Abstract
Raman spectroscopy has become an important tool for biomedical analysis due to its ability to provide label-free, non-destructive molecular fingerprints of biological samples. However, existing deep learning approaches for classifying biological Raman spectra often focus on specific datasets and lack generalizability and interpretability. In this study, BioRamanNet is presented, an interpretable and generalizable deep learning framework designed for classifying a wide range of biological Raman spectra. The model integrates adaptive one-dimensional convolutional layers and squeeze-and-excitation (SE) blocks within a residual network architecture to enhance feature extraction. BioRamanNet was evaluated using four representative Raman spectral datasets—breast cells, extracellular vesicles and particles (EVPs), viruses, and bacteria—achieving classification accuracies of 99.5%, 100%, 99.8%, and 85.3%, respectively. To improve model interpretability, a perturbation-based analysis using Voigt noise was introduced to identify key wavenumber regions influencing classification. These regions were found to correspond closely with known Raman biomarkers, validating their biological significance. The results of this work demonstrate that BioRamanNet is a powerful and interpretable tool for analyzing diverse biological Raman spectra and holds promise for advancing machine learning-assisted biomedical diagnostics.
1. Introduction
Accurate classification of biological samples is a fundamental challenge in biomedical diagnostics and research. Traditional identification methods, such as culture-based assays [1,2] or polymerase chain reaction (PCR) [3], remain reliable but are often time-consuming, labor-intensive, and require specialized laboratory infrastructure. These limitations underscore the demand for rapid, scalable, and label-free techniques capable of characterizing diverse biological systems.
Raman spectroscopy has emerged as a powerful tool for non-destructive biochemical analysis, offering molecular-level insights into cells, viruses, and other biological components. By probing vibrational modes of biomolecules, Raman spectroscopy can generate unique spectral fingerprints that reflect cellular composition, physiological state, and metabolic activity [4,5,6]. The sensitivity of Raman signals is significantly enhanced using surface-enhanced Raman scattering (SERS), which can amplify signal intensity by several orders of magnitude [7,8]. However, SERS suffers from reproducibility issues, especially in complex biological samples like single cells due to variations in local enhancement factors and substrate inhomogeneity [9,10].
The integration of machine learning, particularly deep learning, has substantially improved the analysis and classification of Raman spectral data [11,12]. Deep neural networks (DNNs) excel at automatically learning complex, discriminative features directly from raw spectra, thereby reducing reliance on manual preprocessing or handcrafted feature extraction. A variety of neural architectures have been explored for biological Raman spectroscopy; each tailored to specific sample types and diagnostic goals. For example, PCA combined with support vector machine (PCA-SVM) has been used to classify extracellular vesicles and particles (EVPs) [13]. Other machine learning models, when applied to EV datasets derived from cancer cells using label-free or labeled optical techniques integrated with nanostructured sensors, have shown improved sensitivity and strong potential for early cancer detection [14]. Transformer networks—originally developed for natural language processing—have been successfully adapted to Raman spectral sequences, achieving high accuracy in distinguishing normal and cancerous breast cell lines [15]. More specialized approaches have also emerged. A self-attention mechanism (SAM)-guided convolutional neural network (CNN) enabled single-cell Raman spectral classification of Nosema bombycis spores with high precision and interpretability, identifying key biomolecular markers [16]. Modified ResNet architectures, adapted from computer vision, have been used to classify 30 bacterial isolates and assess their antibiotic resistance [17], while traditional CNNs incorporating segmented spectral processing have improved the classification of bacterial endotoxins [18]. In virology, the CoVari deep neural network demonstrated high classification accuracy in detecting SARS-CoV-2 variants and predicting viral concentrations from Raman spectra [19]. Furthermore, the MultiplexCR algorithm achieved outstanding performance in identifying co-infections using SERS spectra of 11 respiratory viruses, including binary and ternary virus mixtures in saliva samples [20].
Despite these advancements, most existing deep learning models are tailored to specific datasets and lack generalizability across diverse biological Raman spectra. Moreover, their “black-box” nature raises concerns about interpretability—an essential factor for clinical deployment and biological understanding [21,22]. Several methods have been proposed to enhance model transparency, such as dimensionality reduction (PCA, t-SNE, UMAP) [23,24], gradient-based saliency maps (e.g., Grad-CAM) [25], perturbation analysis [26], and comprehensive gradient attribution techniques [27]. In the broader context of optical spectroscopy, recent advances have further enriched the toolbox for interpretability. These include the use of full-gradient saliency maps with biomarker matching in neural networks for Raman spectroscopy [28], feature importance analysis from SVM coefficients for liquid biopsy applications [29], and the development of domain-aware algorithms like peak-sensitive logistic regression (PSE-LR) for generating interpretable feature maps [30]. The integration of attention mechanisms within CNN architectures has been demonstrated to highlight decisive, structure-relevant spectral features, providing sub-molecular level insight into the classification of highly similar compounds [31].
Another challenge is that most deep learning model in this field is rooted in computer vision, where models are designed for 2D image data. Raman spectra, in contrast, are one-dimensional, non- temporal sequences. While 1D convolutional models have seen success in other domains like speech or text processing, Raman spectral data pose unique challenges due to their non-temporal and highly overlapping nature. Current models often fail to address this structural mismatch, limiting their ability to generalize.
To overcome these challenges, BioRamanNet (v1.0, https://github.com/Penguin-Marsfield/BioRamanNet, accessed on 10 November 2025) is proposed, a novel, interpretable deep learning architecture specifically designed for 1D Raman spectral data. In contrast to models that are designed for a single type of biological Raman data and lack interpretability, this architecture is specifically optimized for 1D Raman spectral data, enabling multi-scale learning, improved interpretability, and robust performance across varied biological Raman datasets. This work’s approach combines an adaptive 1D convolutional backbone with squeeze-and-excitation (SE) [32] modules and residual connections, enabling both robust feature extraction and model transparency. BioRamanNet is evaluated on four biologically diverse Raman spectral datasets, breast cells, EVPs, viruses, and bacteria, demonstrating high classification accuracy and strong generalizability. Furthermore, to probe the interpretability of the model, a perturbation-based analysis using Voigt noise is implemented, identifying critical wavenumber regions and validating their biological relevance through comparison with known spectral biomarkers. This work bridges the gap between high-performance classification and interpretability in Raman spectroscopy-based bioanalytic, offering a generalizable and explainable framework for real-world biomedical applications.
2. Materials and Methods
2.1. Spectra Analysis Strategy
Accurate classification of biological samples constitutes a critical task in biomedical research. Given the increasing diversity of spectroscopic analysis techniques, this study utilized a framework integrating deep neural networks with Raman spectroscopy to achieve precise biological sample classification. The methodological workflow is presented below (Figure 1).
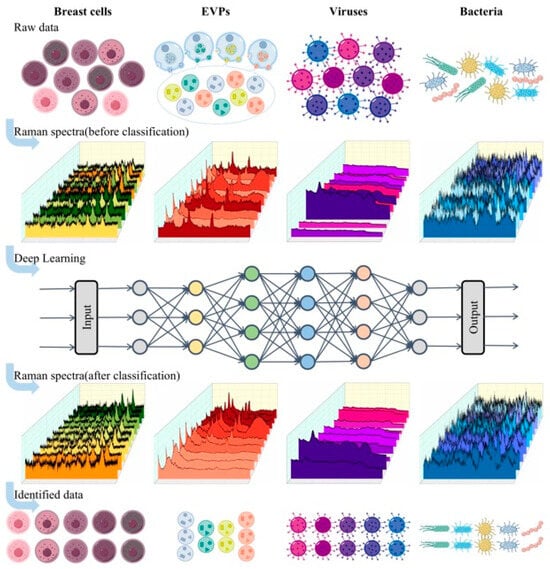
Figure 1.
The schematic outlines the workflow for classifying biological Raman spectral data including breast cells, EVPs, viruses, and bacteria. The process begins with the acquisition of Raman spectra from raw biological samples, resulting in the construction of a structured Raman spectral dataset. A specialized deep neural network is then applied to classify the spectral data, enabling the distinction of different biological samples.
The classification workflow initiates with the batch acquisition of Raman spectra from biological samples. Raw spectral data subsequently undergoes standardized preprocessing, including baseline correction, noise filtration, and normalization. Processed datasets are then randomly partitioned into training, validation, and test subsets. A deep neural network is subsequently implemented to establish spectral classification models, ultimately enabling the automated categorization of biological samples. This standardized protocol ensures both the reliability and reproducibility of classification outcomes.
Current research in the field of spectral analysis focuses on innovations in neural network architecture, and this study is also dedicated to this direction. The core objective of this work is to develop a deep learning framework for biological samples, named BioRamanNet, which is capable of high-precision classification and interpretable analysis of various types of biological Raman spectral data.
2.2. Dataset
Four biological Raman spectra datasets were utilized in this experiment: the breast cell, EVP, virus, and Bacteria-ID datasets. The four categories of biological Raman spectra were selected to rigorously evaluate the generalizability of BioRamanNet. This combination was chosen because it provides a wide range of biological and data-driven challenges. The samples include a large variety of biological structures, from cells to bacteria, ensuring spectral diversity. Furthermore, the datasets vary in the number of classes (from 4 to 30) and the number of samples per class, testing the model’s performance under both data-scarce and data-sufficient conditions. Therefore, achieving high accuracy across this diverse set proves the model’s robustness and utility as a general-purpose tool. The detailed composition, partition description, wavenumber range, and acquisition of each dataset are presented below.
The breast cell dataset consists of five distinct cell lines: MCF-10A, MDA-MB-231, BT-474, SK-BR-3, and T-47D. The numbers of spectra for each cell line are 201 for MCF-10A, 207 for MDA-MB-231, 185 for BT-474, 210 for SK-BR-3, and 237 for T-47D. The dataset was partitioned into a training set (831 spectra) used for model training and optimization, and a test set (209 spectra) employed to assess the model’s performance and generalization ability. During the training stage, the training set was further divided into 80% for training and 20% for validation. All spectra were measured within the wavenumber range of 560 to 1880 cm−1, capturing key spectral features.
The EVP dataset comprises four distinct types: Normal, COLO, DU145, and THP, with 120, 130, 125, and 110 samples. This was divided into a training set containing 339 spectra and a test set containing 146 spectra. During the training stage, the training set was further divided into 80% for training and 20% for validation. All spectra were measured within the wavenumber range of 1000 to 1800 cm−1.
The virus dataset includes 11 distinct types: Ad5, CoV229E, CoVNL63, CoVOC43, FluB, H1N1, H3N2, HMPVA, HMPVB, RSVA2, and RSVB1. Each virus subset originally contained measurements across 12 concentration levels (50, 100, 195, 391, 781, 1562, 3125, 6250, 12,500, 25,000, 50,000, and 100,000). Data from these concentration levels were initially combined to form a unified dataset for each virus type. Subsequently, the 11-virus datasets were merged into a comprehensive dataset. This dataset was partitioned into a training set and a test set. During the training stage, the training set was further divided into 80% for training and 20% for validation. All spectra were measured within the wavenumber range of 401 to 2000 cm−1.
The Bacteria-ID dataset consists of three key subsets: reference, fine-tuning, and test. The reference subset includes 2000 spectra per isolate, providing extensive data for initial model training. The fine-tuning subset, containing 100 spectra per isolate, is used to adjust the model parameters for improved performance on specific isolates. The test subset, also comprising 100 spectra per isolate, is used to evaluate the model’s generalization ability. Additionally, two clinical datasets are included, each featuring 25 patient isolates from five bacterial species. The 2018 clinical subset contains 400 spectra per isolate, whereas the 2019 subset comprises 100 spectra per isolate. For the pre-training and fine-tuning stages, the reference and fine-tuning subsets were further split into training and validation sets with a ratio of 90% to 10%. All spectra were measured within the wavenumber range of 381.98 to 1792.4 cm−1.
The average Raman spectra and standard deviation of 4 different biological datasets are shown in Figure 2. Furthermore, to visualize the inherent feature distribution of each dataset, t-SNE was applied to the Raman spectral data from all four datasets; the resulting projections are presented in Supplementary Figures S1–S4. Through these two approaches, the inter-class differences and intra-class variability within the data can be intuitively observed. The complete dataset partitioning methodology and corresponding sample sizes are fully detailed in Supplementary Table S1. The referenced publications contain comprehensive documentation for all experimental datasets (breast cell, EVP, virus and Bacteria-ID). These documents provide detailed descriptions of data collection protocols, experimental parameters, and preprocessing methodologies. For specific technical details like data sources, analysis methods, and quality checks, please refer to the methods section of the related papers [13,15,17,33].
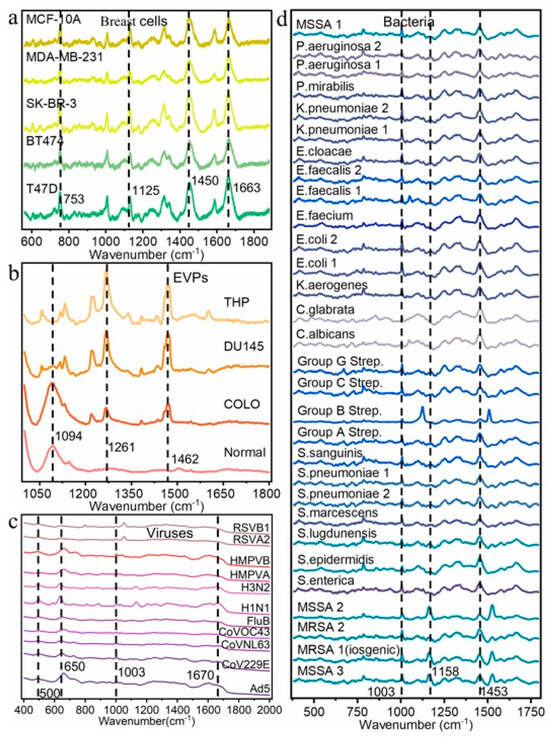
Figure 2.
Raman spectra of 4 different biological datasets. (a) Mean Raman spectra of 5 distinct classes of breast cells. (b) Mean Raman spectra of 4 distinct classes of EVPs. (c) Mean Raman spectra of 11 distinct classes of viruses. (d) Mean Raman spectra of 30 distinct classes of bacteria.
2.3. Construction of the BioRamanNet
The architecture of BioRamanNet was developed through a structured, iterative process to optimize its performance on biological Raman spectra. We began by evaluating several common models, including Support Vector Machine (SVM), Convolutional Neural Networks (CNN), and Recurrent Neural Networks (RNNs). A Residual Network (ResNet) was chosen as the baseline architecture because it demonstrated superior overall accuracy in our preliminary tests, consistent with its proven effectiveness in related tasks such as bacterial classification based on spectral data. However, while this baseline ResNet model performed well on datasets of EVPs, viruses, and bacteria, its performance on the breast cell data was suboptimal, being surpassed by both SVM and RNN. To enhance the model’s ability to capture the defining features of Raman spectra, we introduced an adaptive convolutional layer in place of the standard convolution. This layer employs multiple convolutional kernels of different sizes in parallel, allowing the network to adaptively integrate multi-scale information from the spectral peaks and more effectively model variations in both sharp and broad Raman bands. Furthermore, we incorporated Squeeze-and-Excitation (SE) blocks after key convolutional stages. This allows the model to dynamically recalibrate feature maps, emphasizing the most discriminative spectral channels and suppressing less relevant ones.
As illustrated in Figure 3, the proposed BioRamanNet is a 1D convolutional neural network designed to robustly extract features from biological Raman spectral data. Its architecture is inspired by the ResNet framework and integrates adaptive convolution layers with channel attention mechanisms to enhance feature representation critical for biological spectral discrimination. At the input stage, a custom adaptive convolution layer (AdaptiveConv1d) with kernel size 5 processes raw spectra. This layer employs dual-path processing: (1) a standard 1D convolution captures localized features, and (2) a compressed filter generated through kernel summation extracts global spectral trends. These dual outputs are fused with a bias term, dynamically balancing multi-scale feature extraction. Initialization uses the Kaiming uniform method for gradient stability, followed by batch normalization and ReLU activation.
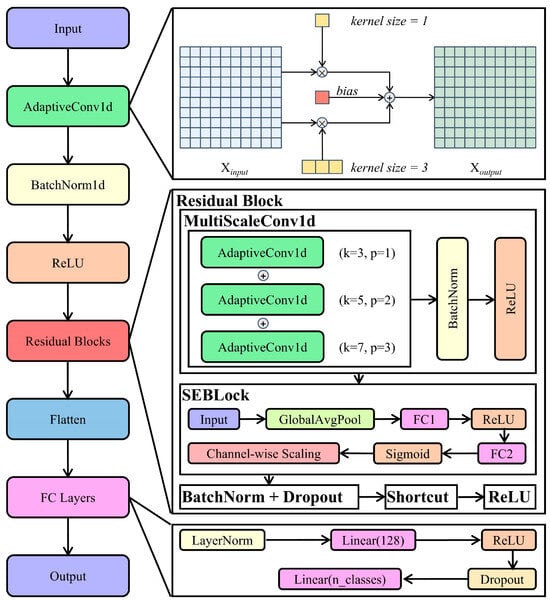
Figure 3.
Architecture of BioRamanNet. The network processes input spectra first through an AdaptiveConv1d layer to integrate local and global features, followed by batch normalization and ReLU activation. Subsequently, stacked residual blocks perform feature extraction: each block employs a MultiScaleConv1d module utilizing parallel AdaptiveConv1d layers with kernel sizes 3, 5, and 7, combined with an SE block for channel-wise attention. Outputs are processed through batch normalization and dropout, while adaptive residual connections maintain gradient flow. Finally, the extracted features pass through layer normalization, a ReLU-activated dense layer (128 units), dropout regularization, and a linear classifier. This architecture enables multi-scale learning and robust performance for biological Raman spectral analysis.
The network core contains stacked residual blocks for hierarchical feature learning. Each block begins with a multi-scale convolution module executing three parallel adaptive convolutions (kernels: 3/5/7). Their combined outputs undergo batch normalization and ReLU activation. Subsequently, a squeeze-and-excitation (SE) block enhances discriminative power—first aggregating channel statistics via global average pooling (squeeze step), then generating channel weights through fully connected layers with bottleneck reduction (ratio = 16) (excitation step), and finally rescaling features (reweighting step). A residual shortcut connection facilitates gradient propagation, while dropout regularizes activations.
Following feature extraction, high-level features are flattened and processed through the classification pathway: layer normalization projects feature to normalized space, a fully connected layer reduces dimensionality to 128 units, ReLU applies nonlinearity, dropout mitigates overfitting, and a final linear layer maps to output classes. The integration of adaptive convolutions, multi-scale fusion, and channel attention enables BioRamanNet to effectively model complex spectral variations in biological Raman data.
2.4. Training and Testing Details
In the training process, the hybrid neural network BioRamanNet, designed for classifying high-dimensional biological Raman spectra, was used for all experiments. First, a grid search was performed to identify optimal hyperparameters (hidden size and hidden layers) for each dataset, training models for 40 epochs to select configurations maximizing validation accuracy: for breast cell data, hidden size 64 with 2 hidden layers; for EVP and virus data, hidden size 128 with 2 layers; for Bacteria-ID data, hidden size 64 with 4 layers.
For breast cell, EVP, and virus datasets, single-step training was applied using a batch size of 5. The larger Bacteria-ID dataset required a different approach: two-phase training with pretraining on reference data followed by fine-tuning on a separate set. Both phases for Bacteria-ID used a batch size of 10. Adamax optimization was employed across all datasets due to its effectiveness with sparse gradients in spectral data. A learning rate of 10−4 was used for breast cell, EVP, and virus classifications. For Bacteria-ID, pretraining used a learning rate of 10−3, reduced to 10−4 during fine-tuning. All optimizations maintained consistent beta parameters (0.5, 0.999). Cross-entropy loss was applied given its suitability for multi-class spectral classification. To avoid overfitting, we employed early stopping, a widely adopted and effective technique used in previous deep learning studies. Training stopped if no improvement occurred for 10 consecutive epochs, with the best-performing model checkpoint preserved.
During evaluation, trained models predicted class labels on held-out test sets. Prediction accuracy and confusion matrices were systematically logged to assess computational efficiency and classifier performance across all datasets.
3. Results
3.1. Classification Performance of the BioRamanNet
Following the description of BioRamanNet, its classification performance was presented across four distinct Raman spectral datasets: breast cell, EVP, virus, and Bacteria-ID. Classification tasks for these biological Raman spectra were conducted independently. BioRamanNet was trained and tested five times on all four datasets. Accuracy and confusion matrices were mainly used to evaluate classification performance. To further demonstrate the robustness of BioRamanNet, precision, recall, and F1 score were also used as evaluation metrics. The Breast Cell, EVP, and Virus datasets are self-collected and were partitioned into training and test sets using identical protocols. In contrast, the Bacteria-ID dataset is a publicly available resource comprising the Reference, Finetune, Test, 2018Clinical, and 2019Clinical subsets. Consequently, the original authors’ multi-stage classification strategy was adopted, differing from the approach used for other datasets. This strategy required pre-training on the Reference subset, fine-tuning on the Finetune subset, testing on the Test subset, and final evaluation on the 2018Clinical and 2019Clinical subsets. Thus, performance is reported separately for the breast cell, EVP, and virus datasets and the Bacteria-ID dataset.
For the breast cell, EVP, and virus datasets, classification involved training and prediction phases. During training, the training set was further split into training and validation subsets. Early stopping was applied, and model weights achieving the highest validation accuracy were retained for final test-set prediction. The BioRamanNet achieved the overall highest test accuracy of 99.5% (Figure 4a) on the 5-class breast cell Raman spectral dataset. For the 4-class EVP dataset, the model attained a test accuracy of 100% (Figure 4b). On the 11-class virus dataset, the model reached the highest test accuracy of 99.8% (Figure 4c). All accuracy values were calculated by first determining the classification accuracy for each individual class and then averaging these values to ensure balanced evaluation across classes.
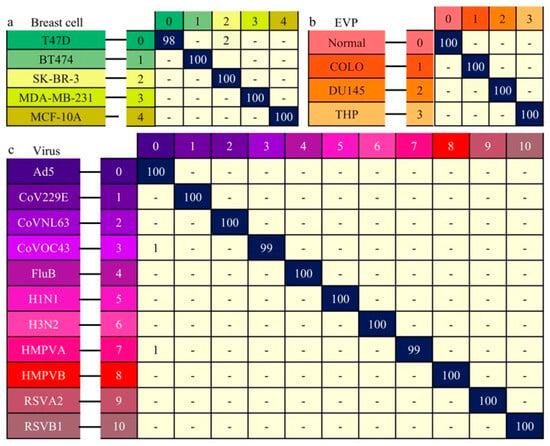
Figure 4.
Classification performance of the neural network for Raman spectra data from breast cells, EVPs, and viruses. Predicted results are presented as confusion matrices. True labels are shown on the left axis, predicted labels on the top axis. Each color-coded label corresponds to a distinct data type. Matrix values represent the probability of true data being classified into each category; values below 0.5% are indicated with “-”. (a) Confusion matrix for classification of five breast cell types (prediction accuracy: 99.5%). (b) Confusion matrix for classification of four EVP types (prediction accuracy: 100%). (c) Confusion matrix for classification of eleven virus types (prediction accuracy: 99.8%).
For the Bacteria-ID dataset, comprising 30 distinct classes. Unlike the other datasets, the training process for this dataset included two stages: pre-training on the reference subset and fine-tuning on the finetune subset. The model achieving the highest validation accuracy during pretraining was selected for fine-tuning, and the best performing finetuned model was then evaluated on the test subset. This model achieved the highest classification accuracy of 85.3% (Figure 5a). The accuracy was computed by averaging the classification accuracies for all 30 classes, providing a comprehensive assessment of the BioRamanNet performance on this dataset. Furthermore, the 30 bacterial isolates can be organized into eight groups based on the recommended empiric treatments for their corresponding species. By mapping the 30-class model outputs to these empiric treatment groups, the model was directly applied to an 8-class empiric treatments task. In this task, the model achieved an accuracy of 97.1% (Figure 5b), reflecting its ability to deliver the correct recommended empiric treatment reliably. To further assess the generalizability of our model, experiments were conducted on two clinical subsets from the Bacteria-ID dataset: the 2018 clinical and 2019 clinical datasets. Notably, the model achieved an accuracy of 100% (Figure 5c,d) on the two clinical datasets.
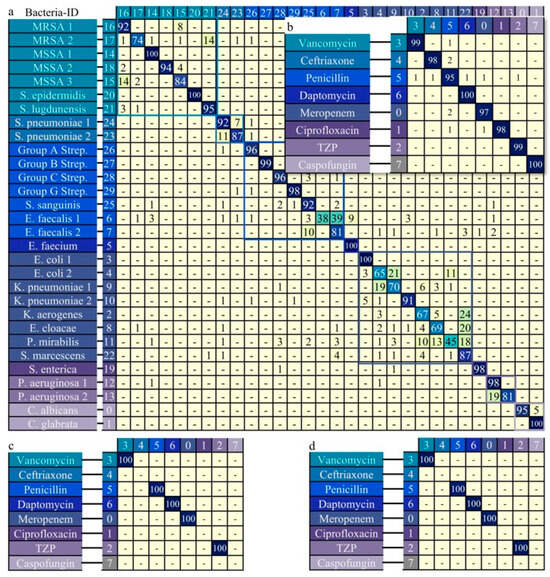
Figure 5.
Classification performance of the neural network for Raman spectra data from the Bacteria-ID dataset. Bacterial isolates (n = 30) were grouped into 8 empiric antibiotic treatment classes based on therapeutic relevance; isolates treated with the same antibiotic share a color and matching color border. Using antibiotic classes as labels aligns with clinical treatment practices. (a) Confusion matrix for the 30-isolate dataset (prediction accuracy: 85.3%). (b) Confusion matrix after regrouping into 8-empiric treatment classes (prediction accuracy: 97.1%). (c) Confusion matrix for the 2018 clinical dataset (prediction accuracy: 100%). (d) Confusion matrix for the 2019 clinical dataset (prediction accuracy: 100%).
To further validate the robustness of BioRamanNet, precision, recall, and F1 score were employed as evaluation metrics. The results, presented as the mean and standard deviation from five runs, are detailed in Table 1. Overall, the model demonstrated exceptional and consistent performance across most classes, with near-perfect scores for breast cells, EVPs, and viruses. The slightly lower, yet still high, scores for the more complex 30-bacteria classification further confirm the generalizability and robustness of the proposed deep learning algorithm for multi-class biological Raman spectral analysis. Additionally, Supplementary Table S2 records the time required for BioRamanNet to perform training and testing on the four datasets. All experiments were performed on a standard workstation with a single NVIDIA GeForce RTX 3080 GPU.

Table 1.
The classification performance of BioRamanNet on the four datasets: breast cell, EVP, virus, and bacteria. BioRamanNet was trained and tested five times on four datasets.
3.2. Comparative Analysis and Ablation Study
To validate the superior classification performance of BioRamanNet, three alternative algorithms were employed: Support Vector Machine (SVM), Recurrent Neural Network (RNN), and Residual Network (ResNet). To ensure a fair comparison, all models were independently optimized via a grid search on a dedicated validation set for key hyperparameters (e.g., regularization parameter C and kernel for SVM; hidden size and layer depth for RNN, ResNet, and BioRamanNet). These algorithms were applied to the four biological Raman spectral datasets (breast cells, EVPs, viruses and bacteria), and their best performance was compared against BioRamanNet’s highest accuracy. The specific outcomes are summarized in Table 2. SVM exhibited the poorest performance among the compared methods, showing the lowest accuracy in three out of the four datasets. RNN achieved good results on datasets with limited categories but demonstrated the worst performance on the virus dataset. ResNet underperformed on the breast cell dataset while showing competitive accuracy on the other datasets, particularly those with richer category diversity. The method used in this work achieved the highest accuracy across all four datasets, consistently outperforming SVM, RNN, and ResNet. These results demonstrate that BioRamanNet possesses superior generalization capability compared to other models when applied to various biological samples.

Table 2.
Comparison of BioRamanNet’s classification performance with previously reported methods for biological Raman spectroscopy.
To assess the contributions of each component in BioRamanNet, an ablation study was conducted to evaluate their individual and combined effects on model performance. An ablation study is a systematic experimental approach that evaluates the impact of removing or modifying specific components in a model to assess their necessity and causal relationships with overall performance. The network was based on a ResNet baseline, augmented with an AdaptiveConv1D module and SE Block. The experiments compared four combinations: (1) the ResNet baseline, (2) the ResNet with the AdaptiveConv1D, (3) the ResNet with the SE Block, and (4) the full model incorporating both modules. Table 3 presents the detailed results. As shown in the results, the addition of AdaptiveConv1D and SE Block modules consistently improved the baseline model’s performance, achieving accuracy enhancements across all four experimental datasets.

Table 3.
Impact assessment of AdaptiveConv1D and SEBlock modules in BioRamanNet through an ablation study.
3.3. Analysis of the Spectral Interpretability
Many biomolecules present in cells share common Raman bands, making single-cell Raman spectra within the fingerprint region (400–1800 cm−1) typically intricate, particularly in complex, mixed biological samples [34]. Similarly, the Raman spectra of viruses, EVPs, and bacteria also exhibited substantial band overlap due to the presence of similar biomolecules, such as proteins, lipids, and nucleic acids. Therefore, translating Raman spectra into meaningful biological insights is crucial for explaining the functions and states of breast cells, viruses, EVPs, and bacteria within complex biological systems.
To enhance the interpretability of the neural network in classifying these biological Raman spectra, a perturbation-based analysis was employed. The core objective was to identify critical wavenumber regions for classification and explore their potential biological significance by systematically evaluating model sensitivity to noise introduced at specific spectral locations using the Voigt profile.
The analysis involved two sequential stages. In Stage 1, each spectrum was divided into continuous segments, and Voigt noise was applied to each segment to measure the impact on classification accuracy, generating accuracy curves. In Stage 2, these accuracy curves were analyzed within sliding windows to compute a sensitivity metric (mean squared error, MSE). Regions with high MSE indicated critical wavenumbers significantly impacting classification. A window size of 6 was selected for Stage 2 as it aligns with the typical half-to-full width range of Raman peaks, optimizing the detection of key spectral features (Supplementary Figures S5–S12 present detailed information across both stages, including the selection of window size and differences in results under various parameters.). This process identified critical spectral regions visualized in heatmaps (Figure 6), where red denotes high-impact areas.
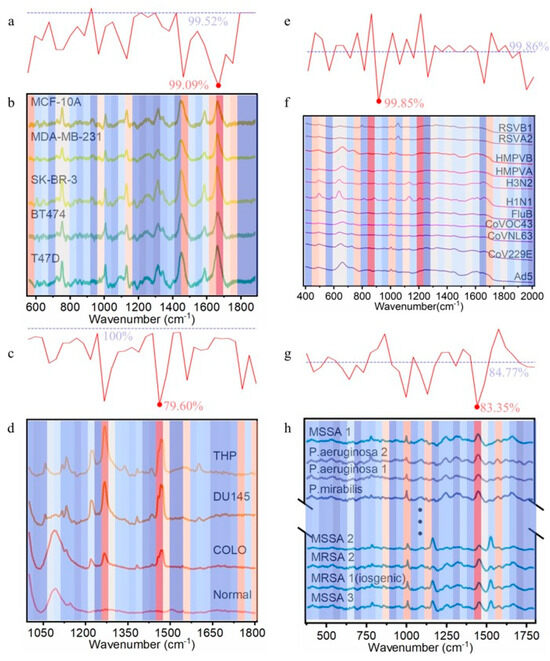
Figure 6.
Classification accuracy responses to Voigt perturbation in the four biological datasets (window size = 6). The intensity of the red color corresponds to the level of importance, whereas the intensity of the blue indicates a lower degree of significance. (a) Mean accuracy change curve after applying Voigt perturbation noise to breast cell data wavenumbers. (b) Heatmap quantifying perturbation-induced accuracy variations for breast cell data. (c) Mean accuracy change curve after applying Voigt perturbation noise to EVP data wavenumbers. (d) Heatmap quantifying perturbation-induced accuracy variations for EVP data. (e) Mean accuracy change curve after applying Voigt perturbation noise to virus data wavenumbers. (f) Heatmap quantifying perturbation-induced accuracy variations for virus data. (g) Mean accuracy change curve after applying Voigt perturbation noise to bacterial data wavenumbers. (h) Heatmap quantifying perturbation-induced accuracy variations for bacterial data.
The top 10 most impactful spectral windows were identified across the four datasets (the data details associated with the entire analysis process are provided in Supplementary Tables S3–S26). Crucially, these algorithmically determined critical regions showed strong correspondence with characteristic Raman peaks previously assigned to biomolecules in breast cells, EVPs, viruses, and bacteria (Table 4), effectively bridging computational findings with biological interpretation.

Table 4.
Critical spectral peaks exhibiting significant sensitivity to Voigt perturbation across biological datasets, with proposed molecular vibration assignments and literature references.
4. Discussion
BioRamanNet, integrating adaptive convolution layers and SE blocks, demonstrated exceptional performance (>99.5% accuracy) on breast cell, EVP, and virus datasets. This confirms its effectiveness in extracting both local spectral features and global channel relationships, making it highly suitable for complex biological Raman spectra.
For the challenging Bacteria-ID dataset, the two-stage strategy proved successful. The primary reason for the comparatively lower accuracy (85.3%) for distinguishing 30 isolates lies in the inherently high spectral overlap due to the biological similarities among the bacterial species, as visualized in the t-SNE plot and average spectra. This highlights the difficulty in capturing extremely fine-grained spectral differences at the isolate level. Despite this challenge, the high accuracy in mapping to empiric treatment groups (97.1%) and perfect performance on clinical datasets (100%) underscore the model’s strong potential for clinical diagnostics.
Ablation studies validated the necessity of both core modules (AdaptiveConv1D and SE blocks) for optimal performance across datasets, with BioRamanNet consistently outperforming other machine learning algorithms.
The novel Voigt perturbation method provides a valuable tool for identifying critical spectral regions, enhancing model interpretability. However, the biological implications of the identified wavenumber ranges could be expanded, particularly regarding unexpected or novel Raman peaks and their connections to cellular or microbial phenotypes. While the current interpretability analysis confirms that BioRamanNet relies on biologically meaningful regions, a deeper exploration of biomarker discovery remains a promising direction for future work.
Key limitations include the model’s computational intensity, which may hinder time-sensitive applications, along with the complexity of the interpretability method and the need for further refinement in fine-grained bacterial classification. In addition, the relatively poor performance of the recurrent neural network (RNN) on the virus dataset—likely due to an architecture mismatch, as viral spectral features may be more localized rather than temporally dependent—warrants further investigation.
Looking forward, several promising research directions emerge. First, model compression techniques, such as pruning redundant network weights and quantization to reduce the numerical precision of parameters, are crucial for future edge-device deployment. Second, incorporating a spectral attention mechanism could be a powerful way to enhance performance on challenging fine-grained classification tasks by allowing the model to focus on the most discriminative spectral regions. Third, the core “perturb-and-measure” concept of our Voigt perturbation approach shows potential for extension to other spectral modalities, such as infrared spectroscopy or mass spectrometry, which represents a valuable future endeavor. Finally, a more complete biological interpretation of salient spectral regions to uncover novel phenotype-related insights would add greater scientific value, necessitating further dedicated experiments.
5. Conclusions
In conclusion, BioRamanNet was developed, a novel hybrid deep learning model integrating adaptive convolutions and SE blocks, which significantly advances the classification accuracy of biological Raman spectra across diverse datasets including breast cells, EVP, viruses, and bacteria. BioRamanNet achieved exceptional performance (>99.5%) on several benchmark datasets and demonstrated robust clinical utility (100% accuracy) on independent clinical bacterial samples. While classification accuracy among 30 distinct bacterial isolates (85.3%) requires further enhancement, the model’s high accuracy (97.1%) in predicting empiric treatment groups underscores its immediate clinical relevance. The ablation studies in this work confirm the critical role of both the AdaptiveConv1D and SE modules. A novel Voigt noise-based perturbation method was introduced to enhance model interpretability by identifying key spectral regions. Future work will focus on optimizing model architecture for efficiency, simplifying the interpretability approach, and refining feature extraction to further improve bacterial isolate classification and solidify BioRamanNet’s utility in real-world diagnostic laboratories.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/aichem1010003/s1.
Author Contributions
Conceptualization, P.Y.; methodology, P.Y., X.L. and Y.L. (Yuxuan Lv); software, X.L. and Y.L. (Yan Li); validation, P.Y., Y.L. (Yan Li) and Y.Z.; resources, P.Y., Y.Z. and B.H.; writing—original draft preparation, X.L. and P.Y.; writing—review and editing, Y.L. (Yuxuan Lv), Y.Z. and B.H.; visualization, X.L. and P.Y.; supervision, P.Y., Y.Z. and B.H.; project administration, P.Y., Y.Z. and B.H.; funding acquisition, P.Y. All authors have read and agreed to the published version of the manuscript.
Funding
We gratefully acknowledge the financial support by the Yanzhao Golden Terrace Talent Initiative of Hebei Province (HJYB202518) and the China Scholarship Council.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Data of breast cells, EVPs, and viruses underlying the results presented in this paper can be obtained from the authors upon reasonable request. Data of Bacteria-ID underlying the results presented in this paper are available in Ref. [17].
Acknowledgments
During the preparation of this manuscript, the authors used DeepSeek R1 for the purposes of improving the readability and language of the manuscript. The authors have reviewed and edited the output and take full responsibility for the content of this publication.
Conflicts of Interest
The funding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
Abbreviations
The following abbreviations are used in this manuscript:
| t-SNE | t-distributed Stochastic Neighbor Embedding |
| UMAP | Uniform Manifold Approximation and Projection |
| ResNet | Residual Network |
| AdaptiveConv1D | Adaptive Convolutional 1D |
| ReLU | Rectified Linear Unit |
References
- Dellinger, R.P.; Levy, M.M.; Rhodes, A.; Annane, D.; Gerlach, H.; Opal, S.M.; Sevransky, J.E.; Sprung, C.L.; Douglas, I.S.; Jaeschke, R.; et al. Surviving Sepsis Campaign: International Guidelines for Management of Severe Sepsis and Septic Shock: 2012. Crit. Care Med. 2013, 41, 580–637. [Google Scholar] [CrossRef]
- Chaudhuri, A.; Martin, P.M.; Kennedy, P.G.E.; Andrew Seaton, R.; Portegies, P.; Bojar, M.; Steiner, I. EFNS guideline on the management of community-acquired bacterial meningitis: Report of an EFNS Task Force on acute bacterial meningitis in older children and adults. Eur. J. Neurol. 2008, 15, 649–659. [Google Scholar] [CrossRef]
- Gouel-Cheron, A.; Lumbard, K.; Hunsberger, S.; Arteaga-Cabello, F.J.; Beigel, J.; Belaunzarán-Zamudio, P.F.; Caballero-Sosa, S.; Escobedo-López, K.; Ibarra-González, V.; Nájera-Cancino, J.G.; et al. Serial real-time RT-PCR and serology measurements substantially improve Zika and Dengue virus infection classification in a co-circulation area. Antivir. Res. 2019, 172, 104638. [Google Scholar] [CrossRef]
- Qian, Y.; Fan, T.; Yao, Y.; Shi, X.; Liao, X.; Zhou, F.; Gao, F. Label-free and Raman dyes-free surface-enhanced Raman spectroscopy for detection of DNA. Sens. Actuators B Chem. 2018, 254, 483–489. [Google Scholar] [CrossRef]
- Barucci, A.; D’Andrea, C.; Farnesi, E.; Banchelli, M.; Amicucci, C.; de Angelis, M.; Hwang, B.; Matteini, P. Label-free SERS detection of proteins based on machine learning classification of chemo-structural determinants. Analyst 2021, 146, 674–682. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chang, K.; Ogunlade, B.; Herndon, L.; Tadesse, L.F.; Kirane, A.R.; Dionne, J.A. From Genotype to Phenotype: Raman Spectroscopy and Machine Learning for Label-Free Single-Cell Analysis. ACS Nano 2024, 18, 18101–18117. [Google Scholar] [CrossRef]
- Langer, J.; Jimenez de Aberasturi, D.; Aizpurua, J.; Alvarez-Puebla, R.A.; Auguié, B.; Baumberg, J.J.; Bazan, G.C.; Bell, S.E.J.; Boisen, A.; Brolo, A.G.; et al. Present and Future of Surface-Enhanced Raman Scattering. ACS Nano 2020, 14, 28–117. [Google Scholar] [CrossRef]
- Moskovits, M. Persistent misconceptions regarding SERS. Phys. Chem. Chem. Phys. 2013, 15, 5301–5311. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Samuelson, D.R.; Xu, Y.; Zhang, H.; Wang, S.; Rasco, B.A.; Xu, J.; Konkel, M.E. Detecting and Tracking Nosocomial Methicillin-Resistant Staphylococcus aureus Using a Microfluidic SERS Biosensor. Anal. Chem. 2013, 85, 2320–2327. [Google Scholar] [CrossRef]
- Butler, H.J.; Ashton, L.; Bird, B.; Cinque, G.; Curtis, K.; Dorney, J.; Esmonde-White, K.; Fullwood, N.J.; Gardner, B.; Martin-Hirsch, P.L.; et al. Using Raman spectroscopy to characterize biological materials. Nat. Protoc. 2016, 11, 664–687. [Google Scholar] [CrossRef] [PubMed]
- Herndon, L.K.; Zhang, Y.; Safir, F.; Ogunlade, B.; Balch, H.B.; Boehm, A.B.; Dionne, J.A. Bacterial Wastewater-Based Epidemiology Using Surface-Enhanced Raman Spectroscopy and Machine Learning. Nano Lett. 2025, 25, 1250–1259. [Google Scholar] [CrossRef]
- Sahli, C.; Kenry. Enhancing Nanomaterial-Based Optical Spectroscopic Detection of Cancer through Machine Learning. ACS Mater. Lett. 2024, 6, 4697–4709. [Google Scholar] [CrossRef]
- Yin, P.; Li, G.; Zhang, B.; Farjana, H.; Zhao, L.; Qin, H.; Hu, B.; Ou, J.; Tian, J. Facile PEG-based isolation and classification of cancer extracellular vesicles and particles with label-free surface-enhanced Raman scattering and pattern recognition algorithm. Analyst 2021, 146, 1949–1955. [Google Scholar] [CrossRef] [PubMed]
- del Real Mata, C.; Jeanne, O.; Jalali, M.; Lu, Y.; Mahshid, S. Nanostructured-Based Optical Readouts Interfaced with Machine Learning for Identification of Extracellular Vesicles. Adv. Heal. Mater. 2023, 12, 2202123. [Google Scholar] [CrossRef]
- Yu, Q.; Shen, X.; Yi, L.; Liang, M.; Li, G.; Guan, Z.; Wu, X.; Castel, H.; Hu, B.; Yin, P.; et al. Fragment-Fusion Transformer: Deep Learning-Based Discretization Method for Continuous Single-Cell Raman Spectral Analysis. ACS Sens. 2024, 9, 3907–3920. [Google Scholar] [CrossRef]
- Xue, M.; Hu, J.; He, X.; Hu, J.; Li, Y.; Wang, G.; Huang, X.; Yuan, Y. Advanced Nosema bombycis Spore Identification: Single-Cell Raman Spectroscopy Combined with Self-Attention Mechanism-Guided Deep Learning. Anal. Chem. 2024, 96, 20255–20266. [Google Scholar] [CrossRef]
- Ho, C.-S.; Jean, N.; Hogan, C.A.; Blackmon, L.; Jeffrey, S.S.; Holodniy, M.; Banaei, N.; Saleh, A.A.E.; Ermon, S.; Dionne, J. Rapid identification of pathogenic bacteria using Raman spectroscopy and deep learning. Nat. Commun. 2019, 10, 4927. [Google Scholar] [CrossRef]
- Yang, Y.; Xu, B.; Haverstick, J.; Ibtehaz, N.; Muszyński, A.; Chen, X.; Chowdhury, M.E.H.; Zughaier, S.M.; Zhao, Y. Differentiation and classification of bacterial endotoxins based on surface enhanced Raman scattering and advanced machine learning. Nanoscale 2022, 14, 8806–8817. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Cui, J.; Luo, D.; Murray, J.; Chen, X.; Hülck, S.; Tripp, R.A.; Zhao, Y. Rapid Detection of SARS-CoV-2 Variants Using an Angiotensin-Converting Enzyme 2-Based Surface-Enhanced Raman Spectroscopy Sensor Enhanced by CoVari Deep Learning Algorithms. ACS Sens. 2024, 9, 3158–3169. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Cui, J.; Kumar, A.; Luo, D.; Murray, J.; Jones, L.; Chen, X.; Hülck, S.; Tripp, R.A.; Zhao, Y. Multiplex Detection and Quantification of Virus Co-Infections Using Label-free Surface-Enhanced Raman Spectroscopy and Deep Learning Algorithms. ACS Sens. 2025, 10, 1298–1311. [Google Scholar] [CrossRef]
- Wong, F.; Omori, S.; Li, A.; Krishnan, A.; Lach, R.S.; Rufo, J.; Wilson, M.Z.; Collins, J.J. An explainable deep learning platform for molecular discovery. Nat. Protoc. 2024, 20, 1020–1056. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Yang, G. Interpretability research of deep learning: A literature survey. Inf. Fusion 2025, 115, 102721. [Google Scholar] [CrossRef]
- Laurens, V.D.M.; Hinton, G. Visualizing Data using t-SNE. J. Mach. Learn. Res. 2008, 9, 2579–2605. [Google Scholar]
- McInnes, L.; Healy, J.; Saul, N.; Grossberger, L. UMAP: Uniform Manifold Approximation and Projection. J. Open Source Softw. 2018, 3, 861. [Google Scholar] [CrossRef]
- Deng, L.; Zhong, Y.; Wang, M.; Zheng, X.; Zhang, J. Scale-Adaptive Deep Model for Bacterial Raman Spectra Identification. IEEE J. Biomed. Health Inform. 2021, 26, 369–378. [Google Scholar] [CrossRef]
- Safir, F.; Vu, N.; Tadesse, L.F.; Firouzi, K.; Banaei, N.; Jeffrey, S.S.; Saleh, A.A.E.; Khuri-Yakub, B.T.; Dionne, J.A. Combining Acoustic Bioprinting with AI-Assisted Raman Spectroscopy for High-Throughput Identification of Bacteria in Blood. Nano Lett. 2023, 23, 2065–2073. [Google Scholar] [CrossRef]
- Ye, J.; Yeh, Y.-T.; Xue, Y.; Wang, Z.; Zhang, N.; Liu, H.; Zhang, K.; Ricker, R.; Yu, Z.; Roder, A.; et al. Accurate virus identification with interpretable Raman signatures by machine learning. Proc. Natl. Acad. Sci. USA 2022, 119, e2118836119. [Google Scholar] [CrossRef]
- Yousuf, S.; Karukappadath, M.I.; Zam, A. Differentiation of Healthy Ex Vivo Bovine Tissues Using Raman Spectroscopy and Interpretable Machine Learning. Lasers Surg. Med. 2025, 57, 517–527. [Google Scholar] [CrossRef]
- Ranasinghe, J.C.; Wang, Z.; Huang, S. Unveiling brain disorders using liquid biopsy and Raman spectroscopy. Nanoscale 2024, 16, 11879–11913. [Google Scholar] [CrossRef]
- Wang, Z.; Ranasinghe, J.C.; Wu, W.; Chan, D.C.Y.; Gomm, A.; Tanzi, R.E.; Zhang, C.; Zhang, N.; Allen, G.I.; Huang, S. Machine Learning Interpretation of Optical Spectroscopy Using Peak-Sensitive Logistic Regression. ACS Nano 2025, 19, 15457–15473. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Li, W.; Liu, Y.; Wang, Y.; Dou, X. Deep-Learning-Assisted Raman Spectral Analysis for Accurate Differentiation of Highly Structurally Similar CA Series Synthetic Cannabinoids. Anal. Chem. 2025, 97, 10812–10820. [Google Scholar] [CrossRef]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-Excitation Networks. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 7132–7141. [Google Scholar]
- Yang, Y.; Xu, B.; Murray, J.; Haverstick, J.; Chen, X.; Tripp, R.A.; Zhao, Y. Rapid and quantitative detection of respiratory viruses using surface-enhanced Raman spectroscopy and machine learning. Biosens. Bioelectron. 2022, 217, 114721. [Google Scholar] [CrossRef]
- Li, M.; Xu, J.; Romero-Gonzalez, M.; Banwart, S.A.; Huang, W.E. Single cell Raman spectroscopy for cell sorting and imaging. Curr. Opin. Biotechnol. 2012, 23, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Sajan, D.; Binoy, J.; Pradeep, B.; Venkata Krishna, K.; Kartha, V.B.; Joe, I.H.; Jayakumar, V.S. NIR-FT Raman and infrared spectra and ab initio computations of glycinium oxalate. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2004, 60, 173–180. [Google Scholar] [CrossRef]
- Stone, N.; Kendall, C.; Smith, J.; Crow, P.; Barr, H. Raman spectroscopy for identification of epithelial cancers. Faraday Discuss. 2004, 126, 141–157. [Google Scholar] [CrossRef]
- Lakshmi, R.J.; Kartha, V.B.; Murali Krishna, C.; Solomon, J.G.R.; Ullas, G.; Uma Devi, P. Tissue Raman Spectroscopy for the Study of Radiation Damage: Brain Irradiation of Mice. Radiat. Res. 2002, 157, 175–182. [Google Scholar] [CrossRef]
- McNesby, K.; Pesce-Rodriguez, R. Handbook of Vibrational Spectroscopy; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2006. [Google Scholar]
- Feng, S.; Chen, R.; Lin, J.; Pan, J.; Chen, G.; Li, Y.; Cheng, M.; Huang, Z.; Chen, J.; Zeng, H. Nasopharyngeal cancer detection based on blood plasma surface-enhanced Raman spectroscopy and multivariate analysis. Biosens. Bioelectron. 2010, 25, 2414–2419. [Google Scholar] [CrossRef]
- Yan, B.; Li, B.; Wen, Z.; Luo, X.; Xue, L.; Li, L. Label-free blood serum detection by using surface-enhanced Raman spectroscopy and support vector machine for the preoperative diagnosis of parotid gland tumors. BMC Cancer 2015, 15, 650. [Google Scholar] [CrossRef]
- Feng, S.; Chen, R.; Lin, J.; Pan, J.; Wu, Y.; Li, Y.; Chen, J.; Zeng, H. Gastric cancer detection based on blood plasma surface-enhanced Raman spectroscopy excited by polarized laser light. Biosens. Bioelectron. 2011, 26, 3167–3174. [Google Scholar] [CrossRef] [PubMed]
- Rygula, A.; Majzner, K.; Marzec, K.M.; Kaczor, A.; Pilarczyk, M.; Baranska, M. Raman spectroscopy of proteins: A review. J. Raman Spectrosc. 2013, 44, 1061–1076. [Google Scholar] [CrossRef]
- Kneipp, K.; Wang, Y.; Kneipp, H.; Perelman, L.T.; Itzkan, I.; Dasari, R.R.; Feld, M.S. Single Molecule Detection Using Surface-Enhanced Raman Scattering (SERS). Phys. Rev. Lett. 1997, 78, 1667–1670. [Google Scholar] [CrossRef]
- Latteyer, F.; Peisert, H.; Göhring, N.; Peschel, A.; Chassé, T. Vibrational and electronic characterisation of Staphylococcus aureus wall teichoic acids and relevant components in thin films. Anal. Bioanal. Chem. 2010, 397, 2429–2437. [Google Scholar] [CrossRef] [PubMed]
- Uchiyama, T.; Sonoyama, M.; Hamada, Y.; Komatsu, M.; Dukor, R.K.; Nafie, L.A.; Oosawa, K. Raman spectroscopic study on the L-type straight flagellar filament of Salmonella. Vib. Spectrosc. 2006, 42, 192–194. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.