Feature Papers in Assessing Environmental Health and Function
Share This Topical Collection
Editor
 Dr. David J. Beale
Dr. David J. Beale
 Dr. David J. Beale
Dr. David J. Beale
E-Mail
Website
Collection Editor
Environmental Metabolomics and Proteomics, Land & Water, Commonwealth Scientific and Industrial Research Organization (CSIRO), Ecoscience Precinct, Dutton Park, QLD 4160, Australia
Interests: metabolomics; environmental multi-omics; pesticide analysis; systems biology; environmental science
Special Issues, Collections and Topics in MDPI journals
Topical Collection Information
Dear Colleagues,
The emergence of novel surveillance techniques and environmental monitoring approaches that utilize metabolomics (and other omics technologies) alongside traditional methodologies is increasing. While traditional environmental and organism health monitoring techniques (e.g., chemical monitoring and bioassays) are highly suited to acute exposure, they are less able to detect subtle shifts in ecosystem function and health. Metabolomics-based approaches have the potential to provide deeper insight into these complex interactions between biological systems and the impacts resulting from anthropogenic environmental changes.
This Special Collection on assessing environmental health and function aims to attract high-quality manuscripts in the field of environmental metabolomics. We encourage researchers from all domains of environmental metabolomics research, integrated with traditional monitoring data and/or other omics datasets (i.e., multi-omics approaches) to contribute manuscripts highlighting novel approaches to ecosystem surveillance.
Dr. David J. Beale
Guest Editor
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 250 words) can be sent to the Editorial Office for assessment.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Metabolites is an international peer-reviewed open access monthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript.
The Article Processing Charge (APC) for publication in this open access journal is 2700 CHF (Swiss Francs).
Submitted papers should be well formatted and use good English. Authors may use MDPI's
English editing service prior to publication or during author revisions.
Keywords
- Environmental metabolomics
- Ecometabolomics
- Systems biology
- Ecosurveillance
- Environmental model organisms
- Wild-caught organisms
- Microbial metabolomics
- Community metabolomics
- Ecosystem health
- Ecosystem function
- Environmental exposure
- Ecotox
- Environmental contaminants
- Metabolomic profiling
- Biological perturbation
Published Papers (4 papers)
Open AccessEditor’s ChoiceArticle
Decoding Fish Origins: How Metals and Metabolites Differentiate Wild, Cultured, and Escaped Specimens
by
Warda Badaoui, Kilian Toledo-Guedes, Juan Manuel Valero-Rodriguez, Adrian Villar-Montalt and Frutos C. Marhuenda-Egea
Cited by 1 | Viewed by 879
Abstract
Background: Fish escape events from aquaculture facilities are increasing and pose significant ecological, economic, and traceability concerns. Accurate methods to differentiate between wild, cultured, and escaped fish are essential for fishery management and seafood authentication. Methods: This study analyzed muscle tissue from
Sparus
[...] Read more.
Background: Fish escape events from aquaculture facilities are increasing and pose significant ecological, economic, and traceability concerns. Accurate methods to differentiate between wild, cultured, and escaped fish are essential for fishery management and seafood authentication. Methods: This study analyzed muscle tissue from
Sparus aurata,
Dicentrarchus labrax, and
Argyrosomus regius using a multiomics approach. Heavy metals were quantified by ICP-MS, fatty acid profiles were assessed via GC-MS, and metabolomic and lipidomic signatures were identified using 1H NMR spectroscopy. Multivariate statistical models (MDS and PLS-LDA) were applied to classify fish origins. Results: Wild seabream showed significantly higher levels of arsenic (9.5-fold), selenium (3.5-fold), and DHA and ARA fatty acids (3.2-fold), while cultured fish exhibited increased linoleic and linolenic acids (6.5-fold). TMAO concentrations were up to 5.3-fold higher in wild fish, serving as a robust metabolic biomarker. Escaped fish displayed intermediate biochemical profiles. Multivariate models achieved a 100% classification accuracy across species and analytical techniques. Conclusions: The integration of heavy metal analysis, fatty acid profiling, and NMR-based metabolomics enables the accurate differentiation of fish origin. While muscle tissue provides reliable biomarkers relevant to human exposure, future studies should explore additional tissues such as liver and gills to improve the resolution of traceability. These methods support seafood authentication, enhance aquaculture traceability, and aid in managing the ecological impacts of escape events.
Full article
►▼
Show Figures
Open AccessEditor’s ChoiceArticle
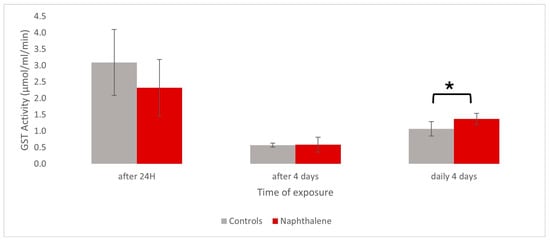
Metabolomic Prediction of Naphthalene Pneumo-Toxicity in the Snail Helix aspersa maxima
by
Aude Devalckeneer, Marion Bouviez and Jean-Marie Colet
Viewed by 1027
Abstract
Background: Polluted soils represent a major problem in many industrialized countries that urgently requires appropriate health risk assessment. The One Health concept that considers a close relationship between human and animal health and ecosystems relies, among other techniques, on continuous monitoring through the
[...] Read more.
Background: Polluted soils represent a major problem in many industrialized countries that urgently requires appropriate health risk assessment. The One Health concept that considers a close relationship between human and animal health and ecosystems relies, among other techniques, on continuous monitoring through the use of animal species as bioindicators. In this context, terrestrial gastropods, already recognized as relevant indicators due to their anatomo-physiology, provide a reliable model to study the pneumotoxic effects of pollutants. On the other hand, risk assessment is based on multi-biomarker studies. Therefore, omic approaches seem particularly useful since they can simultaneously detect numerous early biological changes.
Methods: In this study,
Helix aspersa maxima was exposed to naphthalene, a highly volatile aromatic hydrocarbon responsible for numerous respiratory disorders. Pulmonary membrane extracts and hemolymph samples were analyzed by
1H-NMR spectroscopy after single or repeated exposures to naphthalene.
Results: Numerous metabolic changes were observed, which could be related to membrane lesions, energy, anti-inflammatory, and tumorigenesis pathways.
Conclusions: Our findings highlight the potential of combining animal indicator and omics techniques to predict respiratory health risks in cases of exposure to polluted soils.
Full article
►▼
Show Figures
Open AccessReview
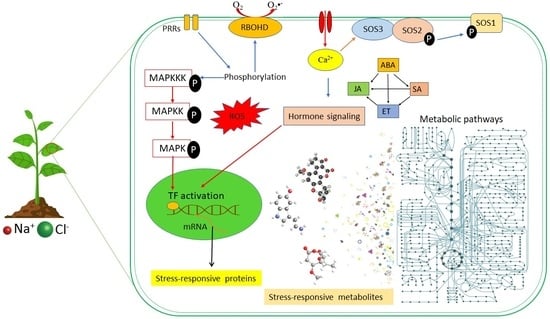
Soil Salinity, a Serious Environmental Issue and Plant Responses: A Metabolomics Perspective
by
Kekeletso H. Chele, Morena M. Tinte, Lizelle A. Piater, Ian A. Dubery and Fidele Tugizimana
Cited by 125 | Viewed by 8183
Abstract
The effects of global warming have increasingly led to devastating environmental stresses, such as heat, salinity, and drought. Soil salinization is a serious environmental issue and results in detrimental abiotic stress, affecting 7% of land area and 33% of irrigated lands worldwide. The
[...] Read more.
The effects of global warming have increasingly led to devastating environmental stresses, such as heat, salinity, and drought. Soil salinization is a serious environmental issue and results in detrimental abiotic stress, affecting 7% of land area and 33% of irrigated lands worldwide. The proportion of arable land facing salinity is expected to rise due to increasing climate change fuelled by anthropogenic activities, exacerbating the threat to global food security for the exponentially growing populace. As sessile organisms, plants have evolutionarily developed mechanisms that allow ad hoc responses to salinity stress. The orchestrated mechanisms include signalling cascades involving phytohormones, kinases, reactive oxygen species (ROS), and calcium regulatory networks. As a pillar in a systems biology approach, metabolomics allows for comprehensive interrogation of the biochemistry and a deconvolution of molecular mechanisms involved in plant responses to salinity. Thus, this review highlights soil salinization as a serious environmental issue and points to the negative impacts of salinity on plants. Furthermore, the review summarises mechanisms regulating salinity tolerance on molecular, cellular, and biochemical levels with a focus on metabolomics perspectives. This critical synthesis of current literature is an opportunity to revisit the current models regarding plant responses to salinity, with an invitation to further fundamental research for novel and actionable insights.
Full article
►▼
Show Figures
Open AccessArticle
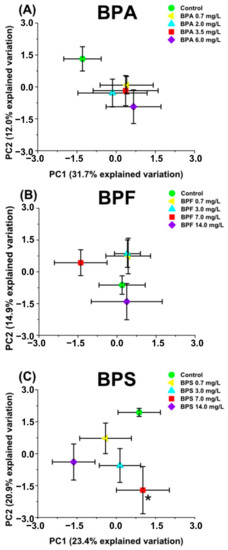
Metabolomics Reveals That Bisphenol Pollutants Impair Protein Synthesis-Related Pathways in Daphnia magna
by
Erico A. Oliveira Pereira, Lisa M. Labine, Sonya Kleywegt, Karl J. Jobst, André J. Simpson and Myrna J. Simpson
Cited by 18 | Viewed by 4603
Abstract
Bisphenols are used in the production of polycarbonate plastics and epoxy resins. Bisphenol A (BPA) has been widely studied and is believed to act as an endocrine disruptor. Bisphenol F (BPF) and bisphenol S (BPS) have increasingly been employed as replacements for BPA,
[...] Read more.
Bisphenols are used in the production of polycarbonate plastics and epoxy resins. Bisphenol A (BPA) has been widely studied and is believed to act as an endocrine disruptor. Bisphenol F (BPF) and bisphenol S (BPS) have increasingly been employed as replacements for BPA, although previous studies suggested that they yield similar physiological responses to several organisms.
Daphnia magna is a common model organism for ecotoxicology and was exposed to sub-lethal concentrations of BPA, BPF, and BPS to investigate disruption to metabolic profiles. Targeted metabolite analysis by liquid chromatography-tandem mass spectrometry (LC-MS/MS) was used to measure polar metabolites extracted from
D. magna, which are linked to a range of biochemical pathways. Multivariate analyses and individual metabolite changes showed similar non-monotonic concentration responses for all three bisphenols (BPA, BPF, and BPS). Pathway analyses indicated the perturbation of similar and distinct pathways, mostly associated with protein synthesis, amino acid metabolism, and energy metabolism. Overall, we observed responses that can be linked to a chemical class (bisphenols) as well as distinct responses that can be related to each individual bisphenol type (A, F, and S). These findings further demonstrate the need for using metabolomic analyses in exposure assessment, especially for chemicals within the same class which may disrupt the biochemistry uniquely at the molecular-level.
Full article
►▼
Show Figures