Abstract
Across the world, asthma is a prominent and widespread respiratory disorder that has a substantial clinical and socioeconomic influence. The classification of asthma subtypes should be performed precisely and effectively, with objectives such as personalized treatments, improved rehabilitation outcomes, and preventing tragic exacerbations. Typical screening approaches are primarily based on spirometry measures, immunologic assessments, and individual clinical diagnoses, and they are commonly affected by limitations such as uncertainty, crossover disparities, and restricted generalizability among various groups of patients. This study utilizes machine learning (ML) methodologies as a Data-Driven Approach (DDA)-based framework for asthma classification to overcome the mentioned challenges. Methodically constructed and evaluated classifiers, such as Random Forest and XGBoost, use the Asthma Disease Dataset from Kaggle, which consists of demographic data, lung function metrics (FEV1, FVC, FEV1/FVC ratio, and PEFR), and immunoglobulin E (IgE) biomarkers. A wide range of metrics such as accuracy, precision, recall, F1-score, receiver operating characteristic area under the curve (ROC-AUC), and average precision (AP) are used exhaustively to assess the performance of the model. The results indicate that though each model exhibits outstanding forecasting abilities, XGBoost has an enhanced classification capability, especially in recall and AP, which minimizes the proportion of false negatives, resulting in a clinically noteworthy result. The significance of the FEV1/FVC ratio, IgE levels, and PEFR as key indicators is recognized by feature interpretability analysis. These results emphasize the ability of ML-powered evaluation in advancing personalized healthcare and revolutionizing the clinical management of asthma.
1. Introduction
As mentioned by the WHO, in 2022, over 262 million people globally struggled with asthma, the most widespread chronic respiratory disorder. The WHO also mentioned that nearly half a million deaths are caused by this disease. The disease is generally described by bronchial hyperreactivity, unstable, blocked airways, and airflow inflammation, and it places a heavy load on patients’ daily activities and a remarkable economic burden on healthcare systems [1]. Asthma is a chronic disease that includes a variety of phenotype versions, such as allergic, non-allergic, occupational, and exercise-related categories. So, asthma is not considered a typical illness. It is crucial to correctly identify the phenotype of asthma and its severity range to customize personalized treatment strategies, prevent sudden flare-ups, and achieve enhanced results that last for longer periods of time [2].
Standard diagnostic methods primarily rely on clinical history, pulmonary function tests such as spirometry (forced expiratory volume in one second [FEV1], forced vital capacity [FVC], and FEV1/FVC ratio), and biomarker indices like immunoglobulin E (IgE). These procedures, although clinically applicable, are mostly restricted by measuring variations, interpretation biases, and intersecting biomarker characteristics in different medical conditions. Spirometry results, for instance, may vary with patient activities, while IgE levels are not specific to asthma. Likewise, typical statistical structures are not well suited to deal with a combined perspective of a diverse number of data, which is essential for correct asthma phenotyping [3].
With the power to detect hidden patterns and find unpredictable relationships in huge datasets, machine learning (ML) represents a feasible solution for boosting diagnostic accuracy [4]. Earlier research has illustrated the importance of machine learning in asthma exacerbation prediction, differentiating phenotypes, and recognizing recurring respiratory problems. However, there still exists an inadequate amount of comprehensive research on solid ensemble methods such as Random Forest and XGBoost using a combination of demographic, spirometry, and immunological data [5].
By developing ML-driven models for asthma classification, this study aims to fill this gap. This work consists of four steps: The first step performs preprocessing and organizing the Asthma Disease Dataset from Kaggle for analysis purposes. The second builds and applies Random Forest and XGBoost classifiers to pulmonary, demographic, and biomarker fields. The third step is the use of assessment metrics such as ROC-AUC, average precision (AP), accuracy, and recall for performance comparison. The study ends with the use of feature importance analysis to recognize the key factors that support the diagnosis of asthma.
By utilizing such measures, this research encourages the formation of reliable, comprehensive decision support systems designed to enhance clinical proficiency and facilitate more accurate asthma management techniques.
2. Background
Various approaches have contributed to the prediction, classification, and management of asthma. Integration of artificial intelligence with machine learning can generate a profound effect and it is used by the authors of [6]. Studies have also used deep neural networks and compared traditional approaches with DL, SVM, and RF to obtain outcomes based on diverse datasets. In [7], the authors use machine learning algorithms on Electronic Health Records (EHR) datasets from clinical data. The results show that the ensemble method used in their research improves asthma exacerbation prediction accuracy by 10–15% compared to traditional regression methods. The challenges are data privacy in individuals’ records and difficulty in data fragmentation. So, here, federated learning and advanced data preprocessing can overcome these challenges.
A combination of data augmentation, feature selection, and XGBoost classification is proposed in [8], where the research denotes the effectiveness of feature engineering in reducing overfitting and improving generalizability. But at the same time, the computational cost of high-quality feature selection is very high. The study conducted in [9] focuses on hyperparameter tuning and optimization algorithms in predictive modeling of asthma exacerbation. The study examines optimization methods such as Genetic Algorithms, Grid Search, and Bayesian Optimization applied to asthma prediction frameworks. The analysis demonstrates a 5–10% improvement in model performance when optimization techniques are incorporated. The limitations are high computational complexity and limited datasets for model training and validation.
Table 1 shows a summary and comparative analysis of machine learning approaches in asthma classification and prediction. Some of these approaches are in collaboration with deep learning, IoT, and sensor technology or with genetic algorithms.

Table 1.
Comparative analysis of machine learning approaches in asthma classification and prediction.
3. Methodology
3.1. Dataset Description
The dataset used in this study was assembled from public repositories, clinical datasets, and custom-acquired data. This dataset was specifically curated to facilitate the exploration of computational methods for asthma prediction and classification, integrating demographic, spirometric, and immunological parameters that reflect both clinical practice and research-oriented perspectives. The Asthma Disease Dataset from Kaggle [19] holds a total of 7425 masked patient records, each associated with an individual clinical profile and 29 columns. Each record consists of demographic attributes and statistical medical results, which are essential to asthma diagnosis and phenotyping. The dataset is organized in a tabular format (CSV), in which rows represent patients and columns represent features. The target variable is Diagnosis, encoded as a binary outcome:
1: Asthmatic Patient;
0: Non-asthmatic Patient.
Table 2 denotes the features that are included in this study from the given dataset. Each feature in the dataset was selected due to its direct clinical association with asthma pathophysiology; for example, spirometry indices (FEV1, FVC, FEV1/FVC, PEFR) are widely used gold-standard diagnostic tools to evaluate lung function, detect airflow limitations, and monitor disease progression. IgE levels serve as an immunological biomarker; they are often elevated in allergic asthma phenotypes and are useful for distinguishing asthma from other respiratory conditions. Demographics (age, gender) captures population-level trends, since asthma prevalence and severity vary by age group and gender (e.g., there is a higher prevalence in young males but a greater severity in adult females).

Table 2.
Features included.
3.2. DDA-Based Framework
The DDA-based framework (Figure 1) delineates the complete workflow for asthma classification using machine learning, beginning with data collection and progressing through preprocessing, feature selection, model training, evaluation, interpretability, and final prediction.
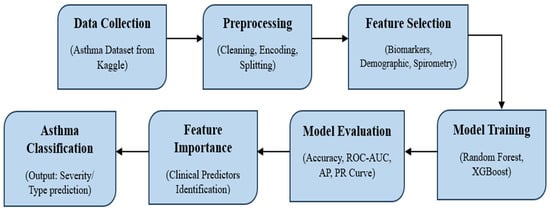
Figure 1.
Data-driven approach-based framework for asthma classification.
3.2.1. Data Collection
The foundation of this framework is the Asthma Disease Dataset from Kaggle, which integrates demographic attributes such as age and gender with clinical measurements, including spirometry indices (FEV1, FVC, FEV1/FVC ratio, PEFR) and biomarkers such as Immunoglobulin E (IgE). This heterogeneous dataset mirrors real-world diagnostic practice, where physicians combine multiple information sources to make informed decisions.
3.2.2. Preprocessing
As clinical datasets in raw form are rarely used for analysis purposes, numerous data preprocessing steps must be performed. Data cleansing involved assigning missing values and addressing outliers to ensure statistical validity. The process of normalization was performed on spirometry and biomarker values to maintain balanced scales, while encoding was performed on categorical variables like gender in a numerical manner. By applying proportional sampling and class proportions for unbiased evaluation, the dataset was then further split into subsets for training (80%) and testing (20%).
3.2.3. Feature Selection
After preprocessing, the next step to perform was selecting the exact features that have clinical significance. The FEV1/ FVC ratio, typically known as spirometry index, indicates airway congestion and persisting diagnostic foundations. IgE level is a biomarker that shows asthma phenotypes related to allergies, while PEFR measures provide lung function. Considering all these features ensured that operational and clinical aspects of asthma were modeled. Feature selection is also important for generating more transparent results that can be used in clinical decisions and for better interpretability.
3.2.4. Model Training
Random Forest (RF) and XGBoost were the two (ensemble) methods used for model development. Construction of various decision trees was carried out on bootstrap data, and their results were aggregated by the Random Forest method, which offers built-in feature importance analysis and robustness against noise. Meanwhile, the XGBoost method incrementally constructs trees to correct errors, captures nonlinear relationships, and, at the same time, incorporates regularization to prevent overfitting, which is a common issue found with medical datasets. Both models were selected for their balance of predictive accuracy, robustness, and clinical applicability.
3.2.5. Model Evaluation
Various supplementary metrics were used to evaluate the model. While accuracy provided a broad measure of performance, ROC-AUC captured the trade-off between sensitivity and specificity, with higher AUC values indicating stronger discriminative ability. Precision–recall curves and the average precision (AP) score were emphasized to account for the clinical consequences of false negatives, where undetected asthma can lead to severe exacerbations. Confusion matrices provided further insights into correct and incorrect classifications, highlighting areas of model strength and weakness.
3.2.6. Feature Importance
By using feature importance analysis, interpretability can be guaranteed. Whereas XGBoost measures feature gain, Random Forest uses Gini-based importance. In line with accepted medical wisdom and bolstering clinical confidence, both consistently found that the most significant predictors were FEV1/FVC ratio, IgE, and PEFR [20].
3.2.7. Asthma Classification
Eventually, the trained models were used for classification of asthma, producing results on the probability of whether the categorized patients had asthma or non-asthma conditions. As per the clinical requirements, the threshold can be adjusted for giving priority to sensitivity or specificity. These results demonstrate how machine learning can evolve into a decision support tool, complementing physician expertise and enabling earlier, more objective, and data-driven asthma management.
4. Experimental Results
A Data-Driven Approach (DDA)-based framework for asthma classification was trained on the ensemble methods of Random Forest and XGBoost by utilizing machine learning methodologies. The purpose of this study was to assess how well a machine learning model (DDA) classifies asthma detection. The model was trained on a dataset of different asthma parameters using the mentioned model training approaches. The findings demonstrated that, particularly in low-label situations, DDA-pretrained models continuously outperformed supervised baselines. An extensive evaluation of the model’s predictive capabilities for the two classes, non-asthmatic (0) and asthmatic (1), was given in the confusion matrix derived for Random forest and XGBoost. Figure 2 shows the confusion matrix for Random Forest; it worked on 300 samples of testing data and classified 179 records as non-asthmatic and 121 as asthmatic.
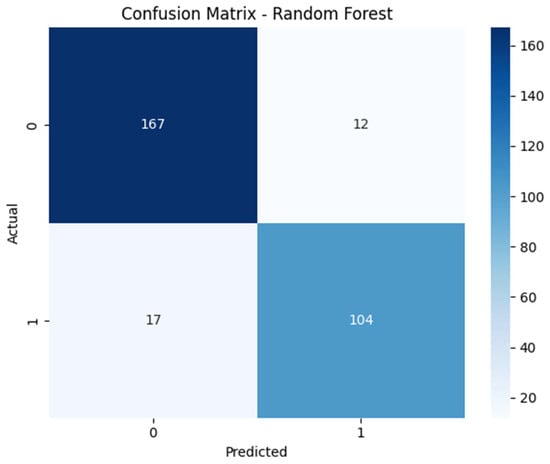
Figure 2.
Confusion matrix illustrating classification performance of Random Forest.
Similarly, Figure 3 denotes the confusion matrix for XFBoost, which was similarly employed on 300 samples and which classified 179 records as non-asthmatic and 121 as asthmatic. In the context of the confusion matrix, both ensemble methods achieved superior performance in terms of classification, but XGBoost produced slightly more true positives, which is crucial to clinical acceptance. Here, both methods performed well in terms of classification, but by analyzing the confusion matrix, it is visible that XGBoost slightly outperformed Random Forest by minimizing false negatives.
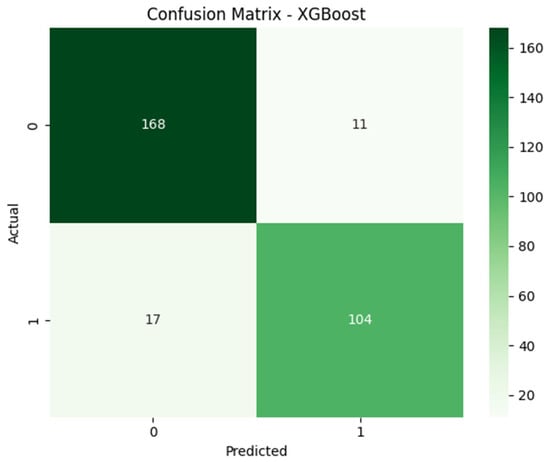
Figure 3.
Confusion matrix illustrating classification performance of XGBoost.
The accuracy analysis (AUC) and the receiver operating characteristic (ROC) curves are highlighted in Figure 4 for both methods. The ROC-AUC curve is used to visualize each model’s potential to achieve both sensitivity and specificity. By observing the visualization, it can be found that both methods achieved 0.95 accuracy in classification, which is a similarity. But the curve for XGBoost increases slightly at the top left side, ensuring more true positives, and lowers slightly in the top right corner, generating fewer false negatives. This confirms that it has slightly superior performance in the classification of asthmatic and non-asthmatic data.
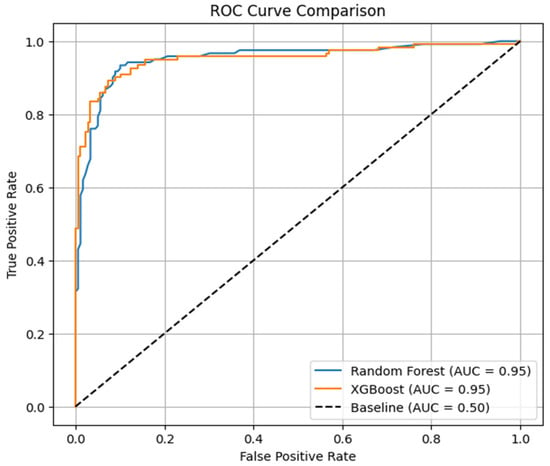
Figure 4.
ROC curve comparison of Random Forest and XGBoost.
To evaluate the classification model’s performance when dealing with an imbalanced dataset (179—non-asthmatic; 121—asthmatic), a precision–recall curve with average precision was used, as shown in Figure 5. Random Forest achieved a score of 0.94 in average precision; it performed well at lower recall, but as recall increased, it lost precision slightly. On the other hand, XGBoost scored an average precision score of 0.95, achieving higher and more stable precision despite variations in recall thresholds. These outcomes denote the ability of XGBoost to handle imbalanced classes and classifications with precision.
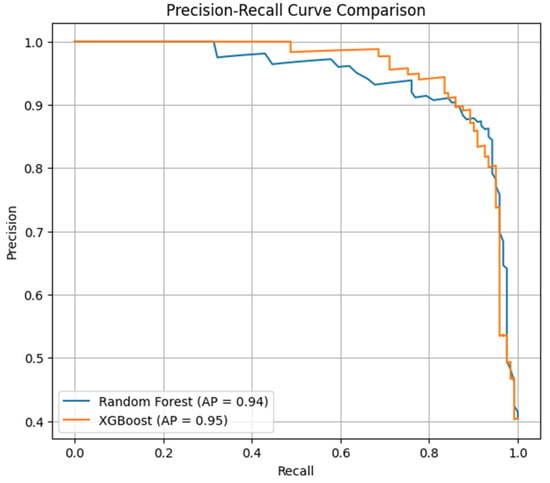
Figure 5.
Comparison of Random Forest and XGBoost with average precision score using precision–recall curve.
Overall, both the Random Forest and XGBoost ensemble methods performed efficiently. XGBosst continuously outperformed Random Forest in ROC-AUC, recall, and average precision. Its potential to minimize false negatives while retaining transparency makes it a versatile tool to deploy in clinical decision-making systems, as it offers reliable, trustworthy, and data-driven insights for personalized asthma management strategies.
5. Conclusions
By employing demographic, spirometry, and biomarker data, this work highlights the ways in which machine learning might serve as an effective tool for improving the classification of asthma. This study also shows that ensemble approaches can identify complicated patterns and increase diagnostic accuracy by employing the Random Forest and XGBoost algorithms. As per the experimental results, XGBoost continuously outperformed Random Forest, particularly in recall and average precision via reducing false negatives, which is a crucial element in clinical settings. In summary, this work sets the foundation for data-driven decision support systems that can supplement medical expertise, enabling early detection, personalized treatment, and better patient outcomes. Future developments focused on feature importance analysis may help to confirm the clinical validity of spirometric indices and IgE levels as key predictors which are highly accepted in medical practice. Meanwhile, the use of huge datasets, using more biomarkers, and using explainable AI approaches may further boost the clinical applicability of such models.
6. Future Scope
Although this study illustrates the effectiveness of machine learning in asthma classification, several approaches or areas are still open for further investigation. Feature importance analysis may confirm the clinical validity of spirometric indices and IgE levels as key predictors which are highly accepted in medical practice. Broad and multifaceted datasets integrating continuous data could improve the generalization of model use across demographic and age categories. Incorporating some more biomarkers, like eosinophil counts, exhaled nitric oxide, and genetic profiles, may help to enhance the identification of asthma phenotypes over those obtained by spirometry and IgE levels. Meanwhile, broadening the process of asthma classification from one-dimensional diagnosis to multi-class allocation such as allergic vs. non-allergic, severity levels, etc., will provide more clinical perspectives. Real-time deployment via mobile health applications and wearable devices could also enable continuous tracking, predictive notifications, and personalized management tactics. Subsequently, to narrow down the divide between computational forecasting and feasible medical decision-making processes, explainable AI frameworks should be utilized to enhance accountability and nurture clinician trust.
Author Contributions
Conceptualization, B.S.P. and A.D.K.; methodology, B.S.P.; software, P.P.P.; validation, B.S.P., A.D.K. and P.P.P.; formal analysis, B.S.P.; investigation, B.S.P.; resources, B.S.P.; data curation, B.S.P.; writing—original draft preparation, B.S.P.; writing—review and editing, B.S.P.; visualization, P.P.P.; supervision, A.D.K.; project administration, B.S.P.; funding acquisition, B.S.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Dr. D. Y. Patil School of Science and Technology, Dr. D. Y Patil Vidyapeeth, Pune, India grant number [DPU/ 1212(1)/ 2024] and the APC was funded by [DPU/ 1212(1)/ 2024].
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in Kaggle at https://www.kaggle.com/datasets/rabieelkharoua/asthma-disease-dataset (accessed on 1 October 2025) [19].
Acknowledgments
The authors gratefully acknowledge the financial support provided by the Dr. D.Y. Patil School of Science and Technology, Dr. D.Y Patil Vidyapeeth, Pune, India, through the Seed Money Project. We also extend our thanks to Dr. D.Y. Patil Vidyapeeth for providing the necessary facilities to conduct this research.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Budiarto, A.; Tsang, K.C.H.; Wilson, A.M.; Sheikh, A.; Shah, S.A. Machine learning–based asthma attack prediction models from routinely collected electronic health records: Systematic scoping review. JMIR AI 2023, 2, e46717. [Google Scholar] [CrossRef] [PubMed]
- Romero Tapia, S.J.; Becerril Negrete, J.R.; Castro Rodriguez, J.A.; Del Río Navarro, B.E. Early prediction of asthma. J. Clin. Med. 2023, 12, 5404. [Google Scholar] [CrossRef] [PubMed]
- Al Meslamani, A.Z. How AI is advancing asthma management? Insights into economic and clinical aspects. J. Med. Econ. 2023, 26, 1489–1494. [Google Scholar] [CrossRef] [PubMed]
- Tsang, K.C.H.; Pinnock, H.; Wilson, A.M.; Shah, S.A. Application of machine learning algorithms for asthma management with mHealth: A clinical review. J. Asthma Allergy 2022, 15, 855–873. [Google Scholar] [CrossRef] [PubMed]
- Tong, Y.; Lin, B.; Chen, G.; Zhang, Z. Predicting continuity of asthma care using a machine learning model: Retrospective cohort study. Int. J. Environ. Res. Public Health 2022, 19, 1237. [Google Scholar] [CrossRef] [PubMed]
- Alkobaisi, S.; Safdar, M.F.; Pałka, P.; Abu Ali, N.A. Artificial intelligence algorithms in asthma management: A review of data engineering, predictive models, and future implications. Appl. Sci. 2025, 15, 3609. [Google Scholar] [CrossRef]
- Molfino, N.A.; Turcatel, G.; Riskin, D. Machine learning approaches to predict asthma exacerbations: A narrative review. Adv. Ther. 2024, 41, 534–552. [Google Scholar] [CrossRef] [PubMed]
- Lee, Z.-J.; Yang, M.-R.; Hwang, B.-J. A sustainable approach to asthma diagnosis: Classification with data augmentation, feature selection, and boosting algorithm. Diagnostics 2024, 14, 723. [Google Scholar] [CrossRef] [PubMed]
- Adamu Aliyu, D.; Akhir, E.A.P.; Saidu, Y.; Adamu, S.; Umar, K.I.; Bunu, A.S.; Mamman, H. Optimization techniques for asthma exacerbation prediction models: A systematic literature review. IEEE Access 2024, 12, 110862–110890. [Google Scholar] [CrossRef]
- Almuhanna, H.; Alenezi, M.; Abualhasan, M.; Alajmi, S.; Alfadhli, R.; Karar, A.S. AI asthma guard: Predictive wearable technology for asthma management in vulnerable populations. Appl. Syst. Innov. 2024, 7, 78. [Google Scholar] [CrossRef]
- Pawan, P.; Singh, R.; Kumar, M.; Goyal, D.; Neeraj, N.; Yadav, D. An effective and comparative analysis of asthma prediction using machine learning algorithms. Int. J. Intell. Syst. Appl. Eng. 2024, 12, 851–856. [Google Scholar]
- Gunawardana, J.; Viswakula, S.D.; Rannan-Eliya, R.P.; Wijemunige, N. Machine learning approaches for asthma disease prediction among adults in Sri Lanka. Health Inform. J. 2024, 30, 14604582241283968. [Google Scholar] [CrossRef] [PubMed]
- Tomita, K.; Yamasaki, A.; Katou, R.; Ikeuchi, T.; Touge, H.; Sano, H.; Tohda, Y. Construction of a diagnostic algorithm for diagnosis of adult asthma using machine learning with random forest and XGBoost. Diagnostics 2023, 13, 3069. [Google Scholar] [CrossRef] [PubMed]
- Aulia, D.; Sarno, R.; Hidayati, S.C.; Rivai, M. Optimization of the electronic nose sensor array for asthma detection based on genetic algorithm. IEEE Access 2023, 11, 74924–74935. [Google Scholar] [CrossRef]
- Xiong, S.; Chen, W.; Jia, X.; Jia, Y.; Liu, C. Machine learning for prediction of asthma exacerbations among asthmatic patients: A systematic review and meta-analysis. BMC Pulm. Med. 2023, 23, 278. [Google Scholar] [CrossRef] [PubMed]
- Awal, M.A.; Hossain, M.S.; Debjit, K.; Ahmed, N.; Nath, R.D.; Habib, G.M.; Khan, M.S.; Islam, M.A.; Mahmud, M.P. An early detection of asthma using BOMLA detector. IEEE Access 2021, 9, 58403–58420. [Google Scholar] [CrossRef]
- Kuo, H.-C.; Lin, B.-S.; Wang, Y.-D.; Lin, B.-S. Development of automatic wheeze detection algorithm for children with asthma. IEEE Access 2021, 9, 126882–126890. [Google Scholar] [CrossRef]
- Singh, O.P.; Palaniappan, R.; Malarvili, M. Automatic quantitative analysis of human respired carbon dioxide waveform for asthma and non-asthma classification using support vector machine. IEEE Access 2018, 6, 55245–55256. [Google Scholar] [CrossRef]
- Kaggle. Asthma Disease Dataset. Available online: https://www.kaggle.com/datasets/rabieelkharoua/asthma-disease-dataset (accessed on 1 October 2025).
- Kim, D.; Cho, S.; Tamil, L.; Song, D.J.; Seo, S. Predicting asthma attacks: Effects of indoor PM concentrations on peak expiratory flow rates of asthmatic children. IEEE Access 2020, 8, 8791–8797. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).