- Article
Assessing Modern AI-Driven Protein-Ligand Modeling with Phenethylamine and Tryptamine Psychedelics
- Benjamin R. Cummins and
- Charles D. Nichols
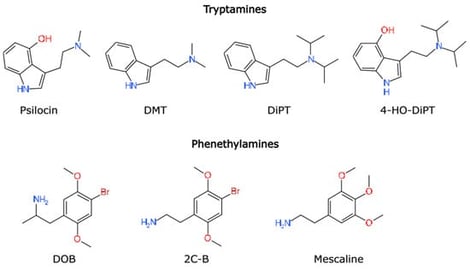
Modern advances in artificial intelligence have accelerated the development of computational tools for protein–ligand structure prediction, yet their real-world performance remains uneven across receptor classes and ligand chemotypes. Recently published cryo-EM structures of several different psychedelics bound to the serotonin 5HT2A receptor provide a unique opportunity to explore how modern AI-based modeling performs in a pharmacologically important GPCR system. Here, we compare three major approaches: AI-based protein–ligand cofolding (Boltz-2), a leading AI-driven docking module (Uni-Mol Docking v2), and a widely used classical physics-based docking pipeline (AutoDock Vina) across a series of tryptamine and phenethylamine psychedelics. Predicted binding poses were comparatively assessed through structural alignment with these newly available cryo-EM complexes. Additionally, calcium-mobilization assays were performed to provide a coarse functional readout for comparison with computationally predicted binding affinities. This study integrates methodological review with exploratory benchmarking to illustrate how different modeling paradigms behave on a shared receptor–ligand test set. Our results highlight substantial variation between modeling strategies, with AI-based cofolding often producing global binding orientations more closely resembling experimental structures, and classical docking showing greater variability across ligands, while still outperforming AI-driven docking on average. These observations underscore both the growing utility and current limitations of AI-assisted structure prediction in serotonergic drug discovery, and emphasize the importance of careful, experimentally anchored evaluation as such tools continue to advance.
10 February 2026



![Overview of the current workflow taking advantage of a recent graph-based force field generator [15] for routine conversion of 2D molecular diagrams to low-energy crystal structures.](https://mdpi-res.com/cdn-cgi/image/w=281,h=192/https://mdpi-res.com/aichem/aichem-01-00002/article_deploy/html/images/aichem-01-00002-g001-550.jpg)
![Evolution of descriptors across GAELLE generated conformation with a genetic algorithm. (Top) Electrophilicity index (in blue)
ω
[eV] plotted for each conformer ranked by increasing reactivity (i.e., decreasing
ω
). (Bottom) Global hardness
η
(in violet) [eV] for the same set of conformers, ranked in ascending order. Each point demonstrates the search for the lowest electrophilicity and higher hardness; hence, the algorithm will find the most reactive structure after 15 evolutions.](https://mdpi-res.com/cdn-cgi/image/w=281,h=192/https://mdpi-res.com/aichem/aichem-01-00001/article_deploy/html/images/aichem-01-00001-g001-550.jpg)