples in ICUs in Europe. In our study, we identified 128 distinguishable pulsotypes and 17 clusters that indicated extended dissemination of CPKP within the hospital ICU setting throughout the study period. Among the clinical isolates, 122 harbored KPC genes (74%), 2 harbored KPC+NDM (1.2%), 38 harbored NDM (23%), 1 harbored NDM+OXA-48 (0.6%), 1 harbored NDM+VIM (0.6%) and 1 harbored the VIM (0.6%) gene. Multiple CPKP strains in our hospital have achieved sustained transmission. The polyclonal endemicity of CPKP presents a further threat for the selection of pathogens resistant to last-resort antimicrobial agents.
Keywords:
Klebsiella pneumoniae; carbapenemases; NDM; KPC; VIM; OXA-48; molecular epidemiology; PFGE 1. Introduction
In recent years, hospital-acquired infections caused by carbapenem-resistant Gram negative bacteria, especially carbapenem-resistant Klebsiella pneumoniae (CRKP), have been observed worldwide causing important public health problems and posing serious infection control issues. CRKP are opportunistic pathogens that cause infections with high morbidity and mortality mainly in hospitalized patients [1,2]. In Europe, the burden of CRKP predominantly affects the south and the east. According to the annual report of the European Centre for Disease Prevention and Control on antimicrobial resistance in Europe, 66.3% of the reported invasive K. pneumoniae isolates in Greece during 2020 were resistant to carbapenems.
Relatively high carbapenem resistance rates were also observed in Romania (48%), Italy (29%) and Bulgaria (28%), while, in the majority of the EU countries, this proportion was below 10% [3]. High carbapenem resistance trends were also observed in other non-EU neighboring countries: Bosnia, Herzegovina, Georgia, the Russian Federation, Serbia and Turkey reported proportions between 25% and 50% whereas, Belarus, the Republic of Moldova and the Ukraine reported proportions exceeding 50% [4].
The most common mechanism of carbapenem resistance among Enterobacterales and, thus, K. pneumoniae is the production of carbapenemases. Carbapenemases are β-lactamases able to hydrolyze all β-lactams, including carbapenems, and are categorized into several types. The carbapenemases most commonly encountered in Greece belong in three classes: class A K. pneumoniae carbapenemase (KPC), class B Verona imipenemase (VIM) and New Delhi metallo-β-lactamase (NDM) and class D oxacillinase-48 (OXA-48) [5].
Carbapenemase-producing K. pneumoniae (CPKP) emerged in Greece in 2002; they were of VIM-type, were involved in various outbreaks and soon became endemic in many hospitals all over the country. VIM-type carbapenemase-producers often belonged to different clones with ST147 being the predominant multi-locus sequence type (MLST) [6]. In 2007, KPC-producing K. pneumoniae isolates were introduced in Greek hospitals and rapidly dominated [7].
Greek KPC-CPKP mostly belonged to the worldwide successful hyperepidemic clone ST258, often associated with multi-drug resistant (MDR) phenotype [8]. The emergence of NDM in CPKP strains in Greece took place in 2011; the majority of them belonged to ST11 and were involved in oligoclonal outbreaks or sporadic cases [9]. OXA-48 type carbapenemases are the most prevalent class D enzymes identified in CPKP strains. The first OXA-48 was detected in Athens, Greece in 2012 and belonged to ST11 [10].
Carbapenemase-encoding genes spread fast via horizontal gene transfer together with other resistance determinants within the K. pneumoniae species in hospital settings, thus, dramatically restricting the available treatment options [11]. Moreover, the local epidemiology and the limited availability for isolation of affected patients in separate rooms in Greek hospitals undermine the efforts for effective infection control strategies.
K. pneumoniae is characterized by a high variety of antimicrobial resistance genes as well as a wide ecological distribution. Thus, in addition to its significance as a nosocomial pathogen (especially the hypervirulent phenotype), K. pneumoniae is considered as one of the most important bacterial species contributing in the dissemination of antimicrobial resistance genes to other human pathogens [12].
K. pneumoniae has the ability to colonize various mucosal surfaces, including the upper respiratory and the gastrointestinal gut. Among hospitalized patients, colonization rates in the nasopharynx are up to 19%, while it can reach as high as 77% in the gastrointestinal tract. Gut colonization often precedes and serves as a reservoir for transmission to other body sites resulting in the development of subsequent infections [13]. The duration of gut colonization with multi-drug resistant (MDR) bacteria, such as carbapenem-resistant K. pneumoniae varies from 43 to 387 days [14].
Driven by a notable variability in the phenotypic testing results for carbapenemase production and the types of carbapenemases present in K. pneumoniae isolates in our hospital, we performed a study to assess the molecular epidemiology of CPKP isolated between 2016 and 2019. Additionally, we investigated the molecular relatedness among strains recovered by rectal surveillance cultures and by respective subsequent infections due to CPKP in the same individual. In order to put our findings in context, we also performed a systematic review regarding the clonality of CPKP isolated from clinical samples originating in intensive care units (ICUs) in Europe.
2. Results
2.1. Carbapenemase Detection
During the study period (January 2016–June 2019) 165 single-patient clinical CRKP were analyzed; the isolates were recovered from 115 patients hospitalized in ICU 1, 6 patients in ICU 2 and 44 patients in ICU 3. Clinical samples included blood (n = 36), central venous catheters (n = 27), bronchial secretions (n = 51), urine (n = 20), pus (n = 13), wound swabs (n = 12), pleural fluid (n = 1), peritoneal fluid (n = 2), nasal swab (n = 1) and cerebrospinal fluid (n = 2).
Phenotypic and molecular testing revealed that all CRKP isolates harbored at least one carbapenemase often combined with ESBL activity. Among the clinical isolates, 122 harbored the KPC gene (73.95%), 2 KPC + NDM (1.21%), 38 NDM (23.04%), 1 NDM + OXA-48 (0.60%), 1 NDM + VIM (0.60%) and 1 VIM (0.60%).
2.2. Pulse-Field Gel Electrophoresis
All the 165 clinical CRKP isolates were typeable by pulse-field gel electrophoresis (PFGE) following digestion by restriction enzyme XbaI, revealing 128 distinguishable pulsotypes (P1-P128; Figure 1).
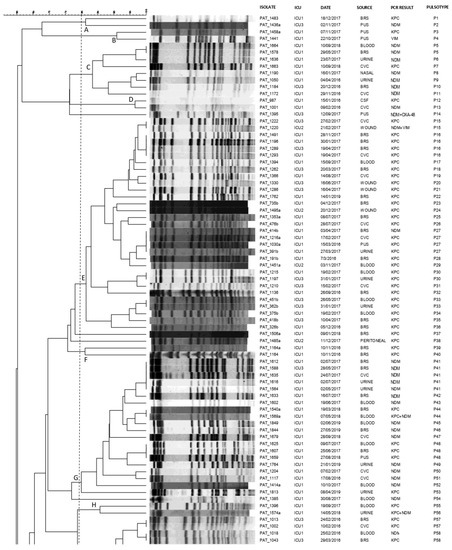
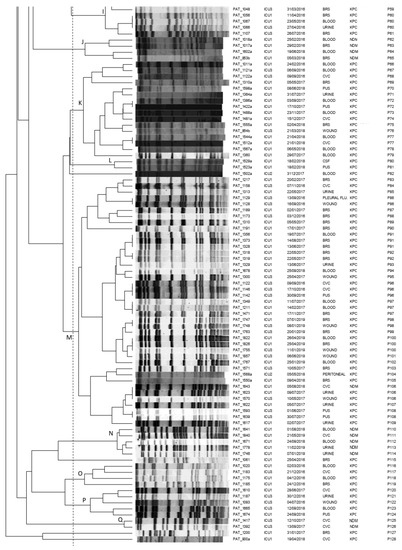
Figure 1.
Pulsotypes of the 165 clinical carbapenemase-producing K. pneumoniae isolates. Clusters (A–Q) were defined at a similarity level of 80%.PAT: patient; ICU: intensive care unit; CVC: central venous catheter; BRS: bronchial secretions; CSF: cerebrospinal fluid; P: pulsotype.
At a similarity level of 80% or above, the majority of CPKP isolates (95.6%, 158/165) were assigned into 17 clusters (A-Q), demonstrating multiclonal dissemination. The remaining seven genomes resulted to be unrelated and were consequently classified as sporadic isolates. KPC as well as NDM genetic determinants demonstrated polyclonal dissemination being present in 14 and 11 distinct clones, respectively.
In more detail, four predominant clusters E, G, K, and M consisting of 31, 19, 16 and 41 CRKP isolates, respectively, were identified: isolates of cluster G were almost exclusively obtained from ICU 1 (18 of 19), while clusters E, K, M consisted of clinical isolates from all ICUs under study. Interestingly, within the above clusters, indistinguishable pulsotypes shared by isolates from different patients and different ICUs were identified. Furthermore, looking at indistinguishable pulsotypes, we could identify common pulsotypes among isolates obtained from different patients during different time periods of the study (P5, P27, P48 and P106).
We also used PFGE analysis for revealing the genetic association among CRKP strains from rectal and clinical samples of 48 representative patients. According to PFGE, in the majority of the cases (81.3%, 39 of 48), the clinical and rectal strains from the same patients were identical (Figure 2). Different pulsotypes were observed for pairs PAT_1422a/b, PAT_1386a/b, PAT_854a/b, PAT_476a/b, PAT_326a/b, PAT_1529a/b, PAT_735a/b, PAT_191a/b and PAT_1216a/b.
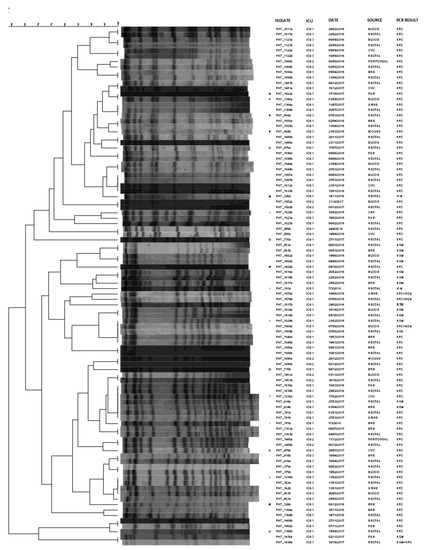
Figure 2.
Pulsotypes of the 48 rectal-clinical carbapenemase-producing K. pneumoniae isolate pairs. Pairs with different pulsotypes are marked with the same symbol. PAT: patient; ICU: intensive care unit; CVC: central venous catheter; BRS: bronchial secretions; CSF: cerebrospinal fluid.
Six clinical-rectal surveillance pairs presented different PCR results (PAT_1017a/b, PAT_1436a/b, PAT_1529a/b, PAT_1569a/b, PAT_191a/b and PAT_326a/b). Among them, PAT_1017a/b, PAT_1529a/b, PAT_191a/b and PAT_326a/b showed also different PFGE profiles. Overall, different pulsotypes were observed for pairs PAT_1422a/b, PAT_1386a/b, PAT_854a/b, PAT_476a/b, PAT_1529a/b, PAT_735a/b, PAT_191a/b and PAT_1216a/b even though the PCR results were identical among the clinical and the rectal samples.
2.3. Non-Susceptibility Rates of CRKP Isolates in the Hospital
The non-susceptibility rates of CRKP isolates in our institution’s ICUs for amikacin, aztreonam, colistin, fosfomycin, gentamicin, piperacillin/tazobactam and tigecycline in isolates during the study period are shown in Table 1 and Figure 3. CRKP isolates presented with high level of resistance to both meropenem and imipenem (MIC50 ≥ 16 mg/L) throughout the study years. On the other hand, a significant increase in resistance was observed for gentamycin ranging from 15% in the first semester of 2016 to 66.7% in the first semester of 2019. In regard to tigecycline, an increase was also observed (23.5% in the 2016a semester to 61.1% in the 2019a semester). As for colistin, an increase was observed (15.8% in the 2016a semester to 27.8% in the 2019a semester).

Table 1.
Imipenem resistant K. pneumoniae susceptibility rates per semester among single patient isolates recovered from the hospital’s ICUs during the study period.
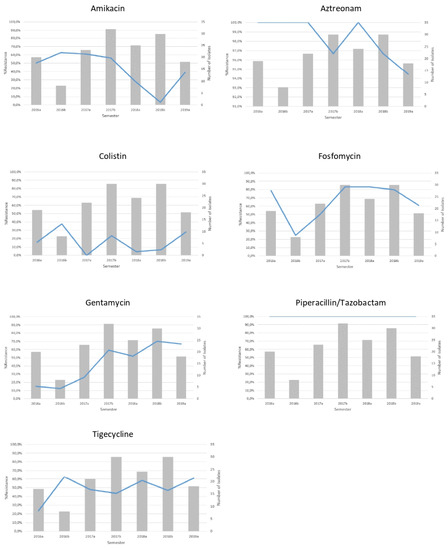
Figure 3.
Non-susceptibility rates (line) and number (bars) of single patient imipenem non-susceptible K. pneumoniae isolates recovered from the hospital’s ICUs per semester.
2.4. Systematic Review Results
Our systematic review search strategy yielded 290 results. After implementation of the exclusion criteria, 39 studies remained. Fourteen studies reported monoclonal dissemination. Data extracted from the remaining 25 studies are reported in Table 2.

Table 2.
Studies reporting non-monoclonal dissemination of carbapenem-resistant Klebsiella pneumoniae clinical strains, including ICU populations.
3. Discussion
The present study evaluated the type of carbapenemases and the molecular epidemiology of K. pneumoniae strains circulating in the ICUs of a tertiary hospital in Thessaloniki, Greece between 2016 and 2019. During the study years, KPC was the predominant carbapenemase; NDM were also present, and a few double-carbapenemase-producers were isolated. On the basis of PFGE, a total of 17 different CRKP transmission clusters were identified.
CRKP isolates have been introduced in our hospital since 2004. At that time, no phenotypic or molecular testing was performed to reveal the type of carbapenemase up to 2010. From 2010 to 2014, phenotypic testing among CRKP isolates revealed that the majority carried KPC enzymes (60%), 25% produced MBL, and 7% co-produced KPC and MBL enzymes. In the time period of 2011–2014, we also observed the first OXA-48 producers in our hospital at a rate of 1.7%. Molecular testing showed that KPC positive strains harbored the blaKPC-2, while MBL-positive strains harbored blaVIM-1.
During 2013–2015, an oligoclonal outbreak caused by 45 CRKP occurred in our hospital [40]. All the patients were hospitalized in the three intensive care units of the hospital, and 17 (68%) of them developed bloodstream infections; the overall mortality of the patients involved in the outbreak was 48% (12/25). Molecular testing verified that all 45 K. pneumoniae isolates co-harbored blaKPC-2 and blaVIM-1 and were associated with OmpK35 deficiency and OmpK36 porin loss. PFGE clustered all isolates into a single clonal type, and multi-locus sequence typing (MLST) assigned them to the emerging high-risk ST147 clonal lineage.
Starting from 2014 until 2016, while KPC producers still prevailed (approximately 75% of the CRKP), we observed a shift among MBL producers from blaVIM-1 towards blaNDM-1. More specifically, VIM-type carbapenemases decreased from 17.6% in 2014 to 6.7% in 2016, whereas NDM-type increased from 1.2% in 2014 to 19.4% in 2016 [41]. In the years 2013–2015, the co-production of KPC and MBL enzymes corresponded to approximately 10% of all CRKP strains [35]. Finally, in 2019, we isolated a strain carrying both NDM-1 and OXA-48 genes classified as ST-11 [42].
In our study, PFGE analysis identified 128 distinguishable pulsotypes and 17 clusters indicating an extended dissemination of CRKP within the hospital setting. Moreover, the dissemination took place over a long-time frame since we included in our study isolates recovered during a 3.5-year period. The presence of identical isolates in all three ICUs highlights their successful dissemination through different hospital wards. More worryingly, the persistence of certain strains throughout the whole study period, despite the various infection control measures that were applied, reflects the difficulties that undermine the efforts for their eradication once they are well-established in a certain geographical area. Indeed, such strains may have persisted in the hospital or/and may have been re-introduced by carrier admissions.
In fact, most of the CRKP carriers that later presented a CRKP infection had identical pulsotypes between their rectal and clinical isolates. The most probable explanation for this finding is that gut colonization preceded infection. In some cases, however, the pulsotypes of rectal and clinical isolates were different indicating that the infection was caused by another nosocomial K. pneumoniae.
There were also two clinical-rectal pairs (PAT_1436a/b and PAT_1569a/b) that harbored different carbapenemase-encoding genes according to PCR results but had identical PFGE pulsotypes. This could be explained by the mobilization of mobile genetic elements, most likely by the loss and acquisition of plasmids [43] even though an infection by a different isolate of the same pulsotype harboring different resistance determinants could not be excluded. PFGE studies in our hospital performed in several CRKP strains during the period of 2011 to 2013 revealed that KPC strains prevailed and that the majority of them belonged to two distinct clones (unpublished data).
A similar pattern of carbapenemases was observed in Hippokration General Hospital of Thessaloniki, Greece where KPC carbapenemases have prevailed among CRKP since 2009 outnumbering the VIM-type carbapenemases that predominated previously [44]. In the same report, KPC-producers belonged to two distinct clones, the predominant of which correspond to the hyperepidemic Greek clone. In a multicenter nationwide surveillance study conducted in several Greek hospitals for CRKP from 2014 to 2016, NDM-producing isolates belonged mainly to one clone, whereas KPC, VIM, OXA-48 and double carbapenemase-producers were mainly categorized in three clones [45]. On the contrary, in our study, both KPC and NDM CRKP isolates showed extremely multiclonal profiles.
In our study, we also observed a rise in the resistance rates of tigecycline, gentamicin and colistin. This is in accordance with other studies reporting elevated resistance rates to last resort antibiotics driven, among other factors, by a vicious cycle of increased last resort antibiotic consumption and subsequent resistance [46].
Our systematic review results showed that non-monoclonal dissemination of K. pneumoniae strains in ICU settings has been described before in countries participating in the EARS-Net (Table 2). Of note, such observations have been reported almost exclusively by Mediterranean countries, mainly from Greece and Italy, and this is in accordance with the epidemiological situation of the region regarding carbapenem-resistance determinants. However, and despite the heterogeneity of settings and methods used, most studies reported rather oligoclonal transmission with further identification of sporadic cases.
In our study, multiple clones circulating simultaneously achieved sustainable dissemination and, according to our knowledge and our systematic review results, this is the first time that such polyclonal dissemination has been observed in Europe. This multi-clonal PFGE observation highlights a possible additional reason for their endemic persistence in our hospital even though infection control measures, including hand hygiene, surveillance for colonization among high-risk patients and contact precautions have been established.
In this context, active surveillance with rectal swab cultures is of outmost importance to control the spread of these pathogens by isolation or cohorting of the colonized patients [29]. However, the spread of CRKP in an endemic environmental niche is a dynamic and multifaceted phenomenon that involves many variables. In a similar situation, more than one CRKP clone may be simultaneously present in the hospital; whereas new admissions may be CRKP carriers most likely by previous hospitalizations in the same or other hospitals. Consequently, a multi-clonal spread is very likely to occur and, even when a previously colonized subject presents a CRKP infection, it is not certain that this infection is directly related to the strain that colonized the patient upon admission.
Our study has several limitations. A multi-centric study would be able to evaluate whether the epidemiological pattern that we observed in our single center study was an isolated phenomenon or more widespread. Including non-ICU along with ICU K. pneumoniae strains would yield a more complete picture for their dissemination. In our analysis, we did not include clinical patient level information, and this limits our ability to draw conclusions with regard to precipitating factors. Finally, we were not able to employ sequencing-based methods to better characterize the molecular epidemiology of the strains included in our study.
4. Materials and Methods
4.1. Study Design
This was a retrospective study that was carried out at AHEPA University Hospital, a 700-bed institution with three ICUs, a central surgical and medical ICU (8 beds, ICU 1), a surgical ICU (4 beds, ICU 2) and a cardiosurgical ICU (5 beds, ICU 3) as well as surgical and internal medicine departments. The study was approved by the institutional medical scientific board. Sample related patient data were retrieved from the laboratory database.
CRKP clinical isolates, recovered in the aforementioned ICUs between January 2016 and June 2019 from 165 single patients, were included in the study. In 48 cases, a rectal isolate (isolated upon admission in ICU for infection control purposes) and a subsequent clinical isolate (isolated by an infection that occurred during ICU stay) were considered, thus forming 48 pairs of surveillance-clinical isolates. Isolates taken from the remaining 117 patients were all recovered from clinical specimens only.
Rectal swabs taken from ICU patients upon admission were inoculated on MacConkey agar plates supplemented with meropenem and ceftazidime discs. All Gram-negative colonies that grew after 24 h of incubation near the discs were further identified and K. pneumoniae isolates were tested for carbapenemase production with phenotypic and molecular techniques. For clinical specimens, standard laboratory procedures were followed depending on each specimen source.
Bacterial identification and antimicrobial susceptibility testing were performed with the Vitek2 automated system (Biomerieux, Marcy-l’Étoile, France). Furthermore, the minimum inhibitory concentration of tigecycline was determined by E-test (Liofilchem, Roseto degli Abruzzi, Italy) and for colistin using the broth microdilution method (Liofilchem, Roseto degli Abruzzi, Italy). The results of all antimicrobial testing were interpreted in accordance with the CLSI criteria. For tigecycline, the breakpoints recommended by the United States Food and Drug Administration were used (susceptible: MIC ≤ 2 mg/L; resistant: MIC ≥ 8 mg/L).
4.2. Carbapenemase Detection
All isolates were phenotypically screened for carbapenemase production with the Modified Hodge Test (MHT), [47] while the type of MBL or KPC was assessed with the Combined Disk Test (CDT) [48]. Following phenotypic identification, PCR assays were performed for carbapenemase-encoding genes using specific primers for blaKPC, blaVIM, blaIMP, blaNDM and blaOXA-48 (Appendix A) [49].
4.3. Pulse-Field Gel Electrophoresis
The genetic relationship among the CPKP isolates was determined by PFGE according to standardized protocol [50] with the XbaI endonuclease (New England Biolabs, Beverly, MA, USA) by using a CHEF-DR III apparatus (Bio-Rad Laboratories Inc., Hercules, CA, USA) for the separation of DNA fragments. XbaI-digested DNA from Salmonella enterica serotype Braenderup H9812 was used as a reference size standard, while PFGE patterns were digitally analyzed using the FPQuest (Bio-Rad Laboratories Pty Ltd., Hercules, CA, USA) software package.
PFGE profiles were compared using the Dice correlation coefficient with a maximum position tolerance of 1.5% and an optimization of 1.5%. Similarity clustering analysis was performed by using the Unweighted Pair Group Method using Averages (UPGMA), and a dendrogram was generated. Two PFGE profiles were classified as indistinguishable if the DNA fragment patterns matched each other completely, while clusters were selected using a cutoff at the 80% level of genetic similarity.
4.4. Non-Susceptibility Rates of CRKP Isolates
For every semester of the study period, we determined the K. pneumoniae non-susceptibility rates of amikacin, aztreonam, colistin, fosfomycin, gentamicin, piperacillin/tazobactam and tigecycline in imipenem non-susceptible single patient isolates from the ICUs using the CLSI 2020 breakpoints.
4.5. Systematic Review
In order to evaluate the extent of non-monoclonal transmission of CRKP strains in the intensive care environment in countries participating in the European Antimicrobial Resistance Surveillance Network (EARS-Net), we undertook a systematic review of the recent literature. We searched MEDLINE via PubMed from 1 January 2000 to 28 April 2021, implementing the search strategy described in Appendix B. Titles and abstracts were screened for studies, which incorporated a molecular epidemiology investigation (PFGE or sequencing methods) of CRKP strains, including samples obtained from ICU patients.
We excluded reviews, case reports, studies restricted to environmental samples, studies that did not explicitly include any ICU clinical samples and studies not conducted in EARS-Net participating countries. No language and patient age restrictions were applied. Eligibility assessment was conducted in duplicate (D.P. and G.M.) and discrepancies were adjudicated by a third author (E.P.).
Next, studies reporting monoclonal transmission were excluded, and, from the remaining studies, we extracted the following data: study setting and eligible samples, sample type (clinical versus surveillance and infection versus colonization), the number of carbapenem-resistant isolates and mechanism of resistance detected, the method used to investigate molecular epidemiology and the number and size of clusters involving ICU patients. If ICU-level data were not available, hospital-wide data were reported. Data extraction was conducted by A.T., G.M., T.P., D.P. and E.P. All steps were conducted in duplicate.
5. Conclusions
In our study, we demonstrated that CPKP in our hospital belonged to a great variety of pulsotypes and clusters, thus, indicating their extended dissemination within the hospital ICU settings. Among them, KPC carbapenemase predominated. The presence of multiple clones harboring variable resistance-determinants poses additional challenges. Further studies are required to identify suitable infection control strategies in a setting of polyclonal dissemination within a context of carbapenem resistance endemicity. Our results highlight the need to urgently reinforce infection-control measures along with antimicrobial stewardship and together with a generous increase in the nosocomial budget in order to contain the transmission of antibiotic resistant organisms.
Author Contributions
Conceptualization, E.P. and L.S.; methodology, E.P., G.M., D.P. (Dimitrios Pilalas), C.K. and T.P.; investigation, E.P., P.M., A.T., D.P. (Dimitra Papadopoulou) C.K., T.P., M.P. and S.M.; resources, D.P. (Dimitra Papadopoulou) and C.K.; software, G.M., D.P. (Dimitrios Pilalas), C.K. and M.P.; validation, E.P., G.M., C.K., formal analysis, G.M., D.P. (Dimitrios Pilala) and M.P.; data curation, E.P., G.M., D.P. (Dimitrios Pilalas), A.T., D.P. (Dimitra Papadopoulou), C.K., M.P.; writing-original draft preparation, E.P., G.M., D.P. (Dimitris Pilalas), A.T., C.K.; writing—review and editing,, E.P., G.M., D.P. (Dimitrios Pilalas), C.K., T.P., L.S.; visualization, E.P., G.M., D.P. (Dimitrios Pilalas) and L.S.; supervision, E.P. and L.S.; project administration, E.P. and L.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We wish to express our gratitude to the laboratory technicians of the Microbiology Laboratory for the performance of the phenotypic and antimicrobial susceptibility tests. We are also grateful to the nurses of the Infection Control Unit of AHEPA Hospital for assistance in collecting the rectal samples.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A. Primers for Carbapenemase Detection
| Gene | Primers (5′–3′) |
| blaKPC | TGTCACTGTATCGCCGTC |
| TATTTTTCCGAGATGGGTGAC | |
| blaIMP | CTACCGCAGCAGAGTCTTTG |
| AACCAGTTTTGCCTTACCAT | |
| blaVIM | TCTACATGACCGCGTCTGTC |
| TGTGCTTTGACAACGTTCGC | |
| blaNDM-1 | GGTTTGGCGATCTGGTTTTC |
| CGGAATGGCTCATCACGATC | |
| blaOXA-48 | TTGGTGGCATCGATTATCGG |
| GAGCACTTCTTTTGTGATGGC |
Appendix B. Systematic Review Search Strategy
- #1
- (critical care OR ICU OR intensive care OR critical ill OR critical illness OR critically ill OR “Intensive Care Units”[Mesh] OR “Critical Care”[Mesh] OR “Critical Illness”[Mesh])
- #2
- carbapenem* OR meropenem OR imipenem OR ertapenem
- #3
- Klebsiella
- #4
- (epidem* OR outbreak OR clon* OR strain*)
- #5
- (PFGE OR puls* OR genom* OR typing OR sequenc* OR MLST OR NGS OR WGS OR cgMLST or wgMLST OR MLVA)
- #6
- (“1 January 2000”[Date–Publication]: “28 April 2021”[Date–Publication])
Final search(29 April 2021): #1 AND #2 AND #3 AND #4 AND #5 AND #6 → 290 results
References
- Falcone, M.; Russo, A.; Iacovelli, A.; Restuccia, G.; Ceccarelli, G.; Giordano, A.; Farcomeni, A.; Morelli, A.; Venditti, M. Predictors of outcome in ICU patients with septic shock caused by Klebsiella pneumoniae carbapenemase–producing K. pneumoniae. Clin. Microbiol. Infect. 2016, 22, 444–450. [Google Scholar] [CrossRef] [PubMed]
- Palacios-Baena, Z.R.; Oteo, J.; Conejo, C.; Larrosa, M.N.; Bou, G.; Fernández-Martínez, M.; Oliver, A.; Zamorano, L.; Alba Rivera, M.; Bautista, V. Comprehensive clinical and epide-miological assessment of colonisation andinfection due to carbapenemase-producing Enterobacteriaceae in Spain. J. Infect. 2016, 72, 152–160. [Google Scholar] [CrossRef] [PubMed]
- WHO Regional Office for Europe and European Centre for Disease Prevention and Control. Surveillance of Antimicrobial Resistance in Europe, 2020 Data. Executive Summary; WHO Regional Office for Europe: Copenhagen, Denmark, 2021. [Google Scholar]
- World Health Organization; Regional Office for Europe. Central Asian and European Surveillance of Antimicrobial Resistance: Annual Report 2020; WHO Regional Office for Europe: Copenhagen, Denmark, 2020. [Google Scholar]
- Queenan, A.M.; Bush, K. Carbapenemases: The Versatile Beta-Lactamases. Clin. Microbiol. Rev. 2007, 20, 440–458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karampatakis, T.; Antachopoulos, C.; Iosifidis, E.; Tsakris, A.; Roilides, E. Molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae in Greece. Futur. Microbiol. 2016, 11, 809–823. [Google Scholar] [CrossRef]
- Giakkoupi, P.; Papagiannitsis, C.C.; Miriagou, V.; Pappa, O.; Polemis, M.; Tryfinopoulou, K.; Tzouvelekis, L.S.; Vatopoulos, A.C. An update of the evolving epidemic of blaKPC-2-carrying Klebsiella pneumoniae in Greece (2009–2010). J. Antimicrob. Chemother. 2010, 66, 1510–1513. [Google Scholar] [CrossRef]
- Pournaras, S.; Protonotariou, E.; Voulgari, E.; Kristo, I.; Dimitroulia, E.; Vitti, D.; Tsalidou, M.; Maniatis, A.N.; Tsakris, A.; Sofianou, D. Clonal spread of KPC-2 carbapenemase producing Klebsiella pneumoniae strains in Greece. J. Antimicrob. Chemother. 2009, 64, 348–352. [Google Scholar] [CrossRef]
- Voulgari, E.; Gartzonika, C.; Vrioni, G.; Politi, L.; Priavali, E.; Levidiotou-Stefanou, S.; Tsakris, A. The Balkan region: NDM-1-producing Klebsiella pneumoniae ST11 clonal strain causing outbreaks in Greece. J. Antimicrob. Chemother. 2014, 69, 2091–2097. [Google Scholar] [CrossRef]
- Voulgari, E.; Zarkotou, O.; Ranellou, K.; Karageorgopoulos, D.E.; Vrioni, G.; Mamali, V.; Themeli-Digalaki, K.; Tsakris, A. Outbreak of OXA-48 carbapenemase-producing Klebsiella pneumoniae in Greece involving an ST11 clone. J. Antimicrob. Chemother. 2012, 68. [Google Scholar] [CrossRef] [Green Version]
- Meletis, G. Carbapenem resistance: Overview of the problem and future perspectives. Ther. Adv. Infect. Dis. 2015, 3, 15–21. [Google Scholar] [CrossRef] [Green Version]
- Wyres, K.; Holt, K. Klebsiella pneumoniae as a key trafficker of drug resistance genes from environmental to clinically important bacteria. Curr. Opin. Microbiol. 2018, 45, 131–139. [Google Scholar] [CrossRef]
- Chang, D.; Sharma, L.; Cruz, C.S.D.; Zhang, D. Clinical Epidemiology, Risk Factors, and Control Strategies of Klebsiella pneumoniae Infection. Front. Microbiol. 2021, 12, 750662. [Google Scholar] [CrossRef] [PubMed]
- Mo, Y.; Hernandez-Koutoucheva, A.; Musicha, P.; Bertrand, D.; Lye, D.; Ng, O.T.; Fenlon, S.N.; Chen, S.L.; Ling, M.L.; Tang, W.Y.; et al. Duration of Carbapenemase-Producing Enterobacteriaceae Carriage in Hospital Patients. Emerg. Infect. Dis. 2020, 26, 2182–2185. [Google Scholar] [CrossRef] [PubMed]
- Hernández-García, M.; García-Fernández, S.; García-Castillo, M.; Melo-Cristino, J.; Pinto, M.F.; Gonçalves, E.; Alves, V.; Costa, E.; Ramalheira, E.; Sancho, L.; et al. Confronting Ceftolozane-Tazobactam Susceptibility in Multidrug-Resistant Enterobacterales Isolates and Whole-Genome Sequencing Results (STEP Study). Int. J. Antimicrob. Agents 2020, 57, 106259. [Google Scholar] [CrossRef] [PubMed]
- Fontana, C.; Angeletti, S.; Mirandola, W.; Cella, E.; Alessia, L.; Zehender, G.; Favaro, M.; Leoni, D.; Rose, D.D.; Gherardi, G.; et al. Whole genome sequencing of carbapenem-resistant Klebsiella pneumoniae: Evolutionary analysis for outbreak investigation. Future Microbiol. 2020, 15, 203–212. [Google Scholar] [CrossRef]
- Galani, I.; Karaiskos, I.; Souli, M.; Papoutsaki, V.; Galani, L.; Gkoufa, A.; Antoniadou, A.; Giamarellou, H. Outbreak of KPC-2-producing Klebsiella pneumoniae endowed with ceftazidime-avibactam resistance mediated through a VEB-1-mutant (VEB-25), Greece, September to October 2019. Eurosurveillance 2020, 25, 2000028. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, C.; Corbella, M.; Gaiarsa, S.; Comandatore, F.; Scaltriti, E.; Bandi, C.; Cambieri, P.; Marone, P.; Sassera, D. Multiple Klebsiella pneumoniae KPC Clones Contribute to an Extended Hospital Outbreak. Front. Microbiol. 2019, 10, 2767. [Google Scholar] [CrossRef]
- Mavroidi, A.; Katsiari, M.; Likousi, S.; Palla, E.; Roussou, Z.; Nikolaou, C.; Mathas, C.; Merkouri, E.; Platsouka, E.D. Changing Characteristics and In Vitro Susceptibility to Ceftazidime/Avibactam of Bloodstream Extensively Drug-Resistant Klebsiella pneumoniae from a Greek Intensive Care Unit. Microb. Drug Resist. 2020, 26, 28–37. [Google Scholar] [CrossRef]
- Gona, F.; Bongiorno, D.; Aprile, A.; Corazza, E.; Pasqua, B.; Scuderi, M.G.; Chiacchiaretta, M.; Cirillo, D.M.; Stefani, S.; Mezzatesta, M.L. Emergence of two novel sequence types (3366 and 3367) NDM-1- and OXA-48-co-producing K. pneumoniae in Italy. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 1687–1691. [Google Scholar] [CrossRef]
- Karampatakis, T.; Tsergouli, K.; Politi, L.; Diamantopoulou, G.; Iosifidis, E.; Antachopoulos, C.; Karyoti, A.; Mouloudi, E.; Tsakris, A.; Roilides, E. Molecular Epidemiology of Endemic Carbapenem-Resistant Gram-Negative Bacteria in an Intensive Care Unit. Microb. Drug Resist. 2019, 25, 712–716. [Google Scholar] [CrossRef]
- Papadimitriou-Olivgeris, M.; Bartzavali, C.; Spyropoulou, A.; Lambropoulou, A.; Sioulas, N.; Vamvakopoulou, S.; Karpetas, G.; Spiliopoulou, I.; Vrettos, T.; Anastassiou, E.D.; et al. Molecular epidemiology and risk factors for colistin- or tigecycline-resistant carbapenemase-producing Klebsiella pneumoniae bloodstream infection in critically ill patients during a 7-year period. Diagn. Microbiol. Infect. Dis. 2018, 92, 235–240. [Google Scholar] [CrossRef]
- Avgoulea, K.; Di Pilato, V.; Zarkotou, O.; Sennati, S.; Politi, L.; Cannatelli, A.; Themeli-Digalaki, K.; Giani, T.; Tsakris, A.; Rossolini, G.M.; et al. Characterization of Extensively Drug-Resistant or Pandrug-Resistant Sequence Type 147 and 101 OXA-48-Producing Klebsiella pneumoniae Causing Bloodstream Infections in Patients in an Intensive Care Unit. Antimicrob. Agents Chemother. 2018, 62, e02457-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ripabelli, G.; Tamburro, M.; Guerrizio, G.; Fanelli, I.; Flocco, R.; Scutellà, M.; Sammarco, M.L. Tracking Multidrug-Resistant Klebsiella pneumoniae from an Italian Hospital: Molecular Epidemiology and Surveillance by PFGE, RAPD and PCR-Based Resistance Genes Prevalence. Curr. Microbiol. 2018, 75, 977–987. [Google Scholar] [CrossRef] [PubMed]
- Bartolini, A.; Basso, M.; Franchin, E.; Menegotto, N.; Ferrari, A.; De Canale, E.; Andreis, S.; Scaggiante, R.; Stefani, S.; Palù, G.; et al. Prevalence, molecular epidemiology and intra-hospital acquisition of Klebsiella pneumoniae strains producing carbapenemases in an Italian teaching hospital from January 2015 to September 2016. Int. J. Infect. Dis. 2017, 59, 103–109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mavroidi, A.; Katsiari, M.; Likousi, S.; Palla, E.; Roussou, Z.; Nikolaou, C.; Maguina, A.; Platsouka, E.D. Characterization of ST258 Colistin-Resistant, blaKPC-Producing Klebsiella pneumoniae in a Greek Hospital. Microb. Drug Resist. 2016, 22, 392–398. [Google Scholar] [CrossRef] [PubMed]
- Bonura, C.; Giuffrè, M.; Aleo, A.; Fasciana, T.; Di Bernardo, F.; Stampone, T.; Giammanco, A.; Palma, D.M.; Mammina, C. The MDR-GN Working Group an Update of the Evolving Epidemic of blaKPC Carrying Klebsiella pneumoniae in Sicily, Italy, 2014: Emergence of Multiple Non-ST258 Clones. PLoS ONE 2015, 10, e0132936. [Google Scholar] [CrossRef] [Green Version]
- Onori, R.; Gaiarsa, S.; Comandatore, F.; Pongolini, S.; Brisse, S.; Colombo, A.; Cassani, G.; Marone, P.; Grossi, P.; Minoja, G.; et al. Tracking Nosocomial Klebsiella pneumoniae Infections and Outbreaks by Whole-Genome Analysis: Small-Scale Italian Scenario within a Single Hospital. J. Clin. Microbiol. 2015, 53, 2861–2868. [Google Scholar] [CrossRef] [Green Version]
- Parisi, S.G.; Bartolini, A.; Santacatterina, E.; Castellani, E.; Ghirardo, R.; Berto, A.; Franchin, E.; Menegotto, N.; De Canale, E.; Tommasini, T.; et al. Prevalence of Klebsiella pneumoniae strains producing carbapenemases and increase of resistance to colistin in an Italian teaching hospital from January 2012 to December 2014. BMC Infect. Dis. 2015, 15, 244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katsiari, M.; Panagiota, G.; Likousi, S.; Roussou, Z.; Polemis, M.; Vatopoulos, C.A.; Platsouka, D.E.; Maguina, A. Carbapenem-resistant Klebsiella pneumoniae infections in a Greek intensive care unit: Molecular characterisation and treatment challenges. J. Glob. Antimicrob. Resist. 2015, 3, 123–127. [Google Scholar] [CrossRef]
- Mezzatesta, M.L.; Caio, C.; Gona, F.; Cormaci, R.; Salerno, I.; Zingali, T.; Denaro, C.; Gennaro, M.; Quattrone, C.; Stefani, S. Carbapenem and multidrug resistance in Gram-negative bacteria in a single centre in Italy: Considerations on in vitro assay of active drugs. Int. J. Antimicrob. Agents 2014, 44, 112–116. [Google Scholar] [CrossRef]
- Papadimitriou-Olivgeris, M.; Marangos, M.; Christofidou, M.; Fligou, F.; Bartzavali, C.; Panteli, E.S.; Vamvakopoulou, S.; Filos, K.S.; Anastassiou, E.D. Risk factors for infection and predictors of mortality among patients with KPC-producing Klebsiella pneumoniae bloodstream infections in the intensive care unit. Scand. J. Infect. Dis. 2014, 46, 642–648. [Google Scholar] [CrossRef]
- Capone, A.; Giannella, M.; Fortini, D.; Giordano, A.; Meledandri, M.; Ballardini, M.; Venditti, C.; Bordi, E.; Capozzi, D.; Balice, M.P.; et al. High rate of colistin resistance among patients with carbapenem-resistant Klebsiella pneumoniae infection accounts for an excess of mortality. Clin. Microbiol. Infect. 2013, 19, E23–E30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tofteland, S.; Naseer, U.; Lislevand, J.H.; Sundsfjord, A.; Samuelsen, Ø. A Long-Term Low-Frequency Hospital Outbreak of KPC-Producing Klebsiella pneumoniae Involving Intergenus Plasmid Diffusion and a Persisting Environmental Reservoir. PLoS ONE 2013, 8, e59015. [Google Scholar] [CrossRef] [Green Version]
- Mammina, C.; Bonura, C.; Di Bernardo, F.; Aleo, A.; Fasciana, T.; Sodano, C.; Saporito, M.A.; Verde, M.S.; Tetamo, R.; Palma, D.M. Ongoing spread of colistin-resistant Klebsiella pneumoniae in different wards of an acute general hospital, Italy, June to December 2011. Euro Surveill. 2012, 17, 20248. [Google Scholar] [CrossRef] [PubMed]
- Richter, S.N.; Frasson, I.; Franchin, E.; Bergo, C.; Lavezzo, E.; Barzon, L.; Cavallaro, A.; Palù, G. KPC-mediated resistance in Klebsiella pneumoniae in two hospitals in Padua, Italy, June 2009-December 2011: Massive spreading of a KPC-3-encoding plasmid and involvement of non-intensive care units. Gut Pathog. 2012, 4, 7. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Romero, I.; Asensio, Á.; Oteo, J.; Muñoz-Algarra, M.; Isidoro, B.; Vindel, A.; Cuevas, O.; Fernández-Romero, S.; Azañedo, L.; Campos, J.; et al. Nosocomial outbreak of VIM-1-producing Klebsiella pneumoniae isolates of multilocus sequence type 15: Molecular basis, clinical risk factors, and outcome. Antimicrob. Agents Chemother. 2012, 56, 420–427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Souli, M.; Galani, I.; Antoniadou, A.; Papadomichelakis, E.; Poulakou, G.; Panagea, T.; Vourli, S.; Zerva, L.; Armaganidis, A.; Kanellakopoulou, K.; et al. An Outbreak of Infection due to β-Lactamase Klebsiella pneumoniae Carbapenemase 2–ProducingK. pneumoniaein a Greek University Hospital: Molecular Characterization, Epidemiology, and Outcomes. Clin. Infect. Dis. 2010, 50, 364–373. [Google Scholar] [CrossRef] [Green Version]
- Giakkoupi, P.; Xanthaki, A.; Kanelopoulou, M.; Vlahaki, A.; Miriagou, V.; Kontou, S.; Papafraggas, E.; Malamou-Lada, H.; Tzouvelekis, L.S.; Legakis, N.J.; et al. VIM-1 Metallo-β-Lactamase-Producing Klebsiella pneumoniae Strains in Greek Hospitals. J. Clin. Microbiol. 2003, 41, 3893–3896. [Google Scholar] [CrossRef] [Green Version]
- Protonotariou, E.; Poulou, A.; Politi, L.; Sgouropoulos, I.; Metallidis, S.; Kachrimanidou, M.; Pournaras, S.; Tsakris, A.; Skoura, L. Hospital outbreak due to a Klebsiella pneumoniae ST147 clonal strain co-producing KPC-2 and VIM-1 carbapenemases in a tertiary teaching hospital in Northern Greece. Int. J. Antimicrob. Agents 2018, 52, 331–337. [Google Scholar] [CrossRef]
- Protonotariou, E.; Poulou, A.; Vasilaki, O.; Papadopoulou, D.; Politi, L.; Kagkalou, G.; Pilalas, D.; Kachrimanidou, M.; Draganoudis, V.; Tsocha, A.; et al. Emergence of NDM-1 producing Klebsiella pneumoniae clinical isolates in northern Greece: A 34-month epidemiological study in a tertiary university hospital (P0270). In Proceedings of the 27th European Congress of Clinical Microbiology and Infectious Diseases, ECCMID 2017, Vienna, Austria, 22–25 April 2017. [Google Scholar]
- Protonotariou, E.; Meletis, G.; Chatzopoulou, F.; Malousi, A.; Chatzidimitriou, D.; Skoura, L. Emergence of Klebsiella pneumoniae ST11 co-producing NDM-1 and OXA-48 carbapenemases in Greece. J. Glob. Antimicrob. Resist. 2019, 19, 81–82. [Google Scholar] [CrossRef]
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile Genetic Elements Associated with Antimicrobial Resistance. Clin. Microbiol. Rev. 2018, 31, e00088-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zagorianou, A.; Sianou, E.; Iosifidis, E.; Dimou, V.; Protonotariou, E.; Miyakis, S.; Roilides, E.; Sofianou, D. Microbiological and molecular characteristics of carbapenemase-producing Klebsiella pneumoniae endemic in a tertiary Greek hospital during 2004–2010. Euro Surveill. 2012, 17, 20088. [Google Scholar] [CrossRef]
- Galani, I.; Karaiskos, I.; Karantani, I.; Papoutsaki, V.; Maraki, S.; Papaioannou, V.; Kazila, P.; Tsorlini, H.; Charalampaki, N.; Toutouza, M.; et al. Epidemiology and resistance phenotypes of carbapenemase-producing Klebsiella pneumoniae in Greece, 2014 to 2016. Eurosurveillance 2018, 23, 1700775. [Google Scholar] [CrossRef] [Green Version]
- Ghenea, A.; Cioboată, R.; Drocaş, A.; Țieranu, E.; Vasile, C.; Moroşanu, A.; Țieranu, C.; Salan, A.-I.; Popescu, M.; Turculeanu, A.; et al. Prevalence and Antimicrobial Resistance of Klebsiella Strains Isolated from a County Hospital in Romania. Antibiotics 2021, 10, 868. [Google Scholar] [CrossRef]
- Wayne, P.A. Clinical and Laboratory Standards Institute. Performance Standards for antimicrobial susceptibility testing; twenty-fifth in-formational supplement. Document M100-S25. Inform. Suppl. 2011, 31, 100–121. [Google Scholar]
- Tsakris, A.; Poulou, A.; Pournaras, S.; Voulgari, E.; Vrioni, G.; Themeli-Digalaki, K.; Petropoulou, D.; Sofianou, D. A simple phenotypic method for the differentiation of metallo- b -lactamases and class A KPC carbapenemases in Enterobacteriaceae clinical isolates. J. Antibicrob. Chemother. 2010, 65, 1664–1671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meletis, G.; Tzampaz, E.; Sianou, E. Phenotypic and molecular methods for the detection of antibiotic resistance mechanisms in Gram negative nosocomial pathogens. In Trends in Infectious Diseases; IntechOpen: London, UK, 2014; pp. 139–162. [Google Scholar]
- Han, H.; Zhou, H.; Li, H.; Gao, Y.; Lu, Z.; Hu, K.; Xu, B. Optimization of Pulse-Field Gel Electrophoresis for Subtyping of Klebsiella pneumoniae. Int. J. Environ. Res. Public Health 2013, 10, 2720–2731. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).