Progress in Plant Genome Sequencing
Abstract
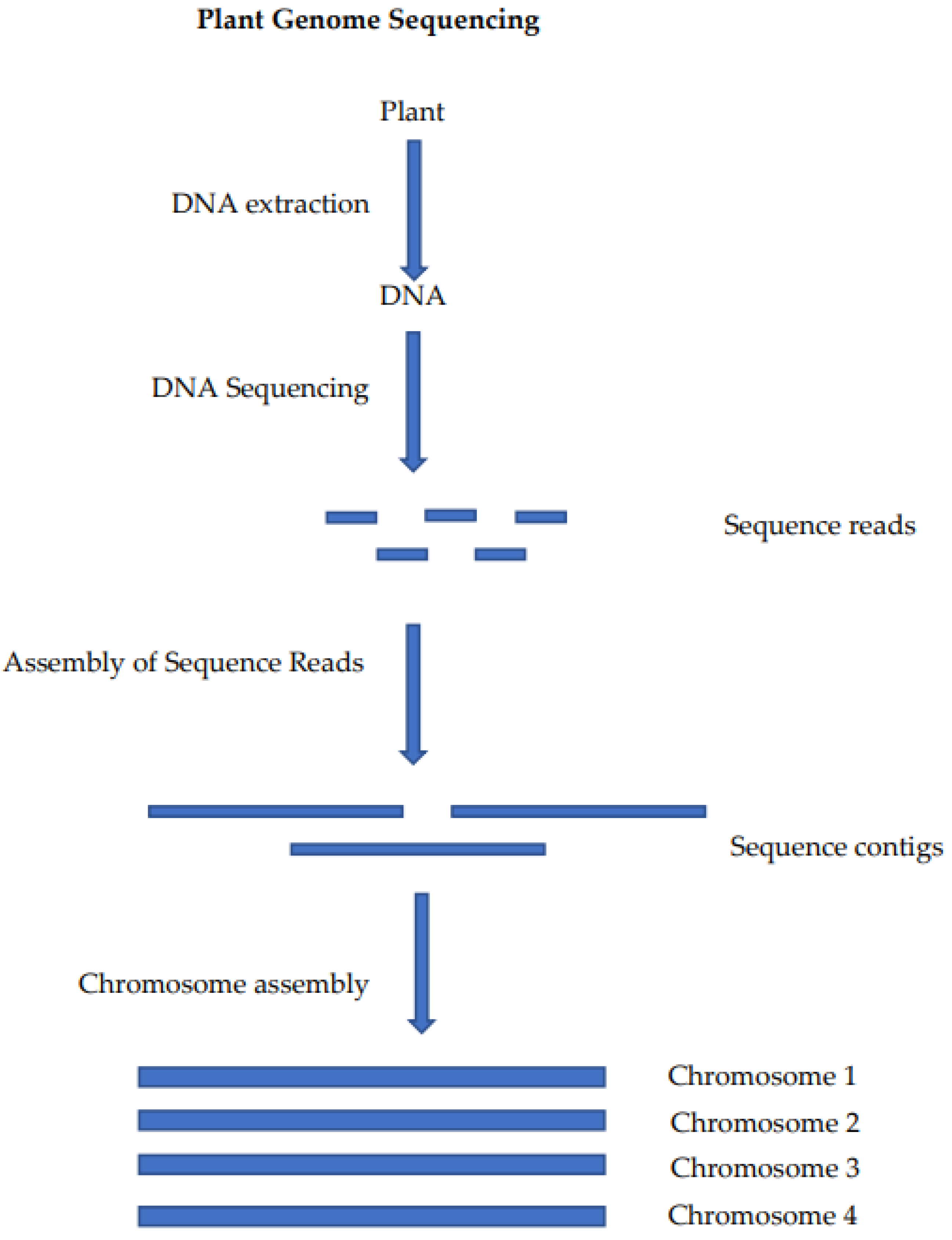
:1. Introduction
2. Diversity of Plant Genomes
3. Applications of Plant Genome Sequencing
3.1. Model Genomes
3.2. Crop Plant Genomes
3.3. Sequencing Plant Biodiversity
3.4. Sequencing Rare and Threatened Species
4. Sequencing Technology
4.1. DNA Isolation
4.2. Sort Read Sequences
4.3. Long Read Sequences
4.3.1. PacBio
4.3.2. ONT
4.3.3. Other
4.3.4. Advances
4.4. Chromosome-Level Assembly
4.5. Haplotype-Resolved Genomes
4.6. Pan-Genomes
4.7. Transcriptomes
4.7.1. RNAseq
4.7.2. Long Read Transcriptomes
4.8. Organelle Genome Sequencing
4.8.1. Chloroplast Genomes
4.8.2. Plant Mitochondrial Genomes
5. Biological Understanding
5.1. Whole Genome Duplications
5.2. Polyploid Challenges
5.3. Genomics of Plants with Diverse Reproductive Biology
5.4. Evolutionary Insights
5.5. Maternal Genome Inheritance
5.6. Importance of Genome Size
6. Enabling Plant Breeding
6.1. Molecular Markers and Plant Selection
6.2. Genetic Manipulation
6.3. Editing Plant Genomes
6.4. Biotechnology Applications (Food, Medicinal and Industrial Crops)
7. IP Issues
8. Future Prospects
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Henry, R.J. Applied Biosciences: Application of Biological Science and Technology. Appl. Biosci. 2022, 1, 38–39. [Google Scholar] [CrossRef]
- Shendure, J.; Balasubramanian, S.; Church, G.M.; Gilbert, W.; Rogers, J.; Schloss, J.; Waterston, R.H. DNA sequencing at 40: Past, present and future. Nature 2017, 550, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Marks, R.A.; Hotaling, S.; Frandsen, P.B.; VanBuren, R. Representation and participation across 20 years of plant genome sequencing. Nat. Plants 2021, 7, 1571–1578. [Google Scholar] [CrossRef]
- Yüksel, B.; Paterson, A.H. Construction and characterization of a peanut HindIII BAC library. Theor. Appl. Genet. 2005, 111, 630–639. [Google Scholar] [CrossRef]
- Garsmeur, O.; Droc, G.; Antonise, R.; Grimwood, J.; Potier, B.; Aitken, K.; Jenkins, J.; Martin, G.; Charron, C.; Hervouet, C.; et al. A mosaic monoploid reference sequence for the highly complex genome of sugarcane. Nat. Commun. 2018, 9, 2638. [Google Scholar] [CrossRef] [PubMed]
- Pucker, B.; Irisarri, I.; de Vries, J.; Xu, B. Plant genome sequence assembly in the era of long reads: Progress, challenges and future directions. Quant. Plant Biol. 2021, 3, 1–14. [Google Scholar] [CrossRef]
- Chen, F.; Dong, W.; Zhang, J.; Guo, X.; Chen, J.; Wang, Z.; Lin, Z.; Tang, H.; Zhang, L. The Sequenced Angiosperm Genomes and Genome Databases. Front. Plant Sci. 2018, 9, 418. [Google Scholar] [CrossRef] [Green Version]
- Sharma, P.A.O.; Alsubaie, B.; Al-Mssallem, I.; Nath, O.; Mitter, N.; Margarido, G.R.A.; Topp, B.; Murigneux, V.; Masouleh, A.K.; Furtado, A.; et al. Improvements in The Sequencing and Assembly of Plant Genomes. Gigabyte 2021. [Google Scholar] [CrossRef]
- Wendel, J.F.; Jackson, S.A.; Meyers, B.C.; Wing, R.A. Evolution of plant genome architecture. Genome Biol. 2016, 17, 37. [Google Scholar] [CrossRef] [Green Version]
- Pellicer, J.; Hidalgo, O.; Dodsworth, S.; Leitch, I.J. Genome Size Diversity and Its Impact on the Evolution of Land Plants. Genes 2018, 9, 88. [Google Scholar] [CrossRef] [Green Version]
- Uddenberg, D.; Akhter, S.; Ramachandran, P.; Sundström, J.F.; Carlsbecker, A. Sequenced genomes and rapidly emerging technologies pave the way for conifer evolutionary developmental biology. Front. Plant Sci. 2015, 6, 970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Wang, X.; Wang, G.; Cui, P.; Wu, S.; Ai, C.; Hu, N.; Li, A.; He, B.; Shao, X.; et al. The nearly complete genome of Ginkgo biloba illuminates gymnosperm evolution. Nat. Plants 2021, 7, 748–756. [Google Scholar] [CrossRef] [PubMed]
- Kaul, S.; Koo, H.L.; Jenkins, J.; Rizzo, M.; Rooney, T.; Tallon, L.J.; Feldblyum, T.; Nierman, W.; Benito, M.I.; Lin, X.Y.; et al. Analysis of The Genome Sequence of The Flowering Plant Arabidopsis Thaliana. Nature 2000, 408, 796–815. [Google Scholar]
- Jackson, S.A. Rice: The First Crop Genome. Rice 2016, 9, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vogel, J.P.; Garvin, D.F.; Mockler, T.C.; Schmutz, J.; Rokhsar, D.; Bevan, M.W.; Barry, K.; Lucas, S.; Harmon-Smith, M.; Lail, K.; et al. Genome Sequencing and Analysis of the Model Grass Brachypodium Distachyon. Nature 2010, 463, 763–768. [Google Scholar]
- Kersey, P.J. Plant genome sequences: Past, present, future. Curr. Opin. Plant Biol. 2018, 48, 1–8. [Google Scholar] [CrossRef]
- Wambugu, P.W.; Henry, R.; Browne, L. Supporting in situ conservation of the genetic diversity of crop wild relatives using genomic technologies. Mol. Ecol. 2022, 31, 2207–2222. [Google Scholar] [CrossRef]
- Wambugu, P.W.; Ndjiondjop, M.-N.; Henry, R.J. Role of genomics in promoting the utilization of plant genetic resources in genebanks. Brief. Funct. Genom. 2018, 17, 198–206. [Google Scholar] [CrossRef]
- Murigneux, V.; Rai, S.K.; Furtado, A.; Bruxner, T.J.C.; Tian, W.; Harliwong, I.; Wei, H.; Yang, B.; Ye, Q.; Anderson, E.; et al. Comparison of long-read methods for sequencing and assembly of a plant genome. GigaScience 2020, 9, giaa146. [Google Scholar] [CrossRef]
- Graham, G.C.; Mayers, P.; Henry, R.J. A simplified method for the preparation of fungal genomic DNA for PCR and RAPD analysis. BioTechniques 1994, 16, 48–50. [Google Scholar]
- Nath, O.; Fletcher, S.J.; Hayward, A.; Shaw, L.M.; Agarwal, R.; Furtado, A.; Henry, R.J.; Mitter, N. A Comprehensive High-Quality DNA and RNA Extraction Protocol for a Range of Cultivars and Tissue Types of the Woody Crop Avocado. Plants 2022, 11, 242. [Google Scholar] [CrossRef] [PubMed]
- Healey, A.; Furtado, A.; Cooper, T.; Henry, R.J. Protocol: A simple method for extracting next-generation sequencing quality genomic DNA from recalcitrant plant species. Plant Methods 2014, 10, 21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guilfoyle, T.J. Chapter 8 Isolation and Characterization of Plant Nuclei. Method Cell Biol. 1995, 50, 101–112. [Google Scholar] [CrossRef]
- Garrido-Cardenas, J.A.; Garcia-Maroto, F.; Alvarez-Bermejo, J.A.; Manzano-Agugliaro, F. DNA Sequencing Sensors: An Overview. Sensors 2017, 17, 588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bundock, P.C.; Eliott, F.G.; Ablett, G.; Benson, A.D.; Casu, R.E.; Aitken, K.S.; Henry, R.J. Targeted single nucleotide polymorphism (SNP) discovery in a highly polyploid plant species using 454 sequencing. Plant Biotechnol. J. 2009, 7, 347–354. [Google Scholar] [CrossRef] [PubMed]
- Brozynska, M.; Furtado, A.; Henry, R.J. Direct Chloroplast Sequencing: Comparison of Sequencing Platforms and Analysis Tools for Whole Chloroplast Barcoding. PLoS ONE 2014, 9, e110387. [Google Scholar] [CrossRef] [Green Version]
- Rhoads, A.; Au, K.F. PacBio Sequencing and Its Applications. Genom. Proteom. Bioinf. 2015, 13, 278–289. [Google Scholar] [CrossRef] [Green Version]
- Hon, T.; Mars, K.; Young, G.; Tsai, Y.-C.; Karalius, J.W.; Landolin, J.M.; Maurer, N.; Kudrna, D.; Hardigan, M.A.; Steiner, C.C.; et al. Highly accurate long-read HiFi sequencing data for five complex genomes. Sci. Data 2020, 7, 399. [Google Scholar] [CrossRef]
- Wang, Y.H.; Zhao, Y.; Bollas, A.; Wang, Y.R.; Au, K.F. Nanopore Sequencing Technology, Bioinformatics and Applications. Nat. Biotechnol. 2021, 39, 1348–1365. [Google Scholar] [CrossRef]
- Wang, J.; Bhakta, N.; Miller, V.A.; Revsine, M.; Litzow, M.; Paietta, E.; Roberts, K.; Gu, Z.; Mullighan, C.; Jones, C.; et al. Nanopore RNA Sequencing as A Diagnostic Tool for Acute Leukemia in Low Resource Settings. Pediatr. Blood Cancer 2021, 68, S107. [Google Scholar]
- Belser, C.; Istace, B.; Denis, E.; Dubarry, M.; Baurens, F.-C.; Falentin, C.; Genete, M.; Berrabah, W.; Chèvre, A.-M.; Delourme, R.; et al. Chromosome-scale assemblies of plant genomes using nanopore long reads and optical maps. Nat. Plants 2018, 4, 879–887. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Pham, L.; Wu, T.-C.; Mo, G.; Xia, Y.; Chang, P.L.; Porter, D.; Phan, T.; Che, H.; Tran, H.; et al. Ultralow-input single-tube linked-read library method enables short-read second-generation sequencing systems to routinely generate highly accurate and economical long-range sequencing information. Genome Res. 2020, 30, 898–909. [Google Scholar] [CrossRef] [PubMed]
- Wang, O.; Chin, R.; Cheng, X.; Wu, M.K.Y.; Mao, Q.; Tang, J.; Sun, Y.; Anderson, E.; Lam, H.K.; Chen, D.; et al. Efficient and unique cobarcoding of second-generation sequencing reads from long DNA molecules enabling cost-effective and accurate sequencing, haplotyping, and de novo assembly. Genome Res. 2019, 29, 798–808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, M.; Al-Eryani, G.; Carswell, S.; Ferguson, J.M.; Blackburn, J.; Barton, K.; Roden, D.; Luciani, F.; Phan, T.G.; Junankar, S.; et al. High-throughput targeted long-read single cell sequencing reveals the clonal and transcriptional landscape of lymphocytes. Nat. Commun. 2019, 10, 3120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, P.; Masouleh, A.K.; Topp, B.; Furtado, A.; Henry, R.J. De novo chromosome level assembly of a plant genome from long read sequence data. Plant J. 2021, 109, 727–736. [Google Scholar] [CrossRef]
- Cheng, H.; Concepcion, G.T.; Feng, X.; Zhang, H.; Li, H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat. Methods 2021, 18, 170–175. [Google Scholar] [CrossRef]
- Healey, A.L.; Shepherd, M.; King, G.J.; Butler, J.B.; Freeman, J.S.; Lee, D.J.; Potts, B.M.; Silva-Junior, O.B.; Baten, A.; Jenkins, J.; et al. Pests, diseases, and aridity have shaped the genome of Corymbia citriodora. Commun. Biol. 2021, 4, 537. [Google Scholar] [CrossRef]
- Harewood, L.; Kishore, K.; Eldridge, M.; Wingett, S.; Pearson, D.; Schoenfelder, S.; Collins, V.P.; Fraser, P. Hi-C as a tool for precise detection and characterisation of chromosomal rearrangements and copy number variation in human tumours. Genome Biol. 2017, 18, 125. [Google Scholar] [CrossRef] [Green Version]
- Chen, P.; Jing, X.; Ren, J.; Cao, H.; Hao, P.; Li, X. Modelling BioNano optical data and simulation study of genome map assembly. Bioinformatics 2018, 34, 3966–3974. [Google Scholar] [CrossRef]
- Shirasawa, K.; Harada, D.; Hirakawa, H.; Isobe, S.; Kole, C. Chromosome-level de novo genome assemblies of over 100 plant species. Breed. Sci. 2021, 71, 117–124. [Google Scholar] [CrossRef]
- Gao, Y.; Zhang, Y.; Feng, C.; Chu, H.; Feng, C.; Wang, H.; Wu, L.; Yin, S.; Liu, C.; Chen, H.; et al. A chromosome-level genome assembly of Amorphophallus konjac provides insights into konjac glucomannan biosynthesis. Comput. Struct. Biotechnol. J. 2022, 20, 1002–1011. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Jiao, C.; Schwaninger, H.; Chao, C.T.; Ma, Y.; Duan, N.; Khan, A.; Ban, S.; Xu, K.; Cheng, L.; et al. Phased diploid genome assemblies and pan-genomes provide insights into the genetic history of apple domestication. Nat. Genet. 2020, 52, 1423–1432. [Google Scholar] [CrossRef] [PubMed]
- Rendón-Anaya, M.; Ibarra-Laclette, E.; Méndez-Bravo, A.; Lan, T.; Zheng, C.; Carretero-Paulet, L.; Perez-Torres, C.A.; Chacón-López, A.; Hernandez-Guzmán, G.; Chang, T.-H.; et al. The avocado genome informs deep angiosperm phylogeny, highlights introgressive hybridization, and reveals pathogen-influenced gene space adaptation. Proc. Natl. Acad. Sci. USA 2019, 116, 17081–17089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Miao, H.; Liu, J.; Xu, B.; Yao, X.; Xu, C.; Zhao, S.; Fang, X.; Jia, C.; Wang, J.; et al. Musa balbisiana genome reveals subgenome evolution and functional divergence. Nat. Plants 2019, 5, 810–821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, T.; Qi, H.; Luan, X.; Xu, W.; Yu, F.; Zhong, Y.; Xu, M. The chromosome-level genome sequence of the camphor tree provides insights into Lauraceae evolution and terpene biosynthesis. Plant Biotechnol. J. 2021, 20, 244–246. [Google Scholar] [CrossRef] [PubMed]
- Iorizzo, M.; Ellison, S.; Senalik, D.; Zeng, P.; Satapoomin, P.; Huang, J.; Bowman, M.; Iovene, M.; Sanseverino, W.; Cavagnaro, P.; et al. A high-quality carrot genome assembly provides new insights into carotenoid accumulation and asterid genome evolution. Nat. Genet. 2016, 48, 657–666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Q.; Yang, J.; Cui, M.-Y.; Liu, J.; Fang, Y.; Yan, M.; Qiu, W.; Shang, H.; Xu, Z.; Yidiresi, R.; et al. The Reference Genome Sequence of Scutellaria baicalensis Provides Insights into the Evolution of Wogonin Biosynthesis. Mol. Plant 2019, 12, 935–950. [Google Scholar] [CrossRef] [Green Version]
- Feng, L.; Lin, H.; Kang, M.; Ren, Y.; Yu, X.; Xu, Z.; Wang, S.; Li, T.; Yang, W.; Hu, Q. A chromosome-level genome assembly of an alpine plant Crucihimalaya lasiocarpa provides insights into high-altitude adaptation. DNA Res. 2022, 29, dsac004. [Google Scholar] [CrossRef]
- Ling, J.; Xie, X.X.; Gu, X.F.; Zhao, J.L.; Ping, X.X.; Li, Y.; Yang, Y.H.; Mao, Z.C.; Xie, B.Y. High-quality chromosome-level genomes of Cucumis metuliferus and Cucumis melo provide insight into Cucumis genome evolution. Plant J. 2021, 107, 136–148. [Google Scholar] [CrossRef]
- Nunn, A.; Rodríguez-Arévalo, I.; Tandukar, Z.; Frels, K.; Contreras-Garrido, A.; Carbonell-Bejerano, P.; Zhang, P.; Cruz, D.R.; Jandrasits, K.; Lanz, C.; et al. Chromosome-level Thlaspi arvense genome provides new tools for translational research and for a newly domesticated cash cover crop of the cooler climates. Plant Biotechnol. J. 2022, 20, 944–963. [Google Scholar] [CrossRef]
- Li, H.L.; Wu, L.; Dong, Z.M.; Jiang, Y.S.; Jiang, S.J.; Xing, H.T.; Li, Q.; Liu, G.C.; Tian, S.M.; Wu, Z.Y.; et al. Haplotype-Resolved Genome of Diploid Ginger (Zingiber Officinale) and Its Unique Gingerol Biosynthetic Pathway. Hortic. Res. 2021, 8, 189. [Google Scholar] [CrossRef] [PubMed]
- Shan, H.; Kong, H. The genome of Ginkgo biloba refined. Nat. Plants 2021, 7, 714–715. [Google Scholar] [CrossRef] [PubMed]
- Al-Dossary, O.; Alsubaie, B.; Kharabian-Masouleh, A.; Al-Mssallem, I.; Furtado, A.; Henry, R.J. The jojoba genome reveals wide divergence of the sex chromosomes in a dioecious plant. Plant J. 2021, 108, 1283–1294. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Murigneux, V.; Haimovitz, J.; Nock, C.J.; Tian, W.; Masouleh, A.K.; Topp, B.; Alam, M.; Furtado, A.; Henry, R.J. The genome of the endangered Macadamia jansenii displays little diversity but represents an important genetic resource for plant breeding. Plant Direct 2021, 5, e364. [Google Scholar] [CrossRef] [PubMed]
- Nock, C.J.; Baten, A.; Mauleon, R.; Langdon, K.S.; Topp, B.; Hardner, C.; Furtado, A.; Henry, R.J.; King, G.J. Chromosome-Scale Assembly and Annotation of the Macadamia Genome (Macadamia integrifolia HAES 741). G3-Genes Genomes Genet. 2020, 10, 3497–3504. [Google Scholar] [CrossRef]
- Peng, X.; Liu, H.; Chen, P.; Tang, F.; Hu, Y.; Wang, F.; Pi, Z.; Zhao, M.; Chen, N.; Chen, H.; et al. A Chromosome-Scale Genome Assembly of Paper Mulberry (Broussonetia papyrifera) Provides New Insights into Its Forage and Papermaking Usage. Mol. Plant 2019, 12, 661–677. [Google Scholar] [CrossRef] [PubMed]
- Lian, X.D.; Zhang, H.P.; Jiang, C.; Gao, F.; Yan, L.; Zheng, X.B.; Cheng, J.; Wang, W.; Wang, X.B.; Ye, X.; et al. De Novo Chromosome-Level Genome of A Semi-Dwarf Cultivar of Prunus Persica Identifies The Aquaporin Pptip2 as Responsible for Temperature-Sensitive Semi-Dwarf Trait And Ppb3-1 for Flower Type And Size. Plant Biotechnol. J. 2022, 20, 886–902. [Google Scholar] [CrossRef]
- Zhuang, W.; Chen, H.; Yang, M.; Wang, J.; Pandey, M.K.; Zhang, C.; Chang, W.-C.; Zhang, L.; Zhang, X.; Tang, R.; et al. The genome of cultivated peanut provides insight into legume karyotypes, polyploid evolution and crop domestication. Nat. Genet. 2019, 51, 865–876. [Google Scholar] [CrossRef]
- Yuan, G.; Tan, S.; Wang, D.; Yang, Y.; Tian, B. Chromosome-Level Genome Assembly of the Rare and Endangered Tropical Plant Speranskia yunnanensis (Euphorbiaceae). Front. Genet. 2022, 12, 755564. [Google Scholar] [CrossRef]
- Xiong, X.; Gou, J.; Liao, Q.; Li, Y.; Zhou, Q.; Bi, G.; Li, C.; Du, R.; Wang, X.; Sun, T.; et al. The Taxus genome provides insights into paclitaxel biosynthesis. Nat. Plants 2021, 7, 1026–1036. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, S.; Shi, L.; Gong, D.; Zhang, S.; Zhao, Q.; Zhan, D.; Vasseur, L.; Wang, Y.; Yu, J.; et al. Haplotype-resolved genome assembly provides insights into evolutionary history of the tea plant Camellia sinensis. Nat. Genet. 2021, 53, 1250–1259. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.; Chen, Y.; Zhang, X.; Feng, Y.; Comes, H.P.; Li, Z.; Zheng, Z.; Yuan, Y.; Wang, L.; Huang, Z.; et al. Genome sequencing and transcriptome analyses provide insights into the origin and domestication of water caltrop ( Trapa spp., Lythraceae). Plant Biotechnol. J. 2022, 20, 761–776. [Google Scholar] [CrossRef] [PubMed]
- Low, W.Y.; Tearle, R.; Liu, R.; Koren, S.; Rhie, A.; Bickhart, D.; Rosen, B.D.; Kronenberg, Z.N.; Kingan, S.B.; Tseng, E.; et al. Haplotype-resolved genomes provide insights into structural variation and gene content in Angus and Brahman cattle. Nat. Commun. 2020, 11, 2071. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Zhao, X.; Mace, E.; Henry, R.; Jordan, D. Exploring and Exploiting Pan-genomics for Crop Improvement. Mol. Plant 2018, 12, 156–169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, B.; Furtado, A.; Henry, R.J. The coffee bean transcriptome explains the accumulation of the major bean components through ripening. Sci. Rep. 2018, 8, 11414. [Google Scholar] [CrossRef] [Green Version]
- Gillies, S.A.; Futardo, A.; Henry, R.J. Gene expression in the developing aleurone and starchy endosperm of wheat. Plant Biotechnol. J. 2012, 10, 668–679. [Google Scholar] [CrossRef]
- Kasirajan, L.; Hoang, N.V.; Furtado, A.; Botha, F.C.; Henry, R.J. Transcriptome analysis highlights key differentially expressed genes involved in cellulose and lignin biosynthesis of sugarcane genotypes varying in fiber content. Sci. Rep. 2018, 8, 11612. [Google Scholar] [CrossRef] [Green Version]
- Nirmal, R.C.; Furtado, A.; Wrigley, C.; Henry, R.J. Influence of Gene Expression on Hardness in Wheat. PLoS ONE 2016, 11, e0164746. [Google Scholar] [CrossRef] [Green Version]
- Rangan, P.; Furtado, A.; Henry, R. Differential response of wheat genotypes to heat stress during grain filling. Exp. Agric. 2019, 55, 818–827. [Google Scholar] [CrossRef] [Green Version]
- Rangan, P.; Furtado, A.; Henry, R.J. The transcriptome of the developing grain: A resource for understanding seed development and the molecular control of the functional and nutritional properties of wheat. BMC Genom. 2017, 18, 766. [Google Scholar] [CrossRef] [Green Version]
- Cuperus, J.T. Single-cell genomics in plants: Current state, future directions, and hurdles to overcome. Plant Physiol. 2021, 188, 749–755. [Google Scholar] [CrossRef] [PubMed]
- Hoang, N.V.; Furtado, A.; Thirugnanasambandam, P.P.; Botha, F.C.; Henry, R.J. De novo assembly and characterizing of the culm-derived meta-transcriptome from the polyploid sugarcane genome based on coding transcripts. Heliyon 2018, 4, e00583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tirumala, R.K.; Subbaiyan, G.K.; Singh, A.K.; Furtado, A.; Henry, R.J. RNA-Seq to Understand Transcriptomes and Application in Improving Crop Quality. In Comprehensive Foodomics, 1st ed.; Cifuentes, A., Ed.; Elsevier: Amsterdam, The Netherlands, 2021; Volume 1, pp. 472–485. [Google Scholar] [CrossRef]
- Margarido, G.R.A.; Correr, F.H.; Furtado, A.; Botha, F.C.; Henry, R.J. Limited allele-specific gene expression in highly polyploid sugarcane. Genome Res. 2022, 32, 297–308. [Google Scholar] [CrossRef] [PubMed]
- Powell, J.J.; Fitzgerald, T.L.; Stiller, J.; Berkman, P.J.; Gardiner, D.M.; Manners, J.M.; Henry, R.J.; Kazan, K. The defence-associated transcriptome of hexaploid wheat displays homoeolog expression and induction bias. Plant Biotechnol. J. 2016, 15, 533–543. [Google Scholar] [CrossRef] [PubMed]
- Cheng, B.; Furtado, A.; Henry, R.J. Long-read sequencing of the coffee bean transcriptome reveals the diversity of full-length transcripts. GigaScience 2017, 6, gix086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, S.; Zhang, J.; Han, H.; Zhang, J.; Ma, H.; Zhang, Z.; Lu, Y.; Liu, W.; Yang, X.; Li, X.; et al. Full-length transcriptome sequences of Agropyron cristatum facilitate the prediction of putative genes for thousand-grain weight in a wheat-A. cristatum translocation line. BMC Genom. 2019, 20, 1025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, H.Z.; Yu, H.M.; Huang, T.; Shen, X.J.; Xia, J.; Pang, F.H.; Wang, J.; Zhao, M.Z. The Complexity of The Fragaria X Ananassa (Octoploid) Transcriptome by Single-Molecule Long-Read Sequencing. Hortic. Res. 2019, 6, 46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoang, N.V.; Furtado, A.; Mason, P.J.; Marquardt, A.; Kasirajan, L.; Thirugnanasambandam, P.P.; Botha, F.C.; Henry, R.J. A survey of the complex transcriptome from the highly polyploid sugarcane genome using full-length isoform sequencing and de novo assembly from short read sequencing. BMC Genom. 2017, 18, 395. [Google Scholar] [CrossRef]
- Hoang, N.V.; Furtado, A.; McQualter, R.B.; Henry, R.J. Next generation sequencing of total DNA from sugarcane provides no evidence for chloroplast heteroplasmy. New Negat. Plant Sci. 2015, 1–2, 33–45. [Google Scholar] [CrossRef] [Green Version]
- Dobrogojski, J.; Adamiec, M.; Luciński, R. The chloroplast genome: A review. Acta Physiol. Plant. 2020, 42, 98. [Google Scholar] [CrossRef]
- Henry, R.; Rice, N.; Edwards, M.; Nock, C. Next-Generation Technologies to Determine Plastid Genome Sequences. In Chloroplast Biotechnology; Maliga, P., Ed.; Humana Press: Totova, NJ, USA, 2014; Volume 1132, pp. 39–46. [Google Scholar]
- Nock, C.J.; Waters, D.L.; Edwards, M.A.; Bowen, S.G.; Rice, N.; Cordeiro, G.M.; Henry, R.J. Chloroplast genome sequences from total DNA for plant identification. Plant Biotechnol. J. 2010, 9, 328–333. [Google Scholar] [CrossRef] [PubMed]
- Guyeux, C.; Charr, J.-C.; Tran, H.T.M.; Furtado, A.; Henry, R.J.; Crouzillat, D.; Guyot, R.; Hamon, P. Evaluation of chloroplast genome annotation tools and application to analysis of the evolution of coffee species. PLoS ONE 2019, 14, e0216347. [Google Scholar] [CrossRef] [PubMed]
- Ananda, G.; Norton, S.; Blomstedt, C.; Furtado, A.; Møller, B.; Gleadow, R.; Henry, R. Phylogenetic relationships in the Sorghum genus based on sequencing of the chloroplast and nuclear genes. Plant Genome 2021, 14, e20123. [Google Scholar] [CrossRef] [PubMed]
- Brozynska, M.; Copetti, D.; Furtado, A.; Wing, R.A.; Crayn, D.; Fox, G.; Ishikawa, R.; Henry, R.J. Sequencing of Australian wild rice genomes reveals ancestral relationships with domesticated rice. Plant Biotechnol. J. 2016, 15, 765–774. [Google Scholar] [CrossRef] [Green Version]
- Healey, A.; Lee, D.J.; Furtado, A.; Henry, R.J. Evidence of inter-sectional chloroplast capture in Corymbia among sections Torellianae and Maculatae. Aust. J. Bot. 2018, 66, 369. [Google Scholar] [CrossRef]
- Hodel, R.G.J.; Zimmer, E.A.; Liu, B.-B.; Wen, J. Synthesis of Nuclear and Chloroplast Data Combined With Network Analyses Supports the Polyploid Origin of the Apple Tribe and the Hybrid Origin of the Maleae—Gillenieae Clade. Front. Plant Sci. 2022, 12, 820997. [Google Scholar] [CrossRef] [PubMed]
- Yoo, K.; Lowry, P.P.; Wen, J. Discordance of chloroplast and nuclear ribosomal DNA data in Osmorhiza (Apiaceae). Am. J. Bot. 2002, 89, 966–971. [Google Scholar] [CrossRef]
- Yu, W.-B.; Huang, P.-H.; Li, D.-Z.; Wang, H. Incongruence between Nuclear and Chloroplast DNA Phylogenies in Pedicularis Section Cyathophora (Orobanchaceae). PLoS ONE 2013, 8, e74828. [Google Scholar] [CrossRef]
- Gualberto, J.M.; Mileshina, D.; Wallet, C.; Niazi, A.K.; Weber-Lotfi, F.; Dietrich, A. The plant mitochondrial genome: Dynamics and maintenance. Biochimie 2014, 100, 107–120. [Google Scholar] [CrossRef]
- Qin, L.; Hu, Y.; Wang, J.; Wang, X.; Zhao, R.; Shan, H.; Li, K.; Xu, P.; Wu, H.; Yan, X.; et al. Insights into angiosperm evolution, floral development and chemical biosynthesis from the Aristolochia fimbriata genome. Nat. Plants 2021, 7, 1239–1253. [Google Scholar] [CrossRef]
- Denoeud, F.; Carretero-Paulet, L.; Dereeper, A.; Droc, G.; Guyot, R.; Pietrella, M.; Zheng, C.; Alberti, A.; Anthony, F.; Aprea, G.; et al. The coffee genome provides insight into the convergent evolution of caffeine biosynthesis. Science 2014, 345, 1181–1184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Julca, I.; Ferrari, C.; Flores-Tornero, M.; Proost, S.; Lindner, A.-C.; Hackenberg, D.; Steinbachová, L.; Michaelidis, C.; Gomes Pereira, S.; Misra, C.S.; et al. Comparative transcriptomic analysis reveals conserved programmes underpinning organogenesis and reproduction in land plants. Nat. Plants 2021, 7, 1143–1159. [Google Scholar] [CrossRef] [PubMed]
- Stein, J.C.; Yu, Y.; Copetti, D.; Zwickl, D.J.; Zhang, L.; Zhang, C.; Chougule, K.; Gao, N.; Iwata, A.; Goicoechea, J.L.; et al. Publisher Correction: Genomes of 13 domesticated and wild rice relatives highlight genetic conservation, turnover and innovation across the genus Oryza. Nat. Genet. 2018, 50, 285–296. [Google Scholar] [CrossRef] [PubMed]
- Park, H.-S.; Lee, W.K.; Lee, S.-C.; Lee, H.O.; Joh, H.J.; Park, J.Y.; Kim, S.; Song, K.; Yang, T.-J. Inheritance of chloroplast and mitochondrial genomes in cucumber revealed by four reciprocal F1 hybrid combinations. Sci. Rep. 2021, 11, 2506. [Google Scholar] [CrossRef] [PubMed]
- Henry, R.J. Genomics strategies for germplasm characterization and the development of climate resilient crops. Front. Plant Sci. 2014, 5, 68. [Google Scholar] [CrossRef]
- Bradbury, L.M.T.; Fitzgerald, T.L.; Henry, R.J.; Jin, Q.; Waters, D.L.E. The gene for fragrance in rice. Plant Biotechnol. J. 2005, 3, 363–370. [Google Scholar] [CrossRef]
- Bradbury, L.M.T.; Henry, R.J.; Jin, Q.; Reinke, R.F.; Waters, D. A Perfect Marker for Fragrance Genotyping in Rice. Mol. Breed. 2005, 16, 279–283. [Google Scholar] [CrossRef]
- Waters, D.L.E.; Henry, R.J.; Reinke, R.F.; Fitzgerald, M.A. Gelatinization temperature of rice explained by polymorphisms in starch synthase. Plant Biotechnol. J. 2006, 4, 115–122. [Google Scholar] [CrossRef]
- Nirmal, R.C.; Furtado, A.; Rangan, P.; Henry, R.J. Fasciclin-like arabinogalactan protein gene expression is associated with yield of flour in the milling of wheat. Sci. Rep. 2017, 7, 12539. [Google Scholar] [CrossRef] [Green Version]
- Furtado, A.; Bundock, P.; Banks, P.M.; Fox, G.; Yin, X.; Henry, R.J. A novel highly differentially expressed gene in wheat endosperm associated with bread quality. Sci. Rep. 2015, 5, 10446. [Google Scholar] [CrossRef]
- Henry, R.J. Plant Genotyping: The DNA Fingerprinting of Plants; CABI Publishing: Oxford, UK, 2001; p. 325. [Google Scholar]
- Henry, R.J.; Edwards, M.; Waters, D.L.E.; Krishnan, S.G.; Bundock, P.; Sexton, T.R.; Kharabian-Masouleh, A.; Nock, C.; Pattemore, J. Application of large-scale sequencing to marker discovery in plants. J. Biosci. 2012, 37, 829–841. [Google Scholar] [CrossRef] [PubMed]
- Krishna, H.; Alizadeh, M.; Singh, D.; Singh, U.; Chauhan, N.; Eftekhari, M.; Sadh, R.K. Somaclonal variations and their applications in horticultural crops improvement. 3 Biotech 2016, 6, 54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henry, R.J. Genomics and Gene-Editing Technologies Accelerating Grain Product Innovation. Cereal Foods World 2019, 64. [Google Scholar] [CrossRef]
- Henry, R.J. Advances in DNA sequencing enabling more rapid development of improved biomass and biofuel conversion technologies. Biofuels 2012, 3, 507–509. [Google Scholar] [CrossRef]
- Sherman, B.; Henry, R.J. Access to biodiversity for food production: Reconciling open access digital sequence information with access and benefit sharing. Mol. Plant 2021, 14, 701–704. [Google Scholar] [CrossRef]
- Sherman, B.; Henry, R.J. The Nagoya Protocol and historical collections of plants. Nat. Plants 2020, 6, 430–432. [Google Scholar] [CrossRef]
- Henry, R. Innovations in Agriculture and Food Supply in Response to the COVID-19 Pandemic. Mol. Plant 2020, 13, 1095–1097. [Google Scholar] [CrossRef]
- Henry, R.J. Innovations in plant genetics adapting agriculture to climate change. Curr. Opin. Plant Biol. 2019, 56, 168–173. [Google Scholar] [CrossRef]
- Abberton, M.; Batley, J.; Bentley, A.; Bryant, J.; Cai, H.; Cockram, J.; De Oliveira, A.C.; Cseke, L.J.; Dempewolf, H.; De Pace, C.; et al. Global agricultural intensification during climate change: A role for genomics. Plant Biotechnol. J. 2015, 14, 1095–1098. [Google Scholar] [CrossRef] [Green Version]
- Henry, R.J. Sequencing of wild crop relatives to support the conservation and utilization of plant genetic resources. Plant Genet. Resour. 2014, 12, S9–S11. [Google Scholar] [CrossRef] [Green Version]
- McCouch, S.; Navabi, Z.K.; Abberton, M.; Anglin, N.L.; Barbieri, R.L.; Baum, M.; Bett, K.; Booker, H.; Brown, G.L.; Bryan, G.J.; et al. Mobilizing Crop Biodiversity. Mol. Plant 2020, 13, 1341–1344. [Google Scholar] [CrossRef] [PubMed]
| Species | Technique * | Reference | |
|---|---|---|---|
| Amorphophallus | Amorphophallas konjac | Hi-C | [41] |
| Apple | Malus domestica | Genetic Map | [42] |
| Avocado | Persea americana | Genetic Map | [43] |
| Banana | Musa balbisiana | Hi-C | [44] |
| Camphor | Cinnamomum camphora | Hi-C | [45] |
| Carrot | Daucus carota | Genetic Map | [46] |
| Chinese Skullcap | Scutellaria baicalensis | Hi-C | [47] |
| Crucihimalaya | Crucihimalaya lasicocarpa | Hi-C | [48] |
| Cucumber | Cucumis metuliferus | Hi-C | [49] |
| Eucalypt | Corymbia citriodora | Genetic Map | [37] |
| Field Pennycresss | Thlaspi arvense | Genetic Map/Hi-C/Bionano | [50] |
| Ginger | Zingiber officinale | Hi-C | [51] |
| Ginkgo | Ginkgo biloba | Hi-C | [52] |
| Jojoba | Simmondsia chinensis | Hi-C | [53] |
| Macadamia | Macadamia jansenii | Hi-C | [54] |
| Macadamia integrifolia | Genetic Map | [55] | |
| Paper Mulberry | Broussonetis papyrifera | Hi-C | [56] |
| Peach | Prunus persica | Hi-C | [57] |
| Peanut | Arachis hypogaea | Hi-C | [58] |
| Speranskia | Speranskia yunnanensis | Hi-C | [59] |
| Taxus | Taxus chinensis | Hi-C | [60] |
| Tea | Camellia sinensus | Hi-C | [61] |
| Water Caltrop | Trapa spp. | Hi-C | [62] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Henry, R.J. Progress in Plant Genome Sequencing. Appl. Biosci. 2022, 1, 113-128. https://doi.org/10.3390/applbiosci1020008
Henry RJ. Progress in Plant Genome Sequencing. Applied Biosciences. 2022; 1(2):113-128. https://doi.org/10.3390/applbiosci1020008
Chicago/Turabian StyleHenry, Robert J. 2022. "Progress in Plant Genome Sequencing" Applied Biosciences 1, no. 2: 113-128. https://doi.org/10.3390/applbiosci1020008
APA StyleHenry, R. J. (2022). Progress in Plant Genome Sequencing. Applied Biosciences, 1(2), 113-128. https://doi.org/10.3390/applbiosci1020008






